Abstract
Background
Control of mRNA translation is fundamentally altered in cancer. Insulin-like growth factor-I (IGF-I) signaling regulates key translation mediators to modulate protein synthesis (e.g. eIF4E, 4E-BP1, mTOR, and S6K1). Importantly the Amplified in Breast Cancer (AIB1) oncogene regulates transcription and is also a downstream mediator of IGF-I signaling.
Materials and Methods
To determine if AIB1 also affects mRNA translation, we conducted gain and loss of AIB1 function experiments in estrogen receptor alpha (ERα)+ (MCF-7L) and ERα- (MDA-MB-231, MDA-MB-435 and LCC6) breast cancer cells.
Results
AIB1 positively regulated IGF-I-induced mRNA translation in both ERα+ and ERα- cells. Formation of the eIF4E-4E-BP1 translational complex was altered in the AIB1 ERα+ and ERα- knockdown cells, leading to a reduction in the eIF4E/4E-BP1 and eIF4G/4E-BP1 ratios. In basal and IGF-I stimulated MCF-7 and LCC6 cells, knockdown of AIB1 decreased the integrity of the cap-binding complex, reduced global IGF-I stimulated polyribosomal mRNA recruitment with a concomitant decrease in ten of the thirteen genes tested in polysome-bound mRNAs mapping to proliferation, cell cycle, survival, transcription, translation and ribosome biogenesis ontologies. Specifically, knockdown of AIB1 decreased ribosome-bound mRNA and steady-state protein levels of the transcription factors ERα and E2F1 in addition to reduced ribosome-bound mRNA of the ribosome biogenesis factor BYSL in a cell-line specific manner to regulate mRNA translation.
Conclusion
The oncogenic transcription factor AIB1 has a novel role in the regulation of polyribosome recruitment and formation of the translational complex. Combinatorial therapies targeting IGF signaling and mRNA translation in AIB1 expressing breast cancers may have clinical benefit and warrants further investigation.
Abbreviations: 4E-BP1, Eukaryotic translation initiation factor 4E-binding protein 1; AIB1, Amplified in Breast Cancer 1; BYSL, Bystin; eIF4E, Eukaryotic translation initiation factor 4E; eIF4G, Eukaryotic translation initiation factor 4 gamma; ERα, Estrogen receptor alpha; HER2, Human epidermal growth factor receptor 2; IGF1R, Type I IGF receptor; IGF-I, Insulin-like growth factor-I; MAPK, Mitogen activated protein kinase; mTORC1, Mammalian target of rapamycin complex 1; S6K1, Ribosomal S6 kinase 1; TNBC, Triple negative breast cancer
Background
The p160 co-activator factor amplified in breast cancer (AIB1) human oncogene, also referred to as SRC3, p/CIP, RAC3, ACTR and TRAM3 has been described as a key pleiotropic “master regulator” of gene transcription in many types of human cancers [1], [2]. Importantly it is over-expressed in up to 64% of all breast cancers and functions as downstream mediator of insulin-like growth factor (IGF-I) tyrosine receptor signaling [3], [4]. AIB1 is phosphorylated on serine and tyrosine residues after the type I IGF receptor (IGF1R) activation by IGF-I, but the effects of IGF1R signaling on AIB1 function require further investigation [3], [5]. The tumorigenic actions of AIB1 are driven by its function as a co-regulator of known transcription factors; estrogen receptor alpha (ERα) in luminal breast cancer or E2F1 in basal triple negative breast cancer (TNBC; defined as ER, progesterone receptor and human epidermal growth factor receptor 2 negative (HER2)) [6]. IGF1R signaling results in activation of downstream effectors of the AKT-pathway and MAPK-pathway that function to control mRNA translation in breast cancer [4], [7]. However, the extent to which AIB1 participates in IGF-I stimulated mRNA translation in breast cancer is not known.
The downstream IGF1R signaling mediator, mammalian target of rapamycin complex 1 (mTORC1) regulates mRNA translation by phosphorylation of S6K1 and 4E-binding protein-1 (4E-BP1) [8], [9], [10]. Cap-dependent translation is a highly regulated cellular process, controlled by the eukaryotic initiation factor 4E (eIF4E), the scaffold protein eIF4G, the ATP-dependent helicase protein eIF4A, and eIF4B to collectively form the eIF4F complex which recognizes the 7-methyl guanosine (m7GTP) cap at the mRNA 5’ terminus [11]. During cellular stress, such as hypoxia and nutrient deprivation, a switch from cap-dependent to cap-independent translation occurs [12]. Hypophosphorylated 4E-BP1 sequesters the rate limiting factor eIF4E (i.e. inactive translation) until it is phosphorylated by mTORC1/S6K1 to allow assembly of the eIF4F complex (i.e. active translation) [13], [14]. Moreover, our laboratory has shown that IGF-I signaling stimulates S6K1 resulting in ERα Ser167 phosphorylation and estradiol-independent gene transcription [15]. S6K1 also enhances eIF4B phosphorylation in breast cancer cells
In addition to IGF-I induced post-translational activation of key translational mediators, assembly of the 43S pre-initiation ribosomal complex and mRNA ribosome recruitment are required for cap-dependent translation to occur [11], [16], [17], [18]. AKT and the transcriptional mediators, c-MYC and E2F1 have all been reported to indirectly regulate mRNA cap-dependent translation by transcriptionally affecting expression of ribosomal RNA required for synthesis of ribosome biogenesis factors [19], [20], [21], [22]. Additionally, the oncogenic actions of Ras and AKT may require increased rates of ribosomal recruitment [23]. However, whether the well characterized transcriptional mediator, AIB1 also possesses similar functions as described for c-MYC, E2F1, Ras and AKT is not well known. Since IGF-I signaling positively regulates mRNA cap-dependent translation and IGF-I can modulate AIB1 phosphorylation, we hypothesized that AIB1 possesses both transcriptional and translational functions in breast cancer.
In this study we examined the functional role of IGF1R signaling and AIB1 on cap-dependent and cap-independent mRNA translation in ERα+ and ERα- breast cancer cells. To determine whether AIB1 is required for ‘active’ mRNA translation we have also studied the regulatory effects of IGF-I and AIB1 on changes to total and ribosome-bound mRNA expression of key cancer-related gene targets associated with breast cancer cell growth (e.g. Ki67 and PCNA [24]), migration (e.g. VEGFA [25]), cell cycle (e.g. p21 [24]), survival (e.g. BCL2 [26]), transcription (e.g. ERα, E2F1 and c-MYC [19]), translational (e.g. eIF4E and 4E-BP1 [11]) and ribosome biogenesis (e.g. BYSL, eIF5 and eIF2α [16], [17], [27], [28]).
