Abstract
Asthma is a heterogeneous clinical syndrome that includes subtypes of disease with different underlying causes and disease mechanisms. Asthma is caused by a complex interaction between genes and environmental exposures; early-life exposures in particular play an important role. Asthma is also heritable, and a number of susceptibility variants have been discovered in genome-wide association studies, although the known risk alleles explain only a small proportion of the heritability. In this review, we present evidence supporting the hypothesis that focusing on more specific asthma phenotypes, such as childhood asthma with severe exacerbations, and on relevant exposures that are involved in gene-environment interactions (GEIs), such as rhinovirus infections, will improve detection of asthma genes and our understanding of the underlying mechanisms. We will discuss the challenges of considering GEIs and the advantages of studying responses to asthma-associated exposures in clinical birth cohorts, as well as in cell models of GEIs, to dissect the context-specific nature of genotypic risks, to prioritize variants in genome-wide association studies, and to identify pathways involved in pathogenesis in subgroups of patients. We propose that such approaches, in spite of their many challenges, present great opportunities for better understanding of asthma pathogenesis and heterogeneity and, ultimately, for improving prevention and treatment of disease.
Keywords: Asthma, Rhinovirus, GWAS, Gene-environment interactions, 17q asthma locus, CDHR3
Introduction
Asthma is a common and complex disorder that shows significant heterogeneity with respect to age of onset, clinical course, and response to therapeutics, as well as a unique age-by-sex interaction, with many more boys diagnosed before puberty but increased incidence and prevalence of diagnosis in girls after puberty and throughout adult life.1 This clinical heterogeneity likely reflects the presence of several disease endotypes (or subtypes of disease defined by a distinct functional or pathobiological mechanism).2 Among the many epidemiologic risk factors for asthma, maternal asthma remains among the most significant predictors of childhood-onset asthma.3 In contrast, growing up on a farm in central Europe provides strong protection against the development of asthma.4 Many other early-life exposures, including viral respiratory tract infections, the composition of the infant’s gut5–7 and airway8 microbiome, intrauterine smoke exposure,9 and maternal diet during pregnancy,10–12 are also associated with risk of asthma. These combined data highlight the important role of environmental exposures on the development of asthma, which is consistent with the observed doubling in prevalence of asthma and allergic diseases in westernized countries over the past 50 years,13 and further suggest that individual risk trajectories are set early and that the critical window of susceptibility might begin in utero and extend to no more than the first few years of life.
In addition to the many environmental exposures implicated in asthma risk,14 twin and family studies suggest that genetic variation also plays an important role. For example, a recent meta-analysis of 71 twin studies comprising 36,903 twin pairs estimated the heritability of asthma to be 0.54 (SE, 0.048).15 Heritability estimates are also higher among boys compared with girls15 and in those with early-onset compared with later-onset asthma.16 Interestingly, despite the dramatic increase in asthma prevalence attributed to environmental factors, increases in heritability estimates over time have also been reported.17 Such increases could be due to changes in the prevalence of environmental factors that increase the penetrance of asthma susceptibility genotypes and manifest their effects through gene-environment interactions (GEIs).
Consistent with estimates of heritability, genome-wide association studies (GWASs) have identified 15 to 20 asthma risk loci that meet stringent thresholds of statistical significance (typically P < 10−8) and replicate across several studies. These loci include genes previously implicated in asthma because of their role in immune and allergy (TH2) pathways (eg, HLA, IL13, IL33, thymic stromal lymphopoietin [TSLP], and IL-1 receptor–like 1 [IL1RL1], which encodes ST2, the receptor for IL-33), transcription factors in many of the known immune pathways associated with asthma (eg, RAR-related orphan receptor A [RORA], SMAD family member 3 [SMAD3], and GATA3), and novel loci (eg, cadherin-related family member 3 [CDHR3] and the 17q12-21 locus encoding ORM1-like 3 [ORMDL3] and gasdermin B [GSDMB], among other genes). Yet despite the many successes of GWASs, the contributions of individual variants to asthma risk are quite small (odds ratios [ORs] of around <1.2), and the combined effect of these loci account for very little of the overall genetic risk for asthma.18 This so-called “missing heritability” has been observed for most complex diseases, and many explanations have been put forth.19,20 Although rare variants, which are typically not well interrogated in GWASs, can influence responses to albuterol21 or long-acting b-agonists,22 a large genome-wide study of low-frequency and rare exonic variants in asthma concluded that variants in this frequency range are unlikely to account for a significant proportion of asthma heritability.23 Rather, we suggest that the discovery of additional risk alleles or those with larger effect sizes has been particularly limited both by the extreme clinical heterogeneity of asthma that is rarely accounted for in the large samples required for GWASs and by our inability to incorporate into GWASs asthma-associated environmental exposures, especially those that occur during critical early-life developmental windows.
In this review we present evidence supporting the hypothesis that focusing on more homogeneous subgroups of asthmatic patients and on relevant exposures involved in GEIs will lead to both the discovery of novel loci and a better understanding of the mechanisms underlying previously discovered asthma genes. In addition, we will discuss the challenges of GEI studies and the advantages of cell models of GEIs for identifying asthma risk loci and the insights that those studies provide into asthma pathogenesis in subgroups of patients. Although we acknowledge the critical role that animal models play in dissecting mechanistic pathways of asthma pathogenesis, a thorough assessment of those models and approaches to asthma gene discovery is beyond the scope of this review, which focuses primarily on human studies.
Challenges of GEI Studies
There are 2 inherent challenges in studies of GEIs. First, unbiased genome-wide studies of interactions, referred to as genome-wide interaction studies (GWISs), require very large sample sizes of at least several thousand cases and control subjects, assuming the most favorable models.24 Whereas GWASs test for association between genotype at each single nucleotide polymorphism (SNP) across the genome with asthma (or other outcome variable), GWISs test for interaction effects between each SNP and a specific exposure on asthma risk (or other outcome variable). Thus, GWISs require even larger sample sizes than GWASs to achieve equivalent power. Unfortunately, most of the large asthma samples assembled for GWASs have limited, if any, information on relevant environmental exposures, with a few exceptions, as discussed below.
Second, exposures are often highly correlated, making it difficult to disentangle the effects of each. This is true for both lifestyle and demographic factors, such as cigarette smoking, activity levels, socioeconomic status and educational levels, and a host of other risk factors for many common diseases, as well as for more general environments, such as growing up on a farm, which captures a complex set of exposures that differ between farm and nonfarm children.
The few GWISs of asthma that have been performed have been generally disappointing, despite sample sizes of more than 1700 children in a study of farm exposures,25 more than 2500 male and more than 2500 female subjects in a study of sex-specific effects,26 and more than 6000 children in a study of in utero and early-life tobacco smoke exposure.27 None of the discovered associations in these 3 relatively large studies met genome-wide criteria for statistical significance, although some interesting candidates were identified and some were replicated within each study. Notably, though, none of variants associated with asthma in GWASs were among the SNPs identified in GWISs, although most SNPs involved in GEIs would not be expected to be significant at the stringent thresholds used in a GWAS because the genotype effects would only be present in the subset of exposed subjects.
However, these studies illustrate general principles that can inform future GEI studies. For example, the most significant association in a sex GWIS in European American subjects in the EVE Consortium was a SNP (rs2549003) near the interferon regulatory factor 1 (IRF1) gene, which was associated with asthma in male (OR, 1.3; 95% CI, 1.18–1.49) but not female (OR, 0.92; 95% CI, 0.82–1.02) subjects.26 A SNP in perfect linkage disequilibrium with this IRF1 SNP was associated with asthma in the large independent GABRIEL GWAS at a P value of .0086, which did not meet genome-wide levels of significance in the GWAS, consistent with a variant that is associated with asthma in only one sex. In fact, it is likely that many SNPs involved in GEIs will show small but not genome-wide significant P values in large GWASs and meta-analyses of GWASs of asthma, and these “midhanging fruit”28 might serve as excellent candidates for future studies of GEIs. Finally, the IRF1 SNP that was associated with asthma in male subjects only or SNPs in strong linkage disequilibrium with the male-associated asthma SNP were reported in previous studies to be cis expression quantitative trait loci (eQTLs) for the IRF1 gene in lymphoblastoid cell lines and sputum and for the nearby genes (RAD50; solute carrier family 22, member 4 [SLC22A4]; and solute carrier family 22, member 5 [SLC22A5]) in lung tissue and lymphoblastoid cell lines (RAD50) or monocytes (SLC22A4 and SLC22A5, see Myers et al26 and the citations therein). These data further suggest that SNPs involved in GEIs are likely to be regulatory in nature, as has been shown in model organisms,29 and that the same SNP can be an eQTL for different genes in different tissues.
