Abstract
Lung cancer continues to be a major public health challenge in the United States despite efforts to decrease the prevalence of smoking; outcomes are especially poor for African-American patients compared to other races/ethnicities. Chronic obstructive pulmonary disease (COPD) co-occurs with lung cancer frequently, but not always, suggesting both shared and distinct risk factors for these two diseases. To identify germline genetic variation that distinguishes between lung cancer in the presence and absence of emphysema, we performed whole-exome sequencing on 46 African-American lung cancer cases (23 with and 23 without emphysema frequency matched on age, sex, histology and pack years). Using conditional logistic regression, we found 6305 variants (of 168 150 varying sites) significantly associated with lung cancer subphenotype (P ≤ 0.05). Next, we validated 10 of these variants in an independent set of 612 lung cancer cases (267 with emphysema and 345 without emphysema) from the same population of inference as the sequenced cases. We found one variant that was significantly associated with lung cancer subphenotype in the validation sample. These findings contribute to teasing apart shared genetic factors from independent genetic factors for lung cancer and COPD.
Summary
Whole-exome sequencing with follow-up genotyping of selected variants identifies an association with lung cancer subphenotyped for emphysema. Our analysis highlights differences in genetic background among lung cancer cases with or without emphysema.
Introduction
Despite declining annual incidence rates in the United States, lung cancer is the third leading form of cancer diagnosed in both men and women, and the leading cause of cancer-related death in the United States, with both higher incidence and mortality rates among African Americans compared to other racial/ethnic groups (1). While chronic obstructive pulmonary disease (COPD) itself is the fourth leading cause of mortality in adults in the United States, it is also considered as an independent risk factor for lung cancer, increasing risk up to 4.5-fold (2). Smoking plays a major role in the development of both diseases; 85–90% of all lung cancers and COPD diagnoses are attributable to cigarette smoking (3). Repeated insult to the lung caused by cigarette smoke triggers a chronic, aberrant inflammatory response that is involved in both airway and lung parenchymal destruction and lung tumorigenesis (4).
Both COPD and lung cancer have been shown to have a genetic component. Large-scale genome-wide association studies (GWAS) and meta-analyses have identified SNPs that confer risk of COPD or related measures of lung function decline; mutations in SERPINA1, leading to alpha1-antitrypsin deficiency, constitute rare but known genetic causal factors (5). The genetic underpinnings of lung cancer have also been studied extensively, from GWAS in population-based samples to whole-exome/whole-genome sequencing in tumor tissue to identify putative driver mutations (summarized in (6)). Given that COPD and lung cancer are etiologically related by smoking-induced inflammation, that a family history of COPD increases risk of lung cancer development, and that they co-occur in only a subset of smokers, suggests a common underlying genetic contribution that is not present in COPD-free lung cancer patients (7). Indeed, there has been a concerted effort to disentangle genetic variants that are jointly associated with lung cancer and COPD from variants that are associated with one or the other disease (reviewed in (8)).
Racial variability adds another layer of complexity to characterizing the smoking-COPD-lung cancer axis. African Americans and whites differ with respect to age at smoking initiation, pack years of exposure, emphysema severity and rates of COPD and related measures of impaired lung function (9–12).
We present a strategy using whole-exome sequence data to evaluate which of these genetic variants distinguish specific lung cancer subphenotypes. We identified sequence variants in a sample of 46 African-American lung cancer cases both with (n = 23) and without (n = 23) CT evidence of a COPD subphenotype, emphysema, to determine whether the genetic profile of lung cancer patients with emphysema is distinct from that of lung cancer patients without emphysema. A set of 12 sequence variants were selected for follow-up genotyping in an independent sample of 612 African-American and white cases, including 267 cases with emphysema and 345 cases without emphysema, to test phenotype–genotype associations with lung cancer in the presence or absence of emphysema.
Materials and methods
Ethics statement
The Wayne State University (WSU) Institutional Review Board approved the procedures used in collecting and processing of participant information, and written informed consent was obtained from all subjects prior to participation.