Methods
Reagents
Cell culture media, trypsin, dimethyl sulfoxide (DMSO) and penicillin and streptomycin (P/S), puromycin and ampicillin antibiotics, HEPES and SuperSignal®West Pico Chemiluminescent substrate were purchased from Thermo Fisher Scientific, (Logan, UT, USA). Fetal bovine serum (FBS) was purchased from Atlas Biologicals (CO, USA), IGF-I from GroPep (Adelaide, AUS) and NVP-AEW541 (AEW; IGF1R inhibitor) from Novartis Pharma, Inc (Basel, Switzerland). Insulin was purchased from Lilly (Indianapolis, IN, USA); complete protease inhibitor cocktail pellets, FuGENE 6 Transfection Reagent and Fast Start Universal SYBR Green Master (Rox) from Roche Biochemicals (Indianapolis, IN, USA); trace elements from Corning, CellGro (Manassas, VA, USA); Dual-Glo luciferase and Lipofectomine 2000 transfection reagents from Promega (Madison, WI, USA); qScript cDNA SuperMix from Quanta Biosciences (Gaithersburg, MD, USA); and 7-Methyl (m7) GTP-Sepharose 4B from GE Healthcare UK Limited.
Antibodies
Primary rabbit polyclonal antibodies SRC-3, total and phospho-AKTSer473, total and phospho-p70 S6 KinaseThr389, eIF4G, eIF4E, total 4E-BP1, total and phospho-ERαSer167, P21, E2F1 and total and phospho-p44/42MAPK Thr202//Tyr204 were purchased from Cell Signaling. Primary mouse monoclonal antibodies PCNA and B-actin were purchased from Cell Signaling and Abcam, respectively and secondary anti-rabbit and anti-mouse IgG horseradish peroxidase-conjugated antibodies from Pierce.
Cell lines and culture
MCF-7L cells, an ER+ cell line, were provided by C. Kent Osborne (Baylor College of Medicine) and ER- MDA-MB-435, LCC6 (a metastatic sub-line of the MDA-MB-435 cells) and MDA-MB-231 cells were provided by Robert Clarke (Lombardi Cancer Center, Georgetown University, Washington, DC, USA). Controversy exists to the lineage origin of the LCC6 cells as to whether they are in fact breast cancer or melanoma cells, although recent evidence suggests that they are of breast origin with a melanocytic phenotype [16], [29], [30]. All cell lines were maintained and cultured at 37°C in 5% CO2, 95% air. MCF-7L wild-type and short hairpin (sh)RNA knockdown cells were grown in DMEM supplemented with 5% FBS plus 11.25 nM insulin, 100 U/ml penicillin, and 100 μg/ml streptomycin. MDA-MB-435 and LCC6 wild-type and shRNA knockdown cells were maintained in DMEM supplemented with 10% FBS plus 11.25 nM insulin, and MDA-MB-231 wild-type and shRNA cells were maintained in DMEM supplemented with 10% FBS plus 100 U/ml penicillin and 100 μg/ml streptomycin (P/S). 293T human kidney cells were cultured in phenol-red DMEM supplemented with 10% FBS plus insulin. Cells were serum deprived in phenol-red free (PRF)-IMEM (supplemented with 20 mM HEPES, 1 × trace elements and 2 μg/ml transferrin) for 24 hours prior to treatment with 5 nM IGF-I or inhibitor (i.e. 0.3 μM AEW). Inhibitor was added to the cells for 4 hours prior to IGF-I exposure. Filter sterilized DMSO was added to the media as a control for the highest concentration used in the inhibitor treatments (i.e. 10 μM).
Preparation and transduction of lentiviral-delivered short-hairpin RNA
AIB1 and scrambled control shRNA was delivered via pLKO.1 lentiviral vectors (RNAi Consortium, Sigma and Plasmid Repository, Addgene), which contain the puromycin resistance gene and drives shRNA expression from the human U6 promoter. The shRNA plasmids (i.e. shCON (scrambled control), shAIB1 and shAIB1 (3′UTR)) and their sequences (Suppl. Table 1) were provided by Dr. Christopher Chien, Department of Oncology, Georgetown University Medical Center, Washington. The shAIB1 sequence was designed to correspond with nucleotides to the coding region (i.e. 564-582 base pairs) and the shAIB1 (3′UTR) sequence to 40 bases after the stop codon in the non-coding untranslated region of the AIB1 mRNA sequence. The shCON scrambled control sequence is not specific for any target mRNA sequence (Qiagen, Valencia, CA). The shRNA was used to create MCF-7L, LCC6, and MDA-MB-231 wild type. shCON, shAIB1, and shAIB1 (3′UTR) stably expressing cells. 2 × 106 293T cells were plated per 10 cm-petri dish, serum deprived for 24 hours and subsequently transfected using FuGENE 6 transfection reagent in PRF-IMEM with 3 μg of shCON and shAIB1 lentiviral vectors combined with 3 μg pCMV-dR8.2 dvpr packaging and 0.375 μg pCMV-VSV-G envelope vectors for 6 hours. Media were replaced and after 48 hours the lentiviral supernatant was centrifuged and filtered sterilized using a 0.22 μm mixed cellulose ester syringe (Thermo Fisher Scientific, USA). 1.0 × 106 cells were plated in 10 cm-petri dishes. The following day the cells were transduced for 24 hours with 1 ml of the lentivirus supernatant added to 9 ml fresh media supplemented with 8 μg/ml polybrene. 2–10 μg/ml of puromycin was added for cell selection.
Dual reporter gene assay
8 × 104 MCF-7L and 6 × 104 MDA-MB-231 and LCC6 cells were serum deprived in PRF-IMEM supplemented with 5% dextran-coated charcoal (DCC)-FBS plus P/S for 24 hours and cultured overnight in 24-well dishes (five replicates per treatment). The media were replaced with fresh PRF-IMEM, and wild-type or stably expressing MCF-7L cells were transiently transfected using Lipofectamine 2000. MDA-MB-231, MDA-MB-435, and LCC6 cells using FuGENE 6 with 250 ng/well of bi-cistronic reporter expression construct (pCDNA3-renilla-LUC-IRES-firefly-LUC), courtesy of V. Polunonvsky (Masonic Cancer Center, University of Minnesota), in addition to 250 ng/well of pCDNA.3-empty vector (i.e., EV) and 250 ng/well of pCDNA.3-AIB1 (i.e., AIB1) expression vectors for 24 hours. The bi-cistronic vector measures cap-dependent translation (renilla luciferase: rLUC) and cap-independent translation (firefly luciferase; fLUC). Post-transfection, the media were replaced with PRF-IMEM +/− 5 nM IGF-I for 24 hours. Luciferase relative light units (RLU) were measured on a Bio-Tek Synergy 2 luminometer and Gen5 software using a Dual-Glo luciferase assay system.