Candidate GEI studies in Asthma
An alternative approach that circumvents the power issues related to genome-wide studies is focusing on candidate genes. Candidate GEI studies have included genes with known roles in immune response to relevant environmental exposures, as well as genes that were identified in GWASs of asthma. Although this approach is limited by our current knowledge and will not detect new genes, it can be quite useful in helping us understand some of the complexities of asthma, defining more homogeneous subgroups of asthmatic subjects, and revealing underlying mechanisms for associations with asthma. As examples of candidate GEIs, we focus the discussion below on a major epidemiologic risk factor for asthma: exposures to microbes (respiratory viruses and bacteria).30–32
The 17q asthma locus and viral wheezing illness in childhood
Wheezing illness with viral respiratory tract infections in infancy and early childhood is a significant risk factor for the eventual diagnosis of asthma by school age,33 and in many children asthma debuts in the first few years of life in the form of respiratory virus– associated wheezing illness.34 For example, moderate-to-severe wheezing illness with respiratory syncytial virus (RSV) or rhinovirus in the first 3 years was associated with risk for asthma by age 6 years, with ORs of 2.6 (95% CI, 1.0–6.3) and 9.8 (95% CI, 4.3–22.0), respectively, in the Childhood Origins of Asthma (COAST) birth cohort of high-risk children.33 In COAST children a SNP at the most significant and highly replicated asthma locus on chromosome 17q12-21 (rs7216389) was associated with asthma by age 6 years, with an OR of 2.3 (95% CI, 1.0–5.2).35 Earlier studies showed that variation at the 17q locus was associated primarily with early onset asthma18,36,37 and that the risk was increased among children exposed to cigarette smoke38,39 and with viral wheezing illness40 in early life. Our groups asked specifically whether the genotype at this locus was associated with RSV- or rhinovirus-associated wheezing illness and whether there was an interaction effect between viral wheezing illness in the first 3 years and genotype on risk for childhood-onset asthma.35
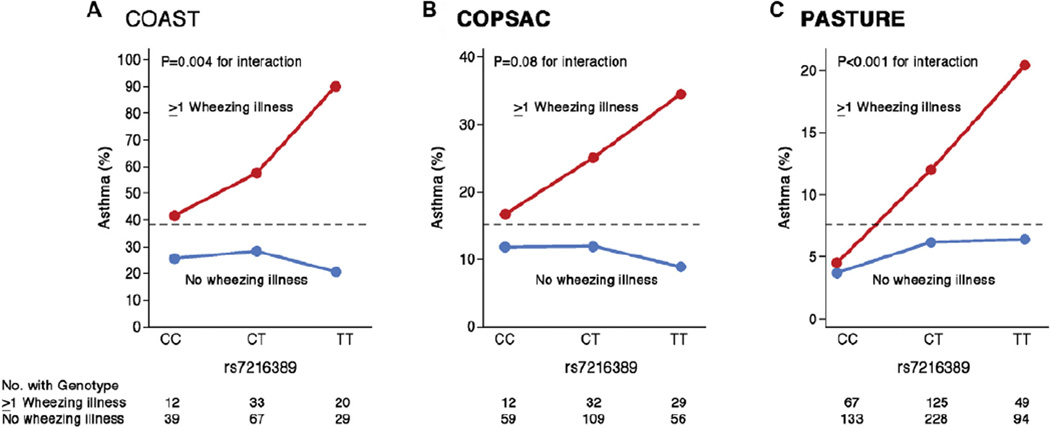
Surprisingly, in the COAST children the 17q genotype at rs7216389 was associated with both the occurrence and number of rhinovirus-associated wheezing illnesses (P =.01 and P <.001, respectively) but not with RSV-associated wheezing illnesses (P =.22 and P =.54, respectively), suggesting that the genotype at this locus was specifically involved in response to rhinovirus or non-RSV viral infection. Additionally, significant interaction effects between rhinovirus-associated wheezing illness and rs7216389 genotype on asthma risk were observed whereby the genotype-specific risk for asthma was present only in the children who experienced a rhinovirus-associated wheezing illness; there was no association with this genotype in the children who did not wheeze with rhinovirus infection (Fig 1, A).35 The OR for asthma among COAST children who wheezed with rhinovirus in early life and had the TT genotype was 26.1 (95% CI, 5.1- 133.0) compared with that for children with neither, and the same pattern of interaction was present in another high-risk birth cohort, the Copenhagen Prospective Studies on Asthma in Childhood–2000 cohort (COPSAC2000; Fig 1, B).
Figure 1. Interaction effects of the 17q genotype and wheezing on asthma risk in 3 birth cohorts.
In all cohorts there is more asthma among children who wheezed in early life (red lines) compared with children who did not wheeze in early life (blue lines), and the associations with 17q genotype are only evident among the children who wheezed. In all cohorts the prevalence of asthma at age 6 years is more than 3-fold higher among TT children who wheezed compared with children who did not wheeze. The dashed line shows the overall prevalence of asthma in each population. Note the different y-axis scales in each panel. A and B, Stratified by rhinovirus-associated wheezing illness in the first 3 years of life. Modified from Caliskan et al.35 C, Stratified by wheezing illness in the first year of life. Modified from Loss et al.41
This finding is remarkable in several respects. First, despite a highly significant main effect on asthma risk in nearly every GWAS of asthma, the association with the 17q locus was present only in children with rhinovirus-associated wheezing illness in early life in 2 independent birth cohorts. This seemingly paradoxical finding can be explained by the ubiquitous nature of rhinovirus infection (ie, all children are exposed) and the fact that the associated alleles at this locus have frequencies of between 30% and 50% in nearly all populations. Thus this is likely an exceptional example of an interaction in which both the exposure and associated allele are very common in the population. It is still unclear whether this interaction reflects a causal role of rhinovirus in asthma development so that prevention of rhinovirus infections in early life would reduce the risk of asthma in children with the 17q risk variants or whether early rhinovirus infections only unmask an underlying asthma propensity with no direct effect on asthma risk. Future studies will also need to address whether this interaction is specific for rhinovirus compared with other, non-RSV respiratory tract viruses because the association between early respiratory tract infections and later asthma might be a general phenomenon for many viruses.42 Nevertheless, this study demonstrates the dramatic effect conditioning on a known risk factor (rhinovirus-associated wheezing illness) can have on genotype-specific effect sizes and illustrates how GEI studies in birth cohorts with longitudinal and extensive assessments of environmental exposures can shed light on pathogenic mechanisms of asthma genes. The children included in this study were ascertained based on having a parent with asthma or allergy (COAST) or a mother with asthma (COPSAC), and therefore it could not be determined whether this interaction is also present in other unselected populations. Indeed, a recent GEI study from the Protection against Allergy Study in Rural Environments (PASTURE) cohort reported similar interaction effects between SNPs at the 17q21 locus and early-life wheezing illness on subsequent development of asthma.41 As in the COAST and COPSAC cohorts, the genotype-specific risks for asthma were present only among the children who wheezed in early life (Fig 1, C).41 Moreover, they reported additional interactions with both risk (presence of older siblings) and protective (exposure to animal sheds) environmental exposures. In particular, they found that the presence of older siblings increased the risk of wheeze and that exposure to animal sheds decreased the risk of wheeze in the first year of life only in children carrying the risk allele (A) at the SNP rs8076131, which is in strong linkage disequilibrium with rs7216389 (P value for genotype-by-exposure interactions with older siblings and animal sheds = .003 and .002, respectively).41 Although specific viral infections were not assessed directly in the study by Loss et al,41 the presence of older siblings is generally thought to increase the risk of early wheeze through increased exposure to viral infections. Overall, this study extends our previous findings to a population-based (unselected) cohort of children and potentially to viruses other than rhinovirus. Importantly, the study by Loss et al now implicates an additional level of interaction with exposures related to animal sheds that harbor high levels of bacteria and fungi and confer protection against the development of asthma only in children with the high-risk (asthma-associated) genotype at the 17q21 locus.
The CDHR3 locus and RV-C infection
A recent GWAS focused on a more specific asthma phenotype characterized by recurrent episodes of severe exacerbations requiring hospitalization between 2 and 6 years of age.43 This study addressed the hypothesis that children with a more severe phenotype represent specific endotypes of asthma and therefore more homogeneous underlying disease mechanisms. As a result, GWASs in these children should provide stronger evidence for genetic association and potentially identify novel loci that confer risk for this more severe phenotype. Indeed, despite the relatively small sample size (1173 cases and 2522 control subjects) compared with other asthma GWASs, this GWAS of recurrent asthma exacerbations in childhood yielded association P values similar to those of previous GWASs in much larger samples18,44 and with considerably larger effects sizes (Table I). This was particularly evident for the children with the largest number of exacerbations (Table I, last column).18,43,44
Table 1.
