Abstract
Summary: Gene Ontology (GO) semantic similarity measures are being used for biological knowledge discovery based on GO annotations by integrating biological information contained in the GO structure into data analyses. To empower users to quickly compute, manipulate and explore these measures, we introduce A-DaGO-Fun (ADaptable Gene Ontology semantic similarity-based Functional analysis). It is a portable software package integrating all known GO information content-based semantic similarity measures and relevant biological applications associated with these measures. A-DaGO-Fun has the advantage not only of handling datasets from the current high-throughput genome-wide applications, but also allowing users to choose the most relevant semantic similarity approach for their biological applications and to adapt a given module to their needs.
Availability and implementation: A-DaGO-Fun is freely available to the research community at http://web.cbio.uct.ac.za/ITGOM/adagofun. It is implemented in Linux using Python under free software (GNU General Public Licence).
Contact: gmazandu@cbio.uct.ac.za or Nicola.Mulder@uct.ac.za
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Recent years have experienced an exponential growth of publicly available and accessible genomics, proteomics and other biological data resulting from high-throughput biology technologies and computational scanning approaches. Retrieving information from these different biological data constitutes an essential step and challenging task which requires the use of computational tools and algorithms for translating these data into different applications. In the context of functional annotation data, the Gene Ontology (GO-Consortium, 2012) provides a way of consistently describing genes and proteins in any organism, producing a well-adapted platform to computationally process data at the functional level. Currently, ∼30 629 514 proteins are already annotated with Gene Ontology (GO) terms in the existing biological databases (see the latest version of GOA UniProt version 143 at http://www.ebi.ac.uk/GOA/uniprot_release, released on 27 May, 2015), thus enabling protein comparisons on the basis of their GO annotations. Several semantic similarity (SS) measures (Mazandu and Mulder, 2013b, 2014) have been suggested to tackle major challenges for knowledge discovery based on these GO annotations. The recent proliferation of these measures in the biomedical and bioinformatics areas was accompanied by the development of tools (http://neurolex.org/wiki/Category:Resource:Gene_Ontology_Tools) that facilitate effective exploration of these measures. These tools include software packages and web-based on-line tools.
None of these tools support all relevant topology-based approaches in the context of GO, except the DaGO-Fun on-line tool (Mazandu and Mulder, 2013a) implementing the GO-universal metric, the Wang et al. (2007) and Zhang et al. (2006) approaches, the G-SESAME on-line tool (Du et al., 2009) and the GOSemSim R package (Yu et al., 2010) which implement the Wang et al. (2007) approach. These tools are often context dependent and only implement SS measures shown to perform well in a specific application. Moreover, these tools work only for proteins contained in the GOA dataset or existing GO-annotated organisms for SS calculations and each has its specific gene or protein identifier (ID) system, making it difficult to meet input requirements of current genome- and proteome-wide applications from high-throughput analysis. Here, we present A-DaGO-Fun (ADaptable Gene Ontology semantic similarity-based Functional analysis), which overcomes these limitations, enabling effective exploration of different protein functional similarity measures, calibrating datasets from high-throughput experiment analyses and providing researchers with the freedom to choose the most relevant measure for their specific applications using their gene or protein ID system and associated GO annotations.
2 Overview of A-DaGO-Fun
A-DaGO-Fun is a repository of python modules for analyzing protein or gene sets at the functional level based on GO annotations using information content-based SS measures. It contains six main functions and implements 101 different functional similarity measures (see Supplementary File). Each of the eight annotation-based and three topology-based approaches, namely Resnik, XGraSM-Resnik, Nunivers, XGraSM-Nunivers, Lin, XGraSM-Lin, Relevance and Li et al. (2010), Wang et al. (2007), Zhang et al. (2006), and GO-universal, is implemented with seven known term pairwise-based functional similarity measures: Avg, Max, ABM, BMA, BMM, HDF and VHDF (see Supplementary File, Appendix 2). A-DaGO-Fun also includes the five IC-based (Information content-based) direct term functional similarity measures: SimGIC, SimDIC, SimUIC and Cosine (SimCOU and SimCOT) for the annotation-based and each of the three topology-based approaches, and the following particular cases: SimUI, SimDB and SimUB, as well as the Normalized Term Overlap (NTO) measure. Depending on the function being used, the user inputs may be two GO terms, a GO term or GO term pair list or file, or proteins and associated GO terms in a dictionary or file. Comprehensive summary reports are generated and made available in a table format. More details are provided in the supplementary File.