Immunoblot
Protein lysates were prepared and concentrations determined as previously described [15]. 40 μg total protein was boiled at 95 °C for 5 minutes and run on SDS-PAGE, transferred to nitrocellulose and probed with antibodies to detect phospho- and/or total-AIB1, AKT, S6K1, MAPK, eIF4G, eIF4E, 4E-BP1, E2F1, ERα, P21, and PCNA steady-state protein levels per the manufacturer’s immunoblot protocol.
m7GTP agarose bead micro-scale affinity chromatography and densitometry
2 × 106 MCF-7L shCON and shAIB1 cells were plated in a 1 × 150-mm-diameter petri dish and grown to approximately 70% confluence in culture medium. The cells were serum starved in PRF-IMEM (plus additives) for 24 hours and subsequently replaced with fresh PRF-IMEM (plus additives) +/− 5nM IGF-I for 24 hours. Protein lysates were prepared and m7GTP cap-binding assay was performed as previously described [15], [31]. 450 μg total-protein was used for the m7GTP agarose bead micro-scale affinity chromatography and 40 μg for separate immunoblot analysis to detect phospho- and/or total-AIB1, AKT, S6K1, MAPK, eIF4G, eIF4E and 4E-BP1 steady-state protein levels. Measurement of m7GTP cap-binding immunoblot protein intensity was performed using densitometry (Bio Rad GS-700 Densitometry Gel Doc) to calculate the eIF4G/4E-BP1 and eIF4E/4E-BP1 ratios as a representation of a relative translational index (arbitrary values) similar to published ratio calculations [32].
Polyribosome preparation and RNA extractions
MCF-7L and LCC6 shCON and shAIB1 cells were used to evaluate the effect of AIB1 on polysome recruitment in ER+ and ER- cells. 2 × 106 cells were plated in 6 × 150-mm-diameter petri dishes and serum deprived for 24 hours in PRF-IMEM (plus additives). Subsequently the medium was either replaced with fresh PRF-IMEM (plus additives) +/− 5nM IGF-I and incubated for an additional 24 hours. Polysome purification was performed as previously described [33], [34]. Total-RNA and RNA from individual polysome fractions were purified using Trireagent (Sigma-Aldrich) following the manufacturer’s protocol. RNA pellets were re-suspended in nuclease-free H2O and concentrations were measured using a plate reader (BioTek, Synergy 2) at a 260/280 nm wavelength absorbance ratio.
Reverse-transcription and quantitative-real-time polymerase chain reaction
Reverse-transcription of 1 μg RNA per individual sample was performed using Transcriptor First Strand cDNA Synthesis Kit (Roche) and qRT-PCR using FastStart Universal SYBr Green Master (Rox) according to manufacturer’s standard protocol. Absolute quantification of RNA copy number was generated using a standard curve of arbitrary concentrations and melting curve analysis using RealPlex4, MasterCycler epgradientS and epMotion 5070 (BioRad). Primer design was performed using http://www.ncbi.nlm.nih.gov/pubmed gene analysis apart from P21 and PCNA were obtained from published research [24] (Suppl. Table 1).
Statistical analysis
A one-way analysis of variance (ANOVA) (GraphPad Prism, version 5.01, GraphPad Software, San Diego, CA, USA) was used to assess statistical significance between treatment samples in the reporter gene assay, polyribosome fractionation assay and qRT-PCR experimental studies. Statistical significance was accepted *P < .05, **P < .01. ***P < .001.
A weighted average calculation was performed to determine the average distribution of RNA across the gradient fractions following IGF stimulation and AIB1 shRNA knock-down. An overall re-distribution of the RNA to the right (or left) indicates a quantitative increase (or decrease) in translation activity.
Results
AIB1 induced cap-dependent and cap-independent mRNA translation
To determine if AIB1 functions as a positive mRNA translational regulator similar to c-MYC, E2F1 and AKT [19], an AIB1 expression vector was transiently transfected into breast cancer cells. ER+ and ER- breast cancer cells were studied as AIB1 is over-expressed in both luminal and basal breast cancer [3], [4]. Cap-dependent and -independent translation was measured using a bi-cistronic reporter construct. IGF-I stimulated cap-dependent (Figure 1A) and cap-independent (Figure 1B) translation in MCF-7L, MDA-MB-231, and MDA-MB-435 cells. In all three cell lines AIB1 expression increased both cap-dependent (Figure 1A, black boxes) and cap-independent translation (Figure 1B, black boxes) in control and IGF-I treated cells compared to empty vector (EV) transfected (empty boxes) cells. IGF-I induced effects were eliminated in all AIB1-transfected cell lines by treatment with an IGF1R tyrosine kinase inhibitor AEW541 (P < .05; data not shown). Immunoblot analysis was performed to show that all cell-lines expressed endogenous AIB1 under basal and IGF-I treated experimental conditions and were therefore suitable for our studies (Figure 1B inset). MCF-7L AIB1-transfected protein cell lysates demonstrated effective AIB1-transfection by an increase in AIB1 steady-state protein expression levels compared to EV-transfected cells (data not shown).
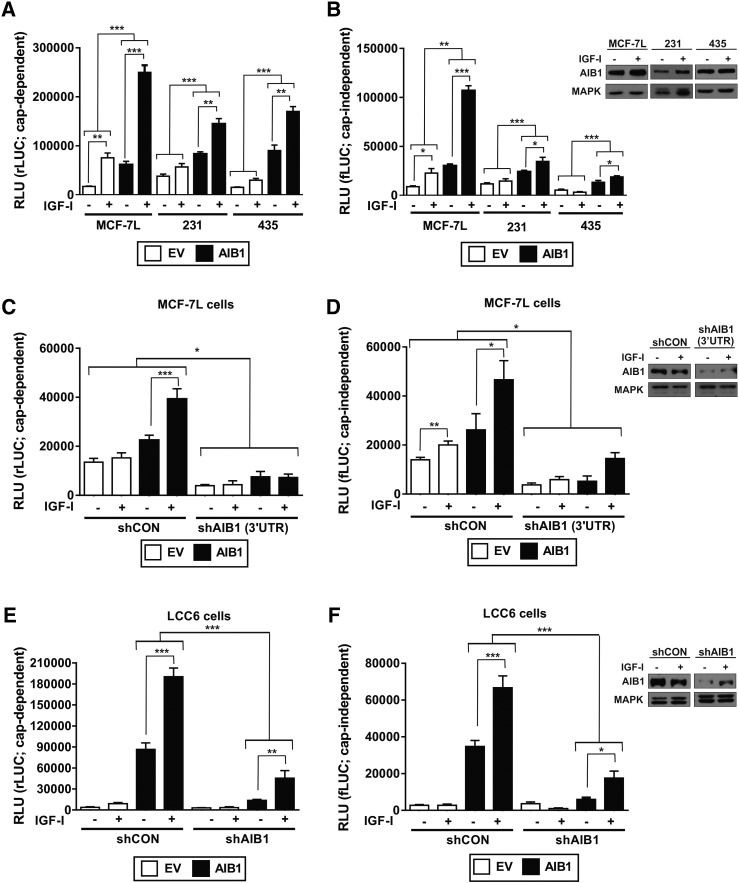
Figure 1.