Association results from 3 large GWAS of asthma: EVE44, GABRIEL18 and COPSACExacerbation43. Of these 3 cohorts, COPSACExacerbation studied a specific phenotype of childhood asthma with severe recurrent exacerbations, whereas the EVE and GABRIEL studies included more heterogeneous asthma populations. Effect sizes (ORs) were consistently larger in the COPSACExacerbation study, and particularly for children with the largest number of exacerbations (last 2 columns in the table). The genome-wide significant loci from each study are shown with mutual replication in the other studies. Replication results were obtained by looking up the associated SNP or one with LD r2 > 0.8 with the associated SNP, either from actual genotyped or imputed data.
| COPSACExacerbation€ |
||||||
|---|---|---|---|---|---|---|
| No. of cases No. of control subjects |
EVE* 5,449 3,385 |
Gabriel§ 10,365 16,110 |
Total 1,173 2,522 |
6 or more exaerbations 358 2,522 |
||
| Locus | Gene | SNP/effect allele | OR (P-value) |
OR (P-value) |
OR (P-value) |
OR (P-value) |
| Top loci in COPSACExacerbation€ | ||||||
| 17q12 | GSDMB | rs2305480/G |
1.26 1.3E-13 |
1.18 9.6E-08 |
2.28 1.3E-48 |
2.72 3.5E-27 |
| 9p24.1 | IL33 | rs928413/G | NA |
1.20 9.2E-10 |
1.50 4.2E-13 |
1.91 6.2E-14 |
| 5q31.1 | RAD50 | rs6871536/C |
1.11 0.06 |
1.14 1.9E-08 |
1.44 1.8E-09 |
1.58 1.3E-06 |
| 2q12.1 | IL1RL1 | rs1558641/G |
1.24 1.2E-04 |
1.12 3.6E-05 |
1.56 6.6E-09 |
2.19 3.2E-08 |
| 7q22.3 | CDHR3 | rs6967330/A | NA |
1.12 5.9E-04 |
1.45 1.4E-08 |
1.63 1.6E-06 |
| Top loci in GABRIEL§ | ||||||
| 6p21.3 |
HLA DQB1 |
rs9273349/C | NA |
1.18 7.0E-14 |
NA | NA |
| 2q12.1 | IL18R1 | rs3771166/G |
1.09 1.3E-06 |
1.15 3.5E-12 |
1.22 2.8E-04 |
1.32 6.3E-04 |
| 9p24.1 | IL33 | rs1342326 /C |
1.19 4.0E-04 |
1.20 8.7E-12 |
1.53 1.1E-10 |
1.77 2.1E-09 |
| 15q22.33 | SMAD3 | rs744910 /G |
1.11 3.7E-03 |
1.12 3.9E-09 |
1.18 0.002 |
1.32 9.5E-04 |
| 22q12.3 | IL2RB | rs2284033 /G |
1.05 0.06 |
1.12 1.2E-08 |
NA | NA |
| 17q12 | GSDMB | rs2305480/G |
1.26 1.3E-13 |
1.18 9.6E-08 |
2.28 1.3E-48 |
2.72 3.5E-27 |
| 17q21.1 | GSDMA | rs3894194/A |
1.10 1.9E-07 |
1.17 4.6E-09 |
1.59 2.6E-21 |
1.82 3.5E-15 |
| Top loci in EVE* | ||||||
| 17q12 | GSDMB | rs11078927/T |
1.27 1.2E-14 |
1.18 9.6E-08 |
2.35 7.6E-57 |
2.90 1.6E-35 |
| 5q22.1 | TSLP | rs1837253/C |
1.19 7.3E-10 |
1.15 3.3E-05 |
1.22 8.8E-04 |
1.33 0.002 |
| 2q12.1 | IL1RL1 | rs10173081/ |
1.20 1.4E-08 |
1.22 1.0E-10 |
1.54 1.8E-07 |
2.16 4.8E-08 |
| 9p24.1 | IL33 | rs2381416/A |
1.18 2.5E-07 |
1.19 1.1E-07 |
1.50 1.1E-13 |
1.84 1.3E-13 |
Associations that met the criteria for genome-wide significance were mainly known asthma loci, with the17q locus as the most significant, but also included one novel susceptibility locus at the CDHR3 gene. CDHR3 is a transmembrane protein and is highly expressed in the human lung and airway epithelium.43,45,46 Its cellular mechanism is unknown, but it belongs to the cadherin family of proteins, which are involved in homologous cell adhesion and several cellular processes, including epithelial polarity and differentiation.47 The most associated variant at this locus was a nonsynonymous SNP (rs6967330) that results in an amino acid change in an interdomain area of the protein, where it could potentially affect protein structure through interference with a disulfide bridge. In support of this, the investigators showed that experimental introduction of the risk variant by means of mutagenesis was associated with increased surface expression of CDHR3.
More recently, it was reported that CDHR3 functions as a receptor for rhinovirus C (RV-C).48 CDHR3 was differentially expressed in epithelial cells susceptible to RV-C infection compared with unsusceptible cells, and its expression on epithelial cells enabled RV-C binding and replication. Importantly, introduction of the risk variant at rs6967330 by means of transfection resulted in 10-fold increased RV-C binding and progeny yield compared with the nonrisk variant. These combined data provide strong evidence that CDHR3 is an RV-C receptor and that the asthma association signal with a SNP in the CDHR3 gene might result from increased susceptibility to RV-C infections. This finding is in line with the exacerbation-related phenotype from the discovery GWAS because RV-C has been reported to be the most common viral trigger of severe asthma exacerbations in children49,50,51 and associated with both more severe disease50 and higher rates of hospital readmissions compared with other viral respiratory tract infections.49 If correct, this would indicate that the CDHR3 variant confers risk to severe childhood asthma through an interaction with RV-C infection, and stronger effect sizes for this risk variant would be expected for RV-C-associated respiratory illness compared with illnesses triggered by other viruses, a hypothesis that is currently being tested. It is notable that the CDHR3 locus was not identified among genome-wide significant SNPs in any GWAS that included all asthmatic patients or even those including only patients with childhood-onset asthma. Only by focusing on children with the extreme phenotype of severe asthma did the evidence for association at this locus rise to genome-wide levels of significance (Table I), demonstrating that performing GWASs on more specific phenotypes can both increase power to detect novel asthma loci and yield greater effect sizes for known variants. Finally, these studies highlight the importance of combining GWAS results with experimental studies on environmental exposures to gain insight into the causal mechanisms of asthma.
Microbial exposures and innate immune genes
The general reduction of exposure to microbes in “westernized societies” is a likely contributor to the increasing prevalence of asthma, allergy, and potentially other immune-mediated diseases.52 In particular, exposure to a farming environment remains one of the strongest protective factors against the development of asthma and allergy, particularly if exposure takes places in utero or in the first year of life.4 The causal factors behind this association are not fully understood but are thought to reflect exposures to microbial products, possibly those related to the ingestion of raw milk and inhalation of microbial components in dust, such as endotoxin (or LPS, a component of the cell wall in gram-negative bacteria). In fact, endotoxin levels in house dust have also been inversely associated with asthma and allergy prevalence in nonfarming environments.53,54
Endotoxin signals through a complex receptor that includes CD14. The first and still most replicated GEI in patients with asthma or allergic disease involves a polymorphism in the promoter of the CD14 gene (rs2569190; 2159/C/T). This SNP was associated with serum levels of CD14,55 suggesting that it regulates expression of the CD14 gene. Studies of interaction effects between this SNP and endotoxin levels on risk for asthma, atopy (specific IgE or skin test), and eczema have yielded remarkably consistent findings in which the T allele is associated with risk for disease only in children with high levels of exposure, but the C allele is associated with disease risk in children with low levels of exposure.54,56–58 SNPs involved in this “flip-flop” type of GEI, in which alleles have opposite effects on risk in different environments, might not be common but would nevertheless be impossible to discover in a GWAS that includes subjects with the full exposure spectrum. Such cryptic confounding would mask this association regardless of sample size or frequency of the associated allele. Moreover, these studies demonstrate the complexity of GEIs in patients with asthma and allergic disease. Even for this single SNP in a gene with a well-defined immune function, GEI effects vary based not just on exposure levels but also on age at exposure, type of microbial exposure,57 and the specific disease phenotype (eg, atopic asthma versus nonatopic wheeze).53,54 Exploring such complex interactions requires extensive clinical studies with longitudinal information on both environmental exposures and disease phenotypes in early life.
In the recent study in the PASTURE cohort described above,41 the authors showed that children carrying the risk allele at the 17q21 locus were both more susceptible to exposure to older siblings and more responsive to the protective effect of animal shed exposure on wheeze in the first year of life. This unexpected interaction between the 17q21 locus and animal shed exposures (likely to bacteria and fungi) demonstrates how carefully designed longitudinal studies of environmental exposures and clinical phenotypes can elucidate novel disease mechanisms and GEIs. In fact, because this interaction was not found in the previous GWIS performed in the same population but using later-onset asthma as the outcome,25 the authors suggest that the interaction with the 17q21 locus highlights the importance of GEI studies focused on early-life events.
Other interactions between microbial/farming exposures and innate immune genes have been reported. These include one SNP in the Toll-like receptor 2 gene (TLR2/ −16934) that was associated with asthma, allergic sensitization, and hay fever in farmers’ children but not in nonfarmers’ children59 and one SNP in the intracellular pattern recognition receptor caspase recruitment domain protein 4 gene (CARD4/ −21596).60 For both of these examples, associations were observed only in children raised on a farm. Overall, these studies highlight the importance of interactions between genetic variation in innate immune genes and early-life exposures to microbes on subsequent risk for asthma and allergic disease, although no studies to date have identified the specific microbes or microbial communities associated with genotype-specific risk or protection in subjects.
