Abstract
Interspecies interactions are essential for the persistence and development of any kind of complex community, and microbial biofilms are no exception. Multispecies biofilms are structured and spatially defined communities that have received much attention due to their omnipresence in natural environments. Species residing in these complex bacterial communities usually interact both intra- and interspecifically. Such interactions are considered to not only be fundamental in shaping overall biomass and the spatial distribution of cells residing in multispecies biofilms, but also to result in coordinated regulation of gene expression in the different species present. These communal interactions often lead to emergent properties in biofilms, such as enhanced tolerance against antibiotics, host immune responses, and other stresses, which have been shown to provide benefits to all biofilm members not only the enabling sub-populations. However, the specific molecular mechanisms of cellular processes affecting spatial organization, and vice versa, are poorly understood and very complex to unravel. Therefore, detailed description of the spatial organization of individual bacterial cells in multispecies communities can be an alternative strategy to reveal the nature of interspecies interactions of constituent species. Closing the gap between visual observation and biological processes may become crucial for resolving biofilm related problems, which is of utmost importance to environmental, industrial, and clinical implications. This review briefly presents the state of the art of studying interspecies interactions and spatial organization of multispecies communities, aiming to support theoretical and practical arguments for further advancement of this field.
Keywords: multispecies biofilms, interspecies interactions, spatial organization
Introduction – Spatial Organization in Biofilms
Microorganisms typically live in dense multispecies communities with distinct patterns of spatial organization, termed biofilms. This specific mode of living generally provides strong fitness advantages to biofilm-associated bacteria compared to their planktonic counterparts (Hoiby et al., 2010; Bjarnsholt et al., 2013; Kragh et al., 2016), making biofilms ubiquitously present in most bacterial habitats including the human body (Burmølle et al., 2010; Bjarnsholt, 2013). Having established the general circumstances for biofilm formation in monocultures, the focus in biofilm research has in recent years been gradually moving toward investigating the complexity and interactions in multispecies biofilms (Zengler and Palsson, 2012; Burmølle et al., 2014).
Observations from numerous studies converge on the finding that bacteria residing in multispecies biofilms show spatial positioning in response to interspecific interactions, which is crucial for community functions (Momeni et al., 2013; Burmølle et al., 2014). For instance, the well-studied oral biofilms show complex organized microbial structures due to interspecies interactions (Zijnge et al., 2010; Ferrer and Mira, 2016). In other cases, bacterial species tend to keep proper distance, thereby avoiding strong substrate competition or toxic compounds secreted by others (Kim et al., 2008), illustrating that different types of interactions can lead to distinct types of spatial organization (Satoh et al., 2007; Momeni et al., 2013). In general, spatially structured environments are believed to facilitate a more effective coexistence of bacterial species, due to a negation of localized competitive interactions as well as the stabilization of beneficial interactions such as co-metabolism and coordinated social activities (Hansen et al., 2007; Pande et al., 2015). This in turn has been illustrated to increase virulence during infections (Harrison et al., 2006).
Visual analysis of biofilm communities is one of the most common tools applied in the study of biofilm model systems as well as natural systems, including in vivo settings. Therefore, the ability to correlate mechanisms of interspecies interactions to the spatial organization of multispecies biofilms is vital for enhancing our understanding of the function and dynamics in these systems. Recent technological advances have resulted in lowered costs and improved resolution. Methods such as next generation sequencing (Medini et al., 2008), advanced mass spectrometry (Park et al., 2014), and high resolution microscopy (Neu and Lawrence, 2015), enable the generation of ample amounts of data to provide more precise analyses of complex communities. However, how interspecies interactions affect spatial organization, and vice versa, are still poorly understood. Therefore, a better understanding of spatial organization based on relevant studies might provide an alternative approach to reveal interspecies interactions of constituent species in multispecies communities.
The main purpose of this review is to establish that there is a link between interspecies interactions and spatial organization of bacterial species in in vitro multispecies biofilms. Species residing in microbial communities with complex structures frequently affect the growth of the others by both cooperative and competitive interactions (Kim et al., 2008; Nadell et al., 2009), and these interplays might change with the development of the community in response to both cell-cell and cell-environment interactions (Coyte et al., 2015). Additionally, the existing methods and the emerging high-resolution techniques, suitable for application in visualizing spatial distribution of bacterial species in complex communities, will be briefly described.