Description of the discovery sample
African-American lung cancer patients were selected from two case–control studies: the Women’s Epidemiology of Lung Diseases (WELD) study which enrolled women with non-small cell lung cancer, and the Exploring Health, Ancestry and Lung Epidemiology (EXHALE) study which focused on African Americans and was not restricted by histologic type of lung cancer. Consequently, the majority of cases selected for the discovery phase of the study were female (39 females, 7 males). All patients had a primary diagnosis of lung cancer. Patients were classified as having emphysema based on radiologist review of computed tomography (CT), using a semi-quantitative approach similar to that developed by the National Emphysema Treatment Trial, and is described in detail in Mina et al. (13). Using this CT-based definition of emphysema to stratify cases, 23 patients without emphysema and 23 patients with emphysema were matched by age, sex and histology (small cell, adenocarcinoma and all other NSCLC). We matched on pack years where possible; however, identifying lung cancer cases without emphysema was a limiting factor, so our discovery sample included four never smoking cases without emphysema, and these cases were matched to the lightest smoking cases with emphysema (range 1–9 pack years).
Description of the validation sample
Six hundred twelve lung cancer cases were selected to validate specific findings in the exome sequencing analysis. Cases were ascertained through the population-based Metropolitan Detroit Cancer Surveillance System, an NCI-funded SEER (Surveillance, Epidemiology and End Results) registry and had participated in one of three studies: the Family History Study (FHS) which enrolled cases diagnosed before age 50 years, WELD or EXHALE (14). The same questionnaire was administered to participants of each study. White cases were frequency matched to African-American cases by self-reported emphysema, age and pack years.
Sample preparation and DNA extraction
The Gentra AutoPure Kit (Qiagen, Valencia, CA) was used to extract DNA from whole blood samples and the Gentra Puregene Kit (Qiagen, Valencia, CA) was used to extract DNA from mouthwash and saliva samples. Sample DNA quantity and quality were assessed using a Nanodrop spectrophotometer and Quantifiler® (Life Technologies) assay, a real-time PCR-based approach for estimation of amplifiable DNA.
Trait definitions
Age was recorded as age at diagnosis. Never smokers were individuals who smoked fewer than 100 cigarettes in their lifetime, while ever smokers included both former and current smokers. Pack years was calculated by multiplying the number of years smoked by the average number of cigarettes smoked per day divided by 20. Family history of lung cancer was recorded as ‘yes’ if the participant reported at least one first-degree relative with a diagnosis of lung cancer. Tumor histology was defined according to clinical assessment (as reported in SEER). Emphysema was classified based on self-reported physician diagnosis in the validation sample participants.
Whole-exome sequencing
Exome sequencing was used to measure all coding genetic variants in the samples. Libraries were prepared per Illumina standard procedures. Briefly, DNA was fragmented using a Covaris S2, the ends repaired with T4 DNA polymerase and Klenow, the 3′ ends adenylated, adapters ligated, with purification on a 2% low range agarose gel followed by excising the 300–400bp fragments and enrichment of samples with 19 cycles of PCR. The libraries were validated using a High Sensitivity DNA chip on an Agilent Technologies 2100 Bioanalyzer. Six libraries were pooled using 500ng of each. The pool was subjected to capture using the Illumina Exome Enrichment kit. The hybridized samples were then purified by hybridization to streptavidin magnetic beads, washing and elution. A second hybridization, washing and elution were followed by 10 cycles of PCR amplification. The amplicons were cleaned using AMPure XP Beads followed by library validation. The exome-enriched, pooled libraries were sequenced on a HiSeq 2500 instrument.
