Abstract
Sequence alterations in the pendrin gene (SLC26A4) leading to functionally affected protein variants are frequently involved in the pathogenesis of syndromic and nonsyndromic deafness. Considering the high number of SLC26A4 sequence alterations reported to date, discriminating between functionally affected and unaffected pendrin protein variants is essential in contributing to determine the genetic cause of deafness in a given patient. In addition, identifying molecular features common to the functionally affected protein variants can be extremely useful to design future molecule-directed therapeutic approaches. Here we show the functional and molecular characterization of six previously uncharacterized pendrin protein variants found in a cohort of 58 Brazilian deaf patients. Two variants (p.T193I and p.L445W) were undetectable in the plasma membrane, completely retained in the endoplasmic reticulum and showed no transport function; four (p.P142L, p.G149R, p.C282Y and p.Q413R) showed reduced function and significant, although heterogeneous, expression levels in the plasma membrane. Importantly, total expression levels of all of the functionally affected protein variants were significantly reduced with respect to the wild-type and a fully functional variant (p.R776C), regardless of their subcellular localization. Interestingly, reduction of expression may also reduce the transport activity of variants with an intrinsic gain of function (p.Q413R). As reduction of overall cellular abundance was identified as a common molecular feature of pendrin variants with affected function, the identification of strategies to prevent reduction in expression levels may represent a crucial step of potential future therapeutic interventions aimed at restoring the transport activity of dysfunctional pendrin variants.
INTRODUCTION
Pendrin (SLC26A4, PDS), first described as the protein encoded by the gene linked to Pendred syndrome (OMIM ID: 274600) (1), is a multifunctional anion exchanger expressed in the inner ear (2) and thyroid (3), among other tissues.
Sequence variations in the pendrin gene (SLC26A4) are found in individuals with syndromic (Pendred syndrome) or nonsyndromic hearing loss, both associated with inner ear malformations such as an enlarged vestibular aqueduct (EVA). Pendred syndrome is an autosomal recessive disease resulting from homozygous or compound heterozygous SLC26A4 sequence variations and characterized by the combination of sensorineural hearing loss and a partial iodide organification defect, clinically revealed by a positive perchlorate discharge test, with or without overt goiter or hypothyroidism (4). In addition, monoallelic (5–7) or biallelic (7) sequence variations in the pendrin gene are found in nonsyndromic EVA (ns-EVA), characterized by deafness without thyroid involvement. On the other hand, monoallelic SLC26A4 sequence variations can be found in deafness not associated with EVA (8). However, whether monoallelic SLC26A4 sequence variations can be regarded as the unique determinant of deafness is currently uncertain.
According to the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) (9), close to 500 different sequence alterations of the pendrin gene have been reported to date, but most of the corresponding protein variants miss a precise functional and molecular characterization (10). In the absence of careful genetic and functional assessments, two factors, i.e., the clinical condition of pseudo-Pendred syndromes (the association of deafness and goiter with no pendrin mutations [11–15]) and the existence of pendrin variants with no functional impairment (10), could lead to an incorrect assignment of the genetic cause of the disease.
Notable effort was devoted to assess the prevalence of pendrin mutations within specific cohorts and the respective molecular defects. Specifically, for the Brazilian population, accounting for 199 million people (according to the World Health Organization estimate for 2012, http://www.who.int/countries/bra/en/), with more than 9 million having hearing problems (http://www.ibge.gov.br/home/estatistica/populacao/censo2010/caracteristicas_religiao_deficiencia/caracteristicas_religiao_deficiencia_tab_xls.shtm, table 1.3.1), and representing the fifth largest population in the world (http://unstats.un.org/unsd/demographic/products/socind/default.htm), former reports described deletions (c.279delT and c.1197delT) found in homozygosity in the pendrin gene in large inbred families, leading to likely nonfunctional proteins caused by frameshift, premature stop codon and truncation (15–17). More recently, we reported the occurrence of 11 known and 2 novel (p.P142L and p.G149R) variants of pendrin in a cohort of 23 unrelated Brazilian patients with nonsyndromic hearing loss and EVA (18,19).
Genetic screening of our cohort of 58 Brazilian patients with a diagnosis of profound deafness (32 without and 26 with EVA) (Table 1) led to identification of SLC26A4 sequence alterations encoding 12 distinct pendrin proteins variants, of which one was novel (p.C282Y) and five were uncharacterized (p.P142L, p.G149R, p.T193I, p.Q413R, p.L445W). These variants, together with a variant (p.R776C) for which previous functional studies led to contradictory results, were characterized in the present study, with the aim of identifying pendrin as the genetic cause of deafness in a given patient by discriminating between functionally affected and unaffected variants and assessing possible correlations between (a) genotype and presence of EVA in these cohorts and (b) function and molecular features of the protein variants.
Table 1.
Genotype and phenotype of patients without (group I) and with (group II) EVA carrying variations in the SLC26A4 sequence.a
| Genotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide change | Amino acid change | Phenotype | |||||||
| Group | Patient ID | Onset of hearing lossb | Allele 1 | Allele 2 | Allele 1 | Allele 2 | EVA | Deafness | Goiter |
| I | C15 | Undetermined | c.412G>T | WT | p.V138F | WT | No | Progressive; profound at 53 years of age | No |
| C26c | 3 | c.845G>A | WT | p.C282Y | WT | No | Profound | No | |
| C01 | 2 | c.1826T>G | WT | p.V609G | WT | No | Profound | No | |
| C04 | 40 | c.1826T>G | WT | p.V609G | WT | No | Profound | No | |
| C09 | Prelingual | c.1826T>G | WT | p.V609G | WT | No | Profound | No | |
| II | 21 | 1 month | c.425C>T | c.279delT | p.P142L | p.S93Rfs3* | Bilateral | Profound | No |
| 16 | Prelingual | c.445G>A | WT | p.G149R | WT | Unilateral | Profound | No | |
| 18 | 5 | c.578C>T | WT | p.T193I | WT | Bilateral | Profound | No | |
| 22 | Prelingual | c.1226G>A | c.1226G>A | p.R409H | p.R409H | Bilateral | Profound | No | |
| 02 | Prelingual | c.1229C>T | c.1707+5G>A | p.T410M | SS | Bilateral | Profound | No | |
| 06 | Prelingual | c.1238A>G | c.412G>T | p.Q413R | p.V138F | Bilateral | Profound | No | |
| 23 | Prelingual | c.1334T>G | c.1001+1G>A | p.L445W | SS | Bilateral | Profound | Yes | |
| L1c | 1 | c.1826T>G | WT | p.V609G | WT | Bilateral | Profound | No | |
| 15 | 5 | c.2326C>T | WT | p.R776C | WT | Bilateral | Profound | No | |
The newly identified pendrin sequence variant is indicated in bold. Light gray cells denote the pendrin variants characterized in the present study; dark gray cells indicate pendrin variants for which the functional test described here could not be applied. SS, splicing site variant; *, stop codon.
Age of onset of hearing loss in years, except when otherwise specified.
Monoallelic mutation (c.35delG) in the connexin GJB2 gene was detected in the same patient.
