The vast majority of biomolecular processes are controlled by the bound water and there is no direct method to address the important biological problems associated with protein–water interactions. In this article, the progress that has been made in synchrotron-based radiolytic labeling and mass spectroscopy techniques for both the identification of bound waters and for characterizing the role of water in protein conformational changes at a high degree of spatial and temporal resolution are summarized.
Keywords: bound water, hydroxyl radical labeling, mass spectrometry, protein conformation, protein modification
Abstract
The vast majority of biomolecular processes are controlled or facilitated by water interactions. In enzymes, regulatory proteins, membrane-bound receptors and ion-channels, water bound to functionally important residues creates hydrogen-bonding networks that underlie the mechanism of action of the macromolecule. High-resolution X-ray structures are often difficult to obtain with many of these classes of proteins because sample conditions, such as the necessity of detergents, often impede crystallization. Other biophysical techniques such as neutron scattering, nuclear magnetic resonance and Fourier transform infrared spectroscopy are useful for studying internal water, though each has its own advantages and drawbacks, and often a hybrid approach is required to address important biological problems associated with protein–water interactions. One major area requiring more investigation is the study of bound water molecules which reside in cavities and channels and which are often involved in both the structural and functional aspects of receptor, transporter and ion channel proteins. In recent years, significant progress has been made in synchrotron-based radiolytic labeling and mass spectroscopy techniques for both the identification of bound waters and for characterizing the role of water in protein conformational changes at a high degree of spatial and temporal resolution. Here the latest developments and future capabilities of this method for investigating water–protein interactions and its synergy with other synchrotron-based methods are discussed.
1. Introduction
Water plays a substantial role in the fundamental events of biological processes and regulates biomolecular structure and dynamics in living cells (Ball, 2008 ▸). Interactions with water are demonstrated to be critical for protein structure, flexibility and folding (Williams et al., 1994 ▸; Renthal, 2008 ▸; Chaplin, 2006 ▸; Angel, Chance et al., 2009 ▸; Jiang & Brünger, 1994 ▸). It has also become increasingly clear that water molecules play an active role in protein–ligand and protein–protein interactions (Bhat et al., 1994 ▸; Braden et al., 1995 ▸; Papoian et al., 2003 ▸; Rispens et al., 2014 ▸; Levinson & Boxer, 2014 ▸; Breiten et al., 2013 ▸), protein activity and catalysis (Nibali & Havenith, 2014 ▸; Royer et al., 1996 ▸; Hare et al., 2010 ▸; Yoon et al., 2004 ▸; Cui et al., 2014 ▸), electron and proton transfer (Garczarek & Gerwert, 2006 ▸; Lin et al., 2005 ▸; Rich & Maréchal, 2013 ▸), transmembrane signaling in receptors (Yuan et al., 2014 ▸; Angel, Chance et al., 2009 ▸) and substrate transfer by ion channels and transporters (Gupta, Chai et al., 2014 ▸; Ostmeyer et al., 2013 ▸). Because proteins are the central players in cellular sensory cascades, antibody-antigen recognition and metabolite transport, delineating protein–water interactions in these systems is of prime importance in understanding these pharmacological targets. Waters in proteins are also an important component of the dynamic aspect of photosynthetic and photoregulatory protein systems (Kerfeld et al., 2003 ▸; Frankel et al., 2013 ▸). Therefore, elucidation of their functional mechanisms, which is important for bioenergy applications, relies on our detailed understanding of protein–water interactions.
X-ray crystallography is one of the tools used to determine the positions of water molecules within proteins at atomic resolution. With the continuous evolution of X-ray sources, detectors, software and protein engineering techniques, very high resolution crystal structures are often obtainable at 2 Å or higher resolution. These structures typically show one to two waters per residue (Carugo & Bordo, 1999 ▸), and analysis of structures from the Protein Data Bank (PDB) has shown a threefold higher concentration of water in cavities and grooves over water at protein surfaces (Williams et al., 1994 ▸; Kuhn et al., 1992 ▸). In contrast to protein surfaces, water in cavities shows a preference for polar side chains over backbone amide bonds when forming hydrogen-bonding networks (Kuhn et al., 1992 ▸). Such water–side-chain hydrogen-bonding networks in protein cavities are formed to a much greater extent than between surface-exposed residues and bulk water (Williams et al., 1994 ▸). Waters in cavities are commonly referred to as bound water (Fig. 1a ▸), which predominantly forms polar contacts with residues rather than with other waters in proximity. Buried water is a form of bound water which is apparently isolated from the bulk and often conserved, forming hydrogen-bonding networks with conserved and functionally important side chains (Angel, Chance et al., 2009 ▸; Halle, 2004 ▸; Renthal, 2008 ▸; Carugo & Bordo, 1999 ▸). The chemical nature of the solvent component used for protein stabilization and crystal formation can affect the occupancy of bound water molecules. The degree to which these solvent components, relative to water, are attracted to or repelled from hydrophobic or polar regions may therefore cause the protein molecule to stabilize preferentially with certain inter- and intra-molecular interactions. Hence, protein crystals might not give structural information on bound water locations that matches the native environment in solution phase and/or physiological conditions. Moreover, while high-resolution crystal structures provide a detailed structural view of the bound water, they also necessarily lack information on the dynamic properties related to protein function.
Figure 1.
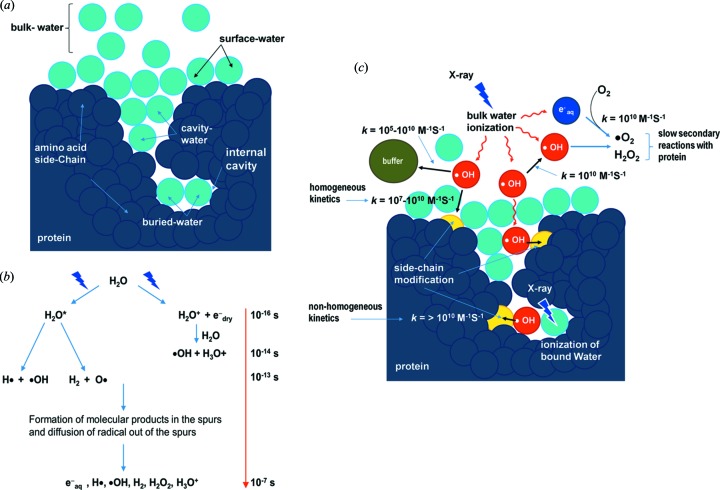
Major reactions scenarios for X-ray radiolysis in dilute protein samples. (a) Schematic showing the position of bulk-, surface- and bound-water (cavity- and buried-water) (light blue) in a protein molecule (dark blue). (b) Radiolysis of water and the timescale of sequence of events. Reproduced from Gupta et al. (2014 ▸) and Liljenzin (2002 ▸). (c) The location of hydroxyl radicals (red) generated in situ from ionization or activation of water by X-ray irradiation. The  OH radicals react with nearby side chains in close proximity, and yield covalent modifications on the protein side chains (yellow). Radiolysis of bulk water starts with the ionization of water on the time scale of 10−16 s. The key product,
OH radicals react with nearby side chains in close proximity, and yield covalent modifications on the protein side chains (yellow). Radiolysis of bulk water starts with the ionization of water on the time scale of 10−16 s. The key product,  OH, diffuses (10−7 s) out in the bulk (red arrows) and undergoes reactions with other
OH, diffuses (10−7 s) out in the bulk (red arrows) and undergoes reactions with other  OH, buffer molecules and protein side chains (bimolecular reactions are indicated by black arrows, and approximate values of the rate constants for different reactions are shown). The rapid counterproductive reactions, such as
OH, buffer molecules and protein side chains (bimolecular reactions are indicated by black arrows, and approximate values of the rate constants for different reactions are shown). The rapid counterproductive reactions, such as  OH—
OH— OH recombination, as well as various other reactions, scavenge
OH recombination, as well as various other reactions, scavenge  OH and reduce the concentration of
OH and reduce the concentration of  OH in the bulk. Thus a sufficient X-ray dose or high-flux-density beam is needed to maintain a steady concentration of
OH in the bulk. Thus a sufficient X-ray dose or high-flux-density beam is needed to maintain a steady concentration of  OH that will lead to a detectable yield of side-chain modification on the protein in solution. In contrast,
OH that will lead to a detectable yield of side-chain modification on the protein in solution. In contrast,  OH radicals formed from activation of a bound water can react faster with side chains in proximity because of the translational and rotational ordering of water and because fewer scavenging reactions by other
OH radicals formed from activation of a bound water can react faster with side chains in proximity because of the translational and rotational ordering of water and because fewer scavenging reactions by other  OH or highly reactive buffer constituents are available. Radiolysis of water also produces electrons, which rapidly become solvated and react with O2 to produce superoxide radicals. In general, the reactivity of side chains to solvated electrons is lower than to hydroxyl radicals (Xu & Chance, 2007 ▸). Peroxides and superoxides undergo slow reactions with protein side chains and are quenched as described in §2.1 and §2.3. Solvated electrons consume O2, which is also required for side-chain labeling by
OH or highly reactive buffer constituents are available. Radiolysis of water also produces electrons, which rapidly become solvated and react with O2 to produce superoxide radicals. In general, the reactivity of side chains to solvated electrons is lower than to hydroxyl radicals (Xu & Chance, 2007 ▸). Peroxides and superoxides undergo slow reactions with protein side chains and are quenched as described in §2.1 and §2.3. Solvated electrons consume O2, which is also required for side-chain labeling by  OH radicals (Fig. 2 ▸); thus short irradiation time as well as high flux density are the key factors for success of the XF-MS experiment. Reproduced in part from Gupta et al. (2014 ▸).
