Abstract
Light is one of the strongest environmental time cues for entraining endogenous circadian rhythms. Emerging evidence indicates that CREB-regulated transcription co-activator 1 (CRTC1) is a key player in this pathway, stimulating light-induced Period1 (Per1) transcription in mammalian clocks. Here, we demonstrate a light-independent role of Drosophila CRTC in sustaining circadian behaviors. Genomic deletion of the crtc locus causes long but poor locomotor rhythms in constant darkness. Overexpression or RNA interference-mediated depletion of CRTC in circadian pacemaker neurons similarly impairs the free-running behavioral rhythms, implying that Drosophila clocks are sensitive to the dosage of CRTC. The crtc null mutation delays the overall phase of circadian gene expression yet it remarkably dampens light-independent oscillations of TIMELESS (TIM) proteins in the clock neurons. In fact, CRTC overexpression enhances CLOCK/CYCLE (CLK/CYC)-activated transcription from tim but not per promoter in clock-less S2 cells whereas CRTC depletion suppresses it. Consistently, TIM overexpression partially but significantly rescues the behavioral rhythms in crtc mutants. Taken together, our data suggest that CRTC is a novel co-activator for the CLK/CYC-activated tim transcription to coordinate molecular rhythms with circadian behaviors over a 24-hour time-scale. We thus propose that CRTC-dependent clock mechanisms have co-evolved with selective clock genes among different species.
Most living organisms have evolved endogenous time-keeping mechanisms known as circadian clocks to anticipate and adapt to daily changes in the environment. External time cues, such as cycles of light, temperature or food availability, entrain the circadian oscillators to sustain 24-hour rhythms. Timing information is subsequently translated into other physiological pathways of the organism, such as sleep, metabolism, immune responses and so forth1,2,3.
At the molecular level, a transcriptional feedback network of circadian transcription factors that regulates daily rhythmic gene expression constitutes a basic framework for cell-autonomous molecular clocks4. In Drosophila, CLK-CYC, a heterodimeric transcription factor composed of CLOCK and CYCLE, binds E-box sequences and activates the transcription of period (per) and timeless (tim) genes from the late afternoon to midnight. PER-TIM complexes accumulate in the cytoplasm at early night and are then translocated to the nucleus, where they inhibit the transcriptional activity of CLK-CYC after midnight. In the morning, light-dependent TIM degradation and cumulative PER phosphorylation destabilize the PER-TIM complex. The consequent de-repression of CLK-CYC activity leads to a new daily cycle5,6,7,8,9. Additional layers of transcriptional feedback loops contribute to the robustness of the molecular clockwork, maintaining high-amplitude rhythms and diverse clock outputs10,11,12,13,14. Moreover, key players in the negative feedback loops are homologous or functionally analogous between flies and mammals15,16,17.
Light stimulates mammalian Per expression via CREB (cAMP response element binding protein)-dependent transcriptional activation, playing important roles in the photic entrainment of mammalian clocks18,19,20. Recent studies have shown that this process requires CRTC (CREB-regulated transcription co-activator)21. In fact, CRTC and its negative regulator SIK1 (salt-inducible kinase 1) constitute a negative feedback loop. Light-activated CRTC induces Sik1 transcription, and then elevated SIK1 feeds back to phosphorylate CRTC proteins, blocking their nuclear entry22,23,24. This mechanism buffers the light-dependent, phase-resetting of clocks such that animals are able to robustly sustain circadian rhythms rather than changing their circadian phase back and forth in response to sudden changes in light regime.
Here, we identify an unexpected role of CRTC in Drosophila clocks and demonstrate that Drosophila CRTC activates tim transcription to align circadian gene expression on a 24-hour time-scale and drive robust free-running rhythms in circadian behaviors. Light-independent effects of Drosophila crtc are evident in the molecular rhythms of both central pacemaker neurons and peripheral clock tissues, implicating an ancestral origin of CRTC-dependent clocks. Given distinct clock functions of CRTC homologs, we suggest a model on how CRTC-dependent clock mechanisms have co-evolved with selective clock targets among different species.
Results
crtc mutation causes long but poor rhythms in circadian behaviors
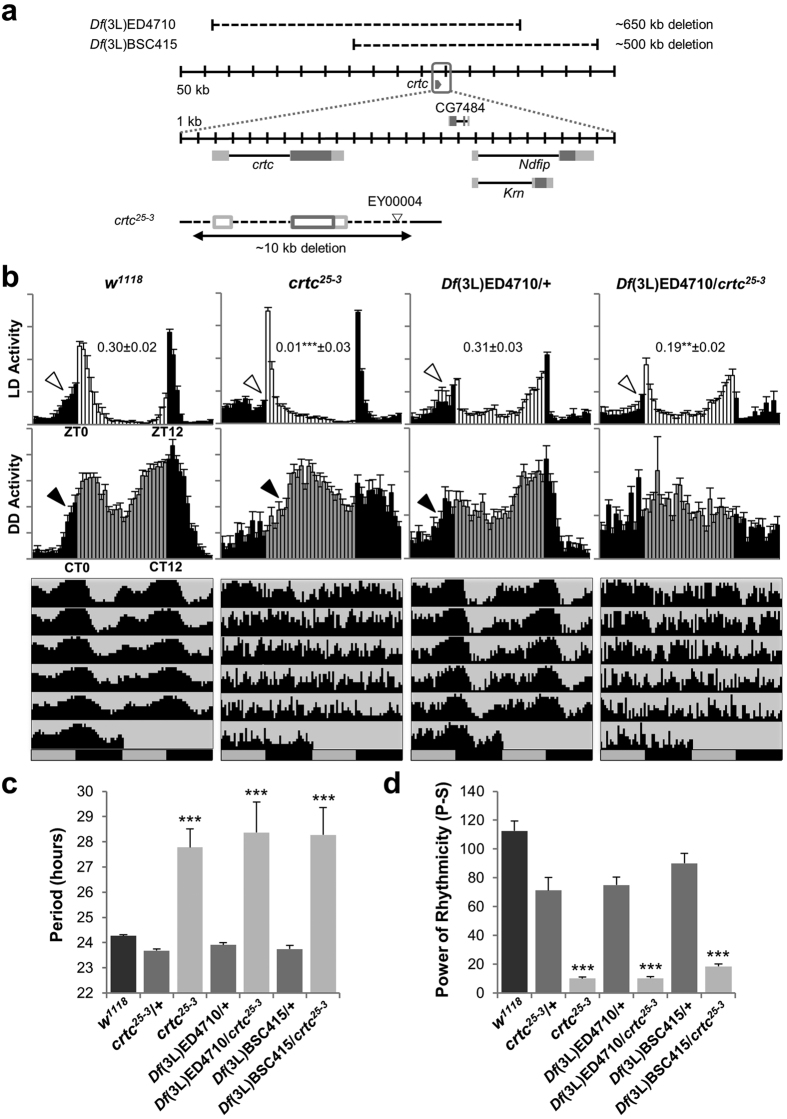
To determine if CRTC regulates circadian rhythms in Drosophila, we first examined how CRTC loss-of-function affects circadian behaviors. We tested a crtc25-3 allele lacking the entire crtc locus as a result of imprecise excision of a transposable element insert (Fig. 1a)25. Wild-type flies showed bimodal peaks of locomotor activity in light: dark (LD) cycles of 12 hours on and 12 hours off (Fig. 1b). They also anticipated the timing of lights-on and -off by gradually increasing locomotor activity around the light transitions. In contrast, crtc mutants displayed compromised morning anticipation as supported by their lower morning index (Fig. 1b, top). A quantitative assessment of circadian periods and rhythmicity in constant dark (DD) following LD entrainment revealed that crtc mutants largely exhibited arrhythmic behaviors with rapid dampening of free-running activity peaks (Fig. 1b–d, Supplementary Table 1). Nonetheless, crtc mutants with detectable rhythmicity showed long-period rhythms (Fig. 1c, Supplementary Table 1, Supplementary Fig. 1). Consistently, we observed a phase delay in the anticipatory morning activity peak of crtc mutants in the first DD cycle (Fig. 1b, middle), suggesting that their morning anticipation in LD cycles was actually masked by a startling response to lights-on. We also noticed that 27% of crtc mutants died during our behavioral tests whereas the majority of control flies survived (Supplementary Fig. 2a). Since crtc mutants are more sensitive to starvation25, we reasoned that 5% sucrose food used in our behavioral tests might partially mimic starvation conditions, thereby lowering the survival rate in crtc mutants. Behavioral tests on 10% sucrose or corn-meal food indeed rescued the lower survival rate in crtc mutants yet they still displayed the poor rhythmicity on the enriched food (Supplementary Fig. 2b, Supplementary Table 2). It is thus unlikely that crtc mutants lose behavioral rhythmicity as they become starved and sick. Finally, chromosomal deletions covering the crtc locus did not complement these circadian defects in trans-heterozygotes with crtc25-3 allele (Fig. 1b–d, Supplementary Table 1), further supporting that CRTC is necessary for sustaining robust circadian behaviors in Drosophila.