A recent study investigated the potentially protective effect of endotoxin and farm dust exposure in a mouse model of house dust mite–sensitized asthma to better understand the mechanism for the farming effect.61 They reported that exposure to endotoxin protected against many of the clinical features of asthma, including house dust mite sensitization, airway eosinophilia, and bronchial hyperreactivity. Moreover, airway epithelial cells from exposed mice produced fewer cytokines that specifically recruit dendritic cells to the lungs. Because of the known role that the A20 protein plays in innate immune response to house dust mites,62 the authors tested the mediating role of this protein by knocking out the gene encoding A20, called TNF-a–induced protein 3 (Tnfaip3), in lung epithelial cells. They found that A20 was an important mediator of the protective effects of endotoxin exposure and validated this finding in human bronchial epithelial cells by showing reduced expression of A20 in cells from asthmatic patients compared with cells from nonasthmatic subjects. Furthermore, a potential modifying effect of A20 was investigated by a “look-up” of 2 SNPs located in or near the human TNF-a–induced protein 3 (TNFAIP3) gene by using data from the earlier GWIS of farming effects on asthma in the PASTURE cohort.25 One of these SNPs (a nonsynonymous variant previously associated with autoimmune disease63) was modestly associated with asthma in this sample (P = .03) and modified the protective farming effect, as evidenced by a stronger genotype-specific effect in children with the nonrisk allele compared with children without the risk allele (OR, 0.14 vs 0.73; P = .03 for interaction). This potential GEI needs to be replicated in additional studies but represents an exciting discovery on a potential mechanism for the protective effect of exposure to microbes. Moreover, this study exemplifies how mechanistic studies targeting specific environmental exposures and the use of mouse models can facilitate identification of genes involved in GEIs that were not identified by using genome-wide approaches (eg, the GWIS in this same population25).
Another important component of microbial exposure is the nature of the commensal microbiomes that populate our bodies, in particular the gut and airways. These communities are potentially modifiable internal environments with putatively important effects on early immune development, allergy, and asthma. For example, the gut microbiome composition in early life has been associated with development of allergic sensitization and asthma,5–7 and the early airway microbiome might be associated with the development of asthma.8 Few GEI studies have been conducted considering the microbiome as a measure of exposure, but in one study an interaction effect between colonization of a specific bacteria, Escherichia coli, and a SNP in the Toll-like receptor 4 (TLR4) gene (rs10759932) on allergic sensitization was reported.64 In this study E coli colonization was associated with protection from allergic sensitization among infants with the TT genotype (OR, 0.31; 95% CI, 0.14–0.68) but not in infants with the C allele (interaction P = .001). Current studies using high-throughput sequencing techniques capture a larger proportion of the microbial diversity and are poised to address the complex relationship between host genotype, the gut and airway microbiome communities, and asthma development. This challenging task will need cross-disciplinary expertise and resources, including development of bioinformatics tools that can handle multiple high-dimensional data sets, approaches for experimental validation, and populations with early-life microbiome and clinical assessments, GWASs, and potentially other omics data. The integration of the microbiome with host genetics is perhaps one of the most formidable yet exciting challenges in the field today.
Moving Forward in the Post-GWAS Era
Two significant challenges in the post-GWAS era are assigning function to variants that are associated with asthma or allergic disease at genome-wide levels of significance and identifying the true associations from among the thousands of SNPs with small P values (eg, P < 10−3) that do not reach genome-wide levels of significance (P < 10−8; ie, the “midhanging fruit”28). Based on data from more than a thousand GWASs of common diseases,65 it is now clear that more than 90% of significant GWAS SNPs reside in noncoding (introns and intergenic) regions of the genome66 and are enriched for variants that are associated with gene expression (ie, eQTLs)67–71 and those that reside in regions involved in gene regulation.72,73 Moreover, gene regulatory architectures differ between tissues, and the enrichments for regulatory variants among GWAS SNPs are more pronounced in disease-relevant tissues.73,74
Finally, assigning an associated SNP to the gene it regulates is not straightforward because assuming it is the nearest gene will be correct only about 65% of the time.74 Thus focusing on SNPs with known regulatory functions in asthma-relevant tissues and cell types, in lieu of the millions of SNPs included in GWASs, would both increase power to detect associations and provide immediate intuition about function and the gene or genes regulated by the associated SNP in relevant cell types. For example, Hao et al75 identified eQTLs in whole lung tissue and then integrated those results withGWAS results from the GABRIEL study18 to identify potentially causal variants at the 17q21 locus, as well as molecular pathways that might underlie asthma risk. Moreover, studying responses to exposures associated with asthma or allergy risk in cell models of GEIs will facilitate the dissection of the contextspecific nature of genotypic risks, the prioritization of variants with small P values in GWASs that do not meet genome-wide levels of significance, and the identification of pathways involved in pathogenesis in subgroups of patients. In the following sections we provide 2 examples that support these contentions and discuss the lessons learned from those studies.
Transcriptional Response of Peripheral Blood Mononuclear Cells (PBMCs) to RV
Because of the important role that rhinovirus plays in both the early inception of asthma and exacerbations of symptoms throughout life,76–78 identifying the genetic variation that influences host response to rhinovirus could identify variants that contribute to both risk for developing asthma and asthma severity. A recent study by Calışkan et al79 addressed this by studying gene expression in PBMCs from 98 subjects before and after rhinovirus infection in vitro. In this model more than half of the nearly 11,000 genes detected as expressed in PBMCs were upregulated (n = 2242) or downregulated (n = 3779) with rhinovirus exposure.
Most of the genetic variation associated with gene expression (eQTLs) was maintained between uninfected and rhinovirusinfected PBMCs, in which 521 and 524 eQTLs, respectively, were detected. However, a small set of SNPs were eQTLs in only one condition (n = 25) or had different genotype-specific effect sizes on gene expression between conditions (n = 13). These 38 SNPs were referred to as response expression quantitative trait loci (reQTLs) and included genes with important known roles in viral function, such as 29,59-oligoadenylate synthetase 1 (OAS1) and interferon regulatory factor 5 (IRF5). Publicly available data on protein-binding sites revealed a 16-fold enrichment of signal transducer and activator of transcription 2 (STAT2)–binding sites near the 38 reQTLs compared with non-reQTLs (P < 10−6). The STAT2 gene itself was 3.7-fold upregulated in rhinovirus-treated cells (P = 1.8 × 10−76), but the eQTL for this gene did not show statistical evidence for an interaction (ie, being an reQTL). Finally, the 38 reQTLs showed a modest enrichment for SNPs associated with asthma in the Genetic Association Database,80 although this difference did not reach statistical significance (7.89% of genes with reQTLs had been associated with asthma compared with 3.76% of all genes detected as expressed, P =.17). However, the studies in the Genetic Association Database are quite heterogeneous with respect to sample composition and evidence for association.
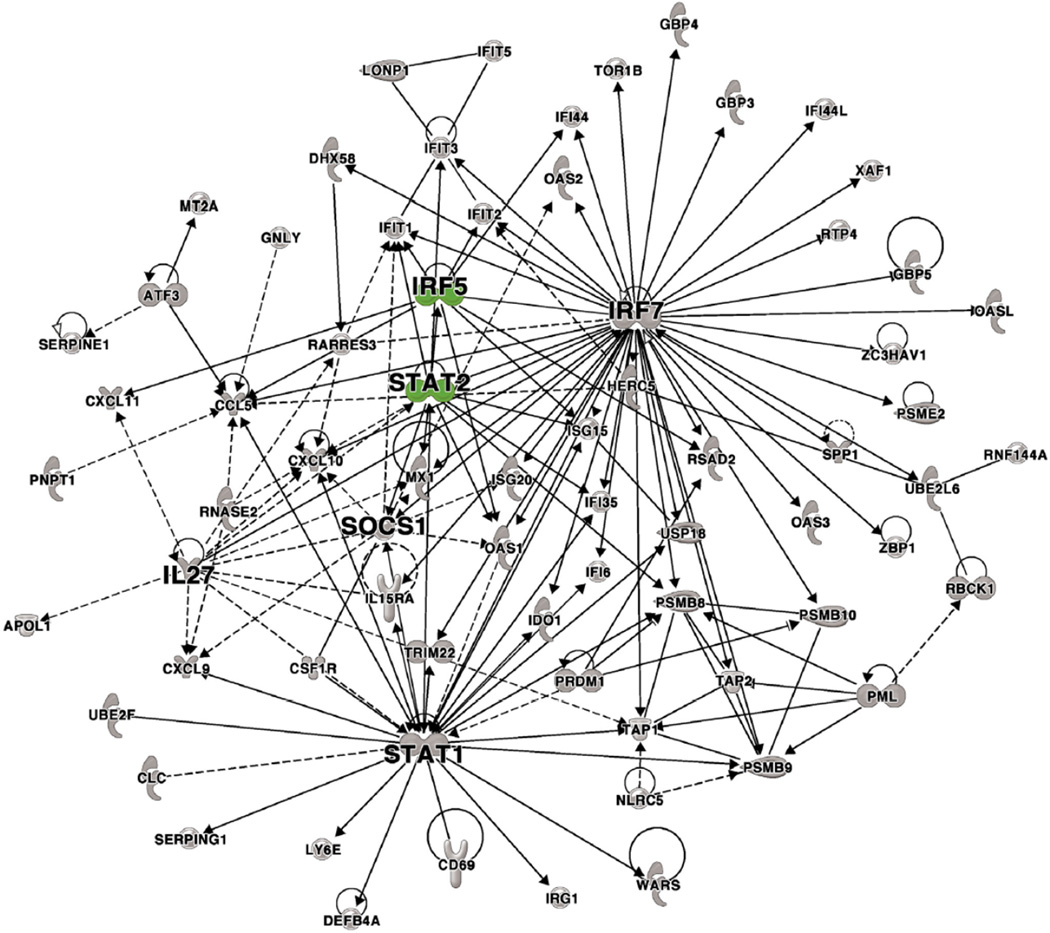
We reasoned that rhinovirus-responsive genes or those with reQTLs might be more associated with asthma exacerbations than with an asthma diagnosis per se. To explore this possibility, we compared the list of rhinovirus-responsive genes in rhinovirustreated cells reported in the study by Calışkan et al79 described above with genes that were differentially expressed in nasal lavage samples collected from children during an acute asthma exacerbation and then again 7 to 14 days later, as reported by Bosco et al.81 We focused on one network from their weighted gene coexpression network analysis that included STAT2 as a hub (Fig 2)79,81 because of the enrichment for STAT2-binding sites near reQTLs. Among the 71 genes in this network, 61 were detected as expressed in the study by Calışkan et al.79 Ninety-five percent (n = 58) of those genes showed significant expression changes in response to rhinovirus compared with 55.3% of all genes tested (P =.0038, Fisher exact test). Overall, the overlapping genes had a median 3.2-fold change in expression response to rhinovirus (median P = 3.4 × 10−58). Moreover, among the 61 genes in this network, 2 (3.3%), IRF5 and OAS1, had reQTLs in the study by Calışkan et al79 compared with 0.035% of all genes tested (P = .026, Fisher exact test). These data suggest that a significant proportion of the transcriptional response in the airways of children with asthma during an exacerbation can be attributed to rhinovirus or other viruses, which is consistent with epidemiologic data, and further suggest that genetic variation between subjects might contribute to a portion of these responses.