OH radicals (Fig. 2 ▸); thus short irradiation time as well as high flux density are the key factors for success of the XF-MS experiment. Reproduced in part from Gupta et al. (2014 ▸).
Other high-resolution methods such as neutron diffraction can provide information on the dynamics of protein surface hydration on a time scale of pico- to nano-seconds and on distance scales of a few angstroms (Tarek & Tobias, 2000 ▸; Nickels et al., 2012 ▸; Frölich et al., 2009 ▸), and high-field nuclear magnetic resonance (NMR) such as NOESY (nuclear Overhauser enhancement spectroscopy) and MRD (magnetic resonance dispersion) methods can also directly determine the dynamics of bound waters in proteins with a high degree of spatial and temporal resolution in solution (Bertini et al., 2000 ▸; Brunne et al., 1993 ▸; Denisov & Halle, 1996 ▸; Denisov et al., 1996 ▸; Nucci et al., 2011 ▸; Otting et al., 1991 ▸; Persson & Halle, 2008 ▸). These methods can determine residence times of individual bound waters on the order of sub-nanoseconds to milliseconds; however, such experimental approaches are limited due to specific sample condition requirements.
The much lower resolution method of vibrational absorption spectroscopy has a high degree of sensitivity to changes in a hydrogen-bonded environment and therefore has proven useful for detecting protein–water interactions. Non-invasive NIR (near infrared at ∼1400 cm−1) spectroscopy has been used as a spectral biomarker to reveal differences in water–protein interactions in protein samples (Gowen et al., 2013 ▸; Tsenkova, 2009 ▸). FTIR (Fourier transform infrared spectroscopy) at O—H and O—D stretching frequencies, together with site-directed mutagenesis, has been used to identify site-specific protein–water interactions, which in turn have revealed important information about the role of internal waters in membrane protein systems (Furutani et al., 2003 ▸; Kandori, 2000 ▸; Garczarek & Gerwert, 2006 ▸; Maréchal & Rich, 2011 ▸; Muroda et al., 2012 ▸). While FITR methods directly determine the location of bound waters, sample preparation requirements and mutagenesis steps often limit the applicability of the technique.
Theoretical predictions from molecular dynamics (MD) simulations provide another important approach that can probe structural, dynamical and thermodynamical properties of protein–water interactions over a broad range of time scales (Beuming et al., 2012 ▸; Tran et al., 2013 ▸; Breiten et al., 2013 ▸; Tarek & Tobias, 2000 ▸; Nibali & Havenith, 2014 ▸). MD results are often unclear due to discrepancies in interpreting experimental results. However, recent microsecond MD simulations have revealed the importance of structural water in both a potassium channel and rhodopsin activation, and the results correlated well with existing biophysical and structural information (Ostmeyer et al., 2013 ▸; Yuan et al., 2014 ▸). It is important that these methods continue to be developed to pinpoint the locations and dynamics of bound water in physiologically relevant samples in order to address a wide range of biological questions.
Recently, the method of synchrotron X-ray radiolytic labeling mass spectrometry (XF-MS) has been shown to be able to directly distinguish the interactions of bulk and bound water with protein side chains in solution. With this technique, reactive hydroxyl radicals ( OH) are generated in situ by high-flux-density X-rays, causing covalent labeling of protein side chains which are detected by mass spectrometry analysis. The in situ covalent labeling approach eliminates most of the complexity associated with sample preparation necessary with other high-resolution techniques and at the same time allows easy comparative structural analysis of protein samples under various physiologically relevant conditions. This review describes the XF-MS method, reviews recent developments and future applications to study water–protein interactions and protein conformational changes, and discusses the synergy of the method with other synchrotron and biophysical methods when a hybrid approach is needed to solve important biological questions.
OH) are generated in situ by high-flux-density X-rays, causing covalent labeling of protein side chains which are detected by mass spectrometry analysis. The in situ covalent labeling approach eliminates most of the complexity associated with sample preparation necessary with other high-resolution techniques and at the same time allows easy comparative structural analysis of protein samples under various physiologically relevant conditions. This review describes the XF-MS method, reviews recent developments and future applications to study water–protein interactions and protein conformational changes, and discusses the synergy of the method with other synchrotron and biophysical methods when a hybrid approach is needed to solve important biological questions.
