Abstract
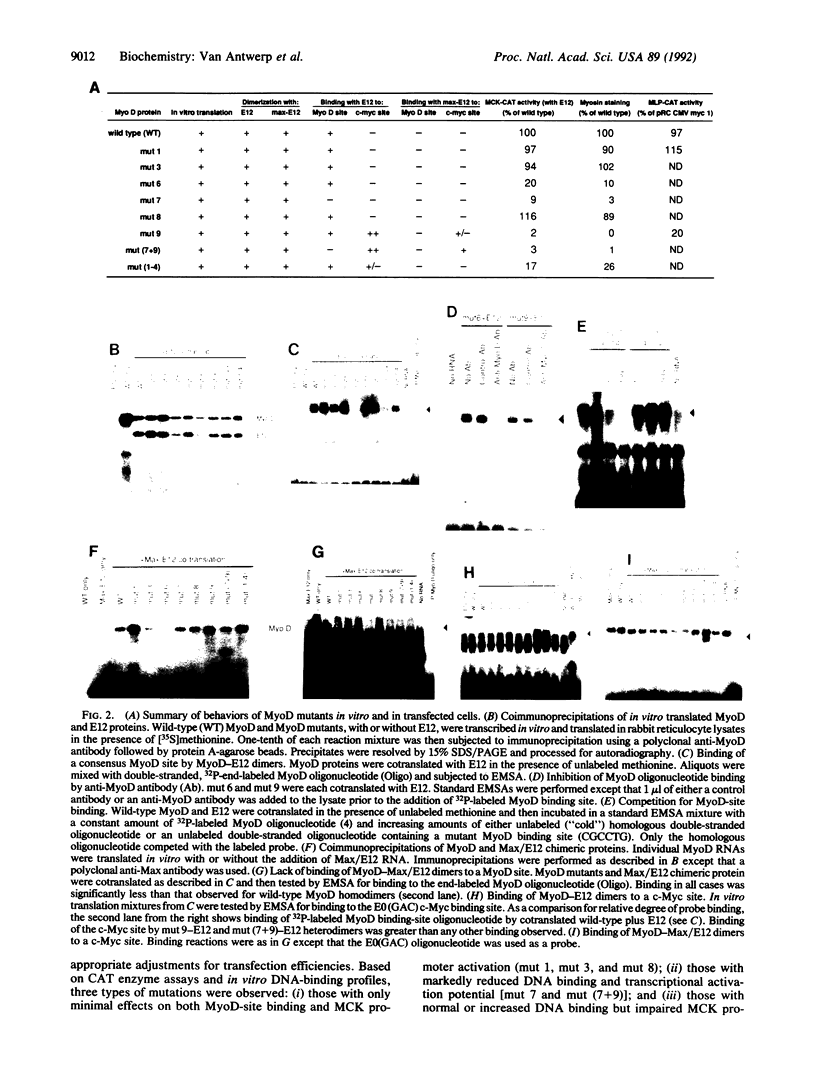
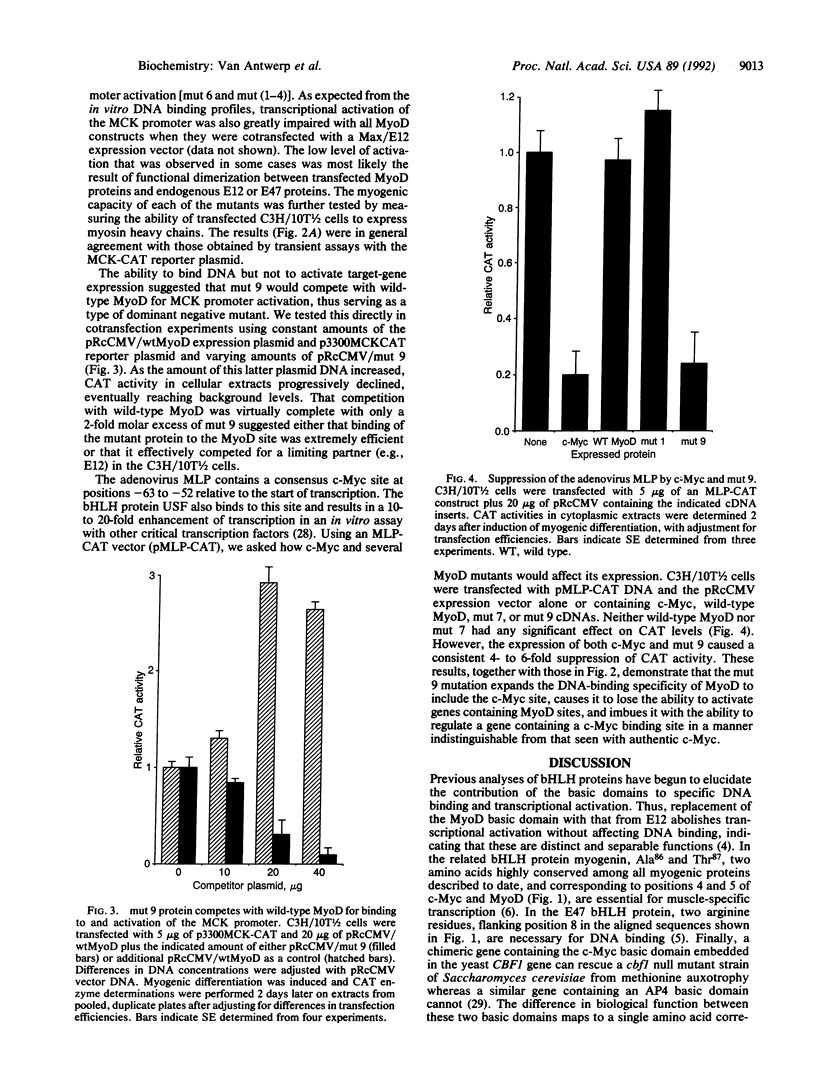
MyoD and c-Myc, members of the large "basic-helix-loop-helix" family of proteins, regulate diverse aspects of both normal and neoplastic growth and specific gene regulation. These two proteins differ at 9 of the 14 amino acids that comprise the basic domains necessary for DNA binding and transcriptional control. Individual amino acids in the MyoD basic domain were mutated to those found at the analogous positions in c-Myc. Four classes of mutants were obtained: (i) those with no effects on MyoD-site binding or activation of MyoD-responsive genes, (ii) those with no effect on MyoD-site binding but with a loss of activation potential, (iii) those with a loss of both DNA binding and activation potential, and (iv) one mutant (mut 9, Leu122----Arg) that left MyoD-site binding unaffected but imparted a new c-Myc-site binding capability. mut 9 competed with wild-type protein for the activation of MyoD-responsive reporter genes but could, like c-Myc, also suppress the adenovirus major-late promoter, which contains a c-Myc binding site. Our studies thus identify specific amino acid residues in the MyoD basic domain that are important for its activity as a DNA-binding transcriptional activator. Most significantly, our results with mut 9 indicate that Leu122 of MyoD is a critical determinant of specific DNA binding and that mutation at this residue can alter this specificity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bader D., Masaki T., Fischman D. A. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982 Dec;95(3):763–770. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Brennan T. J., Chakraborty T., Olson E. N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T. J., Olson E. N. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 1990 Apr;4(4):582–595. doi: 10.1101/gad.4.4.582. [DOI] [PubMed] [Google Scholar]
- Buskin J. N., Hauschka S. D. Identification of a myocyte nuclear factor that binds to the muscle-specific enhancer of the mouse muscle creatine kinase gene. Mol Cell Biol. 1989 Jun;9(6):2627–2640. doi: 10.1128/mcb.9.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J., Rocques P. J., Braun T., Bober E., Arnold H. H., Fisher C., Fletcher C., Brown K., Gusterson B. A., Carter R. L. Expression of members of the myf gene family in human rhabdomyosarcomas. Br J Cancer. 1991 Dec;64(6):1039–1042. doi: 10.1038/bjc.1991.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg C. H., Linkhart T. A., Olwin B. B., Hauschka S. D. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J Cell Biol. 1987 Aug;105(2):949–956. doi: 10.1083/jcb.105.2.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescenzi M., Fleming T. P., Lassar A. B., Weintraub H., Aaronson S. A. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8442–8446. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., Dolde C., Gillison M. L., Kato G. J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C. V., McGuire M., Buckmire M., Lee W. M. Involvement of the 'leucine zipper' region in the oligomerization and transforming activity of human c-myc protein. Nature. 1989 Feb 16;337(6208):664–666. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Denis N., Blanc S., Leibovitch M. P., Nicolaiew N., Dautry F., Raymondjean M., Kruh J., Kitzis A. c-myc oncogene expression inhibits the initiation of myogenic differentiation. Exp Cell Res. 1987 Sep;172(1):212–217. doi: 10.1016/0014-4827(87)90107-8. [DOI] [PubMed] [Google Scholar]
- Dias P., Parham D. M., Shapiro D. N., Webber B. L., Houghton P. J. Myogenic regulatory protein (MyoD1) expression in childhood solid tumors: diagnostic utility in rhabdomyosarcoma. Am J Pathol. 1990 Dec;137(6):1283–1291. [PMC free article] [PubMed] [Google Scholar]
- Endo T., Nadal-Ginard B. Transcriptional and posttranscriptional control of c-myc during myogenesis: its mRNA remains inducible in differentiated cells and does not suppress the differentiated phenotype. Mol Cell Biol. 1986 May;6(5):1412–1421. doi: 10.1128/mcb.6.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone G., Tatò F., Alemà S. Distinctive effects of the viral oncogenes myc, erb, fps, and src on the differentiation program of quail myogenic cells. Proc Natl Acad Sci U S A. 1985 Jan;82(2):426–430. doi: 10.1073/pnas.82.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Predicted structural similarities of the DNA binding domains of c-Myc and endonuclease Eco RI. Science. 1992 Jan 24;255(5043):464–466. doi: 10.1126/science.1734524. [DOI] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff E., Bister K., Klempnauer K. H. Sequence-specific DNA binding by Myc proteins. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4323–4327. doi: 10.1073/pnas.88.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988 Dec 15;336(6200):646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca S. A., Grossi M., Falcone G., Alemà S., Tatò F. Interaction with normal cells suppresses the transformed phenotype of v-myc-transformed quail muscle cells. Cell. 1989 Jul 14;58(1):123–131. doi: 10.1016/0092-8674(89)90409-1. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Lin H., Yutzey K. E., Konieczny S. F. Muscle-specific expression of the troponin I gene requires interactions between helix-loop-helix muscle regulatory factors and ubiquitous transcription factors. Mol Cell Biol. 1991 Jan;11(1):267–280. doi: 10.1128/mcb.11.