Figure 1. Drosophila CRTC is necessary for robust circadian behaviors.
(a) A schematic diagram of crtc mutant alleles. Dark-gray boxes, the translated regions of exons; light-gray boxes, untranslated regions. (b) Male flies homozygous or trans-heterozygous for crtc mutant alleles show long but poor rhythms in circadian behaviors. Normalized activity profiles in LD cycles (top) or on the first day of DD cycles (middle) were averaged from individual flies. Averaged actograms throughout the behavioral analyses were double-plotted (bottom). Anticipatory increase in locomotor activities prior to lights-on (i.e., morning anticipation) was quantified by calculating morning index in individual flies as described in Materials and Methods. Averaged morning index values +/− SEM were shown in the LD activity profiles. White arrow heads, morning anticipation in LD cycles; black arrow heads, morning anticipation in the first DD cycle; white/black bars, LD cycles; gray/black bars, DD cycles; ZT, zeitgeber time; CT, circadian time. Error bars indicate SEM. (c) Circadian periods in DD locomotor rhythms were averaged from rhythmic flies (P-S > 10; see below). ***P < 0.001 to wild-type (w1118) and all heterozygous controls as determined by one-way ANOVA, Tukey post hoc test. Error bars indicate SEM. (d) Rhythmicity in free-running locomotor behaviors was determined by measuring power (P) - significance (S) values from the chi-squared periodograms of individual flies and averaged per each genotype. ***P < 0.001 to wild-type (w1118) and all heterozygous controls as determined by one-way ANOVA, Tukey post hoc test. Error bars indicate SEM.
Drosophila clocks are sensitive to the dosage of CRTC in PDF-expressing neurons
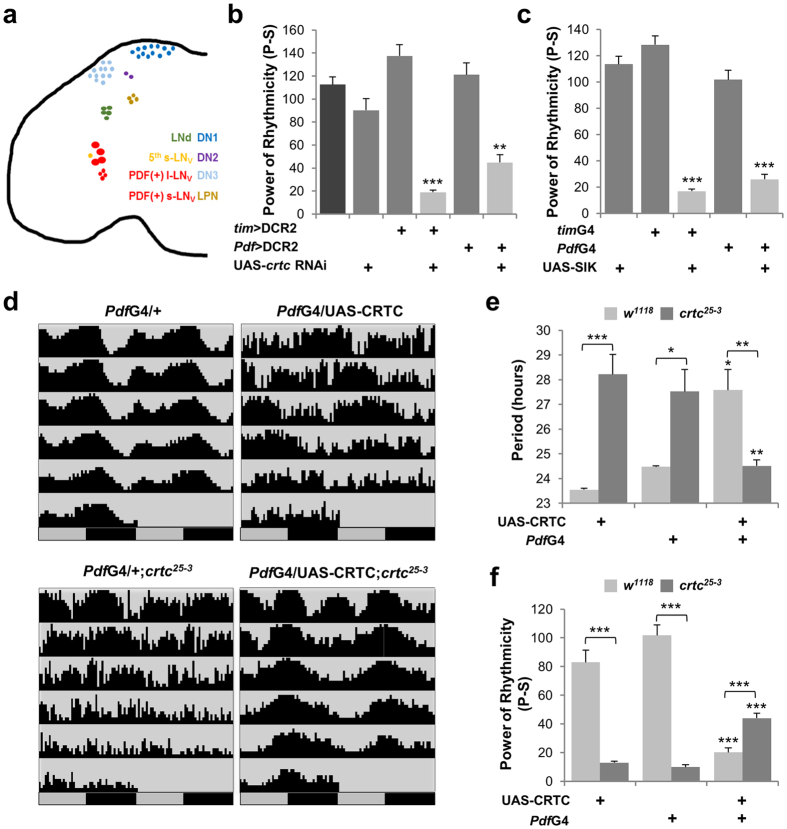
There are ~150 circadian pacemaker neurons that exhibit daily circadian gene expression in the adult fly brain (Fig. 2a). Based on their anatomical location and specific clock gene expression, these clock neurons can be divided into distinct groups that govern different aspects of circadian behaviors26,27,28,29. To map neural loci important for CRTC-dependent circadian behaviors, we examined whether CRTC depletion in specific neurons affected free-running locomotor behaviors. We first validated that our crtc RNA interference (RNAi) transgene decreased endogenous levels of CRTC proteins in fly heads by 30~60% when overexpressed by the pan-neuronal Elav-Gal4 driver or by the clock cell-specific tim-Gal4 driver (Supplementary Fig. 3). As expected, CRTC depletion in all canonical pacemaker neurons led to long but poor behavioral rhythms, phenocopying crtc mutants (Fig. 2b, Supplementary Table 3, tim > DCR2, crtc RNAi). When CRTC depletion was more restricted to large and small ventral lateral neurons (LNv) expressing the circadian neuropeptide PDF (pigment-dispersing factor) (referred to as PDF neurons hereafter)30,31,32, modest but significant decrease in DD rhythmicity was observed as compared to control flies (Fig. 2b, Supplementary Table 3, Pdf > DCR2, crtc RNAi). Consistent with the RNAi phenotype, PDF neuron-specific overexpression of Drosophila SIK2, a negative regulator of CRTC33,34, lengthened circadian periods and dampened behavioral rhythms in DD (Fig. 2c, Supplementary Table 4).
Figure 2. Drosophila clocks are sensitive to the dosage of CRTC in PDF Neurons.
(a) Circadian pacemaker neurons in adult fly brain. tim-Gal4 is expressed in all clock neurons covering lateral neurons (LNs), dorsal neurons (DNs), and lateral posterior neurons (LPNs). Pdf-Gal4 is expressed specifically in PDF-positive large and small ventral LNs (l-LNv and s-LNv, respectively). LNd, dorsal LN; 5th s-LNv, a single s-LNv not expressing a PDF neuropeptide. (b) CRTC depletion in circadian pacemaker neurons leads to arrhythmic circadian behaviors. crtc RNAi transgene was co-expressed along with the RNAi-enhancing DCR2 by tim-Gal4 or Pdf-Gal4 driver. Rhythmicity in free-running locomotor behaviors in DD cycles was measured similarly as in Fig. 1d. **P < 0.01, ***P < 0.001 to controls heterozygous for Gal4 > DCR2 or UAS-crtc RNAi as determined by one-way ANOVA, Tukey post hoc test. Error bars indicate SEM. (c) SIK2 overexpression in circadian pacemaker neurons by tim-Gal4 or Pdf-Gal4 driver leads to arrhythmic circadian behaviors. ***P < 0.001 to controls heterozygous for Gal4 or UAS-SIK as determined by one-way ANOVA, Tukey post hoc test. Error bars indicate SEM. (d) CRTC overexpression in PDF neurons rescues 24-hour behavioral rhythms in crtc mutants. Averaged actograms in DD cycles were double-plotted with their genotypes on top. (e,f) Circadian periods and rhythmicity in DD locomotor behaviors were measured as above. *P < 0.05, **P < 0.01 and ***P < 0.001 to controls heterozygous for Pdf-Gal4 or UAS-CRTC in wild-type (light-gray bars) or crtc mutants (dark-gray bars) as determined by one-way ANOVA, Tukey post hoc test. Error bars indicate SEM.
To determine whether CRTC expression in PDF neurons would be sufficient for robust 24-hour rhythms in DD, we overexpressed CRTC in PDF neurons of crtc mutant flies and compared their circadian behaviors to those in transgenic controls. Indeed, PDF neuron-specific CRTC overexpression restored 24-hour periodicity in circadian behaviors of crtc mutants whereas it partially but significantly rescued the rhythmicity phenotype (Fig. 2d–f, Supplementary Fig. 1, Supplementary Table 5). Unexpectedly, we also found that CRTC overexpression in PDF neurons caused long and weak behavioral rhythms in wild-type flies. These data indicate the presence of a window of CRTC levels or activities effective for the proper control of circadian behaviors, possibly explaining the partial rescue in crtc mutants.