MATERIALS AND METHODS
Patients
A total of 58 Brazilian individuals of undetermined ethnicity (30 females and 28 males aged between 4 and 55 years) diagnosed with profound sensorineural hearing loss were included in the study. These patients were divided in two groups on the basis of the radiological findings, as follows: group I, 32 deaf individuals with no EVA; group II, 26 deaf individuals with EVA (Table 1). In this last group, we included 23 patients from our former study for which we characterized the genotype (18); however, no functional test of the identified pendrin variants was performed. DNA samples of all individuals were obtained from the Clinic of Otorhinolaryngology (State University of Campinas, Brazil). The research was prospectively reviewed and approved by a duly constituted ethics committee (project: numbers 633/2003 and 396/2006). Written informed consent was obtained from all subjects or their legal representatives before blood sampling and genetic testing. For all patients, imaging studies of the inner ear by computer tomography (CT) and magnetic resonance imaging (MRI) of the temporal bones were performed. EVA was defined as an enlargement of the vestibular aqueduct >1.5 mm midway between the endolymphatic sac and the vestibule. The thyroid function of patients was not analyzed (the perchlorate discharge test was not performed); however, it was noticeable that one patient (patient ID 23, group II) presented overt goiter.
Genomic DNA Analysis
Genomic DNA was extracted from blood samples by using a standard phenol–chloroform method. Samples were screened for two common deletions affecting GJB6 [del(GJB6-D13S1830), del(GJB6-D13S1854)] and the m.1555A>G mutation in the mitochondrially encoded 12S rRNA gene MT-RNR1 as previously described (20–22). The entire coding region of GJB2 (exon 2) and the 20 coding exons of SLC26A4 were amplified by polymerase chain reaction (PCR) using primers reported in Supplementary Table S1 and by Everett et al. (1), respectively. The PCR products were sequenced using a 3500 Genetics Analyzer (Life Technologies), with the same primers used for amplification. The resulting sequences were compared against the published NCBI Homo sapiens GJB2 (OMIM ID: 121011, GenBank ID: NG_008358.1) and SLC26A4 (OMIM ID: 605646, GenBank ID: NG_008489.1) DNA sequence reference assembly. Only patients negative for mutations in the above-mentioned genes or harboring a monoallelic SLC26A4 variant were submitted to the genetic analysis for FOXI1 (OMIM ID: 601093, GenBank ID: NG_012068.1) and KCNJ10 (OMIM ID: 602208, GenBank ID: NG_016411.1). The primer pairs used to amplify the coding exons of FOXI1 and KCNJ10 are reported in the Supplementary Table S1. The same primers were also used for sequencing. The newly identified SLC26A4 sequence variants were submitted to the Leiden Open Variation Database (https://grenada.lumc.nl/LOVD2/Usher_montpellier/home.php).
Plasmid Constructs
The pTARGET (Promega Corporation) vector, containing the open reading frame (ORF) of wild-type human pendrin cloned from normal thyroid tissue, was originally provided by P Beck-Peccoz, University of Milan (Italy), and was used for functional tests and Western blot.
For colocalization and determination of pendrin expression levels via imaging, the ORF of wild-type pendrin was subcloned into the pEYFPN1 vector, in frame with the ORF of the enhanced yellow fluorescent protein (EYFP). After transfection of this construct in cells, pendrin is produced with the EYFP fused to its C-terminus (PDS-EYFP). Wild-type PDS-EYFP showed better plasma membrane trafficking than EYFP-PDS (pendrin with the EYFP fused to its N-terminus; data not shown), in agreement with previous observations (5).
The pendrin mutants were made using the QuikChange® site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol, using the primers listed in Supplementary Table S2. All plasmid inserts were sequenced before use in experiments (Microsynth AG, Switzerland) with the primers listed in Supplementary Table S2.
Cell Lines and Transient Transfection
Human embryonic kidney (HEK) 293 Phoenix (23) and HeLa (cervical adenocarcinoma, American Type Cell Culture Collection ATCC-CCL-2) cells were transfected by the calcium phosphate coprecipitation method or with Metafectene Pro (Biontex), following the manufacturer’s instructions. Further details are available in the Supplementary Materials and Methods.
Pendrin Functional Test
For testing the function of pendrin variants, the influx of iodide was measured in wild-type and mutated pendrin-overexpressing and control cells. Because a high transfection efficiency is an essential prerequisite, HEK 293 Phoenix cells, in which transfection efficiencies of ~90% are obtained, are particularly suitable for this approach. Cells were cotransfected with a plasmid encoding for EYFP p.H148Q;I152L (an EYFP variant with substantially improved sensitivity for iodide [24]) and the pTARGET plasmid bearing the cDNA of wild-type or mutated pendrin. Control cells were cotransfected with EYFP p.H148Q;I152L and the empty pTARGET vectors. The functional test was performed as already described (8), with adaptations allowing for the use of a multiplate reader (further details are given in the Supplementary Materials and Methods). Experiments were performed at room temperature. Data are expressed as % fluorescence variations (ΔF%), and a negative ΔF% indicates a flux of iodide from the extracellular to the intracellular milieu.
Colocalization Experiments
Subcellular localization of pendrin variants was determined by colocalization between wild-type or mutant PDS-EYFP and markers of the plasma membrane (PM) (CellMaskTM Deep Red plasma membrane stain, C10046; Invitrogen Molecular Probes) or endoplasmic reticulum (ER) (ER-TrackerTM Red, glibenclamide BODIPY®TR, E34250; Invitrogen Molecular Probes). Being that HEK 293 Phoenix cell morphology is scarcely suitable for colocalization experiments, HeLa cells were used for this purpose.
Imaging was performed by sequential acquisition with a Leica TCS SP5II AOBS confocal microscope (Leica Microsystems) equipped with a HCX PL APO 63×/1.20 Lambda blue water immersion objective and controlled by the LAS AF SP5 software (Leica Microsystems). Further details are given in the Supplementary Materials and Methods.
Semiquantitative Reverse Transcription–PCR for Verification of Equal Transfection Efficacy of the Different Plasmid Constructs
Extraction of total RNA from HEK 293 Phoenix cells transfected with the pTARGET constructs coding for wild-type or mutated pendrin or with the pTARGET empty vector (control) was performed with the All Prep DNA/RNA mini kit (Qiagen). At total of 1 μg total RNA was used for the reverse transcription reaction with the QuantiTect® reverse transcription kit for cDNA synthesis with integrated removal of genomic DNA contamination (Qiagen). For detecting the SLC26A4 transcript, the following primers were used: forward, 5′-TTCCA GCAAC AGCAC GAG-3′, and reverse, 5′-GCCAC TAGCC CAGTA CTAAC TC-3′. The SLC26A4 signal was normalized to the β-actin signal, detected by using the following primers: forward, 5′-GGCAT GGGTC AGAAG GATTC-3′, and reverse, 5′-AGAGG CGTAC AGGGA TAGCA C-3′. These primers span an intron-exon boundary and would disclose an eventual contamination from genomic DNA as a band at 740 bp, which was not detected (Figure 4A). Densitometric analysis was done with the ImageJ 1.46r software (Wayne Rasband, National Institutes of Health, USA).
Figure 4.