2. Synchrotron X-ray radiolytic labeling and mass spectrometry (XF-MS)
XF-MS has developed in the past decade into a nearly routine technique, and has been applied to a diverse range of biological systems, yielding unique structural insights impossible to obtain using only the standard structural methods of crystallography, NMR, electron microscopy and small-angle scattering (Bohon et al., 2008 ▸; Gupta et al., 2007 ▸, 2010 ▸, 2012 ▸; Kiselar & Chance, 2010 ▸; Angel, Gupta et al., 2009 ▸; Chaudhuri et al., 2011 ▸; Kamal et al., 2007 ▸; Kiselar et al., 2003 ▸; Orban et al., 2012 ▸; Padayatti et al., 2013 ▸; Xu & Chance, 2007 ▸; Gupta, Chai et al., 2014 ▸; Leverenz et al., 2015 ▸; Klinger et al., 2014 ▸; Salanga et al., 2014 ▸). The key reaction for XF-MS is the covalent labeling of solvent accessible protein side chains by  OH, which is generated rapidly and isotropically in situ by high-flux-density broad-band synchrotron X-rays in an aqueous buffered solution (Xu & Chance, 2007 ▸; Gupta, Celestre et al., 2014 ▸). Under controlled irradiation and aerobic conditions, labeling results in permanent mass adducts, predominantly +16, +14, +32 and +48 Da, at various side chains. These covalent markers on the protein accessible sites are analyzed by mass-spectrometry-based bottom-up proteomics (Takamoto & Chance, 2006 ▸; Xu & Chance, 2007 ▸). The extent of reaction between protein and
OH, which is generated rapidly and isotropically in situ by high-flux-density broad-band synchrotron X-rays in an aqueous buffered solution (Xu & Chance, 2007 ▸; Gupta, Celestre et al., 2014 ▸). Under controlled irradiation and aerobic conditions, labeling results in permanent mass adducts, predominantly +16, +14, +32 and +48 Da, at various side chains. These covalent markers on the protein accessible sites are analyzed by mass-spectrometry-based bottom-up proteomics (Takamoto & Chance, 2006 ▸; Xu & Chance, 2007 ▸). The extent of reaction between protein and  OH or yield of covalent labeling depends on both the intrinsic reactivity and the solvent accessibility of the residues. However, since most footprinting studies compare two or more states of proteins, the changes in the amount of covalent labeling on the same residue from one state to another depends solely on the solvent accessibility difference between the two states. The information obtained can be interpreted in the context of existing high-resolution structures of one state and compared with that of an unknown state, or incorporated into molecular modeling strategies that provide information about protein or ligand docking and conformational changes (Kamal & Chance, 2008 ▸; Leverenz et al., 2015 ▸; Gupta, Chai et al., 2014 ▸).
OH or yield of covalent labeling depends on both the intrinsic reactivity and the solvent accessibility of the residues. However, since most footprinting studies compare two or more states of proteins, the changes in the amount of covalent labeling on the same residue from one state to another depends solely on the solvent accessibility difference between the two states. The information obtained can be interpreted in the context of existing high-resolution structures of one state and compared with that of an unknown state, or incorporated into molecular modeling strategies that provide information about protein or ligand docking and conformational changes (Kamal & Chance, 2008 ▸; Leverenz et al., 2015 ▸; Gupta, Chai et al., 2014 ▸).
2.1. Chemical basis of radiolysis for XF
A synchrotron bending-magnet beamline can deliver a high flux such as 1015–1016 photon s−1 of continuous X-ray beam with a wide energy range (1–20 keV), and X-rays of this energy range interact with water almost exclusively via the photoelectric effect. The ionization of both bulk and bound water results in formation of  OH radicals. The time scale for the sequence of events in the radiolysis of pure water leading to the generation of the highly reactive but short-lived
OH radicals. The time scale for the sequence of events in the radiolysis of pure water leading to the generation of the highly reactive but short-lived  OH in solution has been discussed previously (Gupta et al., 2007 ▸; Xu & Chance, 2007 ▸) (Fig. 1b
▸). The
OH in solution has been discussed previously (Gupta et al., 2007 ▸; Xu & Chance, 2007 ▸) (Fig. 1b
▸). The  OH undergoes diffusion-controlled recombination reactions with other
OH undergoes diffusion-controlled recombination reactions with other  OH in bulk, and other rapid counterproductive reactions with buffer constituents that lead to scavenging of the radicals as well as generation of secondary radical products. Secondary radical products such as peroxide and superoxide can undergo reactions with proteins by slower kinetic pathways, and are often quenched by methionine amide within milliseconds (Gupta et al., 2007 ▸; Xu & Chance, 2007 ▸) (Fig. 1c
▸). To overcome the loss of
OH in bulk, and other rapid counterproductive reactions with buffer constituents that lead to scavenging of the radicals as well as generation of secondary radical products. Secondary radical products such as peroxide and superoxide can undergo reactions with proteins by slower kinetic pathways, and are often quenched by methionine amide within milliseconds (Gupta et al., 2007 ▸; Xu & Chance, 2007 ▸) (Fig. 1c
▸). To overcome the loss of  OH by the recombination and counterproductive reactions, the synchrotron X-ray beam is focused to deliver flux density of the order of 1016 photon s−1 in a 100 µm spot, such that a sufficient concentration of radicals is generated on a microsecond time scale to label side chains while reducing adverse effects of protein damage by secondary radicals (Gupta, Celestre et al., 2014 ▸). Generation of
OH by the recombination and counterproductive reactions, the synchrotron X-ray beam is focused to deliver flux density of the order of 1016 photon s−1 in a 100 µm spot, such that a sufficient concentration of radicals is generated on a microsecond time scale to label side chains while reducing adverse effects of protein damage by secondary radicals (Gupta, Celestre et al., 2014 ▸). Generation of  OH in situ by ionizing irradiation is a flux-driven process. It is known that in an aqueous environment, and in the presence of other reactants or amino acid side chains (which have bimolecular rate constants for reaction with
OH in situ by ionizing irradiation is a flux-driven process. It is known that in an aqueous environment, and in the presence of other reactants or amino acid side chains (which have bimolecular rate constants for reaction with  OH near diffusion controlled processes),
OH near diffusion controlled processes),  OH is very short-lived and reacts within 1 to 5 molecular diameters of the site of formation (Pryor, 1986 ▸). The intrinsic reactivities of free amino acids towards
OH is very short-lived and reacts within 1 to 5 molecular diameters of the site of formation (Pryor, 1986 ▸). The intrinsic reactivities of free amino acids towards  OH in aqueous solution vary widely; for example, Cys > Met, Trp > Tyr > Phe > His > Iie > Leu > Arg (according to their order of reactivity as measured in aerobic aqueous solution) can react with
OH in aqueous solution vary widely; for example, Cys > Met, Trp > Tyr > Phe > His > Iie > Leu > Arg (according to their order of reactivity as measured in aerobic aqueous solution) can react with  OH extremely rapidly (rate constants 109 to 1010
M
−1 s−1) in contrast to Lys ≃ Val > Ser ≃ Thr ≃ Pro > Gln ≃ Glu > Asp ≃ ASN > Ala > Gly (according to their order of reactivity as measured in aerobic aqueous solution), which react 10- to 1000-fold slower. Thus, the former set of residues generally shows modification in protein samples. The mechanism of the side-chain reaction to
OH extremely rapidly (rate constants 109 to 1010
M
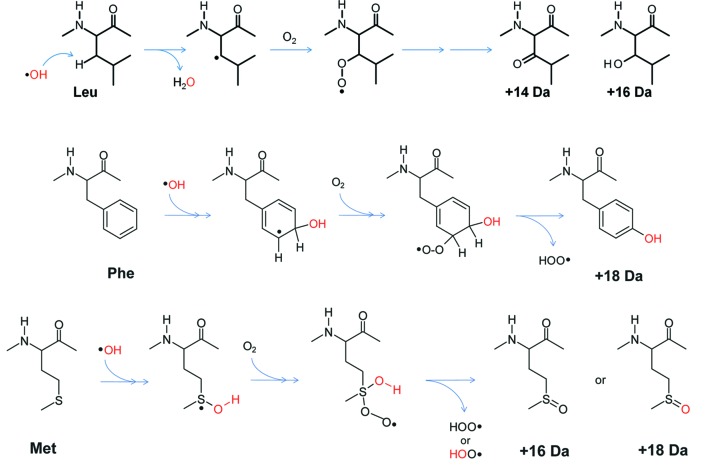
−1 s−1) in contrast to Lys ≃ Val > Ser ≃ Thr ≃ Pro > Gln ≃ Glu > Asp ≃ ASN > Ala > Gly (according to their order of reactivity as measured in aerobic aqueous solution), which react 10- to 1000-fold slower. Thus, the former set of residues generally shows modification in protein samples. The mechanism of the side-chain reaction to  OH can be broadly classified into two major categories: First, hydrogen abstraction from the carbon in a side chain (for most of the aliphatic side chains and Pro), and, second, direct nucleophilic attack on sulfur atoms or conjugated double bonds (Cys, Met, aromatic side chains and His) (Xu & Chance, 2007 ▸). In both cases a protein radical is formed at the side chain, which then undergoes hydroperoxyl elimination in the presence of dissolved molecular oxygen and forms a stable mass adduct (Fig. 2 ▸). Overall, XF-MS can detect modifications in 15 out of 20 standard amino acids, making it a very useful tool for analyzing protein structure at the residue level.