Biological Processes Affecting the Spatial Organization of Microbes in Multispecies Biofilms
A central tenet of biofilm research is that biofilm formation is a dynamic and cyclic process. Up until now, current models describe biofilm formation as a circular process initiated by free-floating microbial cells attaching to a surface, followed by growth into a mature structurally complex biofilm and culminating in the dispersal of detached cells from the biofilm, allowing colonization of new niches (Tolker-Nielsen et al., 2000; Sauer et al., 2002; Hall-Stoodley et al., 2004; Monds and O’Toole, 2009). However, a new study has challenged this model by showing that aggregates have a fitness advantage when initiating biofilms compared to planktonic bacteria (Kragh et al., 2016). Interspecies interactions, including co-metabolism, quorum sensing and production of antimicrobial compounds have been shown to play important roles in regulating microbial activities in vitro (Hojo et al., 2009), and have important functions in shaping the spatial structure of biofilms. It seems that bacterial species organize in three general forms; interspecific segregation, co-aggregation, and/or stratification, based on different types of interspecies interactions. Momeni et al. (2013) have partly verified this by combining computational models and experimental work, indicating that only strong inter-population cooperation leads to partner intermixing in microbial communities. In the following sections, we will discuss the interconnection between different types of interspecies interactions and spatial organization of microbes in multispecies biofilms.
The Role of Metabolic Interactions
Metabolic interactions, leading to cooperation, exploitation, or competition, are ubiquitous in multispecies biofilm and play important roles in maintaining the diversity and stability of microbial communities (Møller et al., 1998; Embree et al., 2015; Zelezniak et al., 2015).
Based on evolutionary theory, it has been suggested that spatial intermixing favors the evolution of metabolic mutualism because it keeps mutualistic partners in close proximity, thereby allowing for stronger reciprocity, which in turn facilitate the exchange of metabolites between partners (Doebeli and Knowlton, 1998; Foster and Wenseleers, 2006). In fact, this was experimentally demonstrated by Pande et al. (2015), who found that cooperative cross-feeding was much more favored in spatially structured environments, such as biofilms, compared to mixed, non-structured environments. In addition, Palmer et al. (2001) studied the interspecific interactions of three pairwise co-aggregating oral bacterial strains composed of Streptococcus gordonii, Streptococcus oralis, and Actinomyces naeslundii. Of these three species, two (S. oralis and A. naeslundii) were incapable of growing as mono-species biofilm, however, both of them showed a luxuriant, intermixing growth pattern when co-cultured on a glass surface using saliva as the sole nutrient source, indicating strong metabolic interdependence between them.
Interestingly, in contrast to intermixing patterns observed in cases of strong metabolic interdependence (mutualism), Estrela and Brown (2013) stated that weak metabolic interdependence results in a spatial structure with initial species segregation. Likewise, exploitative and competitive interactions driven by limited nutrients and space during the development of biofilms lead to greater interspecific segregation. In accordance, Christensen et al. (2002) observed this phenomenon by performing a study showing the shift from weak cooperation to exploitation of two species co-cultured in biofilm. This lead to a layered structure with patchy patterning of species as the biomass increased, with the benefitting species overgrowing the one being benefitted from. Hansen et al. (2007) also found a similar pattern formed between two species, as a result of a recent mutation in the overgrowing strain. This novel exploitative interaction could, however, be transient, since it generates a selective pressure on the producing strain to avoid being exploited.
Competitive metabolic interactions among species often play critical roles in shaping the structure and function of multispecies communities. Scheffer and van Nes (2006) demonstrated that self-organized segregation of species in communities is a direct effect of competitive interactions, using a model based on classical competition theory. Kim et al. (2008) and Momeni et al. (2013) further verified this by performing competitive experiments in mixed species communities, indicating that biofilms with competitive pairs frequently show a spatial structure with interspecific segregation (equal-fitness competition) or one species dominant (unequal-fitness competition).
Chemical Heterogeneity in Multispecies Biofilms
It is well recognized that planktonic, well-mixed conditions result in bacterial cell populations with fairly uniform physiological activity, whereas physiological heterogeneity is common in biofilms due to chemical gradients resulting from the spatial structure (Lee et al., 2005; Gu et al., 2013). Specifically, the extracellular polymeric substances (EPS) provide biofilms a physical structure that segregates microenvironments with different biochemical properties (Lawrence et al., 2007; Madsen et al., 2015), in which bacteria respond and adapt to the local chemical conditions, leading to biological heterogeneity (Xu et al., 1998; Stewart and Franklin, 2008).
As discussed above, metabolic interactions play important roles in the spatial organization of species in multispecies biofilms, which is an indirect reflection of chemical heterogeneity. It has been shown that the layered structure of oral biofilms is a consequence of strong interspecific cooperation among anaerobes and aerobes (Kolenbrander et al., 2006; Mark Welch et al., 2016). Moreover, the chemical heterogeneity is verified in numerous studies using microsensors to detect microscale concentration profiles for a number of compounds in biofilms (Schramm et al., 2000; Chae et al., 2012; Sun et al., 2014), showing that the distribution of these compounds was closely related to that of species in biofilms, which is most likely mechanistically explained by specific metabolic interactions. Therefore, there is potential for elucidating interspecies interactions in detail, by using microelectrodes combined with other techniques, including meta-omics analysis (Embree et al., 2014), fluorescent in situ hybridization (FISH; Almstrand et al., 2013) or stable isotope labeling (Verastegui et al., 2014).