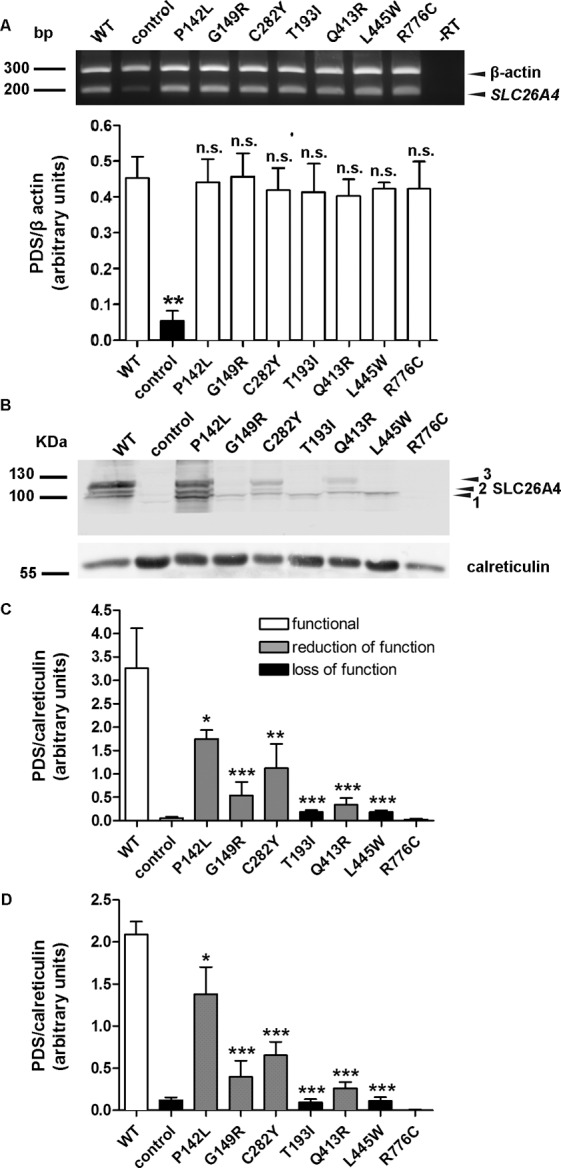
RNA and protein levels of wild-type pendrin and pendrin variants. (A) Top: The levels of the transcripts of wild-type pendrin and its variants, as well as the housekeeping gene β-actin, were detected by reverse transcription PCR in three independent samples of transfected HEK 293 Phoenix cells. In –RT, the PCR was conducted on a sample of total RNA that was not subjected to the reverse transcription reaction, to exclude genomic DNA contamination. For the control, cells were transfected with an empty vector. The presence of a weak band at 200 bp in control corresponds to endogenously expressed pendrin. Bottom: Densitometry of the pendrin signal normalized to the β-actin signal. n.s., Not significant, **p < 0.01 compared with wild-type, one-way ANOVA with Dunnett multiple comparison posttest. (B) Representative immunoblotting on total cellular membrane extracts obtained after expression of untagged wild-type (WT) and mutated pendrin in HEK 293 Phoenix cells. For the control, cells were transfected with an empty vector. 1, 2 and 3 indicate the nonglycosylated, partially glycosylated and maturely glycosylated forms of pendrin, respectively. The expression levels of all (C) or only the maturely glycosylated (D) forms of wild-type (WT) and mutated pendrin were quantified by densitometry and normalized for the housekeeping protein calreticulin. n = 3, ***p < 0.005, **p < 0.01, *p < 0.05 compared with wild-type, one-way ANOVA with Dunnett multiple comparison posttest.
Determination of Wild-Type and Mutant Pendrin Total Expression Levels by Imaging
HeLa cells expressing wild-type or mutant PDS-EYFP were fixed with 3% paraformaldehyde for 15 min, counterstained with 0.1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) for 10 min, washed three times and imaged in Hanks balanced salt solution (HBSS). The nuclear staining with DAPI gives a signal that, expressed as average levels of gray, correlates to the number of cells in the imaged field, therefore giving an indication of cell density. Fluorescence intensity of the whole imaging field (average levels of gray) in the EYFP emission channel was subtracted for the background fluorescence and normalized for the background-subtracted fluorescence intensity in the DAPI emission channel, to obtain wild-type or mutant PDS-EYFP expression levels normalized for the cell density. Imaging was performed by confocal microscopy as described above. Further details are available in the Supplementary Materials and Methods.
Determination of Wild-type and Mutant Pendrin Expression Levels in Total Cell Membranes Lysates by Western Blot
HEK 293 Phoenix cells expressing wild-type or mutant pendrin were collected and centrifuged at 216g for 15 min at 4°C. The extraction of the total membrane protein fraction, including both plasma membrane proteins and membrane proteins from cellular organelles, was obtained with the Plasma Membrane Extraction Kit (MBL International Corporation), according to the manufacturer’s instructions. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot were performed following standard procedures (see Supplementary Materials and Methods). The primary antibodies were rabbit anti-pendrin, 1:10,000, provided by D Eladari (25), or rabbit anti-pendrin, polyclonal, raised against amino acids 586–780 of human pendrin (sc-50346, Santa Cruz Biotechnology) 1:500 and rabbit anti-calreticulin (ab4, Abcam) 1:1,000. Detection of the signal of immunocomplexes was performed with the ODYSSEY infrared imaging system (LI-COR). Blot images were densitometrically analyzed with the ImageJ 1.46r software.
Determination of Wild-type and Mutant Pendrin Expression Levels in the Plasma Membrane Region
HeLa cells expressing wild-type or mutant PDS-EYFP and the enhanced cyan fluorescent protein (ECFP) (pECFPC1 vector, Clontech) were stained with CellMaskTM Deep Red plasma membrane stain, thoroughly washed and imaged in HBSS. EYFP fluorescence intensity of three regions of interest of the plasma membrane of a single cell was subtracted for the background fluorescence, averaged and normalized for the background-subtracted ECFP fluorescence intensity measured in the cytosol of the same cell, to obtain wild-type or mutant PDS-EYFP expression levels normalized for the transfection efficiency of the single cell. Imaging was performed by confocal microscopy as described above. Further details are available in the Supplementary Materials and Methods.
Salts and Chemicals
All salts and chemicals used were per analysis grade.
Statistical Analysis
All data are expressed as arithmetic means ± standard error of the mean (SEM). For statistical analysis, GraphPad Prism software (version 4.00 for Windows, GraphPad Software) was used. Significant differences between datasets were tested by Fisher exact test or one-way analysis of variance (ANOVA) with Bonferroni or Dunnett posttests, as appropriate. Statistically significant differences were assumed at p < 0.05; (n) corresponds to the number of independent measurements.
All supplementary materials are available online at www.molmed.org.
RESULTS
Detection of Sequence Variations in the GJB2, GJB6 and MT-RNR1 Genes
All patients were negative for two common deletions [del(GJB6-D13S1830) and del(GJB6-D13S1854)] affecting GJB6 (encoding connexin 30) and for the m.1555A>G mutation in the mitochondrially encoded 12S rRNA gene MT-RNR1. Two patients (patient ID C26, group I, and L1, group II; Table 1) showed a monoallelic mutation (c.35delG) in the GJB2 gene encoding Connexin 26. All the other patients were homozygous wild-type for GJB2.
Pendrin Allelic Variants in Deaf Brazilian Patients with and without EVA
In the cohort of deaf patients without EVA (group I, Table 1), monoallelic alterations in the sequence of the pendrin gene were found in 5 of 32 subjects. A novel (previously unknown and uncharacterized) pendrin variant (p.C282Y) was detected in this cohort (patient ID C26). All the other patients were homozygous wild-type for the pendrin gene.
In the cohort of deaf patients with EVA (group II, Table 1), as previously described (18), sequence alterations in the pendrin gene were found in 9 of 26 subjects. Of these patients, five carried biallelic and four monoallelic sequence alterations, respectively. All the other subjects were homozygous wild-type for the pendrin gene.
Sequencing of FOXI1 and KCNJ10 Coding Regions
Sequence alterations in the FOXI1 or KCNJ10 coding regions were not detected in these patients.
Function of Pendrin Variants
The impact of the amino acid substitutions p.V138F, p.V609G, p.R409H and p.T410M and the deletion c.279delT (p.S93Rfs3*) on pendrin functionality were previously described by us or others (10; Table 2) and were not characterized further. For the splicing site mutations c.1707+5G>A and c.1001+1G>A (also called IVS15+5G>A and IVS8+1G>A, respectively; evidenced in dark gray in Table 1) most likely leading to an aberrant protein product (26) or no protein product at all (27), the functional test described here could not be applied. Therefore, five previously uncharacterized pendrin variants (p.P142L, p.G149R, p.T193I, p.Q413R, p.L445W), the novel variant p.C282Y and one variant (p.R776C) for which functionality was ambiguously defined (5,14,28) were subjected to a functional test (Figure 1; the pendrin variants were subdivided into two groups for technical reasons).