OH can be broadly classified into two major categories: First, hydrogen abstraction from the carbon in a side chain (for most of the aliphatic side chains and Pro), and, second, direct nucleophilic attack on sulfur atoms or conjugated double bonds (Cys, Met, aromatic side chains and His) (Xu & Chance, 2007 ▸). In both cases a protein radical is formed at the side chain, which then undergoes hydroperoxyl elimination in the presence of dissolved molecular oxygen and forms a stable mass adduct (Fig. 2 ▸). Overall, XF-MS can detect modifications in 15 out of 20 standard amino acids, making it a very useful tool for analyzing protein structure at the residue level.
Figure 2.
Reaction schemes for modification of side chains by  OH. The hydrocarbon side chain (of aliphatic non-polar and polar amino acids) predominantly undergoes hydrogen abstraction by
OH. The hydrocarbon side chain (of aliphatic non-polar and polar amino acids) predominantly undergoes hydrogen abstraction by  OH to give carbon-centered radicals, which then react with molecular O2 under aerobic conditions and subsequently form stable hydroxylated (+16 Da) or carbonylated (+14 Da) products. Aromatics and sulfur-containing side chains directly undergo hydroxylation by
OH to give carbon-centered radicals, which then react with molecular O2 under aerobic conditions and subsequently form stable hydroxylated (+16 Da) or carbonylated (+14 Da) products. Aromatics and sulfur-containing side chains directly undergo hydroxylation by  OH followed by reaction with O2 under aerobic conditions to form stable hydroxylated products (+16 Da) and other oxygen adducts. Aromatics and sulfur-containing side chains can be labeled by 18O (red) from radiolysis of H2
18O water.
OH followed by reaction with O2 under aerobic conditions to form stable hydroxylated products (+16 Da) and other oxygen adducts. Aromatics and sulfur-containing side chains can be labeled by 18O (red) from radiolysis of H2
18O water.
In dilute solutions, amino acids or small polypeptides react with  OH by a homogeneous kinetics process; however,
OH by a homogeneous kinetics process; however,  OH-induced modification in large protein systems follows a complex kinetic pathway, and in many instances does not follow the order of amino acid reactivity listed above. As previously described, protein molecules create distinct hydration environments which are different from that of bulk water. The cavities, grooves and active sites on a protein surface contain hydrogen-bonding networks and ionic interlocks with amino acid side chains that can bind three times as many water molecules as the solvent exposed surface. Low linear energy transfer ionizing radiation, such as 5–20 keV X-ray photon beams, can generate spurs inside such cavities that result in selective modification of local side chains (Fig. 1c
▸). To date there is no report of side-chain reactivity order within a folded protein, and therefore it is possible that the reactivity order is governed by the proximity of the side chain to
OH-induced modification in large protein systems follows a complex kinetic pathway, and in many instances does not follow the order of amino acid reactivity listed above. As previously described, protein molecules create distinct hydration environments which are different from that of bulk water. The cavities, grooves and active sites on a protein surface contain hydrogen-bonding networks and ionic interlocks with amino acid side chains that can bind three times as many water molecules as the solvent exposed surface. Low linear energy transfer ionizing radiation, such as 5–20 keV X-ray photon beams, can generate spurs inside such cavities that result in selective modification of local side chains (Fig. 1c
▸). To date there is no report of side-chain reactivity order within a folded protein, and therefore it is possible that the reactivity order is governed by the proximity of the side chain to  OH, which originates from the ionization of a water molecule that might have strong translational and rotational ordering due to participation in a hydrogen-bonding network. In contrast, a fully exposed side chain has to compete with equally or more reactive
OH, which originates from the ionization of a water molecule that might have strong translational and rotational ordering due to participation in a hydrogen-bonding network. In contrast, a fully exposed side chain has to compete with equally or more reactive  OH or other additives (buffer constituents) in order to yield detectable amounts of modification. Hence, in analyzing XF-MS results it is often not surprising to see limited modification of a fully surface-exposed residue as compared with considerable modification of a similar side chain inside a cavity.
OH or other additives (buffer constituents) in order to yield detectable amounts of modification. Hence, in analyzing XF-MS results it is often not surprising to see limited modification of a fully surface-exposed residue as compared with considerable modification of a similar side chain inside a cavity.
XF-MS has already been shown to label amino acid residues adjacent to bound water in globular proteins (Gupta et al., 2010 ▸). In membrane proteins such as ion channels and receptors, this method detects modification of specific residues inside the transmembrane domain where bound waters are located, in contrast to the nonspecific labeling of the solvent-exposed amino acid residues that are exposed to bulk water (Gupta et al., 2010 ▸; Angel, Gupta et al., 2009 ▸). Thus, in many cases the  OH reaction with amino acid side chains within complex proteins might not strictly follow homogeneous kinetic pathways; instead, the reaction can be strongly affected by the ionization of the local water in the vicinity of the reactive amino-acid side chains (Pearson & Williams, 1987 ▸). We will review recent developments of the XF-MS technique at synchrotron facilities, and experimental results in the context of detecting bound water and protein conformational changes in the following sections.
OH reaction with amino acid side chains within complex proteins might not strictly follow homogeneous kinetic pathways; instead, the reaction can be strongly affected by the ionization of the local water in the vicinity of the reactive amino-acid side chains (Pearson & Williams, 1987 ▸). We will review recent developments of the XF-MS technique at synchrotron facilities, and experimental results in the context of detecting bound water and protein conformational changes in the following sections.
2.2. Synchrotron beamline configuration
XF-MS was originally developed at beamline X9A at the NSLS (Ralston et al., 2000 ▸) and then moved to a similar bending-magnet beamline, NSLS X28C, where it began serving the user community (Gupta et al., 2007 ▸). Beamline X28C was equipped with a palladium-coated toroidal focusing mirror which was used to deliver millisecond X-ray doses to protein samples (Sullivan et al., 2008 ▸). Now the NSLS footprinting program is transitioning to the new XFP beamline at NSLS II. The new XFP beamline will utilize a three-pole wiggler source capable of delivering tenfold-higher flux density than X28C. Recently, XF-MS was also developed at the Advanced Light Source (ALS) using beamlines 5.3.1 and 3.2.1 (Gupta, Celestre et al., 2014 ▸; Gupta et al., 2016 ▸). Beamline 5.3.1 is located on a bending magnet and equipped with a platinum-coated toroidal focusing mirror suitable for focusing a white-light X-ray beam. The broadband X-ray beam (1–13 keV) exits from the beryllium window of the beampipe under ultrahigh vacuum, with a flux of ∼1 × 1016 photon s−1. Focused beam sizes can be set to match 100–535 µm inner-diameter microcapillary tubes, which are used to deliver samples using syringe pumps and allow irradiation on the microsecond timescale. Beamline 3.2.1 is a white-light bending-magnet beamline with similar characteristics to 5.3.1 but without a focusing mirror. The ALS has recently dedicated beamline 3.3.1, from the same bending source as 3.2.1, for development of a dedicated XF-MS facility, which will be the highest-flux-density XF-MS facility in the USA in the coming years, second to the NSLS II XFP beamline. Recently, XF-MS studies were reported using several other low-flux-density synchrotron beamlines (Bohon et al., 2014 ▸). However, the low flux at these beamlines necessitates long irradiation times, resulting in secondary damage, which is time-dependent and results in specific structural perturbations. In contrast, a short pulse of high-flux-density photons produces adequate  OH concentration to overcome scavenging reactions while preserving the structural integrity of complex protein assemblies.