The Connection Between Community Structure and Horizontal Gene Transfer
Biofilm offers an environment with a high cell density, making it well suited for horizontal gene transfer (HGT) (Sørensen et al., 2005). Genes that are transferred horizontally can provide a vast array of new phenotypes, e.g., adjustment of the host metabolic level, resistance toward antibacterial compounds or the ability to form biofilm, thereby facilitating a wide range of adaptions. It has been shown that the presence of conjugative plasmids stimulates biofilm development and modifies the biofilm structure (Ghigo, 2001; Reisner et al., 2006), and the mechanism has been assigned to the plasmid encoded conjugative pili facilitating enhanced attachment and biofilm formation. Reisner et al. (2003) found that cell attachment and microcolony formation were similar for Escherichia coli isolates with and without IncF plasmids, however, expansion of the biofilm structure only occurred for conjugative plasmid-carrying strains resulting in a 70–100 μm thick structure. We observed a different effect when co-culturing Pseudomonas putida, E. coli and Kluyvera sp. in multispecies biofilms; when P. putida harbored the conjugative plasmid pKJK5, the biofilm-attached biomass decreased (Røder et al., 2013). This was also the case when P. putida was grown as single species biofilm with and without the plasmid. In addition, fimbriae encoding genes have been identified on various plasmids and were found to stimulate biofilm formation (Burmølle et al., 2008, 2012).
The studies mentioned above exemplify how biofilm structure can be affected by plasmids; however, biofilm also allows for the possibility of maintaining the plasmids. Biofilms have been proposed to facilitate the maintenance of plasmids over time by allowing plasmids to be retained in the inactive parts of the biofilm, dominated by dormant cells and thus devoid of growth competition (Madsen et al., 2014).
Spatial Organization Reflects and Drives the Function of Multispecies Communities
The development of multispecies biofilms is believed to proceed as a succession of cooperative and competitive events, which are influenced by cell-cell and cell-environment interactions. The continuously changing cell-cell interactions, resulting from the heterogeneity in biofilms, are considered a main force driving the function of multispecies communities (Stewart and Franklin, 2008).
An established example showing this is dental plaque in which species spatially organize in a stratified structure (Marsh et al., 2011; Mark Welch et al., 2016), which is believed to be largely affected by co-aggregation interactions mediated by functional adhesins located on the cell surface (Kolenbrander et al., 2006). Within the structure, individual taxa are located at micron scales in a non-random way suggestive of their functional niches in these communities (Mark Welch et al., 2016). Co-aggregation has been shown to strongly impact the development of oral multispecies communities, because only specific secondary and late colonizers can be adopted into the already attached biofilm (Rickard et al., 2003), which is believed to be important for the proper establishment and function of these communities. However, the effect of co-aggregation on the development and spatial organization of non-oral multispecies communities is still poorly understood.
Spatial organization has been demonstrated to play an important role in reducing the strength of between-species cooperative interactions, contributing to the stability of multispecies communities, such as gut microbiome (Coyte et al., 2015). It is predicted that the host can benefit from zoning species within gut microbiome in order to maintain community stability and species diversity. This strategy, used by the microbes for development of more stable and diverse communities, has similarities to that utilized by plants and animals in nature, which stabilizes ecosystems by branching the food chain into a web with spatial structure (McCann, 2000). Several model investigations have shown that natural food-web structures can, indeed, enhance ecosystem stability (McCann and Hastings, 1997; Huxel and McCann, 1998; Post et al., 2000). Moreover, the specific patterning of microbes leads to increased biomass and enhanced tolerance toward antibiotics compared to their component species individually (Ahmed et al., 2009; Kolenbrander et al., 2010; Lee et al., 2014). Therefore, a dual focus on both spatial organization and multispecies biofilms succession will help identify entry points to uncover the molecular mechanisms of the processes, in order to address biofilm related problems.
Methods to Examine and Link the Spatial Organization to Biological Activities
The key to better understand the forces shaping the structure of biofilms is found in linking the visual observations to specific processes. This can be achieved by selecting appropriate biofilm model systems and high-resolution techniques for analysis (Røder et al., 2016).
Confocal laser scanning microscopy (CLSM) has become a standard technique for structural investigation of hydrated microbiological samples at the microscale. Two- or multiphoton microscopy provides new options for in situ imaging, such as visualizing thick specimens and excitation of ultraviolet fluorochromes. In order to visualize the bacterial cells in biofilms, different probes or differential staining approaches have been applied to distinguish potential interference of fluorochromes (Muffler and Ulber, 2014). Specifically, fluorescent protein, immunofluorescence techniques (Bao et al., 2014) and FISH (Almstrand et al., 2013) are the most commonly used techniques targeting specific bacterial groups or species. Many advanced versions of the FISH technique have been implemented for different purposes, most commonly; catalyzed reporter deposition-FISH (CARD-FISH) for fluorescent signal enhancement (Lupini et al., 2011), peptide nucleic acid-FISH (PNA-FISH) for better penetration and faster hybridization (Werthén et al., 2010) and CLASI-FISH for expanding the number of distinguishable taxa in complex communities (Valm et al., 2012). In addition, an innovative technique, laser ablation electrospray ionization mass spectrometry (LAESI-MS), has been used for studying the spatial distribution of mixed species biofilms and is showing potential advantages contrasted to CLSM experiments (Dean et al., 2015), as LAESI-MS allows for rapid analysis of unfixed and wet biofilms.