Conditional manipulation of CRTC expression in PDF neurons of adult flies is sufficient to affect circadian behaviors
To understand how the genetic manipulation of crtc in PDF neurons affects circadian behaviors, we first tested if crtc mutant flies normally develop circadian pacemaker neurons important for behavioral rhythms. CRTC-depleted or CRTC-overexpressing flies did not show any gross defects in the cell bodies or axonal projections of their PDF neurons (Supplementary Fig. 4). However, crtc mutants constitutively exhibited a higher degree of the axonal arborizations from their small LNv and stronger intensities for anti-PDF staining in the dorsal projections whereas control flies showed rhythmic arborizations and cycling PDF levels in the axonal termini of the pacemaker neurons in LD cycles35,36 (Supplementary Fig. 5). Of note, similar phenotypes have been reported in clock-less tim null mutant flies although tim mutants have more but shorter PDF branches than control flies35,36,37. To further exclude the possibility of crtc effects on the development of PDF neurons, we transiently overexpressed crtc RNAi or CRTC cDNA during the period of behavioral tests. To this end, we employed a Pdf-GeneSwitch-Gal4 driver, which is activated by feeding on mifepristone (RU486)-containing food but not on food containing ethanol (vehicle control), to drive transgenic expression in PDF neurons38. This conditional depletion or overexpression of CRTC in PDF neurons significantly decreased behavioral rhythmicity in DD while modest lengthening of circadian periods was observed in crtc RNAi flies (Supplementary Fig. 6, Supplementary Table 6). Taken together, these data convincingly demonstrate that Drosophila circadian behaviors are sensitive to CRTC levels in PDF neurons of adult fly brains.
Circadian gene expression is phase-delayed by crtc mutation
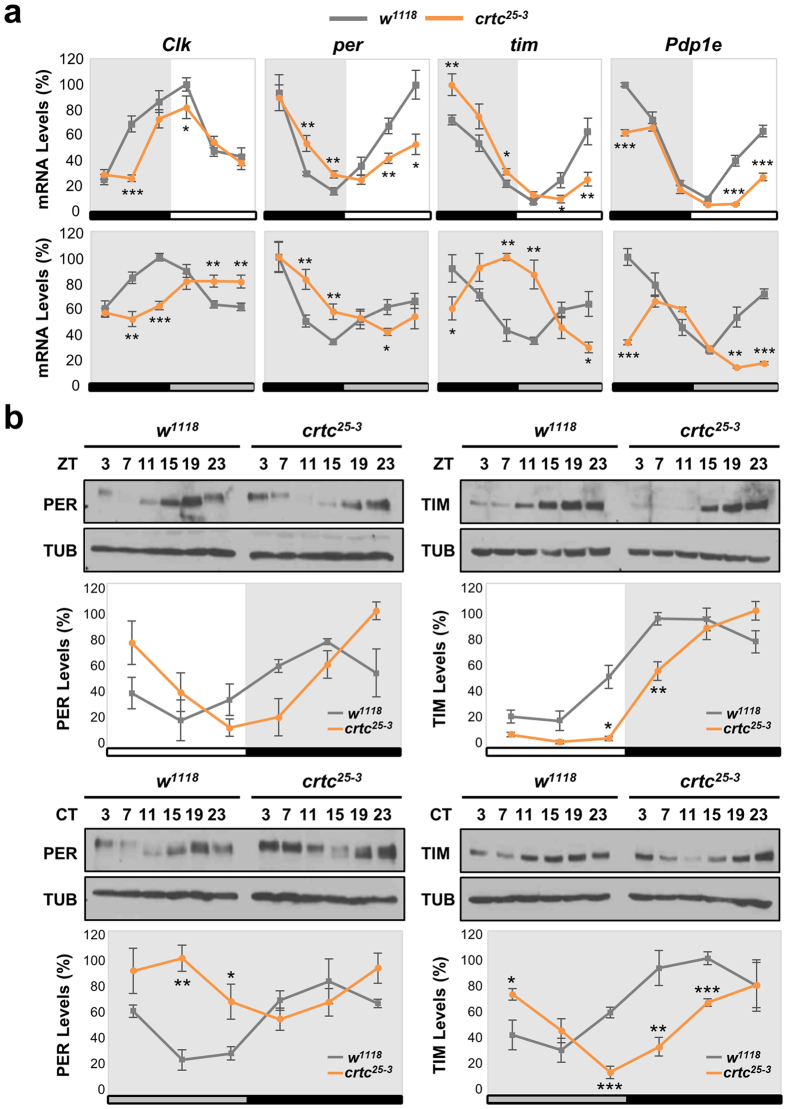
Given the transcriptional co-activator function of CRTC33, we reasoned that CRTC-dependent transcription of clock-relevant genes may explain adult-specific roles of CRTC in sustaining 24-hour behavioral rhythms. We thus examined how CRTC loss-of-function affected the cycling expression of core clock genes in fly heads. Wild-type flies exhibited daily oscillations in abundance of clock mRNAs in both LD and DD cycles (Fig. 3a). The circadian phase of Clk mRNA rhythms was in anti-phase with those of its transcriptional targets per, tim and Pdp1 (Par domain protein 1)10. We found that crtc mutants displayed a ~4-hour delay in the increasing phase of circadian mRNA expression in LD cycles while crtc effects on the declining phase were relatively weak (Fig. 3a, top). The phase delay in mRNA rhythms became more evident during the first-to-second DD cycles following LD cycles (Fig. 3a, bottom), when crtc mutant flies rapidly lost their behavioral rhythms. Accordingly, the circadian expression of PER and TIM proteins in fly head extracts was phase-delayed in crtc mutant flies (Fig. 3b). PER proteins are cumulatively phosphorylated for their ubiquitin-dependent degradation39,40. Wild-type flies thus displayed time-dependent PER phosphorylation in LD cycles, as assessed by shifts in gel mobility (Fig. 3b, top). In crtc mutants, PER phosphorylation was not readily observed until the late D phase of LD cycles, further validating the phase delay in molecular rhythms. crtc mutation had stronger impacts on the oscillating phase of PER and TIM in DD cycles (Fig. 3b, bottom), possibly due to lack of light-induced TIM degradation41,42.
Figure 3. crtc mutation phase-delays circadian gene expression in adult fly heads.
(a) Circadian expression of clock mRNAs in adult fly heads of wild-type (gray lines) and crtc mutants (orange lines). Flies were collected at six different time-points in LD cycles (top) or during the transition from the first to the second DD cycle following LD entrainment (bottom) and total RNAs from heads were purified. Relative levels of Clk, per, tim, and Pdp1 mRNAs were quantified by real-time RT-PCR. X-axis indicates zeitgeber time (ZT) in LD (top) or circadian time (CT) in DD (bottom). Y-axis indicates relative expression levels (%) at each time-point, calculated by normalizing to the peak value (set as 100). White/black bars, LD cycles; gray/black bars, DD cycles. Data represent the average of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 as determined by Student’s t-test. Error bars indicate SEM. (b) Circadian expression of PER and TIM proteins in adult fly heads of wild-type (gray lines) and crtc mutants (orange lines). Head extracts were prepared from flies harvested during LD (top) or the first DD (bottom) cycle and immunoblotted with anti-PER, anti-TIM and anti-TUBULIN (TUB, loading control) antibodies under the same experimental conditions. Images were cropped from full-length blots shown in Supplementary Fig. 12. Protein band intensities in each lane were quantified using ImageJ software and normalized to that of TUB protein. Y-axis indicates the relative expression levels (%) of PER and TIM proteins, calculated by normalizing to the peak value (set as 100). White/black bars, LD cycles; gray/black bars, DD cycles. Data represent the average of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 as determined by Student’s t-test. Error bars indicate SEM.
We next asked if CRTC overexpression has opposing effects on circadian gene expression. That is, we reasoned that CRTC overexpression would activate the rate-limiting circadian transcription, thereby phase-advancing the molecular clocks. To our surprise, however, CRTC overexpression in tim-expressing clock neurons severely dampened the rhythmic amplitude or levels of all clock-relevant mRNAs tested in fly heads (Supplementary Fig. 7). While phase delays were evident in the low-amplitude rhythms of both per and tim mRNAs, non-cycling levels of PDF receptor mRNA decreased by ~50%. These inconsistent effects of CRTC loss-of-function versus CRTC overexpression suggest a possibility that a molar excess of free CRTC proteins might interfere with baseline transcriptional activities in the clock neurons of CRTC-overexpressing flies, likely through the sequestration of general transcription factors. In fact, we observed a similar phenomenon that the transcriptional activation of reporter genes harboring per or tim promoters is rather attenuated by highly overexpressed CLK (Supplementary Fig. 8).