Table 2.
Functional and molecular features of pendrin variants identified patients without (group I) and with (group II) EVA.a
| Group | Patient ID |
Protein variants: Allele 1 Allele 2 |
Function: Allele 1 Allele 2 |
Localization: Allele 1 Allele 2 |
Expression: Allele 1 Allele 2 |
Pendrin-related deafness | Reference: Allele 1 Allele 2 |
|
|---|---|---|---|---|---|---|---|---|
| I | C15 | p.V138F | Lost | ER | ? | Probably not | (44) | |
| WT | WT | PM | WT | — | ||||
| C26b | p.C282Y | Reduced (41%) | PM | Reduced | Probably not | Present work | ||
| WT | WT | PM | WT | — | ||||
| C01 | p.V609G | Reduced (45%) | ? | ? | Probably not | (45) | ||
| WT | WT | PM | WT | — | ||||
| C04 | p.V609G | Reduced (45%) | ? | ? | Probably not | (45) | ||
| WT | WT | PM | WT | — | ||||
| C09 | p.V609G | Reduced (45%) | ? | ? | Probably not | (45) | ||
| WT | WT | PM | WT | — | ||||
| II | 21 | p.P142L | Reduced (76%) | PM | Reduced | Yes | Present work | |
| p.S93Rfs3* | Lost | ? | ? | (46) | ||||
| 16 | p.G149R | Reduced (55%) | PM/ER | Reduced | ? | Present work | ||
| WT | WT | PM | WT | — | ||||
| 18 | p.T193I | Lost | ER | Greatly reduced | ? | Present work | ||
| WT | WT | PM | WT | — | ||||
| 22 | p.R409H | Lost or reduced (33%) | PM/ER | Reduced | Yes | (47) and data not shown | ||
| p.R409H | Lost or reduced (33%) | PM/ER | Reduced | (47) and data not shown | ||||
| 02 | p.T410M | Lost | ER | ? | yes | (44) | ||
| SS (c.1707+5G>A) | Most likely lost | — | Lost | (27) | ||||
| 06 | p.Q413R | Reduced (60%) | ER | Reduced | Yes | Present work | ||
| p.V138F | Lost | ER | ? | (44) | ||||
| 23 | L445W | Lost | ER | Greatly reduced | Yes | Present work | ||
| SS (c.1001+1G>A) | Most likely lost | ? | (26) | |||||
| L1b | p.V609G | Reduced (45%) | ? | ? | ? | (45) | ||
| WT | WT | PM | WT | — | ||||
| 15 | p.R776C | Unaffected | PM | Unaffected | No | Present work | ||
| WT | WT | PM | WT | — |
The magnitude of reduction in pendrin transport activity is indicated in %. Question mark (?) denotes an undetermined feature. SS, splicing site variant; *, stop codon.
Monoallelic mutation (c.35delG) in the connexin GJB2 gene was detected in the same patient.
Figure 1.
Functionality of wild-type pendrin and seven variants identified in Brazilian deaf patients. (A, B) Average intracellular fluorescence intensity measured in HEK 293 Phoenix cells transfected with wild-type (WT) or mutated pendrin and EYFP p.H148Q;I152L or EYFP p.H148Q;I152L alone (control) and bathed in chloride and iodide containing solutions. Arrows indicate the addition of iodide to the extracellular solution. Fluorescence intensity was normalized for the average of fluorescence intensity in high chloride solution. (C, D) Percentage of fluorescence decrease (ΔF%) determined over the experimental period (19 s) for the pendrin variants indicated in (A) and (B). ***p < 0.001, n.s., not statistically significant, one-way ANOVA with Bonferroni multiple comparison posttest.
Iodide flux in cells expressing wild-type pendrin (Figure 1) was significantly higher (p < 0.001) compared with that measured in control cells and confirmed that pendrin is an iodide transporter (29,30), most likely acting as a Cl–/I– anion exchanger in this system (31).
Iodide influx measured in cells expressing pendrin p.T193I and p.L445W (Figures 1A, C) was not significantly different from that measured in control cells, indicating a complete loss of function of these variants. In contrast, iodide influx measured in cells expressing pendrin p.Q413R (Figures 1A, C), p.P142L, p.G149R and p.C282Y (Figures 1B, D) was significantly different from that measured in wild-type and control cells (p < 0.001), indicating that these variants retain residual transport activity. The transport activity of pendrin p.R776C (Figures 1A, C) was indistinguishable from wild-type. The results of the functional test are summarized in Table 2.
Subcellular Localization of Pendrin Variants
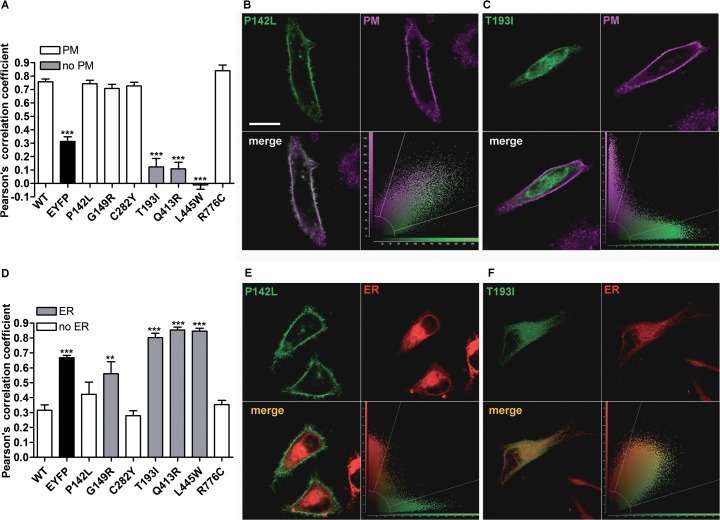
To define their subcellular localization, wild-type or mutated PDS-EYFP were expressed in HeLa cells (Figure 2), and colocalization with the PM or ER was determined. As shown in Figures 2A and D, wild-type pendrin is preferentially targeted to the PM, with poor colocalization with the ER, thereby providing evidence that the C-terminal EYFP tag does not alter proper pendrin trafficking. Similarly, pendrin p.P142L, p.G149R, p.C282Y and p.R776C (Figure 2A) were efficiently transposed to the PM. On the other hand, pendrin p.T193I, p.Q413R and p.L445W failed to reach the PM. Figure 2D shows that pendrin p.G149R, p.T193I, p.Q413R and p.L445W are preferentially located in the ER, whereas pendrin p.P142L, p.C282Y and p.R776C are not preferentially found in this compartment. To conclude, wild-type, p.P142L, p.C282Y and p.R776C pendrin variants show preferential trafficking to the PM, whereas p.T193I, p.Q413R and p.L445W are largely retained in the ER (Table 2). Pendrin p.G149R represents an intermediate situation, since this variant was found to colocalize with both the PM (Figure 2A) and ER (Figure 2D).
Figure 2.