Figure 2. Coexpression network analysis of genes differentially expressed in nasal lavage fluid in children during an asthma exacerbation.
This network includes 58 genes with eQTLs in rhinovirus-treated PBMCs,79 including STAT2 and IRF5 as hubs (shown as green symbols), supporting the hypothesis that genetic variation in response to rhinovirus might modulate response to this virus and to asthma severity more generally. Molecule shapes: oval = transcriptional regulator, diamond = enzyme, dashed line rectangle = channel, up triangle = phosphatase, down triangle = kinase, trapezoid = transporter, circle = other. Modified from Bosco et al.81
The combined results from the studies by Calışkan et al79 and Bosco et al81 demonstrate that cell models of GEIs capture biologically relevant gene regulatory networks and can identify genetic variants that modulate response to an important asthma-associated exposure. Further studies are needed to determine whether the SNPs that are eQTLs or reQTLs in this model are themselves associated with exacerbations or asthma severity. Moreover, because the rhinovirus eQTL study was performed in pooled cells and not isolated peripheral blood immune cells, it is likely that both the number and effect sizes of eQTLs and reQTLs were underestimated.
Epigenetic Responses of Airway Epithelial Cells to IL-13
IL-13 is a TH2 cytokine that plays a central role in asthma pathogenesis by mediating the effects of many asthma-associated exposures, such as allergens, pollution, and viruses, on airway inflammation and remodeling. Although the transcriptional and downstream responses of airway epithelial cells to IL-13 have been well characterized,82,83 little is known about the gene regulatory mechanisms through which IL-13 mediates its effects. A recent study addressed this by focusing on an epigenetic mark of gene regulation, DNA methylation.84
Epigenetics refers to modifications of DNA molecules that do not alter the sequence itself but rather alter the gene regulatory landscape and ultimately the expression of nearby genes. Importantly, DNA methylation levels change in response to many environmental exposures, such as cigarette smoke and pollution,85–87 and likely mediate many downstream effects of these exposures. Finally, DNA methylation levels at many CpG sites vary between subjects, as a result of both individual exposure histories and local genetic variation.88–94 Therefore epigenetic modifications, including DNA methylation, likely play a central role in mediating genotype-specific effects of environmental exposures (ie, GEIs).
In their study, Nicodemus-Johnson et al84 asked 3 central questions: Does exposure to IL-13 alter DNA methylation patterns in cultured airway epithelial cells? Do these changes alter specific genes or pathways? Are IL-13–mediated changes in airway epithelial cells mirrored in freshly isolated cells from asthmatic and nonasthmatic subjects? To answer these questions, the investigators cultured freshly isolated airway epithelial cells from 57 donors in duplicate; 1 culture from each donor was treated with IL-13, and 1 was treated with vehicle only (5% FBS). After just 24 hours in culture, more than 6000 CpG sites (2% of all interrogated sites) were differentially methylated between IL-13–treated and IL-13–untreated cells, and 21% of 8000 differentially expressed genes were near a differentially methylated CpG, suggesting that a significant proportion of the IL-13 transcriptional response is mediated by methylation changes at nearby CpG sites. Further support of this was the observation that the differentially expressed genes and differentially methylated CpGs at these 1590 CpG-gene pairs were overall more correlated compared with all other CpG-gene pairs. Moreover, the genes in the differentially methylated and differentially expressed CpG-gene pairs were enriched in many proinflammatory and profibrotic networks and for genes that were previously associated with asthma, further suggesting the relevance of these epigenetic changes to asthma. These studies answered the first 2 questions in the affirmative: a single 24-hour exposure to IL-13 significantly altered DNA methylation levels in airway epithelial cells, and these changes were associated with both specific genes and pathways previously implicated in asthma pathogenesis.
Studies were performed in freshly isolated airway epithelial cells from 59 asthmatic and 27 nonasthmatic subjects to address the third question and to validate their results ex vivo. Remarkably, more than a third of the IL-13–responsive CpGs in the cell-culture studies were also differentially methylated in airway cells from asthmatic and nonasthmatic subjects, 74% of which showed the same direction of effect: CpGs that became more methylated after IL-13 treatment were more methylated in the asthmatic subjects, and CpGs that became less methylated after IL-13 treatment were less methylated in the asthmatic subjects. This overlap between the 2 studies was significantly higher than expected by chance (P < 10−5), validating the biological relevance of the cell culture of the model and demonstrating the presence of the IL-13–mediated methylation signature in asthmatic subjects.