Image analysis, sequence extraction and base calling were done using Illumina Pipeline v1.6 software. Read mapping was done using BWA which uses the Burrows-Wheeler anti-transform based mapping algorithm. The reference genome used was hg37.61, which was the latest version at the time of analysis. Reads mapped successfully accounted for 86% of the original reads. Multisample variant calling was performed using the SamTools 0.1.12a package and BcfTools with base alignment quality optimization. Realignment near InDels was performed using GATK (15). SnpEff was employed after sequencing to align and call variants on all samples simultaneously. The analysis focuses on those variants which passed quality filtering. (SnpEff: Variant effect analysis. snpeff.sourceforge.net, 2014, version 3.6) (16).
Genotyping of selected variants
The single nucleotide polymorphisms (SNPs) were interrogated with the 5′-nuclease assay using Applied Biosystems TaqMan SNP Genotyping Assays under standard conditions on a Life Technologies QuantStudio 12K Flex. For genotyping quality control, direct repeats, control DNAs and no template controls were used.
Statistical analysis of exome sequence data
Conditional logistic regression was used (gender- and histology-stratified) to identify genetic variants that differentiate between lung cancer in the presence or absence of emphysema. A dominant genetic model was used for each variant, combining all non-wildtype genotypes into a single class and comparing with the wildtype class. For variants where the mutant homozygote genotype was more common than the wildtype genotype, the mutant homozygote served as the ‘wildtype’ genotype and the wildtype genotype served as the ‘mutant’ genotype. A dominant model was chosen to maximize the number of valid tests (e.g. where the minor allele was only present in heterozygotes), while minimizing the degrees of freedom of each test, given the small sample size. As this analysis is exploratory and hypothesis-generating, a nominal significance level of 0.05 was used without consideration of multiple testing.
Selection of sequence variants for genotyping in the validation sample is illustrated in Supplementary Figure 1. Variants with statistically significant effects were further triaged for downstream validation analyses, based on having a quality score (defined as −10*log10(p), where p = probability of base calling error) ≥ 150 (the median among all variants) and located in a gene region previously reported by large-scale genetic association studies (e.g. GWAS) of lung cancer and/or measures of a COPD phenotype. After implementing these filters, 12 variants were chosen that satisfied one of the following criteria: (i) novelty (not previously reported in dbSNP at the time of sequencing), (ii) statistically significant association with lung cancer subphenotype at α = 0.005 (top 10% of significant results) or (iii) location in a gene region with multiple significant non-synonymous SNPs. Custom assays designed for two of these SNPs, rs148827807 and rs143721595, failed and thus were not analyzed further. Detailed information for each of the 10 SNPs used in the validation phase is presented in Supplementary Table 1.
Statistical analysis of genotype (validation) data
Tests of homogeneity of covariates between cases with and without emphysema were performed using chi-squared tests for categorical variables or t-tests for continuous variables. Minor allele relative frequencies for each SNP were estimated separately by race. Logistic regression modeling was used to evaluate associations between each SNP and joint lung cancer/emphysema phenotype in a case-only analysis. As in the exome sequencing analysis, SNPs were coded using a dominant genetic model, where carriers of the minor allele were contrasted with the most frequent homozygote class (the referent genotype). Each SNP effect was adjusted for age, gender, race (in the total sample analysis), pack years, family history of lung cancer and years of education. Since these SNPs were chosen based on prior statistical significance in the exome sequencing analysis (described previously), the modeling results were not adjusted for multiple testing. Tests of Hardy–Weinberg Equilibrium for each of the SNPs were used post-hoc to determine if any of the significant results might be influenced by deviations from HWE (p<0.001).
Results
Description of sequencing sample
The demographic characteristics of the 46 patient samples selected for exome sequencing are described in Supplementary Table 2. There was no significant heterogeneity between lung cancer cases with and without emphysema for sex, age, years of education, family history of lung cancer or pack years of smoking. Only smoking status differed significantly across the two groups, due to the presence of four never smokers among the lung cancer cases without emphysema.