Subcellular localization of wild-type pendrin and its variants. (A) Pearson correlation coefficient referred to the colocalization of wild-type (WT) pendrin and its variants with the PM. ***p < 0.001 compared with wild-type and therefore excluded from the plasma membrane, 9 ≤ n ≤ 25, one-way ANOVA with Bonferroni multiple comparison posttest. Representative examples of pendrin protein variants colocalizing with (B) or excluded from (C) the PM. The corresponding merge images (lower left) and scatterplots (lower right) are shown. Scale bar: 20 μm. (D) Pearson correlation coefficient referred to the colocalization of wild-type pendrin and its variants with the ER. 6 ≤ n ≤ 10, ***p < 0.001 compared with wild-type and therefore colocalizing with the ER, one-way ANOVA with Bonferroni multiple comparison posttest. Representative examples of pendrin protein variants excluded from (E) or colocalizing with (F) the ER. The corresponding merge images (lower left) and scatterplots (lower right) are shown.
Total Expression Levels of Pendrin Variants
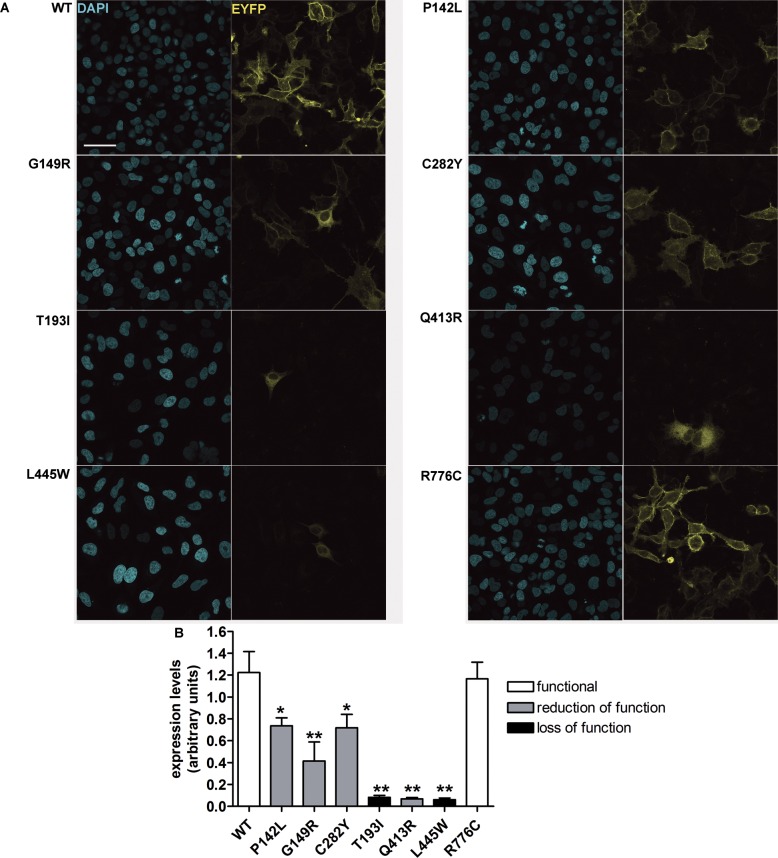
Total expression levels of pendrin variants were evaluated by confocal imaging (Figure 3). Interestingly, the total expression levels of all of the pendrin variants, with the exception of the fully functional variant p.R776C, were substantially reduced with respect to wild-type. A dramatic reduction in the expression levels was observed, especially for those mutants (p.T193I, p.Q413R and p.L445W) entrapped in the ER, whereas the expression levels of those variants (p.P142L and p.C282Y) preferentially targeted to the PM were significantly higher with respect to those of the variants preferentially retained in the ER (p < 0.001).
Figure 3.
Global cellular levels of wild-type pendrin and pendrin variants in intact cells. (A) Wild-type (WT) and mutated pendrin-EYFP (yellow signal) were expressed in HeLa cells. Cells were fixed and nuclei counterstained with DAPI (blue signal) to quantify the cell density. Scale bar: 50 μm. (B) Wild-type (WT) and mutated pendrin total expression levels expressed as fluorescence intensity (levels of gray) normalized for the cell density. n = 16, **p < 0.01, *p < 0.05 compared with wild-type, one-way ANOVA with Dunnett multiple comparison posttest.
Expression levels of pendrin variants in total cellular membrane protein extracts were also evaluated by Western blot after transfection of untagged (that is, with no EYFP fused to the C-terminus) wild-type and mutated pendrin (Figures 4B–D). Bands between 100 and 130 kDa represent the nonglycosylated, partially glycosylated and maturely glycosylated forms of pendrin (Figure 4B) and correspond to what was previously reported (32,33). A signal corresponding to pendrin p.R776C was not detected, most likely due to conformational disruption of the epitope recognized by the antibody used in these experiments (25). Densitometry showed that all functionally impaired pendrin variants exhibit cellular expression levels significantly reduced with respect to the wild-type (Figure 4C) and confirmed the results obtained by imaging, thereby showing evidence that the presence of a C-terminal tag does not significantly influence protein stability. Notably, the pendrin variants with loss of function (p.T193I and p.L445W) only show the nonglycosylated form (Figures 4B, D). Furthermore, the presence of the higher band, corresponding to the maturely glycosylated protein (Figure 4D), correlates with function (Figure 1) and with expression in the plasma membrane region (Figure 5). In these experiments, an antibody directed against the C-terminal 29 amino acids of rat pendrin was used (25). Similar results were obtained with a commercial antibody directed against the amino acids 586–780 of human pendrin (data not shown).
Figure 5.
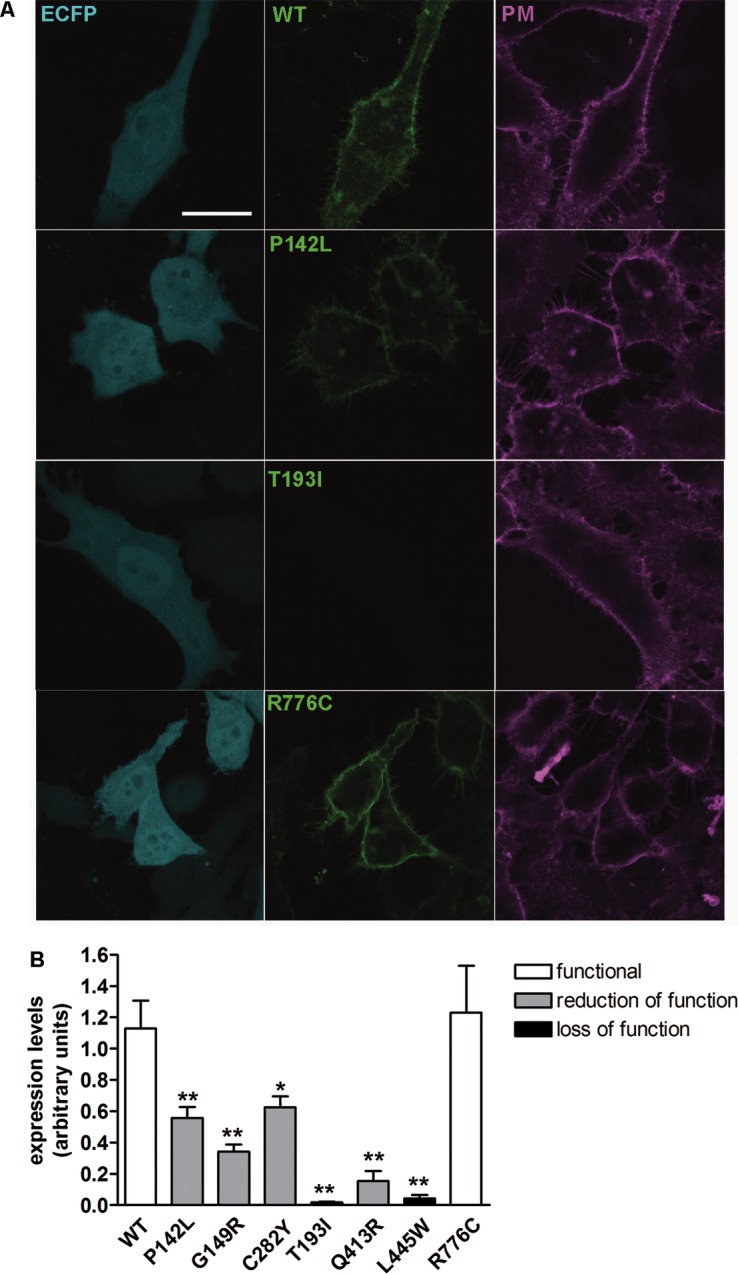
Abundance of wild-type pendrin and pendrin variants in the plasma membrane region of living cells. (A) Wild-type (WT) and mutated pendrin-EYFP (green signal) were expressed in HeLa cells together with ECFP (blue signal) as an indicator of the transfection efficiency of the single cell. Living cells were stained with a suitable dye to clearly identify the plasma membrane (magenta signal). Scale bar: 25 μm. (B) Wild-type (WT) and mutated pendrin fluorescence intensity in three regions of interest of the plasma membrane of a single cell were expressed as levels of gray, averaged and normalized for the fluorescence intensity of ECFP in the cytosol of the same cell. n = 16, **p < 0.01, *p < 0.05 compared with wild-type, one-way ANOVA with Dunnett multiple comparison posttest.