Data processing software, such as Daime (Daims et al., 2006), has been widely used for analyzing co-localization of species in multispecies biofilms (Schillinger et al., 2012; Almstrand et al., 2013), which can be used as an indicator for tracking the shifts of interspecific interactions with the development of the communities. In addition, simple model systems, composed of few species, may be valuable to predict interactions in more complex communities (Ren et al., 2015).
It should be emphasized that the function of a biofilm community cannot be inferred solely from the spatial organization of the sessile organisms without information about their respective metabolites or specific activities. However, recently developed technologies show great promise in providing this additional vital information. As examples, matrix-assisted laser desorption/ionization (MALDI) and laser ablation inductively coupled plasma (LA-ICP) have recently been successfully combined with imaging mass spectrometry (IMS) to visualize proteins and metal distributions within biofilms, demonstrating that metal fluctuations play important roles in microbial community structure (Wakeman et al., 2016). In addition, meta-transcriptomics, in combination with single-cell genome sequencing, has been used to uncover the complicated microbial interactions and metabolic capabilities of an alkane-degrading methanogenic community (Embree et al., 2014). Tracing of isotope labeled substrates can be adapted to reveal the flow of metabolites in complex microbial communities in conjunction with metagenomic analysis, enabling identification of functional genes in different species (Verastegui et al., 2014). Combining such chemical and genetic analysis with detailed information of the spatial organization of multispecies biofilm will facilitate linking the spatial organization to the biological activities.
Conclusion
Space is an important factor that can either facilitate or hamper interactions between cells. This of course is dependent on the physio-chemical nature of the interaction itself, whether e.g., it is expedited via a diffusible molecule or cell-cell contact. However, due to the vast number of interactions occurring in multispecies biofilms it is, in many cases, much cumbersome to try and fully understand each interaction. Moreover, understanding overall mechanisms of interactions and cellular processes that affect spatial organization, and vice versa, can provide a means of associating spatial distribution of different bacteria and how they interact. Therefore, the better understanding of spatial organization based on relevant studies would be an attractive alternative approach that might reveal interspecies interactions of constituent species in multispecies communities. Closing the gap between visual observation and biological processes may become crucial for resolving biofilm related problems, which is of utmost importance to environmental, industrial, and clinical implications.
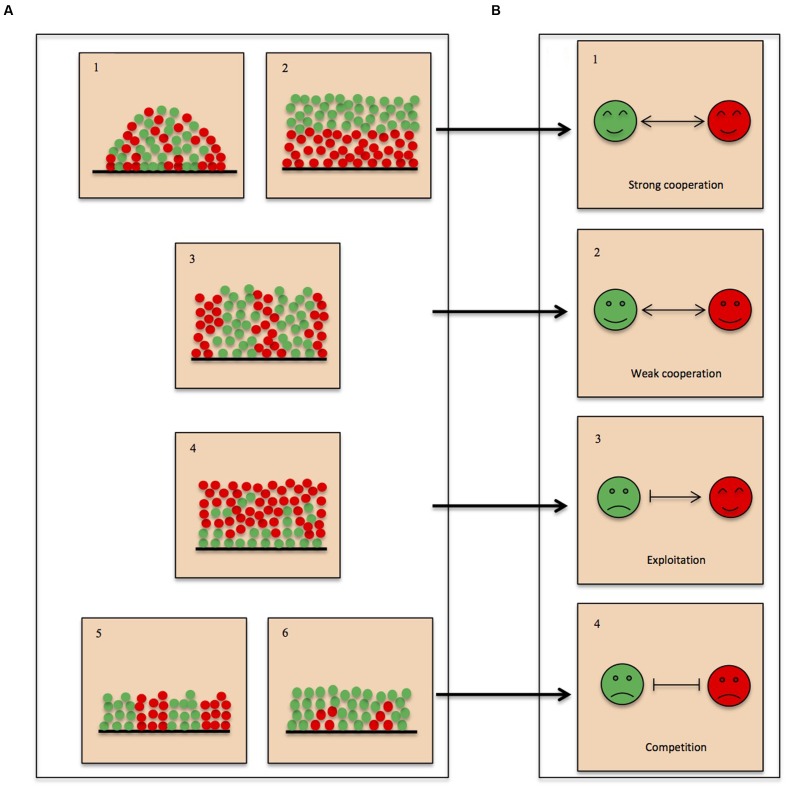
The fitness effects of interspecific interactions are believed to be a major driving force of spatial organization of microbes in multispecies biofilms. Simulations derived from an individual-based fitness model showed that deviations caused by indirect interactions can obscure direct interactions and lead to misinterpretations (Momeni et al., 2013). Additionally, interspecific interactions between competitive pairs could shift to indirect cooperation for coexistence in response to the surrounding species or resources (Wootton, 2002). Even so, there is apparent and predictable correlation between interspecific interactions and spatial organization of microbes in multispecies biofilms, as illustrated in Figure 1: strong interdependence (Figure 1B-1) favors intermixed distribution (Figure 1A-1) or layered structure (Figure 1A-2), mainly dependent on the metabolic properties of the microbes, whereas weak interdependence (Figure 1B-2) is reflected in spatial structures with interspecific segregation when there is no nutrient and space limitation (Figure 1A-3); layered structure with patchy patterning (Figure 1A-4) implying exploitation (Figure 1B-3); mutual inhibition (Figure 1B-4) will result in decreased biomass with patchy patterning (unequal-fitness competition; Figure 1A-5) or interspecific segregation (equal-fitness competition; Figure 1A-6). Hence, detailed description of spatial organization in multispecies biofilm using advanced in situ imaging techniques in combination with recently developed technologies can reveal the molecular mechanisms underpinning interspecies interactions and bring biofilm research an important step forward.