Over-expression and knockdown of AIB1 regulates cap-dependent and cap-independent translation.
Cap-dependent and independent mRNA translation was measured using a bi-cistronic reporter gene assay. A and B) Cap-dependent (i.e. renilla luciferase; rLUC) and cap-independent (i.e. firefly luciferase; fLUC) translation was measured in MCF-7L, MDA-MB-231 (231) and MDA-MB-435 (435) following pCDNA3-EV (EV; empty boxes) and pCDNA3-AIB1 (AIB1; black boxes) vector transfection. Cells were treated with or without IGF-I for 24 hours.
C and D) MCF-7L cells were infected with shRNA scrambled control (shCON) or AIB1 (shAIB1 (3′UTR)) and cap-dependent and cap-independent translation was measured. IGF-I treatment was for 24 hours.
E and F) LCC6 cells were infected with shRNA scrambled control (shCON) or AIB1 (shAIB1) and cap-dependent and cap-independent translation was measured. IGF-I treatment was for 24 hours.
Luciferase expression (rLUC and fLUC) is depicted on the graphs as relative light units (RLU). Error bars are SEM *P < .05, **P < .01. ***P < .001 one-way ANOVA. Experiments were preformed three times. Immunoblot analysis performed on protein lysates from MCF-7L, 231 and 435 cells, and the individual shCON and shAIB1 cell-lines to measure endogenous levels of AIB1 protein levels following IGF-I treatment for 24 hours are shown as insets. All protein samples were run on the same gel.
A significant reduction of both cap-dependent and independent expression was seen when AIB1 levels were suppressed by shRNA in MCF-7L (Figure 1, C and D) and LCC6 cells (Figure 1, E and F) at all levels of AIB1 expression (EV and AIB1-transfected cells). Our findings were further validated using a different shAIB1 knockdown vector in MCF-7L cells and in the MDA-MB-231 shAIB1 cells (P < .01; Suppl. Figure 1, A and C; cap-dependent and Suppl. Figure 1, B and D; cap-independent). Immunoblot analysis confirmed effective shRNA knock-down of AIB1 by decreased levels of AIB1 protein in cell lines infected with the shAIB1 construct (Figure 1, D and F and Suppl. Figure 1, B and D insets). Collectively, these data provide evidence that IGF-I induced mRNA translation is positively modulated by the translational mediator AIB1.
AIB1 suppression altered the integrity of the eIF4F translational complex
To further determine how AIB1 regulates IGF-I induced mRNA cap-dependent translation we measured protein levels of the PI3K and MAPK signaling pathway and key translational mediators. Cells transfected with a scrambled shRNA control (shCON) and shAIB1 MCF-7L and LCC6 cells were treated +/− IGF-I for 24 hours and protein cell lysates were used for immunoblot analysis. In MCF-7L IGF-I induced phosphorylation of key proteins pAKT, pS6K1, pMAPK, and p4E-BP1 within 10 minutes and persisted for up to 24 hours (Figure 2A). This effect was seen for pAKT and p4E-BP1 in the LCC6 cell line (Figure 2B). However, in these cells pS6K1 and pMAPK were constitutively activated even in the absence of growth factor stimulation (Figure 2B). Moreover, following 24 hours of IGF-I treatment, pS6K1 was decreased with an accompanying increase in total-S6K1 protein in LCC6 shAIB1 cells (Figure 2B). Conversely, decreased pAKT and pMAPK protein was seen after 24 hours of IGF-I treatment in the shAIB1 MCF-7L cells (Figure 2A). Most notably and relevant to the translational function of AIB1, the protein levels of hypo-phosphorylated 4E-BP1 were higher in shAIB1 cells compared to control cells in both MCF-7L and LCC6 cells (Figure 2, A and B). Similar changes to 4E-BP1 protein levels were noted using the shAIB1 (3′UTR) MCF-7L and LCC6 cells (Suppl. Figure 2, A and B).
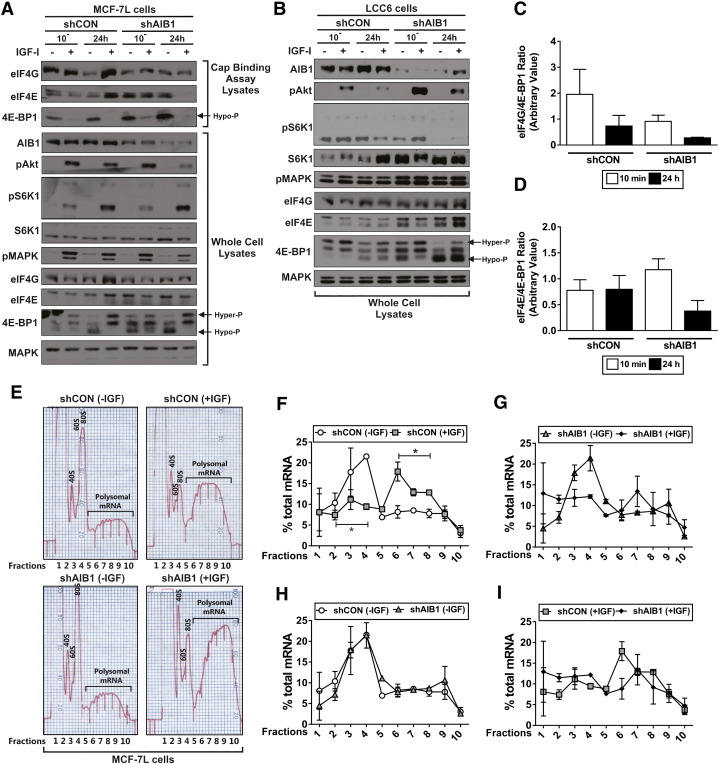
Figure 2.
AIB1 knockdown increases eIF4E and 4E-BP1 complex steady-state protein levels and reduces polyribosome recruitment.
A) Micro-scale affinity chromatography using m7GTP agarose beads (top) was performed using shRNA scrambled control (shCON) or AIB1 (shAIB1) IGF-I treated MCF-7L cell protein lystates. Immunoblot analysis was performed on whole cell protein lysates (bottom). Hyper-phosphorylated (hyper-P; top bands) and hypo-phosphorylated (hypo-P; lower bands) of 4E-BP1 are depicted on the immunoblot images in A and B.
B) Immunoblot analysis was performed on whole cell protein lysates obtained from shCON and shAIB1 LCC6 IGF-I treated cells.
C and D) Densitometry was performed on the m7 GTP cap-binding assay MCF-7L immunoblots to determine a relative translational index by calculating the eIF4G/4E-BP1 and eIF4E/4E-BP1 ratios of control cells for 10 minutes and 24 hours.