Verification of the Transfection Efficacy of the Different Plasmid Constructs
As differences in the protein abundance of the different pendrin variants may potentially arise from differences in the transfection efficacy of the corresponding plasmid constructs, the latter was verified by semiquantitative reverse transcription PCR. Significant differences between the abundance of cDNA (and hence, mRNA) encoding the various pendrin variants in the corresponding batches of transfected cells were not detected (Figure 4A).
Expression Levels of Pendrin Variants in the Plasma Membrane Region
Whereas in Figure 2A we showed the ability of the investigated pendrin variants to reach the PM, the technique used did not allow quantification and comparison of the amount of the different protein variants within this compartment. As shown in Figure 5, the amount of pendrin and its variants in the PM region was evaluated by confocal imaging with a different approach (see Materials and Methods). Expression levels of the different pendrin variants in the PM region mirrored total expression levels (Figures 3, 4). It is important to note that, although the total expression levels of p.Q413R are very low (Figure 3), a residual amount of this variant within the plasma membrane was detected (Figure 5).
DISCUSSION
Genotype–phenotype correlations are difficult to assess for SLC26A4. A recent analysis of six studies with a total enrollment of 769 hearing-impaired probands with EVA revealed that, on average, 45–50% of patients carry at least one SLC26A4 sequence variant. Of these, only 25% carry biallelic SLC26A4 sequence variants, whereas for a notable proportion of deaf individuals with EVA (~55%), SLC26A4 sequence variants were not detected (34). Deafness can unequivocally be assigned to pendrin dysfunction only when both SLC26A4 alleles encode for hypo- or nonfunctional pendrin protein variants. Before the completion of the present study, deafness and EVA could unequivocally be linked to pendrin malfunction in only 2 of 26 individuals in our cohort of deaf patients with EVA (patient IDs 22 and 02, Group II, Table 2), bearing previously characterized pendrin sequence alterations affecting function. The functional assessment of p.P142L, p.Q413R and p.L445W pendrin protein variants presented here allowed for the conclusion that deafness and EVA are most likely due to pendrin dysfunction also for patients 21, 06 and 23 (Table 2). The lack of functional impact of the amino acid substitution p.R776C allowed for the exclusion of pendrin as the genetic cause of deafness and EVA for patient 15 (Table 2).
The functional test used in the present work evaluates the chloride/iodide exchange activity of pendrin (8,31,35,36). Despite that iodide transport may be of no physiological relevance in the inner ear, the chloride/iodide exchange activity generally reflects the chloride/bicarbonate exchange activity (10) and can therefore be used as a tool to discriminate between variants with and without pathogenic potential. Accordingly, when differences between the chloride/iodide and chloride/bicarbonate exchange efficiencies were observed, they were considered to be “moderate” and most likely of no pathophysiological relevance (5).
For patients bearing monoallelic pendrin mutations, interpretation of results is less straightforward. Variants with reduction or loss of function are found both in the group of deaf patients with EVA (p.G149R, p.T193I and p.V609G) and in the group of deaf patients with no EVA (p.V138F, p.C282Y and p.V609G), with no apparent correlation between occurrence of monoallelic, functionally impaired pendrin variants and presence of EVA in these cohorts. In addition, a same variant (that is, p.V609G) could be found in both cohorts with and without EVA. On the basis of these observations, it seems that some defined monoallelic pendrin sequence alterations, although encoding hypofunctional protein variants, as it is the case of p.V609G, are per se not sufficient to cause deafness and EVA, and the association with other genetic, epigenetic and/or environmental factors should be taken into account when attempting to identify the determinant for the deafness phenotype of a given patient. Noteworthy in the present study, the p.V609G amino acid substitution was found in association with a monoallelic GJB2 pathogenic variant in patient L1, possibly leading to EVA. However, the association between monoallelic GJB2 and SLC26A4 sequence alterations affecting functionality does not invariably lead to EVA, since this genetic configuration is also found in patient C26 (carrying the newly identified monoallelic p.C282Y pendrin variant and the c.35delG GJB2 sequence alteration and not showing EVA).
We conclude that the presence of pendrin sequence variants in our cohort of deaf patients without EVA is likely coincidental, and deafness is probably due to other unidentified factors. Pendrin dysfunction could unequivocally be identified as the determinant of deafness in five of nine deaf individuals with EVA bearing pendrin mutations and was excluded in one case, whereas for three cases, the assignment of the genetic cause of deafness and EVA was not conclusive (Table 2).
The identification of pendrin variants with different degrees or no functional impairment led us to attempt correlating transport function and molecular features such as cellular distribution and expression levels of pendrin variants. Pendrin variants showing good colocalization with the PM (p.P142L, p.G149R, p.C282Y; Figure 2A) retain a significant transport activity (Figure 1D), whereas poor colocalization with the PM and retention in the ER (as is the case of variants p.T193I and p.L445W [Figures 2A, D]; the localization of pendrin p.L445W is in agreement with previous findings [5]) correlates with loss of function (Figure 1C).
Retention in the subcellular compartments is an indication of protein misfolding, often leading to degradation (37). This observation prompted us to quantify the cellular expression levels of the different pendrin protein variants, on the basis of the hypothesis that abundance of variants with significant retention in the ER should be reduced with respect to variants with targeting to the PM. Interestingly, however, total (Figures 3 and 4) and PM (Figure 5) expression levels of all of the functionally impaired pendrin variants were significantly reduced with respect to the wild-type, regardless of their cellular localization. Abundance of all protein variants with significant residual activity (p.P142L, p.G149R, p.C282Y and p.Q413R), evaluated in the PM region by means of quantitative imaging (Figure 5), was significantly higher than that of variants with total loss of function (p.T193I and p.L445W). Differences in expression levels of pendrin protein variants did not arise from differences in the transfection efficiency of the corresponding plasmid construct or mRNA abundance (Figure 4A, Supplementary Materials and Methods and Supplementary Table S3).