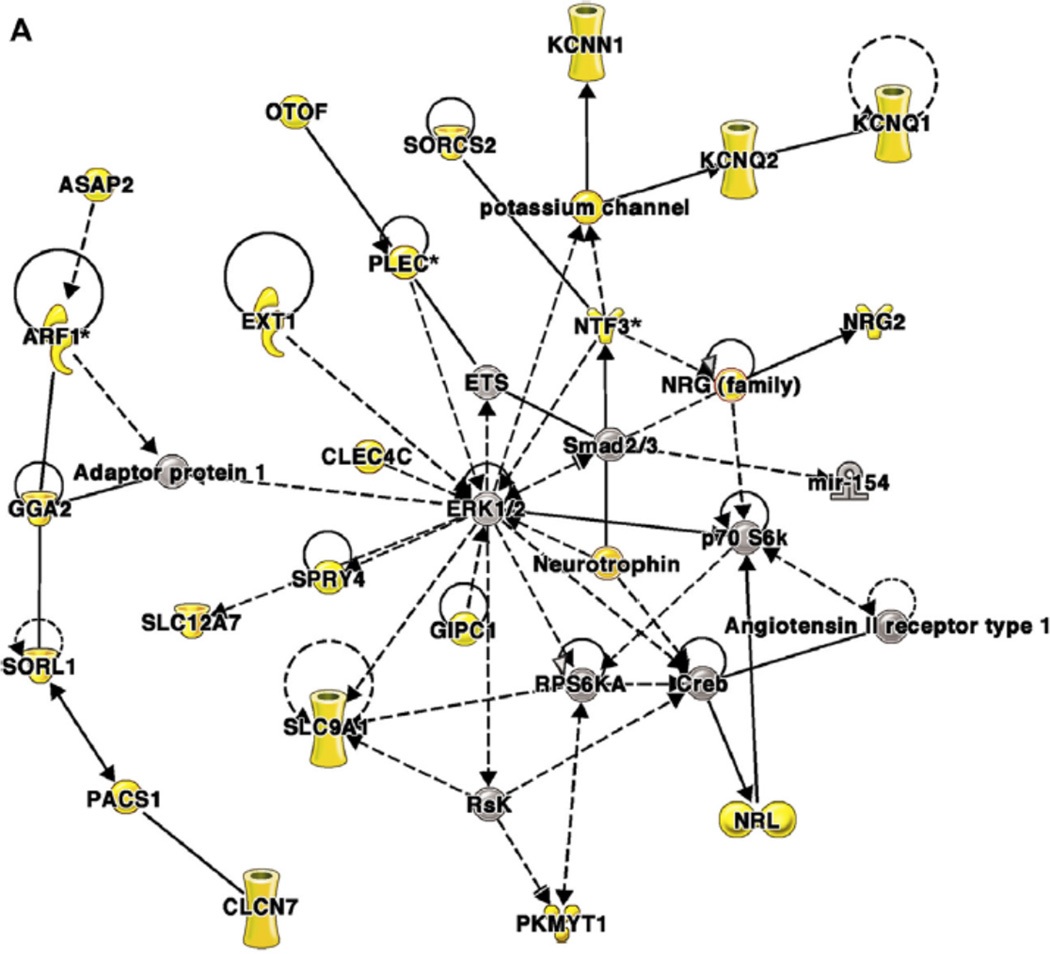
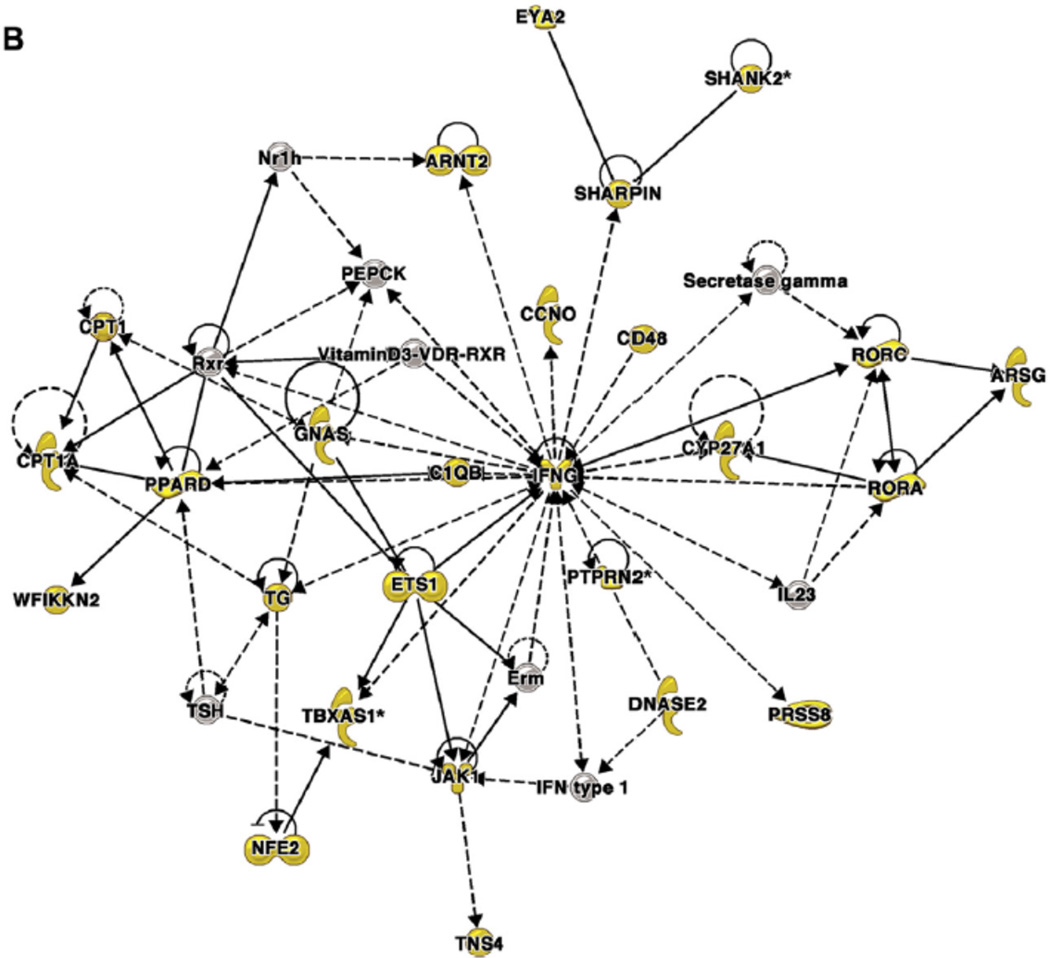
Finally, to elucidate the clinical relevance of these findings, weighted gene coexpression network analysis was used to cluster CpG sites with correlated methylation patterns and then examine associations between the clusters and clinical phenotypes. Overall, 86% of the CpG sites that were differentially methylated both by IL-13 treatment in vitro and asthma status ex vivo clustered into 2 comethylation modules that were each correlated with distinct phenotypes: module 1 was correlated with markers of severity, and module 2 was correlated with eosinophilia (Table II).82,84 The genes near CpGs in module 1 are in networks associated with genes involved in fibrosis, inflammation, and remodeling, and genes near CpGs in module 2 are in networks centered on IFN-g and nuclear factor of kappa light polypeptide gene enhancer in B cells. An example of a network from each module is shown in Fig 3.84
Table 2.
Comethylation modules of correlated methylation patterns at CpG sites in freshly isolated airway epithelial cells with clinical phenotypes in asthmatic subjects.
| Clinical Phenotypes | Co-methylation Module | |
|---|---|---|
| Correlation P-value | ||
| Module 1 | Module 2 | |
| Asthma Severity* | 2.37×10−5 | 0.060 |
| Th2 phenotype+ | 0.79 | 0.54 |
| Steroid resistance╪ | 3.91×10−5 | 0.11 |
| Inhaled corticosteroid usage | 1.04×10−5 | 0.044 |
| Oral steroid usage | 0.050 | 0.12 |
| FEV1 (% predicted) | 7.20×10−5 | 0.020 |
| FeNO | 0.26 | 0.24 |
| IgE | 0.006 | 0.12 |
| Blood eosinophil count | 0.26 | 1.04×10−5 |
| Bronchoalveolar lavage eosinophil count | 0.39 | 1.26×10−6 |
| Atopy§ | 0.0020 | 0.025 |
| Body Mass Index | 3.21×10−6 | 0.15 |
| Gender | 0.87 | 0.81 |
| Current Smoker | 0.45 | 0.57 |
The Bonferroni threshold for significance is .0036. Reprinted with permission from Nicodemus-Johnson et al.84FENO, Fraction of exhaled nitric oxide
Severity was defined using Stepwise classification of asthma per NHLBI guidelines (http://www.nhlbi.nih.gov/health-pro/guidelines/current/asthma-guidelines).
Th2 classifications were determined following published guidelines.82
Defined as an asthma severity score > of 4 or 6.
One or more positive skin prick test.
Figure 3. Coexpression network analysis of genes near differentially methylated CpG dinucleotides in airway epithelial cells from asthmatic and nonasthmatic subjects84.
Networks were generated by using Ingenuity Pathway Analysis (IPA), as previously described.84 Genes near differentially methylated CpGs are shown as yellow symbols; other genes are included to connect the network of differentially methylated CpGs. Molecular shapes (from IPA) were as follows: oval, transcriptional regulator; diamond, enzyme; dashed line rectangle, channel; upward triangle, phosphatase; downward triangle, kinase; trapezoid, transporter; circle, other. A, A network from module 1 that was associated with clinical markers of asthma severity. This network is centered on extracellular signal-regulated kinase 1/2, which was not itself near a differentially methylated CpG. B, A network from module 2 that was associated with eosinophilia. This network is centered on IFN-g, which was near a differentially methylated CpG. Reprinted with permission of the American Thoracic Society from Nicodemus-Johnson et al.84 Copyright 2016 American Thoracic Society.
These studies indicate that exposure to the TH2 cytokine IL-13 elicits coordinated changes in methylation patterns in airway epithelial cells that are correlated with different clinical phenotypes that represent distinct components of asthma pathogenesis. These studies highlight the importance of DNA methylation as a mechanism for a significant proportion of the known IL-13 effects on gene expression in the airways and likely the downstream effects of IL-13 response to many important asthma-associated exposures. Lastly, these data suggest that variation in DNA methylation levels might underlie a significant amount of interindividual differences in response to environmental exposures and therefore to asthma onset and severity, as has been shown for methylation levels in peripheral blood leukocytes and total serum IgE levels.95 The contribution of genetic variation to these differences is yet to be determined in airway epithelial cells, but associations between genotype and methylation levels, referred to as methylation QTLs, are abundant in whole lung tissue and other cell types,88–94 and therefore are also likely to be abundant in these cells as well.
Summary and Conclusions
Asthma is a heterogeneous disease associated with many different causes and pathophysiologic pathways. In this review, we have provided evidence for the important interplay between genes and the environment in setting subjects on specific risk trajectories early in life and the benefits of focusing genetic studies on an extreme asthma subphenotype in early childhood. Associations with the 20 or fewer risk loci identified by means of GWASs might be robust to environmental heterogeneity either because the relevant environmental exposure is sufficiently common among asthmatic subjects, as is the case with the 17q interaction with rhinovirus-induced wheezing illness35 and animal shed exposure,41 or because the associated variants influence asthma risk in the context of many different exposures. However, it is quite likely that many asthma risk loci have been missed and that ORs were underestimated in the large heterogeneous samples included in GWASs. Indeed, only by focusing on an extreme airway phenotype (exacerbations) in children was the association discovered with a variant in CDHR3, a novel asthma susceptibility gene that functions as a receptor for RV-C in airway epithelial cells.43 Based on these observations and other examples reviewed above, we suggest that additional asthma susceptibility loci will be discovered by studies of more homogeneous samples of asthmatic subjects, either by focusing on extreme phenotypes or on asthma-associated environmental exposures (ie, GEIs).