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner J. H., Wold B. J. c-myc inhibition of MyoD and myogenin-initiated myogenic differentiation. Mol Cell Biol. 1991 May;11(5):2842–2851. doi: 10.1128/mcb.11.5.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Sartorelli V., Webster K. A., Kedes L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 1990 Oct;4(10):1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Sejersen T., Sümegi J., Ringertz N. R. Density-dependent arrest of DNA replication is accompanied by decreased levels of c-myc mRNA in myogenic but not in differentiation-defective myoblasts. J Cell Physiol. 1985 Dec;125(3):465–470. doi: 10.1002/jcp.1041250315. [DOI] [PubMed] [Google Scholar]
- Shapiro D. N., Houghton P. J., Hazelton B. J., Germain G. S., Murti K. G., Rahman A., Houghton J. A. Morphological and molecular characterization of spontaneous myogenic differentiation in a human rhabdomyosarcoma cell line. Cancer Res. 1990 Sep 15;50(18):6002–6009. [PubMed] [Google Scholar]
- Smith M. J., Charron-Prochownik D. C., Prochownik E. V. The leucine zipper of c-Myc is required for full inhibition of erythroleukemia differentiation. Mol Cell Biol. 1990 Oct;10(10):5333–5339. doi: 10.1128/mcb.10.10.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino V., Pepperkok R., Davis R. L., Ansorge W., Philipson L. Cell proliferation inhibited by MyoD1 independently of myogenic differentiation. Nature. 1990 Jun 28;345(6278):813–815. doi: 10.1038/345813a0. [DOI] [PubMed] [Google Scholar]
- Tapscott S. J., Lassar A. B., Davis R. L., Weintraub H. 5-bromo-2'-deoxyuridine blocks myogenesis by extinguishing expression of MyoD1. Science. 1989 Aug 4;245(4917):532–536. doi: 10.1126/science.2547249. [DOI] [PubMed] [Google Scholar]
- Tapscott S. J., Weintraub H. MyoD and the regulation of myogenesis by helix-loop-helix proteins. J Clin Invest. 1991 Apr;87(4):1133–1138. doi: 10.1172/JCI115109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M. J., Tapscott S. J., Davis R. L., Wright W. E., Lassar A. B., Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989 Jul 28;58(2):241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- Tonin P. N., Scrable H., Shimada H., Cavenee W. K. Muscle-specific gene expression in rhabdomyosarcomas and stages of human fetal skeletal muscle development. Cancer Res. 1991 Oct 1;51(19):5100–5106. [PubMed] [Google Scholar]
- Voronova A., Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991 Feb 15;251(4995):761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991 Aug;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- Yang B. S., Geddes T. J., Pogulis R. J., de Crombrugghe B., Freytag S. O. Transcriptional suppression of cellular gene expression by c-Myc. Mol Cell Biol. 1991 Apr;11(4):2291–2295. doi: 10.1128/mcb.11.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]