E) Densitometry (O.D.254) traces of polysome preparations. 107 shCON and shAIB1 MCF-7L cells were serum starved in PRF-media (plus additives) for 24 hours and subsequently treated +/− 5 nM IGF-I for 24 hours. Total RNA was isolated, loaded onto sucrose gradients, ultracentrifuged to stratify the ribosome bound mRNA transcripts based on the number of bound ribosomes, and fractionated into 10 fractions. Fraction 10 represents transcripts with the most bound ribosomes. Free ribosomal subunits (40S and 60S), monosomes (80S) and the polysomal mRNA fractions are depicted on the densitometry trace readings.
F-I) Percent mRNA per individual fraction normalized to total polysome bound RNA. Experiments were performed three times with similar results; graphs represent combined data from two experiments. Error bars are SEM *P < .05 one-way ANOVA. A weighted average calculation was performed to demonstrate an overall shift in the average number of bound ribosomes.
m7GTP agarose bead micro-scale affinity chromatography (i.e. cap-binding assay) was employed to determine if AIB1 levels altered recruitment and the assembly of the eIF4F translational complex to the mRNA m7GTP cap required for ‘active’ translation. In the absence of IGF-I we observed an increase in eIF4E-4E-BP1 (hypo-phosphorylated) protein bound to the m7GTP in shAIB1 compared to the shCON cells (Figure 2A) which in effect leads to ‘inactive’ mRNA translation. As expected, IGF-I treatment resulted in decreased levels of 4E-BP1 bound to the m7GTP cap in both the shCON and shAIB1 MCF-7L cells reflecting 4E-BP1 phosphorylation and the release of eIF4E required for ‘active’ mRNA translation. eIF4G was seen to be bound to the m7GTP cap at a higher amount following 24 hours of IGF-I treatment in control cells, compared to the shAIB1 cells suggesting that AIB1 was necessary for optimal recruitment of the eIF4F complex to the m7GTP cap (Figure 2A). More-over basal levels of the eIF4G/4E-BP1 and eIF4E/4E-BP1 ratios were decreased in the shAIB1 compared to shCON cells most notably at 24 hours (Figure 2, C and D). These ratio changes reflect a higher proportion of hypo-phosphorylated 4E-BP1 bound to the m7GTP cap compared to the translational modulators, eIF4E and eIF4G in the shAIB1 cells compared to the shCON cells potentially leading to a ‘brake’ on active mRNA translation. These data suggest AIB1 loss may suppress ‘active’ mRNA translation. Further studies investigating whether AIB1 directly interacts with 4E-BP1, eIF4F or other translational mediators such are required.
Reduction in AIB1 expression reduced IGF-I induced mRNA polyribosomal recruitment
Our initial reporter gene assay findings suggested that AIB1 is an IGF-I downstream translational mediator. To further support this finding, the effect of AIB1 on IGF-I induced mRNA polyribosome recruitment was directly studied [20], [23]. Polyribosomes were isolated from control and 24 hours IGF-I treated MCF-7L and LCC6 shCON and shAIB1 cells to determine changes to total and ribosome-bound mRNA of key translational, transcriptional, migration, cell cycle, proliferation, and ribosome biogenesis mediators.
IGF-I treatment increased the absolute amount of ribosome-bound mRNA in both the shCON and shAIB1 MCF-7L cells (Figure 2E). IGF-treatment led to a strong 0.7 unit left to right shift in the weighted average fraction distribution of ribosome-bound mRNA in the shCON cells, indicating increased global ribosomal loading (P < .05; Figure 2F) but this shift was diminished in the shAIB1 cells (Figure 2G). The overall distribution of ribosome bound mRNA was similar in the absence of IGF-I in the shCON and shAIB1 cells (Figure 2H). Importantly, when AIB1 was suppressed, IGF-I treatment resulted in a 0.5 unit right to left shift of the weighted average fraction distribution of ribosome-bound mRNA, reflecting that AIB1 is necessary for normal translation levels (Figure 2I). We further conducted proof-of-principle experiments using the LCC6 cell line. There were no differences in the amount of ribosome-bound mRNA following IGF-I treatment in either the shCON or shAIB1 LCC6 cells most likely due to the constitutive activation of S6K1 and MAPK in these cells (Figure 2A and Suppl. Figure 3, A–C). However consistent with the MCF-7L cells, AIB1 knockdown reduced the overall transcript ribosome loading, leading to a left to right shift in the weighted average fraction distribution of ribosome-bound mRNA in the absence (i.e. 0.7 unit) and presence (i.e. 0.5 unit) of IGF-treatment (Suppl. Figure 3, D and E).
To evaluate whether AIB1 and IGF-I affected expression of key breast cancer genes we studied mRNA expression levels in the total mRNA and ribosome bound mRNA fractions by quantitative real-time PCR. In MCF-7L cells, shAIB1 decreased AIB1 total mRNA by approximately 50% and ribosome-bound AIB1 mRNA was also reduced (P < .05; Figure 3A). Moreover, we classified genes into specific groups based on total-mRNA expression and identified that the mRNA transcript levels of eIF4E, VEGFA, Ki67, and BYSL were IGF-I- and AIB1-dependent defined by the lack of significant IGF-I induction of gene expression in the shAIB1 cells (P < .05; Figure 3, B–E), E2F1 and PCNA were classified as IGF-dependent and AIB1-independent (P < .05; Figure 3, F and G), ERα and P21 were classified as IGF-independent and AIB1-dependent (P < .05; Figure 3, H and I) and 4E-BP1, BCL2, c-MYC, eIF5 and eIF2α were classified as IGF-1 and AIB1-independent (Figure 4, A–E).
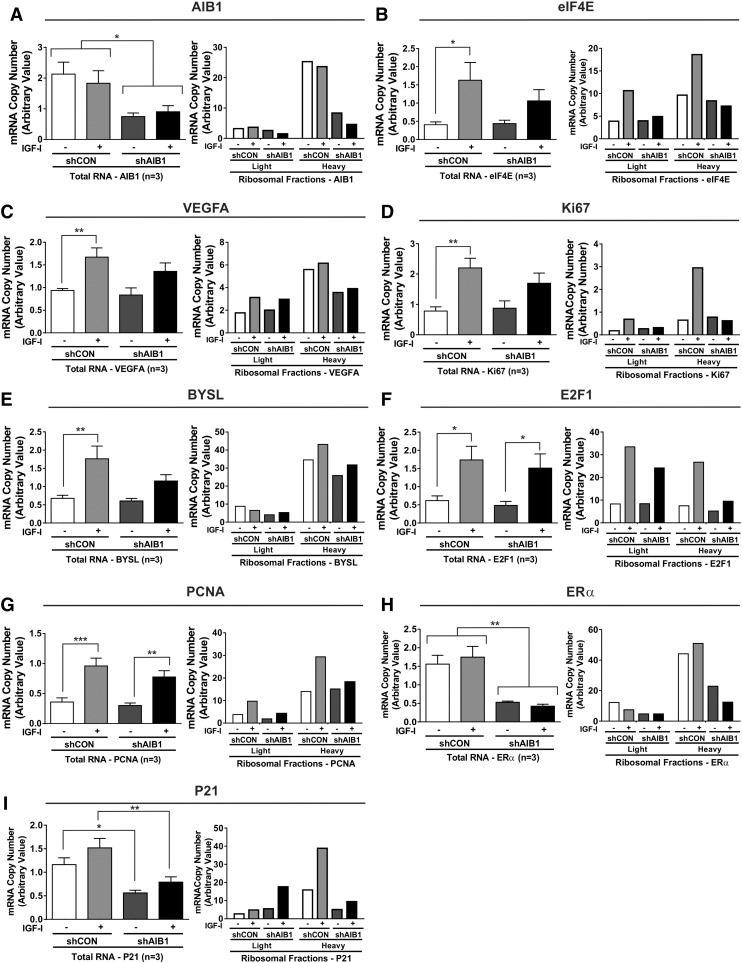
Figure 3.