crtc mutation impacts on PER and TIM oscillations in circadian pacemaker neurons
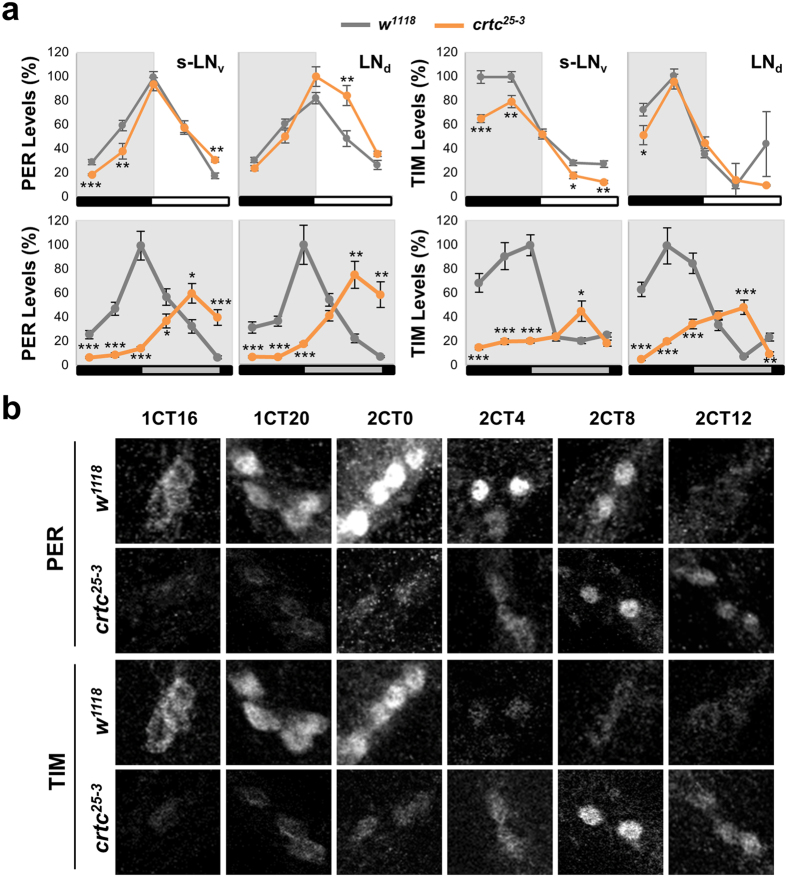
Molecular rhythms in fly head extracts largely reflect the circadian pace of peripheral clock tissues (i.e., photoreceptor neurons in the eye)43 and are not necessarily coupled to rhythmic behaviors in DD. Therefore, we assessed the circadian expression of PER and TIM proteins in behaviorally relevant pacemaker neurons by immunofluorescence assays. These clock neurons include dorsal LN (LNd) and PDF-expressing LNv (see Fig. 2a). In LD cycles, PER and TIM oscillations in the pacemaker neurons were relatively comparable between wild-type and crtc mutant flies (Fig. 4a, top). PER levels were significantly higher in LNd and large LNv of crtc mutants during the early L phase (Supplementary Fig. 9) but the phase difference in PER rhythms was subtle in small LNv. Similarly, there was no apparent phase-delay in TIM cycling yet the overall levels of TIM were modestly reduced in small LNv of crtc mutants. In contrast, ~8-hour phase-delays in PER and TIM rhythms were evident in the pacemaker neurons of crtc mutants during the transition from the first to the second DD cycle (Fig. 4a,b) as comparable to those in head extracts. These phase-delaying effects of crtc seem not due to the failure in the nuclear translocation of PER and TIM proteins (Fig. 4b). In addition, the strongest effects of crtc were observed in small LNv since the peak levels of the delayed TIM cycling substantially dampened in DD, consistent with long but poor rhythmicity in free-running behaviors. Taken together, these data suggest that Drosophila clocks require CRTC to timely induce clock gene expression in both central clock neurons and peripheral clock tissues. Nonetheless, the differential impact of crtc on the robustness of molecular rhythms (e.g., the amplitude of TIM cycling in head extracts versus small LNv) may arise from cell-type specific context of CRTC-dependent transcription.
Figure 4. crtc mutation impacts on PER and TIM oscillations in circadian pacemaker neurons.
(a) Circadian expression of PER and TIM proteins in circadian pacemaker neurons of wild-type (gray lines) and crtc mutants (orange lines). Adult fly brains were dissected at different time-points in LD cycles (top) or during the transition from the first to the second DD cycle following LD entrainment (bottom). Whole-mount immunostaining was performed using anti-PER, anti-TIM, and anti-PDF antibodies. Confocal brain images were obtained from 12–13 hemispheres at each time-point (X-axis). The fluorescence intensity of anti-PER and anti-TIM antibody staining was quantified from individual neurons using ImageJ software and averaged for each group of circadian pacemaker neurons. Y-axis indicates the relative expression levels (%) of PER and TIM proteins, calculated by normalizing to the peak value in each graph (set as 100). White/black bars, LD cycles; gray/black bars, DD cycles. s-LNv, PDF-expressing small ventral lateral neurons; LNd, dorsal LN. *P < 0.05, **P < 0.01 and ***P < 0.001 as determined by Student’s t-test. Error bars indicate SEM. (b) Representative confocal images of small LNv in wild-type (w1118) and crtc mutants (crtc25-3). 1CT/2CT, circadian time in the first/second DD cycle.
CRTC potentiates CLK-CYC–dependent tim transcription
On the basis of gene expression analyses in crtc mutants, we hypothesized that CRTC might facilitate a rate-limiting circadian transcription that coordinates molecular rhythms with 24-hour periodicity. However, the interlocked nature and feedback regulation of circadian transcription factors as well as the possibility of genetic compensation in mutant animals make it difficult to discriminate direct CRTC targets from indirect crtc effects on circadian transcription. To determine if the overall phase-delay in crtc mutants results from the attenuated transcription of general clocks (i.e., CLK-CYC–dependent clock gene expression) or a specific CRTC target, we tested crtc effects on individual clock reporter genes in clock-less Drosophila S2 cells. Of note, S2 cells endogenously express the cyc gene, so CLK overexpression alone is sufficient to support the transcriptional induction of CLK-CYC target reporter genes, such as per-luciferase (luc) or tim-luc44.
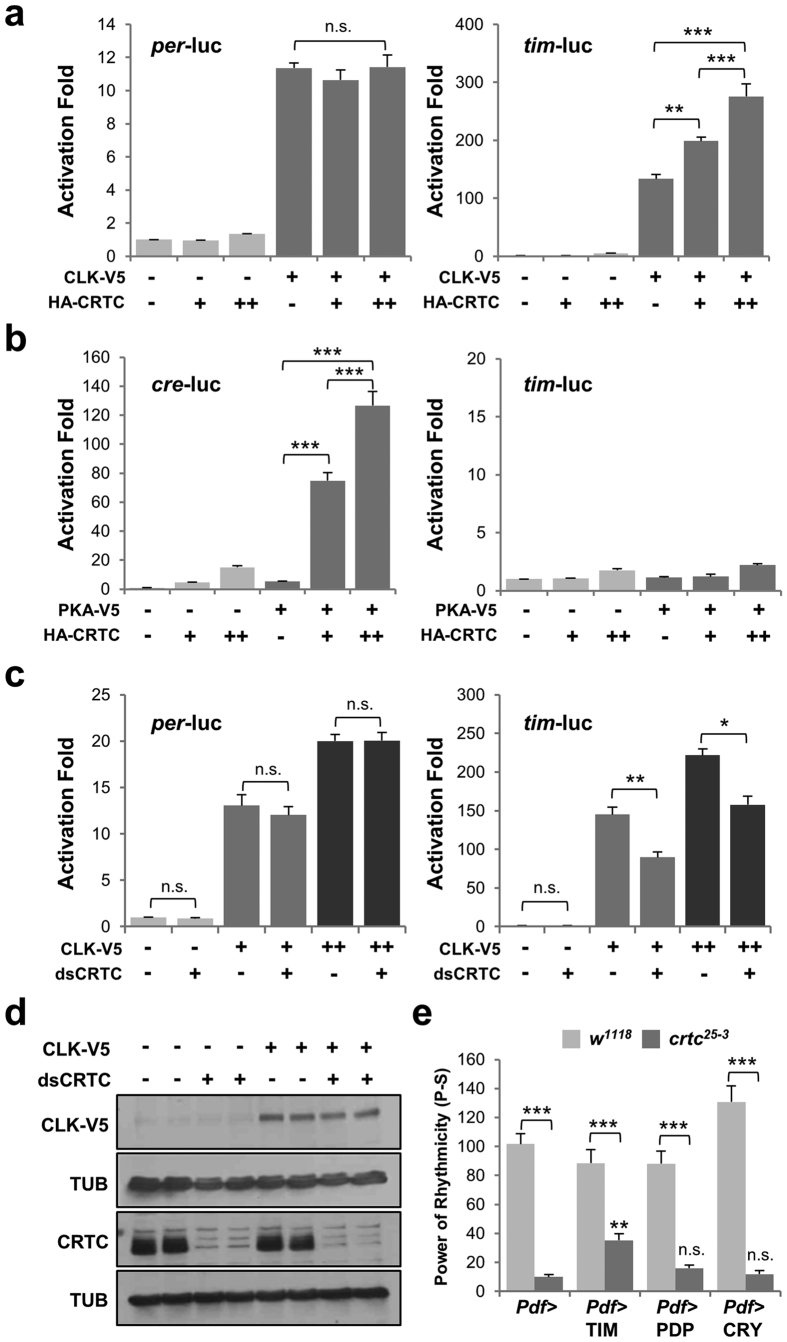
In our S2 reporter assays, CRTC overexpression activated CLK-induced expression of tim-luc, but not per-luc, in a dose-dependent manner (Fig. 5a). Lack of crtc effects on per-luc was not due to the saturating activation of the reporter gene by CLK overexpression (Supplementary Fig. 8). To confirm that crtc effects indeed require CLK, we tested if CREB-dependent transcription could stimulate the expression of tim-luc in the absence of ectopic CLK. We first validated that CRTC and cAMP-dependent protein kinase (PKA) synergistically activated the expression of cre-luc, a reporter gene whose transcription is driven by tandem repeats of cAMP-responsive element (CRE), thus faithfully responding to CREB-dependent transcription in S2 cells (Fig. 5b, left). Under similar conditions, however, CRTC and PKA did not activate the baseline transcription of tim-luc, Clk-luc or per-luc (Fig. 5b, right; Supplementary Fig. 10), verifying that CLK-dependent tim transcription is a prerequisite for crtc effects. Moreover, RNAi-mediated CRTC depletion suppressed CLK-dependent activation of tim-luc but not per-luc (Fig. 5c). We confirmed that CRTC depletion did not affect ectopic CLK expression itself (Fig. 5d), further supporting that CRTC functions as a tim promoter-specific co-activator for CLK-dependent transcription.