Distribution of sequence variant functional effects
Exome sequencing resulted in 168 150 varying sites called among the 46 samples. The frequency of variant sites across the autosomes was fairly homogeneous, accounting for the number of genes per chromosome. The number of variants per gene ranged from 3.1 (chromosome 14) to 5.0 (chromosome 19) across the autosomes, while coverage was lower for the sex chromosomes (2.5 and 0.6 variants/gene for X and Y, respectively). Sampling of the mitochondrial genome was especially dense, with ~10 variants/gene.
Each variant site could be present in multiple transcripts; as such, there were 366 818 functional effects assigned to the 168 150 variable sites using SNPEff. Supplementary Figure 2 describes the distribution of these functional effects; over 80% were assigned within the bounds of a transcriptional region (i.e. 19% were intergenic), and 10% (n = 37 869) of all effects were non-synonymous coding changes. Although they comprise a small fraction of all functional effects, there were 1547 start/stop losses/gains, 433 frameshifts and 392 splice site variants.
Lung cancer subphenotype associated with sequence variants
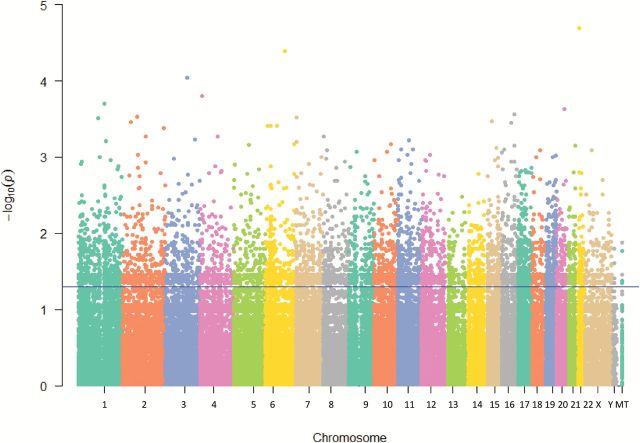
The reference genotype was switched in 2.7% of variant calls (due to relative allele frequency differences in our African-American sample) for use in dominant genetic models. Figure 1 illustrates the results of association testing of lung cancer/emphysema phenotype as compared with lung cancer/no emphysema phenotype with each of the 168 150 variants. Using a conditional logistic regression model, 6305 (3.7%) variants were statistically significant at a nominal α = 0.05 level. The proportion of significant variants (out of all variants) was consistent across chromosomes and the mitochondrial genome, ranging from 3.0% (MT) to 4.5% (chromosome 21).
Figure 1.
Manhattan plot of association between lung cancer subphenotyped for emphysema and individual sequence variants. Results are presented as –log(P value) from a conditional logistic regression model of lung cancer subphenotype and variant genotype (dominant coding), adjusted for age, gender, histology and smoking status.
Description of validation sample
The exome sequencing validation analysis included 612 individuals, described in Table 1. Lung cancer cases were approximately evenly divided among those with a history of emphysema (~44%) and those without a history of emphysema (~56%). Cases with emphysema were significantly older, less educated and had more pack years of smoking than cases without emphysema. There were also significantly higher relative frequencies of whites and those with a family history of lung cancer among cases with emphysema compared to cases without emphysema.
Table 1.
Description of exome sequencing validation sample
| Variable | Lung cancer cases with emphysema (n = 267) | Lung cancer cases without emphysema (n = 345) | P homogeneity |
|---|---|---|---|
| Sex (n, %) | |||
| Male | 78 (29.2) | 111 (32.2) | 0.432 |
| Female | 189 (70.8) | 234 (67.8) | |
| Race (n, %) | |||
| White | 127 (47.6) | 133 (38.6) | 0.025 |
| Black | 140 (52.4) | 212 (61.4) | |
| Age (mean, SD) | 61.8 (9.7) | 58.7 (11.1) | <0.001 |
| Emphysema (n, %) | |||
| No | 0 | 345 (100.0) | — |
| Yes | 267 (100.0) | 0 | |
| Education years (mean, SD) | 12.0 (2.3) | 12.7 (2.6) | <0.001 |
| Family history of lung cancer (n, %) | |||
| No | 219 (82.0) | 275 (79.7) | 0.472 |
| Yes | 48 (18.0) | 70 (20.3) | |
| Smoking status (n, %) | |||
| Never | 0 (0.0) | 29 (8.4) | <0.001 |
| Ever | 267 (100.0) | 316 (91.6) | |
| Packyears (smokers only) (mean, SD) | 48.1 (27.1) | 40.0 (27.9) | 0.001 |
| Histology (n, %) | |||
| Adenocarcinoma | 151 (56.6) | 214 (62.0) | 0.178 |
| Other NSCLCa | 101 (37.8) | 104 (30.1) | |
| Small cell | 11 (4.1) | 17 (4.9) | |
| other | 4 (1.5) | 10 (2.9) | |
a‘other NSCLC’ category includes squamous cell, adenosquamous, large cell and NSCLC not otherwise specified.