OH concentration to overcome scavenging reactions while preserving the structural integrity of complex protein assemblies.
2.3. Liquid-chromatography mass-spectrometry-based quantification, data analysis and recent advancements
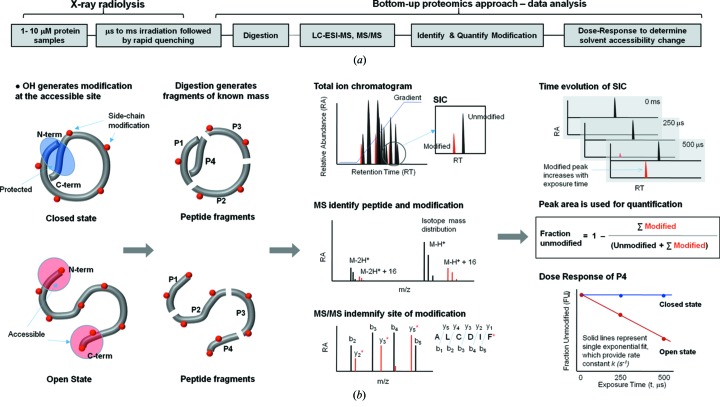
The experimental scheme for a typical XF-MS experiment is shown in Fig. 3 ▸. Dilute buffered protein samples are subjected to a series of irradiations ranging from microseconds to milliseconds (Gupta, Celestre et al., 2014 ▸). The use of microcapillary high-flow-rate sample-handling systems has improved the experiment by allowing short irradiation times, better temperature control, and fast post-exposure quenching by methionine amide to stop any secondary radical reactions (Gupta, Celestre et al., 2014 ▸; Xu & Chance, 2007 ▸). The irradiated samples are subjected to standard mass-spectrometry-based bottom-up proteomics analysis using various protease digestion methods to increase overall sequence coverage. Reverse-phase liquid chromatography coupled to a mass spectrometer generates a mass chromatogram (TIC, total ion chromatogram), where unmodified and modified peptides are separated. We have introduced a new standard-flow liquid-chromatography method with a high degree of chromatographic reproducibility and increased throughput for sample analysis (Gonzalez Fernandez-Nino et al., 2015 ▸). Peak identification and peak area analysis is carried out in a semi-automated fashion using Mascot (Matrix Science Inc), Skyline (Schilling et al., 2012 ▸) and instrument-based software by considering four basic characteristics of unmodified and modified products: (i) accurate m/z (mass to charge ratio); (ii) isotopic m/z distribution of m/z of various charge states; (iii) retention time (RT); and (iv) MS/MS sequencing. Data analysis can also be carried out using the fully automated software ProtMap (Kaur et al., 2009 ▸). The relative abundance of unmodified peptide versus increase in the irradiation time is used to plot dose response curves.
Figure 3.
Schematic of the X-ray footprinting method using synchrotron X-ray radiolysis and mass spectrometry. (a) Overall method, and (b) case study for a protein that undergoes a conformational change from the closed to the open state. The protein is covalently modified after a series of X-ray irradiations of the order of microseconds followed by rapid quenching by methionine amide. Irradiated protein is digested with proteases to generate peptide fragments of known mass. Digested protein is analyzed by reverse-phase liquid chromatography coupled with electrospray mass spectrometry (LC-ESI-MS), in which peptides are separated in the total ion chromatogram (TIC) or mass chromatogram. For any peptide fragment the unmodified and modified m/z is extracted and visualized by selected ion chromatogram (SIC). High-resolution mass spectrometry is used to identify the unmodified and modified peptide fragments by their accurate m/z and isotope distribution. The site of modification is identified from the mass assignment of the y and b fragment ions from the tandem mass spectrometry (MS/MS) of the corresponding peptide. The extent of modification for the series of irradiation points are quantified from the SICs of unmodified and modified pepide fragments. The fraction of unmodified peptide versus exposure time (dose-response or DR-plot) provides site-specific modification rate constants (k s−1). The rates are compared among different sample conditions, and their ratios, which are independent of intrinsic reactivity, account for the degree in solvent accessibility changes due to any conformational transition/interactions. The final results are mapped onto available structures or used as constraints for structural modeling.
A single peptide fragment can have multiple modifications at different residues. Depending on the quality and the resolution of the mass chromatogram, the dose response for each site of modification can be derived to obtain residue speficic data. Otherwise, combined modifications are reported as the dose response for the overall peptide fragment. A pesudo-first-order fit of the dose response plot gives the hydroxyl radical reactivity rate constant, which is a measure of both intrinsic reactivity and solvent accessibility. In a comparative structural analysis, the ratio (R) of rate constants between samples determines the relative solvent accessibility changes, which are independent of any sequence context (Gupta, Chai et al., 2014 ▸; Leverenz et al., 2015 ▸). Finally, the peptide or site-specific R is utilized for structural modeling. Recently, variations in the data-collection method, such as selected reaction monitoring and MS/MS approaches have been undertaken to improve the quality and sensitivity of XF-MS for large protein complexes (Kiselar & Chance, 2010 ▸; Kaur et al., 2015 ▸). In addition, the recent introduction of the protection factor has shown that XF-MS can be used as a tool to quantitatively analyze protein topography (Kaur et al., 2015 ▸; Huang et al., 2015 ▸).
2.4. Comparison with other hydroxyl labeling methods
Chemical generation of hydroxyl radicals is a simple laboratory-based method that can be used to successfully determine protein interactions and dynamics (Xu & Chance, 2007 ▸). However, chemical production of hydroxyl radicals relies on the addition of reagents such as Fe-EDTA and H2O2 that can affect conformation or damage/unfold proteins, as well as remove essential metal ions necessary for protein function. Other methods, such as laser photochemical and electrospray ionization, also generate  OH in protein solutions (Aye et al., 2005 ▸; Hambly & Gross, 2005 ▸; Downard et al., 2012 ▸; Konermann et al., 2013 ▸; Maleknia & Downard, 2012 ▸; Wong et al., 2005 ▸). In the laser photochemical method, a sufficient amount of
OH in protein solutions (Aye et al., 2005 ▸; Hambly & Gross, 2005 ▸; Downard et al., 2012 ▸; Konermann et al., 2013 ▸; Maleknia & Downard, 2012 ▸; Wong et al., 2005 ▸). In the laser photochemical method, a sufficient amount of  OH is produced on the microsecond timescale to conduct footprinting analysis; however, one drawback of this method is that it requires a millimolar concentration of H2O2, which can unfold proteins and/or perturb complex protein assemblies with metal-active centers (Ling et al., 2012 ▸; Shacter, 2000 ▸; Watson et al., 2009 ▸). The electrospray ionization-based hydroxyl radical footprinting method uses a very high electrospray voltage in the presence of O2 to produce
OH is produced on the microsecond timescale to conduct footprinting analysis; however, one drawback of this method is that it requires a millimolar concentration of H2O2, which can unfold proteins and/or perturb complex protein assemblies with metal-active centers (Ling et al., 2012 ▸; Shacter, 2000 ▸; Watson et al., 2009 ▸). The electrospray ionization-based hydroxyl radical footprinting method uses a very high electrospray voltage in the presence of O2 to produce  OH radicals in solutions with a high concentration of volatile buffer components. The radiolytic generation of hydroxyl radicals is a straightforward technique but it is important to control the dose received by samples since both modifications of proteins and secondary radical reactions can lead to unfolding or conformational changes. An alternate approach to X-ray radiolysis is electron beam radiolysis using a van de Graaff generator (Watson et al., 2009 ▸). Data on cyt c and rhodopsin using this approach (Gupta, unpublished data) indicated that the electron pulse delivers high enough flux to probe radiolytic modifications on the sub-microsecond to microsecond time scale. This time scale is fast enough that modifications occur before protein unfolding can take place in simple globular proteins. However, the energy deposition by 2 MeV electrons can be ∼200-fold more than that of a synchrotron X-ray beamline. In this process, direct energized electron-induced protein damage can be a major problem.