Figure 5. timeless is a primary clock target of Drosophila CRTC.
(a) Drosophila S2 cells in 12-well plates were co-transfected with reporter plasmids (50 ng of per-luc or tim-luc; 50 ng of renilla luciferase) and expression vectors for V5-tagged CLK (0.2 ng) and HA-tagged CRTC (0, 50 or 250 ng). Dual luciferase reporter assays were performed 40 hours after transfection. Firefly luciferase activity was first normalized to that of renilla luciferase. Relative fold-activation was then calculated relative to baseline luciferase activity in the absence of any effectors. Data represent the average from three independent experiments. Error bars indicate SEM. n.s., not significant; **P < 0.01, ***P < 0.001 as determined by one-way ANOVA, Tukey post hoc test. (b) Reporter plasmids (50 ng of cre-luc or tim-luc; 50 ng of renilla luciferase) and expression vectors for V5-tagged PKA (5 ng) and HA-tagged CRTC (0, 50 or 250 ng) were cotransfected into S2 cells in 12-well plates. Data represent average +/− SEM (n = 3). ***P < 0.001 as determined by one-way ANOVA, Tukey post hoc test. (c) S2 cells in 12-well plates were pre-incubated with 10 μg of double-stranded RNAs against CRTC (dsCRTC) or EGFP (dsEGFP, control). Forty-eight hours after dsRNA treatment, cells were co-transfected with reporter plasmids (50 ng of per-luc or tim-luc; 50 ng of renilla luciferase) and expression vector for V5-tagged CLK (0.2 and 0.5 ng). Data represent average +/− SEM (n = 3). n.s., not significant, *P < 0.05, **P < 0.01 as determined by Student’s t-test. (d) dsRNA-treated S2 cells were transfected with 10 ng of V5-tagged CLK expression vector. Cell extracts were prepared 2 days after transfection, resolved by SDS-PAGE and immunoblotted for specific proteins under the same experimental conditions. Images were cropped from full-length blots shown in Supplementary Fig. 13. (e) TIM overexpression in PDF neurons rescues circadian behaviors in crtc mutants. Each clock gene was overexpressed in PDF neurons of wild-type or crtc mutants flies. Rhythmicity in DD locomotor behaviors was measured as in Fig. 1d. Data represent average +/− SEM (n = 16–57). n.s., not significant, **P < 0.01 to Pdf-Gal4 controls in crtc mutants as determined by one-way ANOVA, Tukey post hoc test.
These in vitro results are consistent with TIM dampening in the clock neurons of crtc mutants (Fig. 4a, bottom). Given that TIM post-translationally stabilizes PER via a protein-protein interaction45,46, it is likely that the lower amplitude of TIM rhythms could be limiting for robust PER cycling in the pacemaker neurons of crtc mutants. The crtc effects on the end-point measurements of CLK-activated tim-luc expression are also consistent with the phase-delays in tim mRNA cycling in head extracts of crtc mutants (i.e., delays in the full induction of tim) since lack of CRTC failed to potently induce tim transcription within a short window of its increasing phase. We thus reasoned that if tim transcription is rate-limiting in crtc mutants, TIM overexpression would rescue their circadian behaviors. To test this hypothesis, we overexpressed TIM or other clock proteins in PDF neurons of crtc mutants and examined their free-running locomotor behaviors in DD cycles. Indeed, TIM overexpression partially, but significantly, rescued long but weak behavioral rhythms in crtc mutants, whereas PDP1 or CRY (CRYPTOCHROME) overexpression did not show any comparable rescue particularly on the rhythmicity phenotype (Fig. 5e, Supplementary Table 7). Taken together, these data convincingly support our conclusion that CRTC is a novel co-activator for the CLK-activated tim transcription to sustain 24-hour rhythms in Drosophila circadian clocks.
Discussion
CREB-dependent transcription has long been implicated in different aspects of circadian gene expression. In mammalian clocks, light exposure triggers intracellular signaling pathways that activate CREB-dependent Per1 transcription, thereby adjusting the circadian phase of master circadian pacemaker neurons in the suprachiasmatic nucleus (SCN)18,19,20. The phase-resetting process involves the specific CREB coactivator CRTC1 and its negative regulator SIK1, constituting a negative feedback in the photic entrainment via a CREB pathway21,24. In this report, we demonstrated a novel role of Drosophila CRTC that serves to coordinate circadian gene expression with 24-hour locomotor rhythms even in the absence of light. CRTC may regulate several clock-relevant genes, including those clock output genes that might be involved in the rhythmic arborizations and PDF cycling of the circadian pacemaker neurons. However, we identified tim transcription as one of the primary targets of Drosophila CRTC to sustain circadian rhythms in the free-running conditions, thus defining its light-independent clock function.
CREB could employ another transcriptional coactivator CBP (CREB-binding protein) to activate CRE-dependent transcription47,48. In fact, CBP is a rather general coactivator recruited to gene promoters by other DNA-binding transcription factors49. We have previously shown that Drosophila CBP associates with CLK, titrating its transcriptional activity50. Mammalian CBP and the closely related coactivator p300 also form a complex with CLOCK-BMAL1, a homolog of the Drosophila CLK-CYC heterodimer, to stimulate their transcriptional activity51,52. One possible explanation for CRTC-activated tim transcription is that Drosophila CRTC may analogously target the CLK-CYC heterodimer to stimulate CLK-CYC–dependent tim transcription. However, this model does not explain tim-specific crtc effects among other CLK-CYC–induced clock genes. Moreover, CRTC associates with the bZIP domain in CREB protein, whereas CBP/p300 binds CREB through the phosphorylated KID domain53,54, indicating that they might not necessarily target the same transcription factors apart from CREB. Finally, we could not detect a protein complex of CLK and CRTC in Drosophila S2 cells (Supplementary Fig. 11). Thus, it is likely that CRTC and CBP/p300 play unique roles in circadian transcription through their interactions with different DNA-binding transcription factors.
If CRTC augments CLK-CYC–dependent tim transcription indirectly, then why do crtc effects require CLK? A recent study suggested that mammalian CLOCK-BMAL1 may regulate the rhythmic access of other DNA-binding transcription factors to their target promoters in the context of chromatin, acting as a pioneer-like transcription factor55. Given the structural and functional homology between Drosophila CLK-CYC and mammalian CLOCK-BMAL1, the presence of CLK-CYC in the tim promoter might allow the recruitment of additional transcription factors (e.g., CREB) and their co-activators including CRTC for maximal tim transcription (Fig. 6, middle). The transcriptional context of tim promoter might thus define its sensitivity to crtc effects among other clock promoters. In addition, the differential assembly of transcription factors on the tim promoter could explain tissue-specific effects of crtc on TIM oscillations (i.e., circadian pacemaker neurons versus peripheral clock tissues). Interestingly, chromatin immunoprecipitation with V5-tagged CLK protein revealed that CLK-CYC heterodimers associate with both tim and Sik2 gene promoters in fly heads56. In LD cycles, however, their rhythmic binding to the Sik2 promoter is phase-delayed by ~4.5 hours compared with that to the tim promoter. These modes of transcriptional regulation may gate crtc effects on tim transcription in a clock-dependent manner, particularly in the increasing phase of tim transcription.
