Abstract
Culture-based detection of nontuberculous Mycobacteria (NTM) in respiratory samples is time consuming and can be subject to overgrowth by nonmycobacterial bacteria. We describe a single-reaction TaqMan quantitative PCR assay for the direct detection of NTM species in clinical samples that is specific, sensitive, and robust.
TEXT
While rates of infection caused by members of the Mycobacterium tuberculosis complex continue to fall in developed countries (1, 2), disease caused by nontuberculous mycobacteria (NTM) is an area of growing concern (3–6). Pulmonary infection represents more than 90% of NTM cases (7) and has been described in a range of clinical contexts (8–11). Appropriate management of suspected pulmonary NTM infection requires the timely detection and identification of the etiological agent. The current “gold standard” for detection of NTM in respiratory samples relies on protracted in vitro culture, potentially delaying targeted therapy. It also requires samples to undergo decontamination prior to culture to lower levels of commensal microbiota (12) and is associated with variable sensitivity (13). The ability to perform a rapid quantitative screen for the presence of any NTM species would provide an important early indication of mycobacterial involvement and would be informative in cases where samples are culture negative, despite clinical or radiological signs.
To prevent false-positive results arising from the detection of closely related species (14, 15), existing molecular assays target narrow phylogenetic groups or specific pathogens (16–21), require prior mycobacterial isolation by culture (22–24), or are unable to provide accurate species-level NTM identification (25). We describe a TaqMan quantitative PCR (qPCR) assay, based on the single-copy hsp65 gene, for the direct detection of NTM species in respiratory clinical samples.
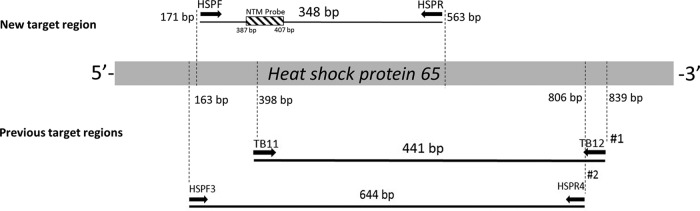
The assay design was based on the full-length hsp65 gene sequences that are available for 116 of the 174 currently described NTM species, including all 56 NTM species reported in respiratory disease (see Fig. S1 in the supplemental material). The PCR primers (forward, HSP171 [5′-CGCCAAGGAGATCGAGCTGG-3′], and reverse, HSP563 [5′-GGACAAGGTCGGCAACGAGGG-3′]) generate a 348-bp hsp65 amplicon and are used in conjunction with a TaqMan probe (5′-FAM-AGAAGGCCGTCGAGAAGGTCA-BHQ-3′ [FAM, 6-carboxyfluorescein; BHQ, black hole quencher]) at an annealing temperature of 60°C (Fig. 1). A detailed description of assay development and methods is provided in the supplemental material.
FIG 1.
Primer and probe target sites. Primer binding sites for previously described NTM detection assay primers are also shown, as follows: #1, Telenti et al. (30); #2, Kim et al. (31).
In silico analysis indicated complete homology to the targeted hsp65 gene region for 77 Mycobacterium species. Fourteen species had ≤2 nucleotide mismatches within the primer binding region, with a corresponding reduction in annealing temperature of up to 4.5°C. However, in all such cases, the corresponding primer binding region showed 100% sequence homology (see Tables S1 and S2 in the supplemental material). Twenty-one mycobacterial species (including M. tuberculosis and Mycobacterium leprae) and 40 assessed nonmycobacterial species had >3 nucleotide mismatches to the primer sequences, requiring an annealing temperature of <55.5°C (see Fig. S2 and Table S3 in the supplemental material).
The assay's performance was assessed using DNA extracts from 15 NTM strains and negative controls that included closely related nonmycobacterial species, common respiratory pathogens, nine M. tuberculosis strains, Mycobacterium bovis, Escherichia coli, and human DNA (Table 1). The assay's sensitivity was assessed using a dilution series of purified Mycobacterium abscessus DNA (selected based both on its clinical importance and its position within NTM phylogeny). The correlation between template concentration and cycle threshold (CT) values was linear between 3.34 × 103 and 2.65 × 108 CFU/ml equivalents (slope, −3.31; R2 = 0.99), with a reaction efficiency of 100%. Analysis using Mycobacterium intracellulare DNA, a species with a single-base primer mismatch, resulted in a linear range of 6.26 × 103 to 4.39 × 108 CFU/ml equivalents (slope, −3.403; R2 = 0.99), with a reaction efficiency of 97%.