OH radicals in solutions with a high concentration of volatile buffer components. The radiolytic generation of hydroxyl radicals is a straightforward technique but it is important to control the dose received by samples since both modifications of proteins and secondary radical reactions can lead to unfolding or conformational changes. An alternate approach to X-ray radiolysis is electron beam radiolysis using a van de Graaff generator (Watson et al., 2009 ▸). Data on cyt c and rhodopsin using this approach (Gupta, unpublished data) indicated that the electron pulse delivers high enough flux to probe radiolytic modifications on the sub-microsecond to microsecond time scale. This time scale is fast enough that modifications occur before protein unfolding can take place in simple globular proteins. However, the energy deposition by 2 MeV electrons can be ∼200-fold more than that of a synchrotron X-ray beamline. In this process, direct energized electron-induced protein damage can be a major problem.
All the radical-generating techniques have unique advantages and disadvantages, and therefore should be applied variously depending on the system under study. The unique advantages of synchrotron-based XF-MS include the following. First, the technique provides a straightforward way to vary the  OH dose from 5- to 20-fold by varying the flow rate of the sample across the fixed size and flux density of a continuous X-ray beam. This simple approach is highly advantageous in terms of generating multiple dose-dependent data to detemine precise hydroxyl radical reactivity rates followed by error analysis for accurate comparisons between multiple states of complex biological samples. Second, in situ
OH dose from 5- to 20-fold by varying the flow rate of the sample across the fixed size and flux density of a continuous X-ray beam. This simple approach is highly advantageous in terms of generating multiple dose-dependent data to detemine precise hydroxyl radical reactivity rates followed by error analysis for accurate comparisons between multiple states of complex biological samples. Second, in situ
 OH generation has the advantage of allowing nearly any type of sample condition without deleterious effects from external reagents. Third, XF-MS can be used to detect bound water, as described below.
OH generation has the advantage of allowing nearly any type of sample condition without deleterious effects from external reagents. Third, XF-MS can be used to detect bound water, as described below.
3. XF distinguishes internal versus bulk water interactions in proteins and probes water dynamics
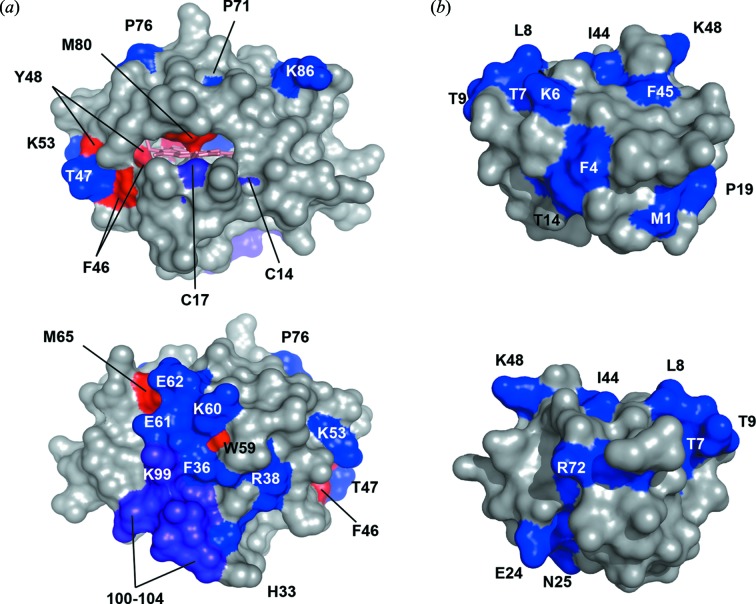
The evidence to support detection of bound water by XF-MS at the protein surface and in the protein interiors first came from temperature-dependent radiolysis of cyt c (Fig. 4 ▸) (Gupta et al., 2012 ▸). Freezing a protein sample will change the properties of diffusible water surrounding a protein and is expected to decrease hydroxyl reactivity. However, in cyt c, upon decreasing the temperature from room temperature to −35°C, the residues in proxmity toi bound water (as determined by high-resolution crystallography) retained their hydroxyl radical reactivity. In contrast, most surface residues showed no observable modification in the frozen state. The modification of residues in the frozen state directly supports the theory that X-rays activate internal waters, which are also shown via NMR to retain their liquid water characteristics at −35°C (Kuntz et al., 1969 ▸; Tompa et al., 2009 ▸). The temperature comparison method was also used to test ubiquitin (which does not have any bound water) and fluorophore (Alexa 488); both showed complete loss of  OH reactivity in the frozen state.
OH reactivity in the frozen state.
Figure 4.
Detection of the site of bound water interaction by temperature-dependent XF-MS. The colored surfaces on the X-ray crystal structures of cyt c (PDB 1HRC) (a) and ubiquitin (PDB 1UBQ) (b) indicate the side-chain residues that are consistently modified at room temperature and/or frozen conditions. Blue indicates the residues that show a 13- to 200-fold decrease in modification upon freezing; violet indicates a 3- to 10-fold decrease; and red indicates minimal to no change (<2-fold) in the modification rate when the sample is frozen as compared with room temperature. In cyt c the residue Y67, which also showed minimal to no change, is completely buried inside the heme cavity and not visible in these orientations. Reproduced from Gupta et al. (2012 ▸).
Following this study, time-resolved (TR) 18O/16O exchange XF-MS was used to determine the dynamics of water exchange. This method is based on the reaction scheme and time-resolved experimental scheme shown in Figs. 2 ▸ and 5(a) ▸, respectively. Only Phe, Tyr, Trp, Met and Cys can be directly labeled by  18OH following irradiation of a protein solution containing H2
18O. The extent of 18O labeling is calculated from the relative isotopic m/z distribution from zoomed scan analysis (Fig. 5b
▸). In a typical water-exchange experiment, 1:1 mixing was carried out between proteins (cyt c and ubiquitin) in H2
18O buffer and regular H2
16O buffer. The temperature of the mixing and irradiation setup was maintained at 4°C to slow the water-exchange process in the proteins. After millisecond delays, the sample was irradiated and immediately quenched. Protease digestion was carried out after completely drying out the sample to eliminate any 18O incorporation from residual H2
18O during the digestion reactions. The extent of water exchange at a particular side-chain residue (the ratio of 18O/16O) versus mixing delays generated site-specific progress curves and determined the residence time of proximal bound water. The temperature-dependent and time-resolved XF-MS methods were able to explicitly probe bound water interactions inside the heme binding pocket of cyt c (Fig. 5d
▸). These results are further supported by the high-resolution crystal structure (Bushnell et al., 1990 ▸), while the millisecond exchange rates are consistent with NMR studies (Bertini et al., 2001 ▸). These two techniques provide a direct and powerful method to follow the structural changes of bound water in enzyme active sites, channels, ion pumps and membrane protein pores. Ultra-fast continuous-flow mixing devices are being developed, and, in combination with the short pulse of intense X-rays available from the focused bending-magnet sources at the ALS or NSLS II, will allow monitoring of even faster time course structural dynamics, which will contribute significantly to our understanding of water-mediated biomolecular processes.