Figure 6. A model for the evolution of CRTC-dependent clocks.
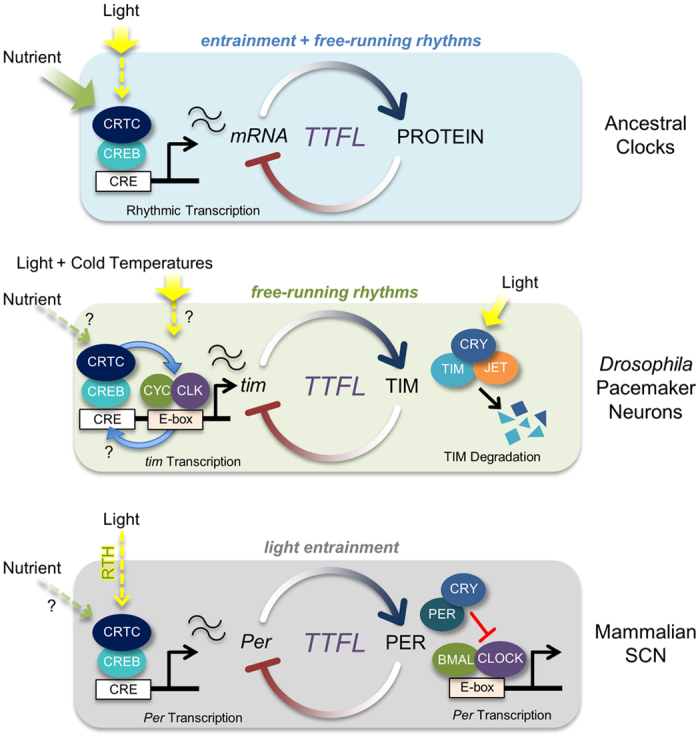
In ancestral organisms, environmental time cues such as light or the availability of nutrients might have been directly accessible to circadian clock cells. Timing information could have converged on the regulation of CRTC through various intracellular signaling pathways to modulate CREB-dependent transcription in the earliest transcription-translation feedback loop (TTFL), thereby entraining and sustaining molecular clocks. In poikilothermic Drosophila, environmental changes in light could be cell-autonomously sensed by circadian pacemaker neurons, whereas metabolic cues are instead provided systemically. Accordingly, post-translational regulation of TIM stability has evolved as the primary strategy for clock entrainment by light since the blue-light photoreceptor CRY and the F box protein JETLAG (JET) trigger light-dependent TIM degradation. CREB-dependent transcriptional regulation of the tim promoter, on the other hand, has been conserved from the original TTFL. Given that Drosophila tim is essential for sustaining molecular rhythms, a transcriptional co-activator function of CRTC in CLK/CYC-induced tim transcription contributes to free-running behaviors in DD. In homeothermic mammals, light cues are indirectly transmitted to master pacemaker neurons in the suprachiasmatic nucleus (SCN) by the synaptic input from the retinohypothalamic tract (RHT). Consequently, the mammalian tim homolog became dispensable for clock function whereas the light-sensing activity of CRY homologs has been replaced by their transcriptional repressor function. Instead, mammalian Per took over a role in light entrainment by retaining the CREB/CRTC-dependent transcriptional regulation from the primitive TTFL.
Transcription from CREB-responsive reporter genes shows daily oscillations, both in Drosophila and mammals19,57,58, implicating this transcriptional strategy in the evolution of molecular clocks. In fact, cAMP signaling and CRE-dependent transcription constitute the integral components of core molecular clocks, serving to regulate daily rhythmic transcription of circadian clock genes57,58,59,60,61. For instance, reciprocal regulation of dCREB2 and per at the transcription level has been reported to sustain free-running circadian rhythms in Drosophila57. During fasting in mammals, a transcriptional program for hepatic gluconeogenesis is induced by CREB phosphorylation and CRTC2 dephosphorylation. Fasting-activated CREB-CRTC2 then stimulates Bmal1 expression61, whereas CLOCK-BMAL1–induced CRY rhythmically gates CREB activity in this process by modulating G protein-coupled receptor activity and inhibiting cAMP-induced CREB phosphorylation62. This molecular feedback circuit thus mutually links mammalian clocks and energy metabolism in terms of CREB-dependent transcription.
On the basis of these observations, we propose a model for the evolution of CRTC-dependent clocks to explain the distinctive circadian roles of CRTC homologs (Fig. 6). CRTC is a transcriptional effector that integrates various cellular signals33. We reasoned that ancestral clocks may have employed CREB-CRTC–mediated transcription to sense extracellular time cues cell-autonomously and integrate this timing information directly into the earliest transcription-translation feedback loop (TTFL). This strategy would have generated simple but efficient molecular clocks to tune free-running molecular rhythms in direct response to environmental zeitgebers, such as light and the availability of nutrients. A circadian role of CRTC then has differentially evolved along with a selective set of clock targets. In poikilothermic Drosophila, light is accessible directly to circadian pacemaker neurons in the adult fly brain. Therefore, TIM degradation by the blue-light photoreceptor CRY63,64 plays a major role in the light entrainment of Drosophila clocks although the photic induction of CLK/CYC-dependent tim transcription has been reported specifically at lower temperatures65. Accordingly, Drosophila CRTC retained a constitutive co-activator function from the ancestral TTFL to support CLK/CYC-activated tim transcription and sustain free-running circadian behaviors. In homeothermic mammals, light input to the SCN is indirectly mediated by neurotransmitter release from presynaptic termini of the retinohypothalamic tract (RHT)66. Intracellular signaling relays in the SCN converge on the dephosphorylation and nuclear translocation of CRTC1 to activate CRE-dependent transcription21,24. Under these circumstances, a circadian role of light-sensitive TIM might have degenerated, while per took over a role in the light-entrainment pathway by retaining CREB-CRTC1–dependent transcriptional regulation from the primitive TTFL. Consequently, mammalian clocks have lost a homolog of the Drosophila-like cry gene family, but instead evolved CRY homologs of the vertebrate-like cry gene family with transcriptional repressor activities in CLOCK-BMAL1–dependent transcription67.
Regulation of metabolism and stress responses by neuronal CREB-CRTC-SIK pathways has been well documented in Drosophila25,34,68. Given our demonstration of a circadian role of CRTC in the pacemaker neurons, it is possible that CRTC might sense metabolic cues in the context of circadian neural circuits to entrain molecular clocks cell-autonomously. Alternatively, but not exclusively, CRTC could participate in the regulation of clock-relevant metabolism as clock outputs from pacemaker neurons69,70. These hypotheses remain to be validated in future studies.
Materials and Methods
Fly Strains
All flies were reared on standard cornmeal-yeast-agar medium at 25 °C under 12 hour:12 hour LD cycles. crtc25-3 and UAS-CRTC (gifts from M. Montminy), as well as UAS-SIK (a gift from J. Chung), Pdf-GeneSwitch-Gal4 (a gift from M. Ceriani), Pdf-Gal4, and tim-Gal4 have been described previously25,32,34,38,71. Df(3L)ED4710, Df(3L)BSC415, UAS-crtc RNAi (stock number 28886), and UAS-DCR2 were obtained from the Bloomington Drosophila Stock Center.
Behavioral Analyses
Individual male flies were placed into glass vials containing 5% sucrose and 2% agar, entrained by three LD cycles at 25 °C and then transferred to constant darkness (DD). Locomotor activities were recorded using the Drosophila Activity Monitor (DAM) system (TriKinetics). Behavioral data were collected from the first to the 6th DD cycle and analyzed to determine the free-running period length and power of rhythmicity in individual flies using the ClockLab analysis software (Actimetrics). The confidence interval of the chi-squared periodogram was set to 0.05 with the testing range of period lengths from 15-hour to 35-hour. The power of rhythmicity (P-S) was calculated by subtracting a Significance value (the minimum measurement considered rhythmic at a given period length with the confidence interval) from a Power value (the observed rhythmicity measurement at the given period length). All the genotypes were tested in multiple behavioral runs. Per each genotype, circadian periods were averaged from rhythmic flies only (which are operationally defined if flies have the power values of rhythmicity greater than 10) whereas the power of rhythmicity values were averaged either from all the flies tested or from rhythmic flies as shown in Supplementary Tables. The averaged locomotor activity profiles were analyzed using Microsoft Excel. To quantify the anticipatory morning activities in LD cycles, we first calculated the normalized total activities from individual flies and then applied them to the equation for the morning index = [(total activity 3 h prior to lights-on)/(total activity 6 h prior to lights-on) − (0.5)].