TABLE 1.
Amplification data for reference and control strains
| Species (strain) | Sourcec | CT value | CFU/ml equivalent |
|---|---|---|---|
| Mycobacterial speciesa | |||
| M. abscessus | ATCC 19977 | 16.9 | 2.65 × 108 |
| M. avium | Clinical strain | 20.1 | 7.56 × 107 |
| M. chelonae | ATCC 35752 | 22.6 | 1.26 × 107 |
| M. flavescens | Collection strain | 28.3 | 2.42 × 105 |
| M. fortuitum | ATCC 9820 | 18.6 | 2.08 × 108 |
| M. goodii | Clinical strain | 21.3 | 3.21 × 107 |
| M. gordonae | Clinical strain | 23 | 9.93 × 106 |
| M. interjectum | Clinical strain | 21.2 | 3.34 × 107 |
| M. intracellulare | Clinical strain | 23 | 9.99 × 106 |
| M. kansasii | Clinical strain | 24.9 | 2.68 × 106 |
| M. lentiflavum | Clinical strain | 21 | 3.81 × 107 |
| M. marinum | Collection strain | 23.3 | 7.72 × 106 |
| M. simiae | Clinical strain | 19 | 1.62 × 108 |
| M. smegmatis | Clinical strain | 20.8 | 4.39 × 107 |
| M. bovis (BCG) | Collection strain | NDd | ND |
| M. tuberculosis (H37Rv) | Clinical strain | ND | ND |
| M. tuberculosis (Uganda 1) | Clinical strain | ND | ND |
| M. tuberculosis (Orygis) | Clinical strain | ND | ND |
| M. tuberculosis (MDR) | Clinical strain | ND | ND |
| M. tuberculosis (LAM/Uganda 1) | Clinical strain | ND | ND |
| M. tuberculosis (EAI) | Clinical strain | ND | ND |
| M. tuberculosis (EAI 1) | Clinical strain | ND | ND |
| M. tuberculosis (BJ Delhi CAS) | Clinical strain | ND | ND |
| M. tuberculosis (BJ Delhi CAS 1) | Clinical strain | ND | ND |
| Nonmycobacterial speciesb | |||
| Rhodococcus equi | Collection strain | ND | ND |
| Nocardia farcinica | Collection strain | ND | ND |
| Corynebacterium glucuronolyticum | Collection strain | ND | ND |
| Staphylococcus aureus | Clinical strain | ND | ND |
| Pseudomonas aeruginosa | Clinical strain | ND | ND |
| Haemophilus influenzae | Clinical strain | ND | ND |
| Escherichia coli | Clinical strain | ND | ND |
| Streptococcus pneumoniae | Clinical strain | ND | ND |
| Human | Placental DNA | ND | ND |
South Australian pathology collection.
Flinders Medical Centre pathology laboratory.
ATCC, American Type Culture Collection.
ND, not detected.
The potential for carryover of clinical sample components to influence assay performance was assessed in three ways. First, the amplification efficiency and dynamic range of M. abscessus DNA were determined following the addition of DNA extracts from culture- and qPCR-negative bronchoalveolar lavage (BAL) and sputum samples. Second, the assay's performance was assessed following the addition of purified human DNA at a concentration that substantially exceeded the levels in respiratory clinical samples. Third, the impact of the addition of horse blood prior to DNA extraction on M. abscessus DNA amplification efficiency was assessed, using a dilution series starting at 50% (vol/vol). In each case, no significant change in assay performance was observed (Mann Whitney test, P > 0.3) (see Fig. S3 to S5 in the supplemental material).
Assay validation was performed using 42 respiratory samples from patients suspected of respiratory NTM infection, including 30 BAL samples and 12 sputum samples (of which 8 were NTM positive according to standard diagnostic testing; see Table S4 in the supplemental material). Positive results from NTM culture were confirmed by qPCR, and species identity was confirmed by DNA sequencing. However, in three cases, samples were NTM culture negative but qPCR positive. Mycobacterium avium was detected in the BAL sample at a concentration of 8.7 × 104 CFU/ml equivalents, while Mycobacterium flavescens and M. avium were detected in the two sputum samples at 2.1 × 104 and 6.4 × 103 CFU/ml equivalents, respectively (Table 2; see also Table S5).