Lung cancer subphenotype associations in validation sample of lung cancer cases
Relative frequencies of 10 SNPs identified by exome sequencing were evaluated to detect differences between lung cancer cases subphenotyped for emphysema in the African-American validation sample using a case–case comparison (N = 352) (Table 2). Odds of the lung cancer/emphysema phenotype were 86% higher in carriers of the minor allele for rs13144371 compared to major allele homozygotes (OR = 1.86 for AA/AG versus GG, P = 0.031).
Table 2.
Association between SNPs and lung cancer cases with emphysema as compared to cases without emphysema, assuming dominant genetic model for each SNPa
| SNP | Nearest gene(s) | Chr | Minor allele | African Americans (N = 352) | Whites (N = 260) | P interaction c | Total sample (N = 612) | ||
|---|---|---|---|---|---|---|---|---|---|
| MARFb | OR (95%CI) | MARFb | OR (95%CI) | OR (95% CI) | |||||
| rs2256721 | CHIA | 1 | T | 0.201 | 0.73 (0.47, 1.15) | 0.265 | 0.70 (0.41, 1.18) | 0.973 | 0.72 (0.51, 1.02) |
| rs6680778 | CHD1L | 1 | T | 0.344 | 0.80 (0.51, 1.24) | 0.178 | 0.70 (0.40, 1.23) | 0.801 | 0.78 (0.55, 1.10) |
| rs13144371 | IBSP | 4 | A | 0.109 | 1.86 (1.06, 3.26) | 0.219 | 1.57 (0.90, 2.72) | 0.523 | 1.60 (1.09, 2.36) |
| rs140750089 | FLJ14186 | 4 | A | 0.255 | 0.71 (0.45, 1.12) | 0.247 | 1.06 (0.63, 1.79) | 0.223 | 0.84 (0.60, 1.18) |
| rs303061 | LINC00518 | 6 | C | 0.216 | 1.29 (0.82, 2.02) | 0.220 | 0.83 (0.49, 1.41) | 0.131 | 1.06 (0.76, 1.49) |
| rs9501572 | HLA-B | 6 | G | 0.369 | 0.75 (0.48, 1.17) | 0.274 | 0.91 (0.54, 1.54) | 0.471 | 0.80 (0.57, 1.12) |
| rs3130071 | SNORA38, PRRC2A | 6 | T | 0.026 | 0.77 (0.27, 2.16) | 0.125 | 1.39 (0.78, 2.46) | 0.335 | 1.23 (0.76, 2.00) |
| rs6944332 | PMS2CL | 7 | G | 0.475 | 1.49 (0.86, 2.60) | 0.905 | 1.22 (0.60, 2.47) | 0.640 | 1.30 (0.85, 2.00) |
| rs199638324 | FAM86B2 | 8 | T | 0.030 | 1.30 (0.42, 4.00) | 0.004 | No test | — | 1.16 (0.38, 3.57) |
| rs1288775 | GATM, GATM-AS1 | 15 | T | 0.205 | 1.22 (0.78, 1.91) | 0.692 | 1.56 (0.48, 5.08) | 0.611 | 1.34 (0.88, 2.03) |
Bold indicates significant result at α = 0.05.
aEach logistic regression model was adjusted for age, race (in total sample), sex, pack years education years and family history of lung cancer (yes/no).
bMinor allele relative frequencies estimated in controls only.
cTest of interaction effects on lung cancer subphenotype between SNP relative frequencies and race, in a logistic regression model adjusted for age, sex, pack years, education years and family history of lung cancer.