FIGURE 1.
Linking interspecific interactions to spatial organizations in multispecies biofilms. (A) Spatial organization of microbes in biofilms. Based on observation, there are five general forms in which the bacteria are organized: (1) intermixing; (2) layered structure without patchy patterning; (3,5) interspecific segregation; (4) layered structure with patchy patterning; (6) patchy patterning structure. Both (4) and (6) represent that one species is dominant in the biofilms. (B) Interspecific interactions. Interspecific interactions are divided into three groups based on whether species residing in multispecies communities benefit or suffer from the specific interaction: (1,2) cooperation; (3) exploitation; (4) competition. Cooperation and exploitation lead to increased biomass of one or all member species in mixed species compared to single-species biofilms (A-1,2,3,4), whereas competition results in decreased biomass of all member species in mixed species compared to single-species biofilms (A-5,6). Respectively, arrows and vertical bars represent growth facilitation and inhibition.
Author Contributions
WL and HR conducted the literature study and wrote the draft manuscript. TB, JM, SS, and MB edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was partly funded by grants from The Danish Council for Independent Research; ref no: DFF-1335-00071 and ref no: DFF-1323-00235 (SIMICOM) and from the Villum Young Investigator Programme (ref no. 10098).
References
- Ahmed N. A., Petersen F. C., Scheie A. A. (2009). AI-2/LuxS is involved in increased biofilm formation by Streptococcus intermedius in the presence of antibiotics. Antimicrob. Agents Chemother. 53 4258–4263. 10.1128/AAC.00546-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almstrand R., Daims H., Persson F., Sörensson F., Hermansson M. (2013). New methods for analysis of spatial distribution and coaggregation of microbial populations in complex biofilms. Appl. Environ. Microbiol. 79 5978–5987. 10.1128/AEM.01727-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K., Belibasakis G. N., Thurnheer T., Aduse-Opoku J., Curtis M. A., Bostanci N. (2014). Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol. 14:258 10.1186/s12866-014-0258-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T. (2013). The role of bacterial biofilms in chronic infections. APMIS Suppl. 136 1–51. 10.1111/apm.12099 [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T., Alhede M., Alhede M., Eickhardt-Sørensen S. R., Moser C., Kühl M., et al. (2013). The in vivo biofilm. Trends Microbiol. 21 466–474. 10.1016/j.tim.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Burmølle M., Bahl M. I., Jensen L. B., Sørensen S. J., Hansen L. H. (2008). Type 3 fimbriae, encoded by the conjugative plasmid pOLA52 enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 154 187–195. 10.1099/mic.0.2007/010454-0 [DOI] [PubMed] [Google Scholar]
- Burmølle M., Norman A., Sørensen S. J., Hansen L. H. (2012). Sequencing of IncX-plasmids suggests ubiquity of mobile forms of a biofilm-promoting gene cassette recruited from Klebsiella pneumoniae. PLoS ONE 7:e41259 10.1371/journal.pone.0041259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmølle M., Ren D., Bjarnsholt T., Sørensen S. J. (2014). Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 22 84–91. 10.1016/j.tim.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Burmølle M., Thomsen T. R., Fazli M., Dige I., Christensen L., Homøe P., et al. (2010). Biofilms in chronic infections – a matter of opportunity – monospecies biofilms in multispecies infections. FEMS Immunol. Med. Microbiol. 59 324–336. 10.1111/j.1574-695X.2010.00714.x [DOI] [PubMed] [Google Scholar]
- Chae K. J., Kim S. M., Oh S. E., Ren X., Lee J., Kim I. S. (2012). Spatial distribution and viability of nitrifying, denitrifying and ANAMMOX bacteria in biofilms of sponge media retrieved from a full-scale biological nutrient removal plant. Bioprocess Biosyst. Eng. 35 1157–1165. 10.1007/s00449-012-0701-9 [DOI] [PubMed] [Google Scholar]
- Christensen B. B., Haagensen J. A. J., Heydorn A., Molin S. (2002). Metabolic commensalism and competition in a two-species microbial consortium. Appl. Environ. Microbiol. 68 2495–2502. 10.1128/AEM.68.5.2495-2502.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte K. Z., Schluter J., Foster K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350 663–666. 10.1126/science.aad2602 [DOI] [PubMed] [Google Scholar]