TABLE 2.
NTM-positive respiratory samples as determined by qPCR, and corresponding diagnostic culture results
| Sample type | CT value | CFU/ml equivalent | Identification obtained with: |
|
|---|---|---|---|---|
| qPCR/sequencing | Diagnostic microbiology | |||
| BAL fluid | 30.5 | 1.36 × 105 | M. avium | M. avium |
| BAL fluid | 30.9 | 1.02 × 105 | M. avium | M. avium |
| BAL fluid | 32.1 | 4.58 × 104 | M. intracellulare | M. intracellulare |
| BAL fluid | 35.4 | 1.13 × 104 | M. avium | M. avium |
| BAL fluid | 34 | 8.70 × 104 | M. avium | —a |
| BAL fluid | 32.7 | 3.02 × 104 | M. avium | M. avium |
| Sputum | 31.1 | 3.22 × 104 | M. massiliense | M. massiliense |
| Sputum | 31.7 | 2.08 × 104 | M. flavescens | —a,b |
| Sputum | 31.5 | 2.44 × 104 | M. abscessus | M. abscessus |
| Sputum | 33.4 | 6.43 × 103 | M. avium | —a |
| Sputum | 35.5 | 1.49 × 103 | M. avium | M. avium |
—, not detected.
Sample was recorded as “insufficient for adequate assessment” for standard diagnostic analysis.
Negative culture results in patients with suspected NTM infection are not uncommon, with NTM isolated from subsequent samples in some instances (26). A number of factors could contribute to discrepancies between culture-dependent and molecular analysis. For example, culture overgrowth by nonmycobacterial species can substantially reduce NTM detection, while sample decontamination techniques used to prevent this can reduce mycobacterial viability (12). In addition, NTM recovery can be reduced in patients receiving commonly used antibiotics, such as macrolides and quinolones (12).
In the case of the culture-negative, qPCR-positive BAL sample, high levels of Haemophilus influenzae growth were reported. While this species is fastidious, the finding suggests the potential presence of other nonmycobacterial species that might have contributed to the failure to isolate NTM through bacterial overgrowth. In the case of the culture-negative sputum sample in which M. flavescens was detected by PCR, M. abscessus had been isolated from this patient on a previous occasion (although definitive typing had not been performed), and the patient had received apparently successful eradication therapy. While the basis for discordance remains unclear, it is important to highlight that, as with all PCR-based assays, a positive result does not rely on the presence of viable bacterial cells (27, 28). DNA in nonviable bacteria or present in the extracellular environment (as might occur following successful antibiotic therapy) can also act as a PCR template (29), a factor that must be taken into account when interpreting disparities between culture and PCR-based results. In the case of the patient in whose sample M. avium was detected by qPCR alone, the corresponding sample was recorded as being “insufficient for adequate assessment” by culture (an outcome that is sometimes interpreted wrongly at the clinical level as a culture-negative result). However, sputum samples collected both prior to and following this sample were also found to be culture negative, suggesting a genuine discrepancy between culture and qPCR results.
Our study was unable to assess the assay's ability to detect a number of rare or recently described NTM species for which high-quality DNA sequence data are not yet available. It was further limited by a requirement for DNA sequencing to identify the source of positive PCR results, a technology that is not available in all laboratories. However, the single-reaction qPCR assay described offers substantial advantages over other molecular assays in terms of time and cost. Importantly, the assay does not amplify DNA from M. tuberculosis, a range of closely related nonmycobacterial species, or common airway bacteria and is unaffected by the presence of high concentrations of human DNA or blood derivatives. Our assay provides a specific, sensitive, and robust means to rapidly screen respiratory clinical samples for the presence of NTM and represents an important adjunct to conventional diagnostic approaches.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to David Gordon, Microbiology and Infectious Diseases, Flinders Medical Centre, who provided clinical isolates and bacterial type strains.
The authors declare no conflict of interest.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit-sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01410-16.