Immunoblotting
Fifty fly heads were homogenized in lysis buffer consisting of 25 mM Tris-Cl pH 7.5, 300 mM NaCl, 10% glycerol, 1 mM PMSF, 1 mM DTT, 0.5% NP-40, and protease/phosphatase inhibitor cocktail. Soluble extracts were clarified by centrifugation, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes (Amersham) for immunoblotting. Membranes were probed with rabbit anti-CRTC (a gift from J. Chung), guinea pig anti-TIM50, rabbit anti-PER (a gift from E.Y. Kim), mouse anti-TUB (Developmental Studies Hybridoma Bank) and anti-V5 (Invitrogen) primary antibodies, and detected using appropriate horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (ECL) reagents (Pierce).
Immunofluorescence Assay
Male flies were entrained in three LD cycles and then transferred to constant darkness. Brains were dissected at six different time-points in LD cycles or from the first to the second DD cycle. Whole-mount immunostaining was performed as described previously72,73. Rabbit anti-PER (a gift from E. Y. Kim), guinea pig anti-TIM50 and mouse anti-PDF (Developmental Studies Hybridoma Bank) were used as primary antibodies, and Alexa Fluor 488-conjugated anti-rabbit IgG, Alexa Fluor 594-conjugated anti-guinea pig IgG, and Alexa Fluor 647-conjugated anti-mouse IgG (Jackson Immuno Research Laboratories, Inc.) were used as secondary antibodies. Brain images were obtained by confocal laser-scanning microscopy (Olympus FV1000). For quantitative analyses of PER and TIM levels in the circadian pacemaker neurons, the fluorescence intensity from each group of clock cells was quantified using ImageJ software, as described previously72. The quantification of the axonal arborizations from small LNv was performed as described previously36. To quantify PDF levels at the axonal termini of small LNv, the intensities of anti-PDF staining were quantified using ImageJ software. Those above a threshold levels were integrated from each z-stacked confocal images and averaged from 16–20 hemispheres per each genotype at ZT0 or ZT12.
Quantitative Real-time PCR
Flies were entrained in three LD cycles and then transferred to constant darkness. For transcription analyses of clock genes, flies were harvested at six different time-points in LD cycles or during the transition from the first to the second DD cycle. Total RNA was extracted from 50 fly heads using TRIzol reagent (Ambion), according to the manufacturer’s instructions. Purified RNA was digested with RQ1 DNase to remove genomic DNAs, extracted with a mixture of phenol:chloroform:isoamyl alcohol (25:24:1), and then reverse-transcribed with M-MLV reverse transcriptase using oligo dT primers, according to the manufacturer’s instructions (Promega). Quantitative polymerase chain reaction (PCR) was performed on a CFX96 system (Bio-Rad) using TOPreal qPCR 2X PreMIX (Enzynomics) and the following specific primer pairs: rp49 (control), 5′-GAA GAA GCG CAC CAA GCA CT-3′ (forward) and 5′-TTG AAT CCG GTG GGC AGC AT-3′ (reverse); tim, 5′-GAC TTG CCA AAT CCC TCA TC-3′ (forward) and 5′-GAA GCA CTG CAA CTC GAT CA-3′ (reverse); per, 5′-GAC CGA ATC CCT GCT CAA TA-3′ (forward) and 5′-GTG TCA TTG GCG GAC TTC TT-3′ (reverse); Clk, 5′-TAC TGC GTG AGG ATA TCG-3′ (forward) and 5′-GTT GTT GTT CTG GTT GC-3′ (reverse); Pdp1e, 5′-GTC GCC CTC CTC CTT GTA TT-3′ (forward) and 5′-CCA ATG AGC ATC ACA ACC AT-3′ (reverse); cry, 5′-CCA CCG CTG ACC TAC CAA A-3′ (forward) and 5′-GGT GGA AGC CCA ATA ATT TGC-3′ (reverse); and pdfr, 5′-CAG CTC GTT AGC ATT GTC CA-3′ (forward) and 5′-ACG TTT AAG TTG GCC ACA GG-3′ (reverse).
S2 Cell Culture, Plasmids, Transfection, and Luciferase Assay
Drosophila S2 cells were cultured in Shields and Sang M3 insect medium (Sigma) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Invitrogen). Transient transfection was performed using Effectene according to the manufacturer’s instructions (Qiagen). crtc cDNA (a gift from J. Chung) was subcloned into a modified pAc vector for expression of HA-tagged CRTC. pAc-CLK-V5, pAc-PKA-V5, per-luc, tim-luc, Clk-luc, cre-luc and renilla-luc vectors have been described previously10,44,50. For S2 RNAi experiments, double-stranded RNAs (dsRNAs) were in vitro-transcribed using a MEGAscript RNAi kit (Ambion). S2 cells were treated with dsRNA against EGFP (control) or CRTC for 2 days before transfection. Luciferase assays were performed 44 hours after transfection using the Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions (Promega). Luminescence was measured using a TriStar2 plate reader (Berthold Technologies).
Additional Information
How to cite this article: Kim, M. et al. CRTC Potentiates Light-independent timeless Transcription to Sustain Circadian Rhythms in Drosophila. Sci. Rep. 6, 32113; doi: 10.1038/srep32113 (2016).
Supplementary Material
Acknowledgments
We thank J. Chung for providing the anti-CRTC antibody, plasmid DNAs and transgenic flies; E.Y. Kim for the anti-PER antibody; and M. Montminy for CRTC transgenic flies. This work was supported by grants from the National Research Foundation (NRF) of Korea (NRF-2016R1A2B4011111) funded by the Ministry of Science, ICT and Future Planning (MSIP), the Republic of Korea (J.C.), and the 2014 Creativity & Innovation Research Fund (project no. 1.140016) of UNIST (Ulsan National Institute of Science and Technology) and Basic Science Research Program through the NRF funded by the MSIP, the Republic of Korea (NRF-2014R1A1A1003184) (C.L.).
Footnotes
Author Contributions J.C. and C.L. conceived the project and supervised the work. M.K., H.L. and C.L. performed the experiments. M.K., H.L., J.-H.H. and C.L. analyzed the data. M.K., J.C. and C.L. wrote the paper.
References
- Bass J. & Takahashi J. S. Circadian integration of metabolism and energetics. Science 330, 1349–1354, 10.1126/science.1195027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G. & Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell metabolism 13, 125–137, 10.1016/j.cmet.2011.01.006 (2011). [DOI] [PubMed] [Google Scholar]
- Curtis A. M., Bellet M. M., Sassone-Corsi P. & O’Neill L. A. Circadian clock proteins and immunity. Immunity 40, 178–186, 10.1016/j.immuni.2014.02.002 (2014). [DOI] [PubMed] [Google Scholar]
- Partch C. L., Green C. B. & Takahashi J. S. Molecular architecture of the mammalian circadian clock. Trends in cell biology 24, 90–99, 10.1016/j.tcb.2013.07.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R. & Chung B. Y. Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 72, 605–624, 10.1146/annurev-physiol-021909-135815 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N. & Helfrich-Forster C. Setting the clock–by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 585, 1435–1442, 10.1016/j.febslet.2011.02.028 (2011). [DOI] [PubMed] [Google Scholar]
- Hardin P. E. Molecular genetic analysis of circadian timekeeping in Drosophila. Advances in genetics 74, 141–173, 10.1016/B978-0-12-387690-4.00005-2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. & Sehgal A. Speed control: cogs and gears that drive the circadian clock. Trends in neurosciences 35, 574–585, 10.1016/j.tins.2012.05.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataroglu O. & Emery P. The molecular ticks of the Drosophila circadian clock. Current opinion in insect science 7, 51–57, 10.1016/j.cois.2015.01.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran S. A. et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112, 329–341 (2003). [DOI] [PubMed] [Google Scholar]
- Lim C. et al. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Current biology: CB 17, 1082–1089, 10.1016/j.cub.2007.05.039 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Stoleru D., McDonald M., Nawathean P. & Rosbash M. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes & development 21, 1675–1686, 10.1101/gad.1552607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A. et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes & development 21, 1687–1700, 10.1101/gad.1552207 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richier B., Michard-Vanhee C., Lamouroux A., Papin C. & Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. Journal of biological rhythms 23, 103–116, 10.1177/0748730407313817 (2008). [DOI] [PubMed] [Google Scholar]
- Brown S. A., Kowalska E. & Dallmann R. (Re)inventing the circadian feedback loop. Dev. Cell 22, 477–487, 10.1016/j.devcel.2012.02.007 (2012). [DOI] [PubMed] [Google Scholar]
- Hardin P. E. & Panda S. Circadian timekeeping and output mechanisms in animals. Current opinion in neurobiology 23, 724–731, 10.1016/j.conb.2013.02.018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. & Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci 16, 1544–1550, 10.1038/nn.3543 (2013). [DOI] [PubMed] [Google Scholar]
- Ginty D. D. et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260, 238–241 (1993). [DOI] [PubMed] [Google Scholar]
- Obrietan K., Impey S., Smith D., Athos J. & Storm D. R. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. The Journal of biological chemistry 274, 17748–17756 (1999). [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z., Cermakian N., Reppert S. M. & Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proceedings of the National Academy of Sciences of the United States of America 99, 7728–7733, 10.1073/pnas.102075599 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K. et al. Clock and light regulation of the CREB coactivator CRTC1 in the suprachiasmatic circadian clock. J. Neurosci. 33, 9021–9027, 10.1523/JNEUROSCI.4202-12.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V. & Cohen S. M. TORCing up metabolic control in the brain. Cell Metab. 7, 357–358, 10.1016/j.cmet.2008.04.006 (2008). [DOI] [PubMed] [Google Scholar]