Extending our analyses to white cases, one SNP was too rare to be analyzed (rs199638324); additionally, the minor allele was different from that in African Americans for rs6944332 and rs1288775 (Table 2). There were no significant associations between lung cancer subphenotype (with and without emphysema) and any of these SNPs among white cases.
Despite observing differences in relative allele frequencies between African Americans and whites, we found no evidence of heterogeneity of SNP effects on lung cancer subphenotype by race (Table 2). When African-American and white lung cancer cases were pooled, we observed a significant association between lung cancer subphenotype and rs13144371, where the relative frequency of AA/AG genotypes was higher among cases with emphysema compared to cases without emphysema (OR = 1.60, P = 0.017). This direction of effect is consistent in both African Americans and whites (OR = 1.80 and 1.57, respectively).
Discussion
COPD has been associated with risk of lung cancer, and susceptibility to both diseases (either separately or in tandem) has been associated with genetic variation, yet little has been done to identify genetic variation associated with lung cancer in the presence or absence of COPD. We present a strategy to identify and validate exome sequence variants in germline DNA associated with lung cancer subphenotyped by emphysema. SNP rs13144371 genotype, a missense in the IBSP gene located on chromosome 4q22.1, was found to distinguish between lung cancer in the presence and absence of emphysema.
Chromosomal region 4q22 has been reported in several GWAS of lung function, lung cancer and a joint COPD/lung cancer phenotype. A meta-analysis of four CHARGE (Cohorts for Heart and Aging Research in Genetic Epidemiology) consortium cohorts of European ancestry found associations between FEV1/FVC and variants in the FAM13A gene region, located ~1Mb downstream from IBSP (17). COPD was associated with variants in FAM13A in a combined analysis of three large white COPD studies and replicated in two additional studies (18).
Evaluating lung cancer and COPD jointly, Young et al. (2011) report that the 4q22 locus, marked by rs7671167, was independently associated with both COPD and lung cancer, as well as a joint COPD/lung cancer phenotype (19,20). Our findings in IBSP are consistent with those of Young et al. and further implicate 4q22 in defining lung cancer subphenotypes.
African Americans exhibit greater genetic diversity than other populations, with an excess of low frequency variants compared to expectations under the neutral model (21). We chose to do exome sequencing in African Americans to maximize identification of variable sites while also evaluating potential race-specific differences in relative frequencies according to lung cancer subphenotype (presence/absence of emphysema). Despite statistically significant differences in relative allele frequencies and covariate distributions (e.g. age, sex, pack years) between African Americans and whites in our sample, when we extended the validation to a sample of white lung cancer cases, we found that SNP effects distinguishing cases with emphysema and cases without emphysema were consistent across African Americans and whites.
The strengths of this study include: whole-exome sequencing in an African-American case group with relatively lower levels of cigarette exposure and greater genetic diversity than seen in other populations, emphysema classification based on radiologic evidence in the discovery sample and selection of a validation set from the same population of inference. There are some limitations to this study. The small number of cases in the discovery set did not allow for the identification of very rare variants (minor allele relative frequencies less than 0.01). We do not correct for multiple testing in our validation set and were limited in the number of SNPs that could be included in the validation phase. Emphysema was based on self-report in the validation set, so there is the possibility of misclassification, especially among African Americans (13). Any misclassification would result in a bias towards the null hypothesis of no difference in relative allele frequencies between cases with and without emphysema.