AIB1 knockdown reduces recruitment of IGF-I induced target mRNA to polysomes.
qRT-PCR analysis was performed on total and fractionated mRNA samples following polysomal fractionation obatined from lentiviral stably expressing shRNA scrambled control (shCON) and AIB1 (shAIB1) MCF-7L cells. Primers were used as probes to measure relative changes in gene expression of total RNA in light (1-6; lower percentage of ribosome-bound mRNA) and heavy (7-10; higher percentage of ribosome mRNA) pooled ribosomal fractions; A) AIB1, B) eIF4E, C) VEGFA, D) Ki67, E) BYSL, F) E2F1, G) PCNA, H) ERα and I) P21. Three replicate experiments were performed with similar results. Graphs for total RNA depict three treatment and technical replicates. Error bars are SEM *P < .05, **P < .01, ***P < .001 one-way ANOVA. Independent examples of a single experiment for each gene are shown of the ribosomal fractions. RPLPO primers were used as a control for each gene examined. The graphs represent the mRNA copy number (arbitrary value) of each reference gene normalized to RPLPO.
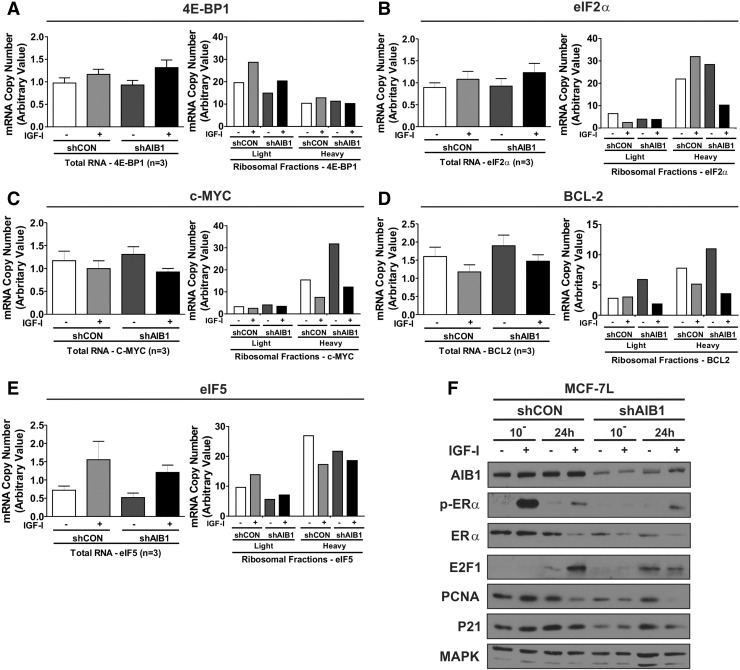
Figure 4.
AIB1 regulates polyribosome recruitment to cancer-related targets.
qRT-PCR analysis was performed on total and fractionated mRNA samples following polysomal fractionation obatined from lentiviral stably expressing shRNA scrambled control (shCON) and AIB1 (shAIB1) MCF-7L cells. Primers were used as probes to measure relative changes in gene expression of total RNA and in light (1-6; lower percentage of ribosome-bound mRNA) and heavy (7-10; higher percentage of ribosome-bound mRNA) pooled ribosomal fractions; A) 4E-BP1, B) eIF2α, C) c-MYC, D) BCL2 and E) eIF5. Graphs for total RNA depict three treatment replicates comprised of three technical replicates. Error bars are SEM. Independent examples of a single experiment for each gene are shown of the ribosomal fractions. RPLPO primers were used as a control for each gene examined. The graphs represent the mRNA copy number (arbitrary value) of each reference gene normalized to RPLPO.
F) Immunoblot analysis was performed on protein lysates obatined from shCON and shAIB1 MCF-7L cells. Total MAPK was used as a loading control.
Importantly, the translational modulation by IGF-I and AIB1 revealed that the majority of target genes, ten of the thirteen measured in our study were positively regulated by IGF-I and AIB1 (i.e. eIF4E, VEGFA, Ki67, BYSL, E2F1, PCNA, ERα, P21; Figures 3, B–I and 4E-BP1 and eIF2α; Figure 4A and B). Further testing of the ten genes revealed that ribosomal loading was reduced in the heavy mRNA fractions of the shAIB1 IGF-I treated cells compared to the shCON cells. Moreover, in the absence of IGF-I, an additional reduction in ribosomal loading was observed in the four genes VEGFA, BYSL, ERα and P21. Of the remaining three genes c-MYC, BCL-2 and eIF5 IGF-I treatment led to a negative regulation of ribosomal recruitment most notably in the heavy fractions (Figure 4, C–E). Immunoblot analysis confirmed our findings by the reduction of total and pERα, E2F1, P21 and PCNA steady-state protein levels in the AIB1 knock-down MCF-7L cells (Figure 4F).
Lastly, to validate our findings we tested five genes using the total and polysome fractionated mRNA samples obtained from shCON and shAIB1 LCC6 cells. Total and ribosome bound AIB1 mRNA was reduced by approximately 80% in the control and IGF-I treated shAIB1 vs. shCON cells (Suppl. Figure 3F). We identified an IGF-I-independent and AIB1-dependent gene transcriptional regulation of 4E-BP1, E2F1 and BYSL, yet eIF4E was not significantly affected by either IGF-I or AIB1 (P < .05; Suppl. Figure 3, G–J). A reduction in the ribosome loading of 4E-BP1 and E2F1 mRNA was evident under control and IGF-I treatment conditions (Suppl. Figure 3G and H), yet eIF4E mRNA was slightly increased by IGF-I treatment following AIB1 knockdown (Suppl. Figure 3J). We observed a reduction of ribosome-bound BYSL mRNA of the heavier fractions in the control AIB1 knockdown cells (Suppl. Figure 3I). Collectively, our findings support our hypothesis that AIB1 influences both gene transcription and mRNA translation and provides evidence that AIB1 potentially modulates ribosomal recruitment on a subset of cancer-related genes in a cell-line specific manner.