3 A-DaGO-Fun and other tools
As mentioned previously, there have been numerous tools developed for producing GO term and protein SS scores and we refer the interested reader to the Supplementary File (Appendix 3) where these tools are described in terms of SS measures and input size that each tool supports. Each of these tools uses its specific gene ID system in integrating and constructing its database. As examples, the three recently introduced tools, KU-GOAL (Jeong and Chen, 2014), GOssTo (Caniza et al., 2014), SML (Harispe et al., 2013) and DaGO-Fun (Mazandu and Mulder, 2013a), use UniProt IDs. Thus, the user’s datasets have to be aligned with tool ID requirements when exploring SS measures from these tools. However, in the ID mapping process, some annotation content may be left out of the SS score retrieval, especially if the user input IDs are redundant or some gene IDs cannot be efficiently mapped to their corresponding annotation content. Another critical issue is that these tools are organism-based tools, except GOSim (Fröhlich et al., 2007) and DaGO-Fun (Mazandu and Mulder, 2013a), and use different information content values for a term, whereas a term in the GO structure is expected to have a unique information content value which should not depend on the corpus under consideration (Mazandu and Mulder, 2013b). Furthermore, regardless of species coverage these tools may not support less popular species and all existing tools are very limited for newly annotated organisms.
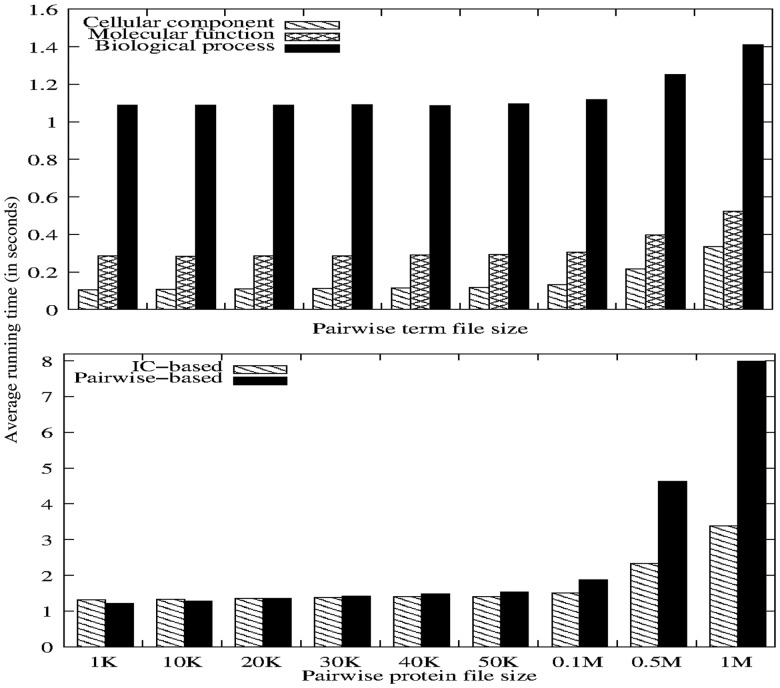
A-DaGO-Fun solves these issues by storing term IC values precomputed using the GO structure and protein GO annotations as provided by the GOA dataset (Huntley et al., 2014). This enables it to handle any dataset under any ID system with their associated GO annotations and to quickly respond to user queries as shown in Fig. 1. These results show that A-DaGO-Fun retrieves SS scores faster than existing software packages (Caniza et al., 2014; Harispe et al., 2013), outputting SS scores in a few seconds. A-DaGO-Fun outputs SS scores of a 100 MB file of protein pairs in 213.77 s (or 3 min 34 s) and 692.24 s (or 11 min 32 s) on average using IC- and pairwise-based functional similarity measures, respectively, and in 34.29, 24.53 and 24.45 s for biological process, molecular function and cellular component term pairwise files respectively, compared to SML producing these scores in 133 min 27 s (Harispe et al., 2013). Furthermore, A-DaGO-Fun implements topology-based approaches producing a fixed and well-defined information content value for a given GO term independent of the corpus under consideration.