Lung cancer continues to be a major public health challenge, and new screening modalities may improve early diagnosis and survival. Lung cancer is 2 to 4-fold more likely to occur in individuals with a history of COPD, yet that history is not yet used as a significant driver for screening. Understanding what differentiates lung cancer patients with and without emphysema will help delineate a highest risk group for screening, while at the same time, point to mechanistic leads for lung cancer development. Results from the present study support previously published associations between genetic variation on 4q22 and lung cancer/COPD. Future work to study joint lung cancer/COPD phenotypes is needed to refine genetic profiles for subsets of patients to optimize prediction of and improve outcomes for lung cancer.
Supplementary material
Supplementary Tables 1 and 2 and Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (R01CA060691 to A.G.S., R01CA087895 to A.G.S., R01CA141769 to A.G.S., HHSN26120100028 to A.G.S. and P30CA022453 to A.G.S.).
Supplementary Material
Acknowledgements
We would like to thank Valerie Ratliff and Susan Santer for preparing and extracting the DNA samples.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- COPD
chronic obstructive pulmonary disease
- CT
computed tomography
- GWAS
genome-wide association studies
- SNP
single nucleotide polymorphism
References
- 1. Howlader N., et al. (2015). SEER Cancer Statistics Review, 1975–2012. National Cancer Institute, Bethesda, MD 2015 http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission, posted 2015. [Google Scholar]
- 2. Punturieri A., et al. (2009) Lung cancer and chronic obstructive pulmonary disease: needs and opportunities for integrated research. J. Natl. Cancer Inst., 101, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sethi J.M., et al. (2000) Smoking and chronic obstructive pulmonary disease. Clin. Chest Med., 21, 67–86, viii. [DOI] [PubMed] [Google Scholar]
- 4. Houghton A.M. (2013) Mechanistic links between COPD and lung cancer. Nature Rev. Cancer, 13, 233–245. [DOI] [PubMed] [Google Scholar]
- 5. Tang W., et al. (2014) Large-scale genome-wide association studies and meta-analyses of longitudinal change in adult lung function. PLoS One, 9, e100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Z., et al. (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. Cancer, 14, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu A.H., et al. (1988) Personal and family history of lung disease as risk factors for adenocarcinoma of the lung. Cancer Res., 48(24 Pt 1), 7279–7284. [PubMed] [Google Scholar]
- 8. Schwartz A.G. (2012) Genetic epidemiology of cigarette smoke-induced lung disease. Proc. Am. Thorac. Soc., 9, 22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatila W.M., et al. ; National Emphysema Treatment Trial Research Group. (2006) Advanced emphysema in African-American and white patients: do differences exist? Chest, 130, 108–118. [DOI] [PubMed] [Google Scholar]
- 10. Chatila W.M., et al. (2004) Smoking patterns in African Americans and whites with advanced COPD. Chest, 125, 15–21. [DOI] [PubMed] [Google Scholar]
- 11. Coughlin S.S., et al. (2014) Opportunities to address lung cancer disparities among African Americans. Cancer Med., 3, 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinch A.G., et al. (2011) The landscape of recombination in African Americans. Nature, 476, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mina N., et al. (2012) The relationship between chronic obstructive pulmonary disease and lung cancer in African American patients. Clin. Lung Cancer, 13, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz A.G., et al. (2009) Racial differences in the association between SNPs on 15q25.1, smoking behavior, and risk of non-small cell lung cancer. J. Thorac. Oncol., 4, 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H., et al. ; 1000 Genome Project Data Processing Subgroup. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cingolani P., et al. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin)., 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hancock D.B., et al. (2010) Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet., 42, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho M.H., et al. (2010) Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet., 42, 200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young R.P., et al. (2011) FAM13A locus in COPD is independently associated with lung cancer - evidence of a molecular genetic link between COPD and lung cancer. Appl. Clin. Genet., 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Young R.P., et al. (2011) Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLoS One, 6, e16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lohmueller K.E., et al. (2010) The effect of recent admixture on inference of ancient human population history. Genetics, 185, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.