- Daims H., Lücker S., Wagner M. (2006). Daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8 200–213. 10.1111/j.1462-2920.2005.00880.x [DOI] [PubMed] [Google Scholar]
- Dean S. N., Walsh C., Goodman H., van Hoek M. L. (2015). Analysis of mixed biofilm (Staphylococcus aureus and Pseudomonas aeruginosa) by laser ablation electrospray ionization mass spectrometry. Biofouling 31 151–161. 10.1080/08927014.2015.1011067 [DOI] [PubMed] [Google Scholar]
- Doebeli M., Knowlton N. (1998). The evolution of interspecific mutualisms. Proc. Natl. Acad. Sci. U.S.A. 95 8676–8680. 10.1073/pnas.95.15.8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embree M., Liu J. K., Al-bassam M. M., Zengler K. (2015). Networks of energetic and metabolic interactions define dynamics in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 112 15450–15455. 10.1073/pnas.1506034112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embree M., Nagarajan H., Movahedi N., Chitsaz H., Zengler K. (2014). Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J. 8 757–767. 10.1038/ismej.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrela S., Brown S. P. (2013). Metabolic and demographic feedbacks shape the emergent spatial structure and sunction of microbial communities. PLoS Comput. Biol. 9:e1003398 10.1371/journal.pcbi.1003398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M. D., Mira A. (2016). Oral biofilm architecture at the microbial scale. Trends Microbiol. 24 246–248. 10.1016/j.tim.2016.02.013 [DOI] [PubMed] [Google Scholar]
- Foster K. R., Wenseleers T. (2006). A general model for the evolution of mutualisms. J. Evol. Biol. 19 1283–1293. 10.1111/j.1420-9101.2005.01073.x [DOI] [PubMed] [Google Scholar]
- Ghigo J. M. (2001). Natural conjugative plasmids induce bacterial biofilm development. Nature 412 442–445. 10.1038/35086581 [DOI] [PubMed] [Google Scholar]
- Gu H., Hou S., Yongyat C., De Tore S., Ren D. (2013). Patterned biofilm formation reveals a mechanism for structural heterogeneity in bacterial biofilms. Langmuir 29 11145–11153. 10.1021/la402608z [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Hansen S. K., Rainey P. B., Haagensen J. A. J., Molin S. (2007). Evolution of species interactions in a biofilm community. Nature 445 533–536. 10.1038/nature05514 [DOI] [PubMed] [Google Scholar]
- Harrison F., Browning L. E., Vos M., Buckling A. (2006). Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 4:21 10.1186/1741-7007-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35 322–332. 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Hojo K., Nagaoka S., Ohshima T., Maeda N. (2009). Bacterial interactions in dental biofilm development. J. Dent. Res. 88 982–990. 10.4161/viru.2.5.16140 [DOI] [PubMed] [Google Scholar]
- Huxel G. R. R., McCann K. (1998). Food web stability: the influence of trophic flows across habitats. Am. Nat. 152 460–469. 10.1086/286182 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Boedicker J. Q., Choi J. W., Ismagilov R. F. (2008). Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl. Acad. Sci. U.S.A. 105 18188–18193. 10.1073/pnas.0807935105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. E., Palmer R. J., Periasamy S., Jakubovics N. S. (2010). Oral multispecies biofilm development and the key role of cell–cell distance. Nat. Rev. Microbiol. 8 471–480. 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E., Palmer R. J., Rickard A. H., Jakubovics N. S., Chalmers N. I., Diaz P. I. (2006). Bacterial interactions and successions during plaque development. Periodontol. 2000 42 47–79. 10.1111/j.1600-0757.2006.00187.x [DOI] [PubMed] [Google Scholar]
- Kragh K. N., Diggle S. P., Allen R. J., Gordon V. (2016). Role of multicellular aggregates in biofilm formation. mBio 7:e00237–16 10.1128/mBio.00237-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. R., Swerhone G. D. W., Kuhlicke U., Neu T. R. (2007). In situ evidence for microdomains in the polymer matrix of bacterial microcolonies. Can. J. Microbiol. 53 450–458. 10.1139/W06-146 [DOI] [PubMed] [Google Scholar]
- Lee B., Haagensen J. A. J., Ciofu O., Bo J., Høiby N., Molin S., et al. (2005). Heterogeneity of biofilms formed by nonmucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J. Clin. Microbiol. 43 5247–5255. 10.1128/JCM.43.10.5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. W. K., Periasamy S., Mukherjee M., Xie C., Kjelleberg S., Rice S. A. (2014). Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 8 894–907. 10.1038/ismej.2013.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupini G., Proia L., Di Maio M., Amalfitano S., Fazi S. (2011). CARD-FISH and confocal laser scanner microscopy to assess successional changes of the bacterial community in freshwater biofilms. J. Microbiol. Methods 86 248–251. 10.1016/j.mimet.2011.05.011 [DOI] [PubMed] [Google Scholar]