- Takemori H. & Okamoto M. Regulation of CREB-mediated gene expression by salt inducible kinase. J. Steroid Biochem. Mol. Biol. 108, 287–291, 10.1016/j.jsbmb.2007.09.006 (2008). [DOI] [PubMed] [Google Scholar]
- Jagannath A. et al. The CRTC1-SIK1 pathway regulates entrainment of the circadian clock. Cell 154, 1100–1111, 10.1016/j.cell.2013.08.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 7, 434–444, 10.1016/j.cmet.2008.02.010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R. & Emery P. A plastic clock: how circadian rhythms respond to environmental cues in Drosophila. Mol. Neurobiol. 38, 129–145, 10.1007/s12035-008-8035-y (2008). [DOI] [PubMed] [Google Scholar]
- Collins B. & Blau J. Even a stopped clock tells the right time twice a day: circadian timekeeping in Drosophila. Pflugers Arch. 454, 857–867, 10.1007/s00424-006-0188-9 (2007). [DOI] [PubMed] [Google Scholar]
- Hermann-Luibl C., Yoshii T., Senthilan P. R., Dircksen H. & Helfrich-Forster C. The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. The Journal of neuroscience: the official journal of the Society for Neuroscience 34, 9522–9536, 10.1523/JNEUROSCI.0111-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith E. J. & Ceriani M. F. Experimental assessment of the network properties of the Drosophila circadian clock. The Journal of comparative neurology 523, 982–996, 10.1002/cne.23728 (2015). [DOI] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J. & Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868, 10.1038/nature02926 (2004). [DOI] [PubMed] [Google Scholar]
- Grima B., Chelot E., Xia R. & Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873, 10.1038/nature02935 (2004). [DOI] [PubMed] [Google Scholar]
- Renn S. C., Park J. H., Rosbash M., Hall J. C. & Taghert P. H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802 (1999). [DOI] [PubMed] [Google Scholar]
- Altarejos J. Y. & Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12, 141–151, 10.1038/nrm3072 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Kim W. & Chung J. Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC). J. Biol. Chem. 286, 2658–2664, 10.1074/jbc.C110.119222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H. et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 97, 3608–3613, 10.1073/pnas.070036197 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M. P., Berni J. & Ceriani M. F. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS biology 6, e69, 10.1371/journal.pbio.0060069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert K. I. et al. Circadian changes in Drosophila motor terminals. Developmental neurobiology 67, 415–421, 10.1002/dneu.20332 (2007). [DOI] [PubMed] [Google Scholar]
- Depetris-Chauvin A. et al. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Current biology: CB 21, 1783–1793, 10.1016/j.cub.2011.09.027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B. et al. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420, 178–182, 10.1038/nature01122 (2002). [DOI] [PubMed] [Google Scholar]
- Garbe D. S. et al. Cooperative interaction between phosphorylation sites on PERIOD maintains circadian period in Drosophila. PLoS genetics 9, e1003749, 10.1371/journal.pgen.1003749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Qian Z., Myers M. P. & Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature 380, 129–135, 10.1038/380129a0 (1996). [DOI] [PubMed] [Google Scholar]
- Peschel N., Chen K. F., Szabo G. & Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Current biology: CB 19, 241–247, 10.1016/j.cub.2008.12.042 (2009). [DOI] [PubMed] [Google Scholar]
- Zeng H., Hardin P. E. & Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J 13, 3590–3598 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington T. K. et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603 (1998). [DOI] [PubMed] [Google Scholar]
- Price J. L., Dembinska M. E., Young M. W. & Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. The EMBO journal 14, 4044–4049 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V., Lanjuin A. & Rosbash M. TIMELESS-dependent positive and negative autoregulation in the Drosophila circadian clock. The EMBO journal 18, 675–686, 10.1093/emboj/18.3.675 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia J. C. et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859, 10.1038/365855a0 (1993). [DOI] [PubMed] [Google Scholar]
- Kwok R. P. et al. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370, 223–226, 10.1038/370223a0 (1994). [DOI] [PubMed] [Google Scholar]
- Goodman R. H. & Smolik S. CBP/p300 in cell growth, transformation, and development. Genes & development 14, 1553–1577 (2000). [PubMed] [Google Scholar]
- Lim C. et al. Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol. Cell Biol. 27, 4876–4890, 10.1128/MCB.02155-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S. et al. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes to cells: devoted to molecular & cellular mechanisms 5, 739–747 (2000). [DOI] [PubMed] [Google Scholar]
- Etchegaray J. P., Lee C., Wade P. A. & Reppert S. M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421, 177–182, 10.1038/nature01314 (2003). [DOI] [PubMed] [Google Scholar]
- Luo Q. et al. Mechanism of CREB recognition and coactivation by the CREB-regulated transcriptional coactivator CRTC2. Proceedings of the National Academy of Sciences of the United States of America 109, 20865–20870, 10.1073/pnas.1219028109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I. et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91, 741–752 (1997). [DOI] [PubMed] [Google Scholar]
- Menet J. S., Pescatore S. & Rosbash M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes & development 28, 8–13, 10.1101/gad.228536.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abruzzi K. C. et al. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes & development 25, 2374–2386, 10.1101/gad.178079.111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin M. P., Zhou H. & Yin J. C. The Drosophila dCREB2 gene affects the circadian clock. Neuron 22, 777–787 (1999). [DOI] [PubMed] [Google Scholar]
- O’Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S. & Hastings M. H. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320, 949–953, 10.1126/science.1152506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. et al. CREB influences timing and entrainment of the SCN circadian clock. Journal of biological rhythms 25, 410–420, 10.1177/0748730410381229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi S. et al. cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. The Journal of biological chemistry 286, 32416–32423, 10.1074/jbc.M111.258970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. et al. Glucagon-CREB/CRTC2 signaling cascade regulates hepatic BMAL1 protein. The Journal of biological chemistry 290, 2189–2197, 10.1074/jbc.M114.612358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E. E. et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature medicine 16, 1152–1156, 10.1038/nm.2214 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busza A., Emery-Le M., Rosbash M. & Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 304, 1503–1506, 10.1126/science.1096973 (2004). [DOI] [PubMed] [Google Scholar]
- Koh K., Zheng X. & Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312, 1809–1812, 10.1126/science.1124951 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. F., Majercak J. & Edery I. Clock-gated photic stimulation of timeless expression at cold temperatures and seasonal adaptation in Drosophila. Journal of biological rhythms 21, 256–271, 10.1177/0748730406289306 (2006). [DOI] [PubMed] [Google Scholar]
- Hannibal J., Moller M., Ottersen O. P. & Fahrenkrug J. PACAP and glutamate are co-stored in the retinohypothalamic tract. The Journal of comparative neurology 418, 147–155 (2000). [PubMed] [Google Scholar]
- Zhu H. et al. The two CRYs of the butterfly. Current biology: CB 15, R953–R954, 10.1016/j.cub.2005.11.030 (2005). [DOI] [PubMed] [Google Scholar]
- Iijima K., Zhao L., Shenton C. & Iijima-Ando K. Regulation of energy stores and feeding by neuronal and peripheral CREB activity in Drosophila. PloS one 4, e8498, 10.1371/journal.pone.0008498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Zheng X. & Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell metabolism 8, 289–300, 10.1016/j.cmet.2008.09.006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAngelo J. R., Erion R., Crocker A. & Sehgal A. The central clock neurons regulate lipid storage in Drosophila. PloS one 6, e19921, 10.1371/journal.pone.0019921 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M. & Hall J. C. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422, 66–94 (2000). [DOI] [PubMed] [Google Scholar]
- Lim C. et al. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature 470, 399–403, 10.1038/nature09728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Sonn J. Y., Oh Y., Lim C. & Choe J. SIFamide and SIFamide receptor defines a novel neuropeptide signaling to promote sleep in Drosophila. Molecules and cells 37, 295–301, 10.14348/molcells.2014.2371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.