18OH following irradiation of a protein solution containing H2
18O. The extent of 18O labeling is calculated from the relative isotopic m/z distribution from zoomed scan analysis (Fig. 5b
▸). In a typical water-exchange experiment, 1:1 mixing was carried out between proteins (cyt c and ubiquitin) in H2
18O buffer and regular H2
16O buffer. The temperature of the mixing and irradiation setup was maintained at 4°C to slow the water-exchange process in the proteins. After millisecond delays, the sample was irradiated and immediately quenched. Protease digestion was carried out after completely drying out the sample to eliminate any 18O incorporation from residual H2
18O during the digestion reactions. The extent of water exchange at a particular side-chain residue (the ratio of 18O/16O) versus mixing delays generated site-specific progress curves and determined the residence time of proximal bound water. The temperature-dependent and time-resolved XF-MS methods were able to explicitly probe bound water interactions inside the heme binding pocket of cyt c (Fig. 5d
▸). These results are further supported by the high-resolution crystal structure (Bushnell et al., 1990 ▸), while the millisecond exchange rates are consistent with NMR studies (Bertini et al., 2001 ▸). These two techniques provide a direct and powerful method to follow the structural changes of bound water in enzyme active sites, channels, ion pumps and membrane protein pores. Ultra-fast continuous-flow mixing devices are being developed, and, in combination with the short pulse of intense X-rays available from the focused bending-magnet sources at the ALS or NSLS II, will allow monitoring of even faster time course structural dynamics, which will contribute significantly to our understanding of water-mediated biomolecular processes.
Figure 5.
Determination of residence time of tightly bound water by XF-MS. (a) Rapid mixing combined with 18O-mediated hydroxyl radical labeling to monitor the time-course of exchange of water in cyt c. LC-ESI-MS is used to identify and isolate the modified peptides, targeted MS/MS is used to identify the sites of 18O labeling, and zoom scans are used to quantify the ratio of 18O- versus 16O-labeling at various mixing delays. (b) Zoom scans for singly protonated peptide 61–72 showing the decrease in the abundance of the 2m/z shifted 18O monoisotopic mass (arrow) that corresponds to the water exchange at M65 and Y67 with increase in the mixing delays. (c) Progress curves (circles and error bars) of water exchange for the 18O labeled side-chain residues. The solid line represents the fit to a single exponential function. Residues W59 and F36 have exchange that is complete at the first measurement, while the rates of exchange of C14, C17, F46, Y48, M65, Y67 and M80 are discretely measured. (d) Sites of 18O-modifications are visualized from the crystal structure 1HRC using PyMOL (DeLano Scientific). The 18O-labeled residues (light blue) in and around the heme (light pink) crevices, and the position of residue T78 (gray) and conserved waters (cyan spheres) HOH112, HOH139 are shown in two orientations of the cyt c molecule. Reproduced from Gupta et al. (2012 ▸).
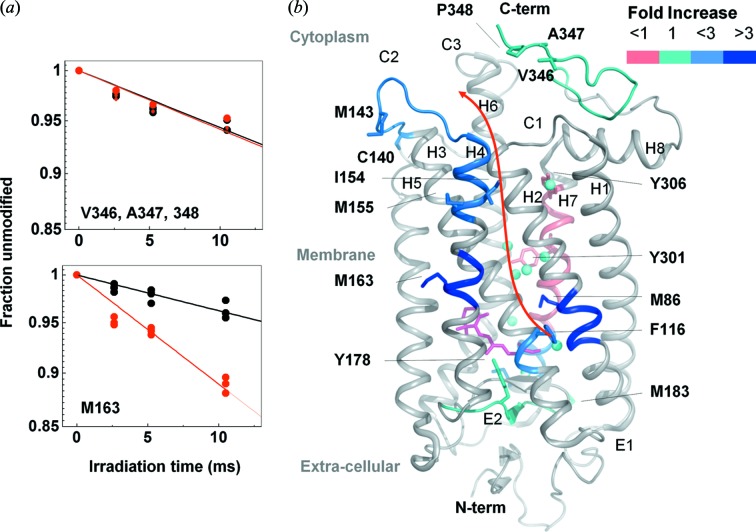
The X-ray crystal structures of rhodopsin and of its photo-intermediates have dramatically increased our understanding of structural rearrangements upon the activation of G-protein-coupled receptors (GPCRs) (Orban et al., 2012 ▸). But it is also increasingly clear that static structures alone are not sufficient to provide a complete understanding of GPCR function, especially given the prominent role that is played by structural waters, which are only visible in high-resolution crystal structures. XF-MS has emerged as a novel approach to study GPCRs by in situ labeling of transmembrane (TM) residues, which are located in proximity to bound water (Angel, Chance et al., 2009 ▸). The first molecular details of the photoactivation process in the solution state came from XF-MS studies of dark, activated (meta II) and opsin states of detergent-solubilized samples (Angel, Gupta et al., 2009 ▸). Results indicated that local conformational changes arising from the isomerization of the covalently bound retinal appeared to be propagated to the cytoplasmic surface by means of water reorganization, and rearrangement of the hydrogen-bonding network between bound water and amino-acid side chains in the TM domain (Fig. 6 ▸). This study allowed XF-MS to be used to structurally validate a homology model for the 5-HT4R receptor, and predict sites for internal water–side-chain interactions necessary for its activation process (Padayatti et al., 2013 ▸).
Figure 6.
XF-MS probes bound water mediated signal transfer pathway in bovine rhodopsin. (a) DR-plots of dark (black) versus meta II (red) for the peptide p333–348 (at the solvent exposed cytoplasmic side) and p160–164 (at the TM region) for modified residues as indicated. (b) Pictorial summary of relative solvent accessibility changes for the photoactivation of dark to meta II state. Residues with rate constants >0.1 s−1 are shown as sticks. The color coding represents the ratio of rate constants between meta II and rhodopsin. Conserved transmembrane waters are shown as cyan spheres. The changes in rates of modification reflect local structural changes inside the TM domain upon formation of meta II. The results demonstrate disruption and reorganization of multiple close-packing interactions, mediated by both side chains and bound waters. The information is transmitted from the chromophore (ligand-binding site) to the cytoplasmic surface for G-protein activation. Results from Angel, Gupta et al. (2009 ▸).
XF-MS combined with hydrogen deuterium exchange mass spectrometry (HDX-MS) studies on rhodopsin, activated rhodopsin and rhodopsin-G-protein complex have elucidated the finer details of the dynamics of the internal water rearrangements that accompany G-protein binding, as well as revealing the location of the protein–protein interactions in both rhodopsin and G-protein (Orban et al., 2012 ▸).