Fig. 1.
Running time for retrieval of term semantic and functional similarity scores versus file size in kilobyte (K) or megabyte (M)
4 Conclusion
A-DaGO-Fun provides a python portable application to the large community of GO users and to a broad computational audience, enabling tool designers or developers and experienced end-users as well as non-programmers to retrieve SS scores. This application can be extended, modified and adapted to a defined user need, ensuring that GO SS data and related biological applications are conveniently accessible to researchers and can effectively be used in their functional analyses based on GO annotations.
Supplementary Material
Funding
Some of the authors are funded in part by Government of Canada via the International Development Research Centre (IDRC) through the African Institute for Mathematical Sciences - Next Einstein Initiative (AIMS-NEI). The remaining authors are fully or part funded by the National Human Genome Research Institute of the National Institutes of Health under Award Number U41HG006941. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest: none declared.
References
- Caniza H., et al. (2014) GOssTo: a user-friendly stand-alone and web tool for calculating semantic similarities on the gene ontology. Bioinformatics, 30, 2235–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., et al. (2009) G-SESAME: web tools for GO-term-based gene similarity analysis and knowledge discovery. Nucleic Acids Res., 37, D345–D349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich H., et al. (2007) GOSim–an R-package for computation of information theoretic GO similarities between terms and gene products. BMC Bioinformatics, 8, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GO-Consortium. (2012) The gene ontology: enhancements for 2011. Nucleic Acids Res., 40, D559–D564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harispe S., et al. (2013) The Semantic Measures library and Toolkit: fast computation of semantic similarity and relatedness using biomedical ontologies. Bioinformatics, 30, 740–742. [DOI] [PubMed] [Google Scholar]
- Huntley R.P., et al. (2014) The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res., 43, D1057–D1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.C., Chen X.W. (2014) A new semantic functional similarity over gene ontology. IEEE/ACM Trans. Comput. Biol. Bioinform., 12, 322–334. [DOI] [PubMed] [Google Scholar]
- Li B., et al. (2010) Effectively integrating information content and structural relationship to improve the go-based similarity measure between proteins. ArXiv e-prints, 1001.0958. [Google Scholar]
- Mazandu G.K., Mulder N.J. (2013a) DaGO-Fun: tool for gene ontology-based functional analysis using term information content measures. BMC Bioinformatics, 14, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazandu G.K., Mulder N.J. (2013b) Information content-based gene ontology semantic similarity approaches: toward a unified framework theory. BioMed Res. Int., 2013, Article ID 292063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazandu G.K., Mulder N.J. (2014) Information content-based gene ontology functional similarity measures: which one to use for a given biological data type? PLoS One, 9, e113859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.Z., et al. (2007) A new method to measure the semantic similarity of GO terms. Bioinformatics, 23, 1274–1281. [DOI] [PubMed] [Google Scholar]
- Yu G., et al. (2010) GOSemSim: an R package for measuring semantic similarity among go terms and gene products. Bioinformatics, 26, 976–978. [DOI] [PubMed] [Google Scholar]
- Zhang P., et al. (2006) Gene functional similarity search tool (GFSST). BMC Bioinformatics, 7, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.