Discussion
Our previous work has shown that IGF-I signaling affects gene transcription in breast cancer cells via its ability to activate PI3K/mTORC1/S6K signaling [15]. These pathways are also involved in enhanced mRNA cap dependent translation [11] and it seemed likely that the IGF-I downstream transcriptional mediator, oncogenic factor AIB1 would stimulate both transcription and translation. Here we show for the first time a role for AIB1 in the control of mRNA cap-dependent and cap-independent in breast cancer cells. While AIB1 clearly participates in IGF-I mediated effects, we also show that AIB1 influences gene expression independently of IGF-I signaling.
The translational machinery (eIF4F) components, 4E-BP1, eIF4E, and eIF4G are over-expressed in many different types of cancer and have been found to be oncogenic [11], [12], [32]. Given these oncogenic properties, it follows that the oncogene AIB1 also participates in enhancing mRNA translation. The translational activator eIF4E has become an attractive cancer therapy target for inhibitor therapies including: the antisense oligonucleotide 4E-ASO, the small molecule inhibitor, 4EGI-l and the antiviral drug ribavirin [11], [32]. An inhibitor of mTORC1 has been approved for the treatment of endocrine resistant breast cancer [35] which certainly influences cap dependent translation. More specific inhibitors of translation are being tested in clinical trials; the translational inhibitor ribavirin is currently being tested as a monotherapy in an ongoing Phase I and II clinical trial (NCT01309490) for solid cancers including breast cancer highlighting the potential therapeutic impact of inhibiting mRNA translation [36]. Recently it has been reported that activated mRNA translation is a promoting factor in driving therapy resistance in melanoma [37].
Targeting of ER with selective ER-modulators (tamoxifen, fulvestrant) to reduce its activity and aromatase inhibitors (letrozole, anastrozole, and exemestane) to reduce estrogen levels has been a successful therapeutic strategy. Given that clinical approaches are determined by ER expression and activity in breast cancer, the novel insights relating to the molecular regulation of ER by AIB1 revealed in this study have important clinical implications for ER-positive disease. In light of our findings, it is plausible that AIB1, like c-MYC, acts as a translational regulator by the formation of a transcriptional complex with ERα on promoter regions of ribosome biogenesis genes required for translation [19], [20], [21], [22] and this crosstalk could serve as an additional therapeutic target that would affect both ERα and ribosomal function. Success of the mTORC1 inhibitor, everolimus, in the treatment of ER + breast cancer supports this possibility [35]. Based on our findings a similar effect may occur in the LCC6 cells by AIB1 forming a transcriptional complex with E2F1. It is tempting to speculate that AIB1 indirectly regulates c-MYC activity via AKT and E2F1 signaling pathways, as recently reported in bladder cancer to aid in its translational regulation in specific molecular subtypes of breast cancer [22], [23], [38].
It is now becoming increasing evident that in conjunction with the PI3K/AKT/mTORC1 and Ras signaling hubs, that transcriptional factors such as C-MYC co-operate in a finely tuned cellular process to regulate protein synthesis via activation of polymerase I and II ribosomal RNA (rRNA) synthesis and subsequent production of ribosome biogenesis factors [20], [39]. Our study demonstrates novel findings that in addition to C-MYC, over-expression of AIB1 may function in a similar manner to fuel cancer cells by transcriptionally increasing rates of rRNA and production of ribosome biogenesis machinery. Clinically our findings are relevant to the development of novel and more effective IGF1R-targeted cancer therapies in line with a recent study that has shown a synergistic effect of co-targeting ribosome biogenesis using a Phase I novel small molecule inhibitor, CX561 and mRNA translation using the mTORC inhibitor, everolimus in MYC-driven lymphoma l [40], [41].
In support of our results, in vitro inhibition of 4E-BP1 function increases the malignant phenotype in breast cancer cells [42] and over-expression of hypo-phosphorylated 4E-BP1 results in a reduction of breast cancer cell proliferation [43]. Furthermore, our findings demonstrating an altered ratio of key eIF4F translational mediators, in a reduced AIB1 protein cellular context, complement reports that a high eIF4E/4E-BP1 ratio is associated with a better clinical prognosis in luminal A breast cancer subtypes [32] suggesting a role for cap dependent translation in the ERα expressing subset of breast cancers. Our findings provide new evidence that AIB1 may function to promote mRNA translation of cancer-associated genes in part by increasing mRNA ribosomal recruitment and thus the rate of translation initiation.
Conclusions
IGF-I signalling in breast cancer has been supported by preclinical and population data but inhibitors of IGF-I action have been disappointing in clinical trials [44]. The inability to develop or test specific biomarkers of “IGF-driven” breast cancers may have contributed to the relatively limited benefit of IGF-I receptor inhibitors. Our data suggest that AIB1 expressing breast cancers may identify a subgroup of breast cancer sensitive to IGF-I signaling inhibition as multiple genes are transcriptionally regulated by the combined effects of enhanced signaling and AIB1 function. Furthermore, combined targeting of IGF-I signaling and AIB1 function might be a useful therapeutic strategy in breast cancer.
Acknowledgements
This work was supported by funding provided by a Susan G. Komen for the Cure Post-doctoral Research and Training Post-doctoral Fellowship KG101465 (ASO and AMO), Komen for the Cure, SAC110039 (DY), NIH R01CA CA074285 (DY), and Cancer Center Support Grant P30 CA 077598. We would like to acknowledge Dr. Christopher Chien for the preparation of the AIB1 knockdown breast cancer cell-lines used in the studies detailed in this manuscript. We acknowledge V. Polunonvsky for providing the pCDNA3-renilla-LUC-IRES-firefly-LUC vector and Dr. Karen Scott for assistance with generation of the plasmid sequence. Thank you to Dr. Scott Dehm and Luke Brand for reporter gene assay technical assistance undertaken for this manuscript.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2016.01.001.
Contributor Information
Aleksandra M. Ochnik, Email: aleksandra.ochnik@sydney.edu.au.
Mark S. Peterson, Email: peter026@umn.edu.
Svetlana V. Avdulov, Email: choch002@umn.edu.
Annabell S. Oh, Email: AOh@komen.org.
Peter B. Bitterman, Email: bitte001@umn.edu.
Douglas Yee, Email: yeexx006@umn.edu.
Appendix A. Supplementary data
Supplementary materials.