The significant residual activity of pendrin p.Q413R appears to be substantiated by detection of a small amount of the protein in the PM region that could not be quantified in colocalization experiments, as mentioned earlier. The low expression levels of pendrin p.Q413R, however, seems to be insufficient to justify its significant transport function. It is possible that the amino acid substitution Q413R leads to protein misfolding and consequent degradation and simultaneously confers an intrinsic gain of transport function, therefore leading to significant iodide intake (Figure 1C) despite dramatically reduced expression levels (Figures 3–5). Expression levels of the variant p.R776C were not reduced with respect to the wild-type, therefore confirming its nature of a benign polymorphism. We conclude that functionally impaired pendrin variants show cellular expression levels reduced with respect to the wild-type and functionally unaffected variants, with mutants with reduced function showing significant, although heterogeneous, expression levels at the PM and mutants with null function being undetectable in the PM and completely retained in the ER.
Misfolded proteins are degraded via a series of pathways collectively denoted as ER-associated degradation (ERAD), requiring ubiquitination by different E3 ubiquitin ligases as an essential step (38). Single amino acid substitutions may impair the ability of the affected polypeptide to reach the functional conformation; consequently, prolonged interaction with molecular chaperones, enhanced proteolytic degradation and reduced cellular half-life of mutant proteins are events occurring in a number of genetic diseases (39). A previous report gathered evidence that wild-type pendrin-GFP colocalizes with ubiquitin in the cell, therefore suggesting a role for polyubiquitination in pendrin degradation (40). Accordingly, ubiquitination prediction programs (http://www.ubpred.org/) identify pendrin lysine residues at positions 632, 647, 734 and 753 as putative ubiquitination sites (Supplementary Figure S1). Recently, Lee et al. (33) identified the specific ER-resident E3 ubiquitin ligase involved in the degradation of wild-type and mutated pendrin. It is therefore likely to hypothesize that the reduced global and PM expression levels of functionally impaired pendrin variants are the consequence of an enhanced degradation by the ubiquitin proteasome system.
The mutations analyzed in the present work do not lie in close proximity of a glycosylation site (Supplementary Figure S1) and most likely do not directly impede glycosylation. Some amino acid substitutions, however, as in the case of p.T193I and p.L445W, may cause protein misfolding and premature degradation, therefore indirectly interfering with or preventing glycosylation (Figure 4D).
Studies on mouse models lacking pendrin expression selectively in the endolymphatic sac (41), or in the cochlea and the vestibular labyrinth (42), showed that pendrin activity is only required during a critical time period of embryonic development and suggested that a temporally and spatially limited therapy directed to the endolymphatic sac and focused on the prenatal phase could restore normal hearing in patients with deafness linked to pendrin mutations (43). On the basis of these considerations and on the findings presented here, we suggest that prevention of reduction of expression levels, together with the assistance of proper protein folding, should be regarded as an essential component of potential therapeutic approaches aimed at restoring or increasing the transport activity of dysfunctional pendrin variants. Further studies are needed to elucidate the molecular mechanism leading to the reduction of cellular abundance of pathogenic pendrin protein variants.
Conclusion
Cellular expression levels of functionally impaired pendrin protein variants are significantly reduced with respect to those of wild-type and fully functional variants. Reduction of protein abundance correlates with reduction of ion transport function, regardless of subcellular localization. Therefore, a molecular feature common to pathogenic pendrin protein variants has been identified and could represent the target of future molecule-directed therapies aimed at restoring pendrin function.
Supplementary Material
Acknowledgments
This work was supported by the Seventh Framework Programme (grant PIRSES-GA-2008-230661) and Fonds zur Förderung der wissenschaftlichen Forschung (FWF) (grant P18608) to M Paulmichl and Fundação de Amparo à Pesquisa do Estado de São Paulo and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior to EL Sartorato. C Nofziger was supported by the Roche Postdoc Fellowship Program (grant 231). The anti-pendrin antibody was a gift from D Eladari, Institut National de la Santé et de la Recherche Médicale, Paris, France. We sincerely thank Elisabeth Mooslechner for her expert secretarial assistance.
Footnotes
Online address: http://www.molmed.org doi: 10.2119/molmed.2015.00226
Disclosure
Disclosure
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: de Moraes VCS, et al. (2016) Reduction of cellular expression levels is a common feature of functionally affected pendrin (SLC26A4) protein variants. Mol. Med. 22:41–53.
REFERENCES
- Everett LA, et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat. Genet. 1997;17:411–22. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- Royaux IE, et al. Localization and functional studies of pendrin in the mouse inner ear provide insight about the etiology of deafness in Pendred syndrome. J. Assoc. Res. Otolaryngol. 2003;4:394–404. doi: 10.1007/s10162-002-3052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royaux IE, et al. Pendrin, the protein encoded by the Pendred syndrome gene (PDS), is an apical porter of iodide in the thyroid and is regulated by thyroglobulin in FRTL-5 cells. Endocrinology. 2000;141:839–45. doi: 10.1210/endo.141.2.7303. [DOI] [PubMed] [Google Scholar]
- Bizhanova A, Kopp P. Genetics and phenomics of Pendred syndrome. Mol. Cell. Endocrinol. 2010;322:83–90. doi: 10.1016/j.mce.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Choi BY, et al. Hypo-functional SLC26A4 variants associated with nonsyndromic hearing loss and enlargement of the vestibular aqueduct: genotype-phenotype correlation or coincidental polymorphisms? Hum. Mutat. 2009;30:599–608. doi: 10.1002/humu.20884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor SP, et al. SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J. Med. Genet. 2005;42:159–65. doi: 10.1136/jmg.2004.024208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur. J. Hum. Genet. 2006;14:773–9. doi: 10.1038/sj.ejhg.5201611. [DOI] [PubMed] [Google Scholar]
- Pera A, et al. Functional assessment of allelic variants in the SLC26A4 gene involved in Pendred syndrome and nonsyndromic EVA. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18608–13. doi: 10.1073/pnas.0805831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossena S, et al. Molecular and functional characterization of human pendrin and its allelic variants. Cell Physiol. Biochem. 2011;28:451–66. doi: 10.1159/000335107. [DOI] [PubMed] [Google Scholar]
- Fugazzola L, et al. Differential diagnosis between Pendred and pseudo-Pendred syndromes: clinical, radiologic, and molecular studies. Pediatr. Res. 2002;51:479–84. doi: 10.1203/00006450-200204000-00013. [DOI] [PubMed] [Google Scholar]
- Kara C, Kilic M, Ucakturk A, Aydin M. Congenital goitrous hypothyroidism, deafness and iodide organification defect in four siblings: Pendred or pseudo-Pendred syndrome? J. Clin. Res. Pediatr. Endocrinol. 2010;2:81–4. doi: 10.4274/jcrpe.v2i2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N, Lunardi C, Shield JP. Sensori-neural deafness and hypothyroidism: autoimmunity causing ‘pseudo-Pendred syndrome.’. Horm. Res. 2006;65:267–8. doi: 10.1159/000092513. [DOI] [PubMed] [Google Scholar]