REFERENCES
- 1.Brode SK, Daley CL, Marras TK. 2014. The epidemiologic relationship between tuberculosis and non-tuberculous mycobacterial disease: a systematic review. Int J Tuberc Lung Dis 18:1370–1377. doi: 10.5588/ijtld.14.0120. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2015. Leprosy. Fact sheet N101. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs101/en/ Accessed 3 May 2016. [Google Scholar]
- 3.Marras TK, Chedore P, Ying AM, Jamieson F. 2007. Isolation prevalence of pulmonary non-tuberculous mycobacteria in Ontario, 1997-2003. Thorax 62:661–666. doi: 10.1136/thx.2006.070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martín-Casabona N, Bahrmand AR, Bennedsen J, Thomsen VO, Curcio M, Fauville-Dufaux M, Feldman K, Havelkova M, Katila ML, Köksalan K, Pereira MF, Rodrigues F, Pfyffer GE, Portaels F, Urgell JR, Rüsch-Gerdes S, Tortoli E, Vincent V, Watt B, Spanish Group for Non-Tuberculosis Mycobacteria. 2004. Non-tuberculous mycobacteria: patterns of isolation. A multi-country retrospective survey. Int J Tuberc Lung Dis 8:1186–1193. [PubMed] [Google Scholar]
- 5.Mirsaeidi M, Machado RF, Garcia JG, Schraufnagel DE. 2014. Nontuberculous mycobacterial disease mortality in the United States, 1999-2010: a population-based comparative study. PLoS One 9:e91879. doi: 10.1371/journal.pone.0091879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson RM, NTM working group at Queensland TB Control Centre and Queensland Mycobacterial Reference Laboratory. 2010. Changing epidemiology of pulmonary nontuberculous mycobacteria infections. Emerg Infect Dis 16:1576–1583. doi: 10.3201/eid1610.091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Brien RJ, Geiter LJ, Snider DE Jr. 1987. The epidemiology of nontuberculous mycobacterial diseases in the United States. Results from a national survey. Am Rev Respir Dis 135:1007–1014. [DOI] [PubMed] [Google Scholar]
- 8.Horsburgh CR Jr, Gettings J, Alexander LN, Lennox JL. 2001. Disseminated Mycobacterium avium complex disease among patients infected with human immunodeficiency virus, 1985-2000. Clin Infect Dis 33:1938–1943. doi: 10.1086/324508. [DOI] [PubMed] [Google Scholar]
- 9.Park IK, Olivier KN. 2015. Nontuberculous mycobacteria in cystic fibrosis and non-cystic fibrosis bronchiectasis. Semin Respir Crit Care Med 36:217–224. doi: 10.1055/s-0035-1546751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prince DS, Peterson DD, Steiner RM, Gottlieb JE, Scott R, Israel HL, Figueroa WG, Fish JE. 1989. Infection with Mycobacterium avium complex in patients without predisposing conditions. N Engl J Med 321:863–868. doi: 10.1056/NEJM198909283211304. [DOI] [PubMed] [Google Scholar]
- 11.Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, Saulson A, Hedberg K. 2010. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features: an emerging public health disease. Am J Respir Crit Care Med 182:977–982. doi: 10.1164/rccm.201003-0503OC. [DOI] [PubMed] [Google Scholar]
- 12.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Diseases Society of America . 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 13.van Ingen J. 2015. Microbiological diagnosis of nontuberculous mycobacterial pulmonary disease. Clin Chest Med 36:43–54. doi: 10.1016/j.ccm.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Dai J, Chen Y, Dean S, Morris JG, Salfinger M, Johnson JA. 2011. Multiple-genome comparison reveals new loci for Mycobacterium species identification. J Clin Microbiol 49:144–153. doi: 10.1128/JCM.00957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radomski N, Lucas FS, Moilleron R, Cambau E, Haenn S, Moulin L. 2010. Development of a real-time qPCR method for detection and enumeration of Mycobacterium spp. in surface water. Appl Environ Microbiol 76:7348–7351. doi: 10.1128/AEM.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foongladda S, Pholwat S, Eampokalap B, Kiratisin P, Sutthent R. 2009. Multi-probe real-time PCR identification of common Mycobacterium species in blood culture broth. J Mol Diagn 11:42–48. doi: 10.2353/jmoldx.2009.080081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung KL, Yip CW, Cheung WF, Lo AC, Ko WM, Kam KM. 2009. Development of a simple and low-cost real-time PCR method for the identification of commonly encountered mycobacteria in a high throughput laboratory. J Appl Microbiol 107:1433–1439. doi: 10.1111/j.1365-2672.2009.04324.x. [DOI] [PubMed] [Google Scholar]