We further propose that cell models of GEIs provide a power tool for identifying GEIs in controlled environments. This approach has several advantages. First, cell models of GEI allow the decoupling of effects of a single exposure from all other correlated exposures and nongenetic factors that confound both genome-wide and candidate gene studies of GEIs in populations.
Second, these studies can be performed in relevant cell types and identify both genetic and epigenetic associations with response to asthma-associated exposures. Because variants involved in GEIs are likely to be regulatory in nature29 and gene regulatory landscapes differ between cell types,73,74 focusing these studies on respiratory cells might identify GEIs that are particularly relevant to asthma.
Third, studies of GEIs with asthma-associated exposures and relevant cell types will identify SNPs for functional GWASs of asthma, as well as sort through associations with small P values that do not reach the criteria for genome-wide significance in asthma GWASs.
Finally, identifying correlated modules of response to exposures in relevant cells will identify pathways that contribute to different arms of asthma pathogenesis and the genetic and epigenetic variation that contributes to these responses.
In summary, we suggest that improved and more comprehensive understanding of the genetic architecture of asthma will require a combination of GWASs focusing on more homogeneous subtypes of disease, GEI studies in birth cohorts and cell models, and ultimate integration with other types of omics data. In particular, elucidating the gene regulatory architecture in asthma-relevant cell types and in response to asthma-associated exposures will both facilitate the discovery of additional asthma risk alleles and provide biological insight into the mechanisms through which host genotype modifies responses to asthma-associated exposures. Such studies present great challenges but also tremendous opportunities to understand asthma pathogenesis and heterogeneity and ultimately to improve prevention and treatment of disease.
Acknowledgments
We thank Minal Calışkan, Jessie-Nicodemus, Johannes Waage, and Anthony Bosco for helpful discussions of their data.
C.O. is supported by R01 HL058197, U19 AI095230, U01 AI106683, P01 HL070831, and R01 HL129735. K.B. is supported by The Lundbeck Foundation grant no. R163-2013-16235 & R16-A1694; The Danish Ministry of Health grant no. 903516; Danish Council for Strategic Research grant no. 0603-00280B; The Danish Council for Independent Research grant no. 10-082884 & 271-08-0815; and The Capital Region Research Foundation (no grant no.).
Abbreviations
- CARD4
caspase recruitment domain protein 4 gene
- CDHR3
cadherin-related family member 3
- COAST
Childhood Origins of Asthma
- COPSAC2000
Copenhagen Prospective Studies on Asthma in Childhood – 2000 cohort
- DMC
differentially methylated CpG
- eQTL
expression quantitative trait locus
- GAD
Genetic Association Database
- GATA3
GATA binding protein 3
- GEI
gene-environment interaction
- GSDMB
gastermin B
- GWAS
genome-wide association studies
- GWIS
genome-wide interaction studies
- HDM
house dust mite
- HLA
human leukocyte antigen
- IFN-γ
interferon gamma
- IL13
interleukin 13
- IL1RL1
interleukin 1 receptor-like 1
- IL33
interleukin 33
- IRF1
interferon regulatory factor 1
- IRF5
interferon regulatory factor 5
- LPS
lipolysaccharide
- meQTL
methylation quantitative trait locu
- NFκβ
nuclear factor of kappa light polypeptide gene enhancer in B cells
- OAS1
2’,5’-oligoadenylate synthetase 1
- OR
odds ratio
- ORMDL3
ORM1-like 3
- PASTURE
Protection against Allergy Study in Rural Environments
- PBMC
peripheral blood mononuclear cell
- reQTL
response expression quantitative trait locus
- RORA
RAR-related orphan receptor A
- RSV
respiratory syncytial virus
- RV
rhinovirus
- SLC22A4
solute carrier family 22, member 4
- SLC22A5
solute carrier family 22, member 5
- SMAD3
SMAD family member 3
- SNPs
single nucleotide polymorphisms
- STAT2
signal transducer and activator of transcription 2
- Th2
T helper 2
- TLR2
toll like receptor 2
- TLR4
toll-like receptor 4
- TSLP
thymic stromal lymphopoietin
- TNFAIP3
tumor necrosis factor, alpha-induced protein 3
- WGCNA
Weighted Gene Co-expression Network Analysis
Glossary
- CpG
Shorthand for cytosine-phosphate-guanine dinucleotide in which a cytosine base is located adjacent to a guanine base on the chromosomal strand (as opposed to cytosine forming chemical bonds to guanine on the opposite DNA strand because of complementarity).
- EFFECT SIZE
A measurement of the magnitude of effect of a genotype (or treatment) on a specific outcome.
- ENDOTYPE
A subtype of disease defined by a distinct functional or pathobiological mechanism.
- HERITABILITY
The proportion of phenotypic variation that can be attributed to genetic variation. Heritability estimates range from 0 (no contribution of genes to phenotypic variation) to 1.0 (phenotypic variation explained entirely by genetic variation).
- LINKAGE DISEQUILIBRIUM
When alleles at linked loci occur together on the same chromosome more frequently than expected by chance (ie, alleles at linked loci that are inherited together).
- MICROBIAL DIVERSITY
A term used to describe bacterial community metrics that include richness (the number of different species represented in the community) and evenness (how equal is the abundance of different species).
- ODDS RATIO
The odds of exposure to a risk factor in cases divided by the odds of exposure to a risk factor in control subjects. The odds ratio is used in case-control studies and is a reasonable estimate of relative risk when the number of patients with the disease is small compared with the number of patients without the disease. An odds ratio of 5 implies that exposure to the risk factor increases the odds of having the disease by 5 times.
- PENETRANCE
The proportion of subjects with a particular genotype who express the phenotype.
- SINGLE NUCLEOTIDE POLYMORPHISM
A type of genetic polymorphism caused by a single base substitution.
Footnotes
Disclosure of potential conflict of interest
C. Ober has received grants from the National Institutes of Health. K. Bønnelykke declares that he has no relevant conflicts of interest.
References
- 1.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372:1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 3.Lim RH, Kobzik L, Dahl M. Risk for asthma in offspring of asthmatic mothers versus fathers: a meta-analysis. PLoS One. 2010;5:e10134. doi: 10.1371/journal.pone.0010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 5.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–652. doi: 10.1016/j.jaci.2011.04.060. e1-5. [DOI] [PubMed] [Google Scholar]
- 7.West CE, Renz H, Jenmalm MC, Kozyrskyj AL, Allen KJ, Vuillermin P, et al. The gut microbiota and inflammatory noncommunicable diseases: associations and potentials for gut microbiota therapies. J Allergy Clin Immunol. 2015;135:3–13. doi: 10.1016/j.jaci.2014.11.012. quiz 4. [DOI] [PubMed] [Google Scholar]
- 8.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bonnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 9.Hylkema MN, Blacquiere MJ. Intrauterine effects of maternal smoking on sensitization, asthma, and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009;6:660–662. doi: 10.1513/pats.200907-065DP. [DOI] [PubMed] [Google Scholar]
- 10.Litonjua AA, Weiss ST. Is vitamin D deficiency to blame for the asthma epidemic? J Allergy Clin Immunol. 2007;120:1031–1035. doi: 10.1016/j.jaci.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Olsen SF, Osterdal ML, Salvig JD, Mortensen LM, Rytter D, Secher NJ, et al. Fish oil intake compared with olive oil intake in late pregnancy and asthma in the offspring: 16 y of registry-based follow-up from a randomized controlled trial. Am J Clin Nutr. 2008;88:167–175. doi: 10.1093/ajcn/88.1.167. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Xun P, He K. Fish and fish oil intake in relation to risk of asthma: a systematic review and meta-analysis. PLoS One. 2013;8:e80048. doi: 10.1371/journal.pone.0080048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 14.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27:107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- 16.Thomsen SF, Duffy DL, Kyvik KO, Backer V. Genetic influence on the age at onset of asthma: a twin study. J Allergy Clin Immunol. 2010;126:626–630. doi: 10.1016/j.jaci.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Skadhauge LR, Backer V. Increase in the heritability of asthma from 1994 to 2003 among adolescent twins. Respir Med. 2011;105:1147–1152. doi: 10.1016/j.rmed.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drake KA, Torgerson DG, Gignoux CR, Galanter JM, Roth LA, Huntsman S, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. 2014;133:370–378. doi: 10.1016/j.jaci.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega VE, Hawkins GA, Moore WC, Hastie AT, Ampleford EJ, Busse WW, et al. Effect of rare variants in ADRB2 on risk of severe exacerbations and symptom control during longacting beta agonist treatment in a multiethnic asthma population: a genetic study. Lancet Respir Med. 2014;2:204–213. doi: 10.1016/S2213-2600(13)70289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igartua C, Myers RA, Mathias RA, Pino-Yanes M, Eng C, Graves PE, et al. Ethnic-specific associations of rare and low-frequency DNA sequence variants with asthma. Nat Commun. 2015;6:5965. doi: 10.1038/ncomms6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 25.Ege MJ, Strachan DP, Cookson WO, Moffatt MF, Gut I, Lathrop M, et al. Gene-environment interaction for childhood asthma and exposure to farming in Central Europe. J Allergy Clin Immunol. 2011;127:138–144. doi: 10.1016/j.jaci.2010.09.041. 44 e1-4. [DOI] [PubMed] [Google Scholar]