- Madsen J. S., Burmølle M., Sørensen S. J. (2014). A spatiotemporal view of plasmid loss in biofilms and planktonic cultures. Biotechnol. Bioeng. 110 3071–3074. 10.1002/bit.25204 [DOI] [PubMed] [Google Scholar]
- Madsen J. S., Lin Y. C., Squyres G. R., Price-Whelan A., Torio A. S., Song A., et al. (2015). Facultative control of matrix production optimizes competitive fitness in Pseudomonas aeruginosa PA14 biofilm models. Appl. Environ. Microbiol. 81 8414–8426. 10.1128/AEM.02628-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch J. L., Rossetti B. J., Rieken C. W., Dewhirst F. E., Borisy G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113 E791–E800. 10.1073/pnas.1522149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P. D., Moter A., Devine D. A. (2011). Dental plaque biofilms: communities, conflict and control. Periodontology 55 16–35. 10.1111/j.1600-0757.2009.00339.x [DOI] [PubMed] [Google Scholar]
- McCann K., Hastings A. (1997). Re-evaluating the omnivory–stability relationship in food webs. Proc. Biol. Sci. 264 1249–1254. 10.1098/rspb.1997.0172 [DOI] [Google Scholar]
- McCann K. S. (2000). The diversity-stability debate. Nature 405 228–233. 10.1038/35012234 [DOI] [PubMed] [Google Scholar]
- Medini D., Serruto D., Parkhill J., Relman D. A., Donati C., Moxon R., et al. (2008). Microbiology in the post-genomic era. Nat. Rev. Microbiol. 6 419–430. 10.1038/nrmicro1901 [DOI] [PubMed] [Google Scholar]
- Møller S., Sternberg C., Andersen J. B., Christensen B. B., Ramos J. L., Givskov M., et al. (1998). In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64 721–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momeni B., Brileya K. A., Fields M. W., Shou W. (2013). Strong inter-population cooperation leads to partner intermixing in microbial communities. Elife 2013:e00230 10.7554/eLife.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds R. D., O’Toole G. A. (2009). The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17 73–87. 10.1016/j.tim.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Muffler K., Ulber R. (2014). Productive Biofilms. Berlin: Springer. [Google Scholar]
- Nadell C. D., Xavier J. B., Foster K. R. (2009). The sociobiology of biofilms. FEMS Microbiol. Rev. 33 206–224. 10.1111/j.1574-6976.2008.00150.x [DOI] [PubMed] [Google Scholar]
- Neu T. R., Lawrence J. R. (2015). Innovative techniques, sensors, and approaches for imaging biofilms at different scales. Trends Microbiol. 23 233–242. 10.1016/j.tim.2014.12.010 [DOI] [PubMed] [Google Scholar]
- Palmer R. J., Kazmerzak K., Hansen M. C., Kolenbrander P. E. (2001). Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69 5794–5804. 10.1128/IAI.69.9.5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S., Kaftan F., Lang S., Svatos A., Germerodt S., Kost C. (2015). Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 10 1413–1423. 10.1038/ismej.2015.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A. J., Murphy K., Krieger J. R., Brewer D., Taylor P., Habash M., et al. (2014). A temporal examination of the planktonic and biofilm proteome of whole cell Pseudomonas aeruginosa PAO1 using quantitative mass spectrometry. Mol. Cell. Proteomics 13 1095–1105. 10.1074/mcp.M113.033985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post D. M., Conners M. E., Goldberg D. S., Jan N. (2000). Prey preference by a top predator and the stability of linked food chains. Ecology 81 8–14. 10.1890/0012-9658(2000)081[0008:PPBATP]2.0.CO;2 [DOI] [Google Scholar]
- Reisner A., Haagensen J. A., Schembri M. A., Zechner E. L., Molin S. (2003). Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48 933–946. 10.1046/j.1365-2958.2003.03490.x [DOI] [PubMed] [Google Scholar]
- Reisner A., Höller B. M., Molin S., Zechner L., Ho B. M., Zechner E. L. (2006). Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 188 3582–3588. 10.1128/JB.188.10.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Madsen J. S., Sørensen S. J., Burmølle M. (2015). High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 9 81–89. 10.1038/ismej.2014.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard A. H., Gilbert P., High N. J., Kolenbrander P. E., Handley P. S. (2003). Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11 94–100. 10.1016/S0966-842X(02)00034-3 [DOI] [PubMed] [Google Scholar]