- Pfarr N, et al. Goitrous congenital hypothyroidism and hearing impairment associated with mutations in the TPO and SLC26A4/PDS genes. J. Clin. Endocrinol. Metab. 2006;91:2678–81. doi: 10.1210/jc.2006-0142. [DOI] [PubMed] [Google Scholar]
- Kopp P, et al. Phenocopies for deafness and goiter development in a large inbred Brazilian kindred with Pendred’s syndrome associated with a novel mutation in the PDS gene. J. Clin. Endocrinol. Metab. 1999;84:336–41. doi: 10.1210/jcem.84.1.5398. [DOI] [PubMed] [Google Scholar]
- Camargo R, et al. Aggressive metastatic follicular thyroid carcinoma with anaplastic transformation arising from a long-standing goiter in a patient with Pendred’s syndrome. Thyroid. 2001;11:981–8. doi: 10.1089/105072501753211073. [DOI] [PubMed] [Google Scholar]
- Lofrano-Porto A, et al. Pendred syndrome in a large consanguineous Brazilian family caused by a homozygous mutation in the SLC26A4 gene. Arq. Bras. Endocrinol. Metabol. 2008;52:1296–303. doi: 10.1590/s0004-27302008000800015. [DOI] [PubMed] [Google Scholar]
- de Moraes VC, et al. Molecular analysis of SLC26A4 gene in patients with nonsyndromic hearing loss and EVA: identification of two novel mutations in Brazilian patients. Int. J. Pediatr. Otorhinolaryngol. 2013;77:410–3. doi: 10.1016/j.ijporl.2012.11.042. [DOI] [PubMed] [Google Scholar]
- Ramos PZ, et al. Etiologic and diagnostic evaluation: algorithm for severe to profound sensorineural hearing loss in Brazil. Int. J. Audiol. 2013;52:746–52. doi: 10.3109/14992027.2013.817689. [DOI] [PubMed] [Google Scholar]
- del Castillo FJ, et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 2005;42:588–94. doi: 10.1136/jmg.2004.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RA, et al. Maternally inherited nonsyndromic hearing loss. Am. J. Med. Genet. 1999;84:369–72. doi: 10.1002/(sici)1096-8628(19990604)84:4<369::aid-ajmg12>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Tamagawa Y, Ocho S, Hoshino T, Kitamura P. Hereditary sensorineural hearing loss of unknown cause involving mitochondrial DNA 1555 mutation. ORL J. Otorhinolaryngol. Relat. Spec. 2000;62:100–3. doi: 10.1159/000027725. [DOI] [PubMed] [Google Scholar]
- DiCiommo DP, Duckett A, Burcescu I, Bremner R, Gallie BL. Retinoblastoma protein purification and transduction of retina and retinoblastoma cells using improved alphavirus vectors. Invest. Ophthalmol. Vis. Sci. 2004;45:3320–9. doi: 10.1167/iovs.04-0140. [DOI] [PubMed] [Google Scholar]
- Galietta LJ, Haggie PM, Verkman AS. Green fluorescent protein-based halide indicators with improved chloride and iodide affinities. FEBS Lett. 2001;499:220–4. doi: 10.1016/s0014-5793(01)02561-3. [DOI] [PubMed] [Google Scholar]
- Knauf F, et al. Identification of a chloride-formate exchanger expressed on the brush border membrane of renal proximal tubule cells. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9425–30. doi: 10.1073/pnas.141241098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogazzi F, et al. A novel mutation in the pendrin gene associated with Pendred’s syndrome. Clin. Endocrinol. (Oxf) 2000;52:279–85. doi: 10.1046/j.1365-2265.2000.00930.x. [DOI] [PubMed] [Google Scholar]
- Ganaha A, et al. Pathogenic substitution of IVS15 + 5G>A in SLC26A4 in patients of Okinawa Islands with enlarged vestibular aqueduct syndrome or Pendred syndrome. BMC Med. Genet. 2013;14:56. doi: 10.1186/1471-2350-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, et al. Molecular epidemiology and functional assessment of novel allelic variants of SLC26A4 in non-syndromic hearing loss patients with enlarged vestibular aqueduct in China. PLoS One. 2012;7:e49984. doi: 10.1371/journal.pone.0049984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, Wang R, Kreman TM, Sheffield VC, Karniski LP. The Pendred syndrome gene encodes a chloride-iodide transport protein. Nat. Genet. 1999;21:440–3. doi: 10.1038/7783. [DOI] [PubMed] [Google Scholar]
- Gillam MP, et al. Functional characterization of pendrin in a polarized cell system: evidence for pendrin-mediated apical iodide efflux. J. Biol. Chem. 2004;279:13004–10. doi: 10.1074/jbc.M313648200. [DOI] [PubMed] [Google Scholar]
- Dossena S, et al. Fast fluorometric method for measuring pendrin (SLC26A4) Cl–/I– transport activity. Cell Physiol. Biochem. 2006;18:67–74. doi: 10.1159/000095164. [DOI] [PubMed] [Google Scholar]
- Yoon JS, et al. Heterogeneity in the processing defect of SLC26A4 mutants. J. Med. Genet. 2008;45:411–9. doi: 10.1136/jmg.2007.054635. [DOI] [PubMed] [Google Scholar]
- Lee K, Hong TJ, Hahn JS. Roles of 17-AAG-induced molecular chaperones and Rma1 E3 ubiquitin ligase in folding and degradation of Pendrin. FEBS Lett. 2012;586:2535–41. doi: 10.1016/j.febslet.2012.06.023. [DOI] [PubMed] [Google Scholar]
- Pique LM, et al. Mutation analysis of the SLC26A4, FOXI1 and KCNJ10 genes in individuals with congenital hearing loss. PeerJ. 2014;2:e384. doi: 10.7717/peerj.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror AA, et al. Calcium oxalate stone formation in the inner ear as a result of an Slc26a4 mutation. J. Biol. Chem. 2010;285:21724–35. doi: 10.1074/jbc.M110.120188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugazzola L, et al. High phenotypic intrafamilial variability in patients with Pendred syndrome and a novel duplication in the SLC26A4 gene: clinical characterization and functional studies of the mutated SLC26A4 protein. Eur. J. Endocrinol. 2007;157:331–8. doi: 10.1530/EJE-07-0263. [DOI] [PubMed] [Google Scholar]
- Tamura T, Sunryd JC, Hebert DN. Sorting things out through endoplasmic reticulum quality control. Mol. Membr. Biol. 2010;27:412–27. doi: 10.3109/09687688.2010.495354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–90. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bross P, et al. Protein misfolding and degradation in genetic diseases. Hum. Mutat. 1999;14:186–98. doi: 10.1002/(SICI)1098-1004(1999)14:3<186::AID-HUMU2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Shepshelovich J, et al. Protein synthesis inhibitors and the chemical chaperone TMAO reverse endoplasmic reticulum perturbation induced by overexpression of the iodide transporter pendrin. J. Cell Sci. 2005;118:1577–86. doi: 10.1242/jcs.02294. [DOI] [PubMed] [Google Scholar]
- Hulander M, et al. Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development. 2003;130:2013–25. doi: 10.1242/dev.00376. [DOI] [PubMed] [Google Scholar]
- Li X, et al. SLC26A4 targeted to the endolymphatic sac rescues hearing and balance in Slc26a4 mutant mice. PLoS. Genet. 2013;9:e1003641. doi: 10.1371/journal.pgen.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangemann P. Mouse models for pendrin-associated loss of cochlear and vestibular function. Cell. Physiol. Biochem. 2013;32:157–65. doi: 10.1159/000356635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Metcalfe RA, Watson PF, Weetman AP, Trembath RC. Mutations of the PDS gene, encoding pendrin, are associated with protein mislocalization and loss of iodide efflux: implications for thyroid dysfunction in Pendred syndrome. J. Clin. Endocrinol. Metab. 2002;87:1778–84. doi: 10.1210/jcem.87.4.8435. [DOI] [PubMed] [Google Scholar]
- Dossena S, et al. Identification of allelic variants of pendrin (SLC26A4) with loss and gain of function. Cell. Physiol. Biochem. 2011;28:467–76. doi: 10.1159/000335108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palos F, et al. Pendred syndrome in two Galician families: insights into clinical phenotypes through cellular, genetic, and molecular studies. J. Clin. Endocrinol. Metab. 2008;93:267–77. doi: 10.1210/jc.2007-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam MP, Bartolone L, Kopp P, Benvenga S. Molecular analysis of the PDS gene in a nonconsanguineous Sicilian family with Pendred’s syndrome. Thyroid. 2005;15:734–41. doi: 10.1089/thy.2005.734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.