- 26.Myers RA, Scott NM, Gauderman WJ, Qiu W, Mathias RA, Romieu I, et al. Genome-wide interaction studies reveal sex-specific asthma risk alleles. Hum Mol Genet. 2014;23:5251–5259. doi: 10.1093/hmg/ddu222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholtens S, Postma DS, Moffatt MF, Panasevich S, Granell R, Henderson AJ, et al. Novel childhood asthma genes interact with in utero and early-life tobacco smoke exposure. J Allergy Clin Immunol. 2014;133:885–888. doi: 10.1016/j.jaci.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ober C. Asthma genetics in the post-genome-wide association study (GWAS) era. ATS Journal. 2016 [Google Scholar]
- 29.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bisgaard H, Hermansen MN, Bonnelykke K, Stokholm J, Baty F, Skytt NL, et al. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ. 2010;341:c4978. doi: 10.1136/bmj.c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. Bmj. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykes A, Johnston SL. Etiology of asthma exacerbations. J Allergy Clin Immunol. 2008;122:685–688. doi: 10.1016/j.jaci.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 33.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995;332:133–138. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 35.Caliskan M, Bochkov YA, Kreiner-Moller E, Bonnelykke K, Stein MM, Du G, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–908. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bisgaard H, Bonnelykke K, Sleiman PM, Brasholt M, Chawes B, Kreiner-Moller E, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179:179–185. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 38.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 39.Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124:605–607. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 40.Smit LA, Bouzigon E, Pin I, Siroux V, Monier F, Aschard H, et al. 17q21 variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36:57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 41.Loss GJ, Depner M, Hose AJ, Genuneit J, Karvonen AM, Hyvarinen A, et al. The Early Development of Wheeze: Environmental Determinants and Genetic Susceptibility at 17q21. Am J Respir Crit Care Med. 2015 doi: 10.1164/rccm.201507-1493OC. [DOI] [PubMed] [Google Scholar]
- 42.Bonnelykke K, Vissing NH, Sevelsted A, Johnston SL, Bisgaard H. Association between respiratory infections in early life and later asthma is independent of virus type. J Allergy Clin Immunol. 2015;136:81–86. doi: 10.1016/j.jaci.2015.02.024. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 44.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 46.Ross AJ, Dailey LA, Brighton LE, Devlin RB. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am J Respir Cell Mol Biol. 2007;37:169–185. doi: 10.1165/rcmb.2006-0466OC. [DOI] [PubMed] [Google Scholar]
- 47.Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–369. doi: 10.1016/j.biocel.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 48.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015;112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cox DW, Bizzintino J, Ferrari G, Khoo SK, Zhang G, Whelan S, et al. Human rhinovirus species C infection in young children with acute wheeze is associated with increased acute respiratory hospital admissions. Am J Respir Crit Care Med. 2013;188:1358–1364. doi: 10.1164/rccm.201303-0498OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, et al. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller EK, Khuri-Bulos N, Williams JV, Shehabi AA, Faouri S, Al Jundi I, et al. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol. 2009;46:85–89. doi: 10.1016/j.jcv.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu AH. Revisiting the hygiene hypothesis for allergy and asthma. J Allergy Clin Immunol. 2015;136:860–865. doi: 10.1016/j.jaci.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347:869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 54.Simpson A, John SL, Jury F, Niven R, Woodcock A, Ollier WE, et al. Endotoxin exposure, CD14, and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med. 2006;174:386–392. doi: 10.1164/rccm.200509-1380OC. [DOI] [PubMed] [Google Scholar]
- 55.Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A Polymorphism* in the 5’ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20:976–983. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- 56.Zambelli-Weiner A, Ehrlich E, Stockton ML, Grant AV, Zhang S, Levett PN, et al. Evaluation of the CD14/−260 polymorphism and house dust endotoxin exposure in the Barbados Asthma Genetics Study. J Allergy Clin Immunol. 2005;115:1203–1209. doi: 10.1016/j.jaci.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, et al. Opposite effects of CD 14/−260 on serum IgE levels in children raised in different environments. J Allergy Clin Immunol. 2005;116:601–607. doi: 10.1016/j.jaci.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 58.Smit LA, Siroux V, Bouzigon E, Oryszczyn MP, Lathrop M, Demenais F, et al. CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults. Am J Respir Crit Care Med. 2009;179:363–368. doi: 10.1164/rccm.200810-1533OC. [DOI] [PubMed] [Google Scholar]
- 59.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, et al. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol. 2004;113:482–488. doi: 10.1016/j.jaci.2003.12.374. [DOI] [PubMed] [Google Scholar]
- 60.Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, et al. Association between exposure to farming, allergies and genetic variation in CARD4/NOD1. Allergy. 2006;61:1117–1124. doi: 10.1111/j.1398-9995.2006.01128.x. [DOI] [PubMed] [Google Scholar]
- 61.Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015;349:1106–1110. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- 62.Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30:383–391. doi: 10.1016/j.it.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 63.Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Penders J, Thijs C, Mommers M, Stobberingh EE, Dompeling E, Reijmerink NE, et al. Host-microbial interactions in childhood atopy: toll-like receptor 4 (TLR4), CD14, and fecal Escherichia coli. J Allergy Clin Immunol. 2010;125:231–236. doi: 10.1016/j.jaci.2009.10.011. e1-5. [DOI] [PubMed] [Google Scholar]
- 65.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjalmsson BJ, Xu H, et al. Partitioning Heritability of Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. Am J Hum Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Torres JM, Gamazon ER, Parra EJ, Below JE, Valladares-Salgado A, Wacher N, et al. Cross-Tissue and Tissue-Specific eQTLs: Partitioning the Heritability of a Complex Trait. Am J Hum Genet. 2014;95:521–534. doi: 10.1016/j.ajhg.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, Laviolette M, et al. Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol. 2010;84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gern JE, Busse WW. Association of rhinovirus infections with asthma. Clin Microbiol Rev. 1999;12:9–18. doi: 10.1128/cmr.12.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jackson DJ, Johnston SL. The role of viruses in acute exacerbations of asthma. J Allergy Clin Immunol. 2010;125:1178–1187. doi: 10.1016/j.jaci.2010.04.021. quiz 88-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caliskan M, Baker SW, Gilad Y, Ober C. Host genetic variation influences gene expression response to rhinovirus infection. PLoS Genet. 2015;11:e1005111. doi: 10.1371/journal.pgen.1005111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 81.Bosco A, Ehteshami S, Panyala S, Martinez FD. Interferon regulatory factor 7 is a major hub connecting interferon-mediated responses in virus-induced asthma exacerbations in vivo. J Allergy Clin Immunol. 2012;129:88–94. doi: 10.1016/j.jaci.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee JH, Kaminski N, Dolganov G, Grunig G, Koth L, Solomon C, et al. Interleukin-13 induces dramatically different transcriptional programs in three human airway cell types. Am J Respir Cell Mol Biol. 2001;25:474–485. doi: 10.1165/ajrcmb.25.4.4522. [DOI] [PubMed] [Google Scholar]
- 84.Nicodemus-Johnson J, Naughton KA, Sudi J, Hogarth DK, Naureckas ET, Nicolae DL, et al. Genome-wide methylation study identifies an IL-13 induced epigenetic signature in asthmatic airways. Am J Resp Care Med. 2016 doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lovinsky-Desir S, Miller RL. Epigenetics, asthma, and allergic diseases: a review of the latest advancements. Curr Allergy Asthma Rep. 2012;12:211–220. doi: 10.1007/s11882-012-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pacheco KA. Epigenetics mediate environment : gene effects on occupational sensitization. Curr Opin Allergy Clin Immunol. 2012;12:111–118. doi: 10.1097/ACI.0b013e328351518f. [DOI] [PubMed] [Google Scholar]
- 87.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Y, Li X, Aryee MJ, Ekstrom TJ, Padyukov L, Klareskog L, et al. GeMes, clusters of DNA methylation under genetic control, can inform genetic and epigenetic analysis of disease. Am J Hum Genet. 2014;94:485–495. doi: 10.1016/j.ajhg.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teh AL, Pan H, Chen L, Ong ML, Dogra S, Wong J, et al. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res. 2014;24:1064–1074. doi: 10.1101/gr.171439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banovich NE, Lan X, McVicker G, van de Geijn B, Degner JF, Blischak JD, et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10:e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lemire M, Zaidi SH, Ban M, Ge B, Aissi D, Germain M, et al. Long-range epigenetic regulation is conferred by genetic variation located at thousands of independent loci. Nat Commun. 2015;6:6326. doi: 10.1038/ncomms7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi J, Marconett CN, Duan J, Hyland PL, Li P, Wang Z, et al. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat Commun. 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liang L, Willis-Owen SA, Laprise C, Wong KC, Davies GA, Hudson TJ, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520:670–674. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]