- 18.Lim SY, Kim BJ, Lee MK, Kim K. 2008. Development of a real-time PCR-based method for rapid differential identification of Mycobacterium species. Lett Appl Microbiol 46:101–106. doi: 10.1111/j.1472-765X.2007.02278.x. [DOI] [PubMed] [Google Scholar]
- 19.Ngan GJ, Ng LM, Jureen R, Lin RT, Teo JW. 2011. Development of multiplex PCR assays based on the 16S-23S rRNA internal transcribed spacer for the detection of clinically relevant nontuberculous mycobacteria. Lett Appl Microbiol 52:546–554. doi: 10.1111/j.1472-765X.2011.03045.x. [DOI] [PubMed] [Google Scholar]
- 20.Richardson ET, Samson D, Banaei N. 2009. Rapid identification of Mycobacterium tuberculosis and nontuberculous mycobacteria by multiplex, real-time PCR. J Clin Microbiol 47:1497–1502. doi: 10.1128/JCM.01868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gopinath K, Singh S. 2009. Multiplex PCR assay for simultaneous detection and differentiation of Mycobacterium tuberculosis, Mycobacterium avium complexes and other Mycobacterial species directly from clinical specimens. J Appl Microbiol 107:425–435. doi: 10.1111/j.1365-2672.2009.04218.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim JU, Cha CH, An HK. 2015. Direct identification of mycobacteria from clinical specimens by multiplex real-time PCR. J Appl Microbiol 118:1498–1506. doi: 10.1111/jam.12780. [DOI] [PubMed] [Google Scholar]
- 23.Kim K, Lee H, Lee MK, Lee SA, Shim TS, Lim SY, Koh WJ, Yim JJ, Munkhtsetseg B, Kim W, Chung SI, Kook YH, Kim BJ. 2010. Development and application of multiprobe real-time PCR method targeting the hsp65 gene for differentiation of Mycobacterium species from isolates and sputum specimens. J Clin Microbiol 48:3073–3080. doi: 10.1128/JCM.00939-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran AC, Halse TA, Escuyer VE, Musser KA. 2014. Detection of Mycobacterium avium complex DNA directly in clinical respiratory specimens: opportunities for improved turn-around time and cost savings. Diagn Microbiol Infect Dis 79:43–48. doi: 10.1016/j.diagmicrobio.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 25.Perry MD, White PL, Ruddy M. 2014. Potential for use of the Seegene Anyplex MTB/NTM real-time detection assay in a regional reference laboratory. J Clin Microbiol 52:1708–1710. doi: 10.1128/JCM.03585-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarand J, Davis JP, Cowie RL, Field SK, Fisher DA. 2016. Long-term follow-up of Mycobacterium avium complex lung disease in patients treated with regimens including clofazimine and/or rifampin. Chest 149:1285–1293. doi: 10.1378/chest.15-0543. [DOI] [PubMed] [Google Scholar]
- 27.Rogers GB, Stressmann FA, Koller G, Daniels T, Carroll MP, Bruce KD. 2008. Assessing the diagnostic importance of nonviable bacterial cells in respiratory infections. Diagn Microbiol Infect Dis 62:133–141. doi: 10.1016/j.diagmicrobio.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy N, Gillespie SH, Saruni AO, Kisyombe G, McNerney R, Ngowi FI, Wilson S. 1994. Polymerase chain reaction for assessing treatment response in patients with pulmonary tuberculosis. J Infect Dis 170:713–716. doi: 10.1093/infdis/170.3.713. [DOI] [PubMed] [Google Scholar]
- 29.Rogers GB, Marsh P, Stressmann AF, Allen CE, Daniels TV, Carroll MP, Bruce KD. 2010. The exclusion of dead bacterial cells is essential for accurate molecular analysis of clinical samples. Clin Microbiol Infect 16:1656–1658. doi: 10.1111/j.1469-0691.2010.03189.x. [DOI] [PubMed] [Google Scholar]
- 30.Telenti A, Marchesi F, Balz MM, Bally F, Bottger EC, Bodmer T. 1993. Rapid identification of Mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol 31:175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, Kim SH, Shim TS, Kim MN, Bai GH, Park YG, Lee SH, Chae GT, Cha CY, Kook YH, Kim BJ. 2005. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int J Syst Evol Microbiol 55:1649–1656. doi: 10.1099/ijs.0.63553-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.