References
- 1.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh AS, Lahusen JT, Chien CD, Fereshteh MP, Zhang X, Dakshanamurthy S, Xu J, Kagan BL, Wellstein A, Riegel AT. Tyrosine phosphorylation of the nuclear receptor coactivator AIB1/SRC-3 is enhanced by Abl kinase and is required for its activity in cancer cells. Mol Cell Biol. 2008;28:6580–6593. doi: 10.1128/MCB.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahusen T, Henke RT, Kagan BL, Wellstein A, Riegel AT. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res Treat. 2009;116:225–237. doi: 10.1007/s10549-009-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sachdev D, Yee D. Disrupting insulin-like growth factor signaling as a potential cancer therapy. Mol Cancer Ther. 2007;6:1–12. doi: 10.1158/1535-7163.MCT-06-0080. [DOI] [PubMed] [Google Scholar]
- 8.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5'TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 12.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 13.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 15.Becker MA, Ibrahim YH, Cui X, Lee AV, Yee D. The IGF pathway regulates ERalpha through a S6K1-dependent mechanism in breast cancer cells. Mol Endocrinol. 2011;25:516–528. doi: 10.1210/me.2010-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maag D, Algire M, Lorsch J. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 17.Jennings M, Pavitt G. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature. 2010;465:378–381. doi: 10.1038/nature09003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 19.Cole MD, Cowling VH. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene. 2009;28:1169–1175. doi: 10.1038/onc.2008.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannan KM, Sanij E, Hein N, Hannan RD, Pearson RB. Signaling to the ribosome in cancer--It is more than just mTORC1. IUBMB Life. 2011;63:79–85. doi: 10.1002/iub.428. [DOI] [PubMed] [Google Scholar]
- 21.Devlin JR, Hannan KM, Ng PY, Bywater MJ, Shortt J, Cullinane C, McArthur GA, Johnstone RW, Hannan RD, Pearson RB. AKT signalling is required for ribosomal RNA synthesis and progression of Emu-Myc B-cell lymphoma in vivo. FEBS J. 2013;280(21):5307–5316. doi: 10.1111/febs.12135. [DOI] [PubMed] [Google Scholar]
- 22.Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci Signal. 2011;4:ra56. doi: 10.1126/scisignal.2001754. [DOI] [PubMed] [Google Scholar]
- 23.Rajasekhar V, Viale A, Socci N, Wiedmann M, Hu X, Holland E. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Chen Q, Li W, Su X, Chen TK, Liu Y, Zhao Y, Yu CT. Overexpression of transcriptional coactivator AIB1 promotes hepatocellular carcinoma progression by enhancing cell proliferation and invasiveness. Oncogene. 2010;10:3386–3397. doi: 10.1038/onc.2010.90. [DOI] [PubMed] [Google Scholar]
- 25.Maae E, Andersen R, Steffensen K, Jakobsen E, Brandslund I, Sørensen F, Jakobsen A. Prognostic impact of VEGFA germline polymorphisms in patients with HER2-positive primary breast cancer. Anticancer Res. 2012;32:3619–3627. [PubMed] [Google Scholar]
- 26.Barillé-Nion S, Bah N, Véquaud E, Juin P. Regulation of cancer cell survival by BCL2 family members upon prolonged mitotic arrest: opportunities for anticancer therapy. Anticancer Res. 2012;32:4225–4233. [PubMed] [Google Scholar]
- 27.Fukuda M, Miyoshi M, Nadano D. The role of bystin in embryo implantation and in ribosomal biogenesis. Cell Mol Life Sci. 2008;65:92–99. doi: 10.1007/s00018-007-7302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyoshi M, Okajima T, Matsuda T, Fukuda M, Nadano D. Bystin in human cancer cells: intracellular localization and function in ribosome biogenesis. Biochem J. 2007;404:373–381. doi: 10.1042/BJ20061597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers AF. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009;69:5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 30.Hollestelle A, Schutte M. Comment Re: MDA-MB-435 and M14 cell lines: identical but not M14 Melanoma? Cancer Res. 2009;69:7893. doi: 10.1158/0008-5472.CAN-09-2396. [DOI] [PubMed] [Google Scholar]
- 31.Polunovsky VA, Gingras AC, Sonenberg N, Peterson M, Tan A, Rubins JB, Manivel JC, Bitterman PB. Translational control of the antiapoptotic function of Ras. J Biol Chem. 2000;275:24776–24780. doi: 10.1074/jbc.M001938200. [DOI] [PubMed] [Google Scholar]
- 32.Pettersson F, Yau C, Dobocan MC, Culjkovic-Kraljacic B, Retrouvey H, Puckett R, Flores LM, Krop IE, Rousseau C, Cocolakis E. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin Cancer Res. 2011;17:2874–2884. doi: 10.1158/1078-0432.CCR-10-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Takasu T, Perlman DM, Peterson MS, Burrichter D, Avdulov S, Bitterman PB, Polunovsky VA. Translation factor eIF4E rescues cells from Myc-dependent apoptosis by inhibiting cytochrome c release. J Biol Chem. 2003;278:3015–3022. doi: 10.1074/jbc.M208821200. [DOI] [PubMed] [Google Scholar]
- 34.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci U S A. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baselga J, Campone M, Piccart M, Burris HA, III, Rugo HS, Sahmoud T, Noguchi S, Gnant M, Pritchard KI, Lebrun F. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101:18105–18110. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro N, Thuaud F. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513:105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 38.Tong ZT, Wei JH, Zhang JX, Liang CZ, Liao B, Lu J, Fan S, Chen ZH, Zhang F, Ma HH. AIB1 predicts bladder cancer outcome and promotes bladder cancer cell proliferation through AKT and E2F1. Br J Cancer. 2013;108:1470–1479. doi: 10.1038/bjc.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devlin JR, Hannan KM, Ng PY, Bywater MJ, Shortt J, Cullinane C, McArthur GA, Johnstone RW, Hannan RD, Pearson RB. AKT signalling is required for ribosomal RNA synthesis and progression of Emu-Myc B-cell lymphoma in vivo. FEBS J. 2013;280:5307–5316. doi: 10.1111/febs.12135. [DOI] [PubMed] [Google Scholar]
- 40.Devlin JR, Hannan KM, Hein N, Cullinane C, Kusnadi E, Ng PY, George AJ, Shortt J, Bywater MJ, Poortinga G. Combination therapy targeting ribosome biogenesis and mRNA translation synergistically extends survival in MYC-driven lymphoma. Cancer Discovery. 2016;6(1):59–70. doi: 10.1158/2159-8290.CD-14-0673. [DOI] [PubMed] [Google Scholar]
- 41.Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 42.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 43.Pons B, Peg V, Vazquez-Sanchez MA, Lopez-Vicente L, Argelaguet E, Coch L, Martinez A, Hernandez-Losa J, Armengol G, Ramon YCS. The effect of p-4E-BP1 and p-eIF4E on cell proliferation in a breast cancer model. Int J Oncol. 2011;39:1337–1345. doi: 10.3892/ijo.2011.1118. [DOI] [PubMed] [Google Scholar]
- 44.Yee D. Insulin-like growth factor receptor inhibitors: baby or the bathwater? J Natl Cancer Inst. 2012;104:975–981. doi: 10.1093/jnci/djs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.