- Røder H. L., Hansen L. H., Sørensen S. J., Burmølle M. (2013). The impact of the conjugative IncP-1 plasmid pKJK5 on multispecies biofilm formation is dependent on the plasmid host. FEMS Microbiol. Lett. 344 186–192. 10.1111/1574-6968.12175 [DOI] [PubMed] [Google Scholar]
- Røder H. L., Sørensen S. J., Burmølle M. (2016). Studying bacterial multispecies biofilms: where to start? Trends Microbiol. 24 503–513. 10.1016/j.tim.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Satoh H., Miura Y., Tsushima I., Okabe S. (2007). Layered structure of bacterial and archaeal communities and their in situ activities in anaerobic granules. Appl. Environ. Microbiol. 73 7300–7307. 10.1128/AEM.01426-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Sauer K., Camper A. K., Camper A. K., Ehrlich G. D., Ehrlich G. D., et al. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184 1140–1154. 10.1128/JB.184.4.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer M., van Nes E. H. (2006). Self-organized similarity, the evolutionary emergence of groups of similar species. Proc. Natl. Acad. Sci. U.S.A. 103 6230–6235. 10.1073/pnas.0508024103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillinger C., Petrich A., Lux R., Riep B., Kikhney J., Friedmann A., et al. (2012). Co-localized or randomly distributed? pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis. PLoS ONE 7:e37583 10.1371/journal.pone.0037583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A., DeBeer D., Gieseke A., Amann R. (2000). Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2 680–686. 10.1046/j.1462-2920.2000.00150.x [DOI] [PubMed] [Google Scholar]
- Sørensen S. J., Bailey M., Hansen L. H., Kroer N., Wuertz S., Sorensen S. J., et al. (2005). Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3 700–710. 10.1038/nrmicro1232 [DOI] [PubMed] [Google Scholar]
- Stewart P. S., Franklin M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6 199–210. 10.1038/nrmicro1838 [DOI] [PubMed] [Google Scholar]
- Sun J., Hu S., Sharma K. R., Ni B. J., Yuan Z. (2014). Stratified microbial structure and activity in sulfide- and methane- producing anaerobic sewer biofilms. Appl. Environ. Microbiol. 80 7042–7052. 10.1128/AEM.02146-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolker-Nielsen T., Brinch U. C., Ragas P. C., Andersen J. B., Jacobsen C. S., Molin S. (2000). Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182 6482–6489. 10.1128/JB.182.22.6482-6489.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm A. M., Mark Welch J. L., Borisy G. G. (2012). CLASI-FISH: principles of combinatorial labeling and spectral imaging. Syst. Appl. Microbiol. 35 496–502. 10.1016/j.syapm.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verastegui Y., Cheng J., Engel K. (2014). Multisubstrate isotope labeling and metagenomic analysis of active soil bacterial communities. MBio 5 1–12. 10.1128/mBio.01157-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman C. A., Moore J. L., Noto M. J., Zhang Y., Singleton M. D., Prentice B. M., et al. (2016). The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Commun. 7:11951 10.1038/ncomms11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werthén M., Henriksson L., Jensen P. Ø., Sternberg C., Givskov M., Bjarnsholt T. (2010). An in vitro model of bacterial infections in wounds and other soft tissues. APMIS 118 156–164. 10.1111/j.1600-0463.2009.02580.x [DOI] [PubMed] [Google Scholar]
- Wootton J. T. (2002). Indirect effects in complex ecosystems: recent progress and future challenges. J. Sea Res. 48 157–172. 10.1016/S1385-1101(02)00149-1 [DOI] [Google Scholar]
- Xu K. D., Stewart P. S., Xia F., McFeters G. (1998). Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64 4035–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelezniak A., Andrejev S., Ponomarova O., Mende D. R., Bork P., Patil K. R. (2015). Correction for Zelezniak et al, metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. U.S.A. 112 6449–6454. 10.1073/pnas.1522642113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengler K., Palsson B. O. (2012). A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol. 10 366–372. 10.1038/nrmicro2763 [DOI] [PubMed] [Google Scholar]
- Zijnge V., Van Leeuwen M. B. M., Degener J. E., Abbas F., Thurnheer T., Gmür R., et al. (2010). Oral biofilm architecture on natural teeth. PLoS ONE 5:e9321 10.1371/journal.pone.0009321 [DOI] [PMC free article] [PubMed] [Google Scholar]