4. Studying gating in a K+ channel by XF-MS
Ion channels are excellent targets for XF-MS-based solution structural studies since activation of these channels is in general associated with ‘gating’ events, which usually result in dramatic changes in water accessibility of the conserved transmembrane pore regions (Aryal et al., 2015 ▸). These proteins also respond to a variety of regulatory stimuli by allosteric conformational regulation in distal domains (Swartz, 2004 ▸). Crystallography has proved to be challenging as the closed or inactive form of ion channels appears to be energetically favorable, resulting in channels preferentially crystallizing in the closed state (Tao et al., 2009 ▸; Uysal et al., 2009 ▸), while their size and complexity precludes NMR analysis, which has left the question of structural transitions during gating unanswered for a number of years.
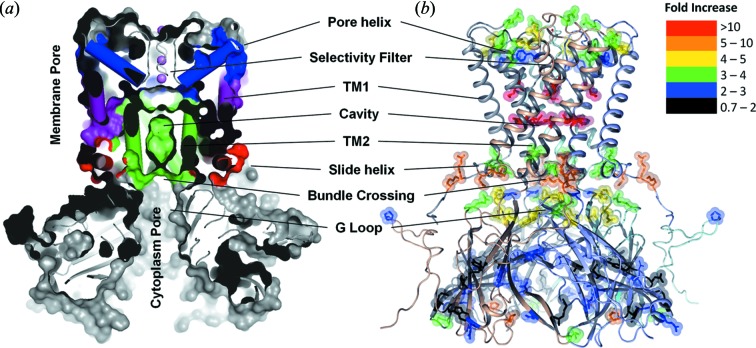
The first XF-MS studies on ion channels were carried out on a detergent reconstituted full length bacterial inwardly rectifying ion channel, KirBac3.1 (Gupta et al., 2010 ▸). Comparative XF-MS labeling between open and closed states revealed that, upon opening of the KirBac3.1 channel, there was a significant increase in the solvent accessibility near the selectivity filter, along the central cavity, and at the interface between the TM and the cytoplasmic regulatory domain of the channel (Fig. 7 ▸). This study not only supports the existence of a potential gate inside the channel but also identifies functionally important amino acid residues. In particular, the highest accessibility changes (<30-fold) were observed at the hydrophobic residue L124, which is located inside the central cavity. A few years later, these observations were confirmed in full by the crystal structure of the open state of the channel (Bavro et al., 2012 ▸). Notably, although the existence of such hydrophobic gating for K-channels had been suggested much earlier (Zimmerberg & Parsegian, 1986 ▸), the XF-MS directly visualized the role of L124 during the transition. This finding provides a basis for understanding the dynamics at the side-chain level for a wide variety of bacterial and eukaryotic channels in the presence of ligands and effectors that affect the gating process.
Figure 7.
XF-MS identifies conformation changes during gating of a K+ ion channel, KirBac3.1. (a) Cross-sectional view showing multiple surface-exposed and buried cavities in close KirBac3.1 (PDB-1XL4). TM1 (purple) and TM2 (green) denote transmembrane helix-1 (outer) and transmembrane helix-2 (inner), respectively. Pore helix and side helix are colored blue and red, respectively. (b) Solvent accessibility changes from the closed to the open conformation in KirBac3.1 are visualized on the structure of closed KirBac3.1, where the subunits are represented by different colors. The modified residues are shown by sticks. Color coding indicates the changes in the modification rates or solvent accessibilities upon transition from the closed to the open state. Residues that undergo increased interactions with water due to changes in the structure of the channel in the open state show dramatic increase in labeling efficiency. The results support the proposed existence of three potential gates within the channel. Results from Gupta et al. (2010 ▸).
More recently, XF-MS studies on the pH-dependent bacterial K+ channel, KcsA (Gupta, unpublished data), showed modification at side chains that are in proximity to structural waters inside the TM domains. Comparison of the XF-MS data of low (closed/inactivated) and high pH (open/activate) states of the wild-type and E71A mutant revealed a dramatic difference in the solvent accessibility of the selectivity filter. These findings are consistent with the recent suggestion that structured/bound water plays an important role in stabilizing the selectivity filter conformation, and underscores the potential of XF-MS for the study of these systems (Ostmeyer et al., 2013 ▸).
5. Identifying conformational hot-spots in a Zn2+ transporter
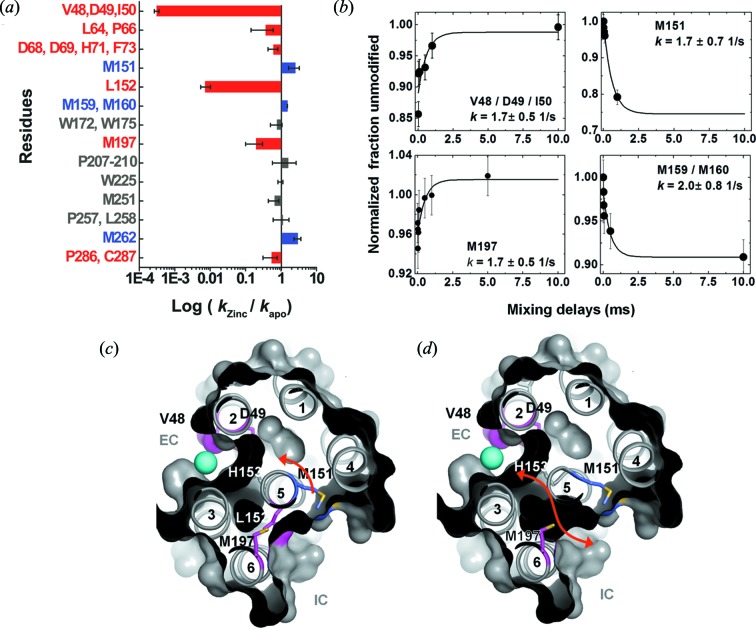
Prospects for further developing XF-MS approaches for following water accessibility changes in TM domains of membrane proteins came from the study of the prototypical proton-coupled Zn2+ transporter YiiP found in the cytoplasmic membrane of E. coli (Fig. 8 ▸) (Gupta, Chai et al., 2014 ▸). This transporter has the unique characteristic of transporting Zn2+ against the concentration gradient with the exchange of protons (Chao & Fu, 2004 ▸). The crystal structure of Zn-YiiP is available and showed the absence of any polar residues that could carry protons to the Zn binding or transport site, and showed the presence of a hydrophobic barrier that divides the transport pathway between the intra- and extra-cellular cavity (Lu & Fu, 2007 ▸; Lu et al., 2009 ▸). A comparative XF-MS analysis of Zn2+-YiiP and Apo-YiiP identified specific and reciprocal solvent accessibility changes in the residues adjacent to these cavities and within the hydrophobic barrier. At the extracellular side, Zn2+ binding residue D49 (TM2, at the active site) and L152 (TM5, at the hydrophobic gate) showed a significant decrease in water accessibility upon zinc binding which is consistent with the available crystal structure of Zn-YiiP (Figs. 9c and 9d ▸). Residue M197, which is at the entrance of the intracellular surface near L152, also showed a decrease in water accessibility. L152 and M197 interact with residues from TM3 and TM6, and this cluster of residues forms a highly conserved TM5 → TM3-TM6 packing core that stabilizes the TM helical arrangment and hydrophobic barrier. Solvent accessiblity data suggest that Zn2+ access to the transport site shuts off water access to L152, suggesting that L152 on TM5 may function as an inter-cavity gate that controls alternating access of zinc ions and water molecules to the transport site. In contrast, increased water accessibility was observed in several methionine residues located on TM5 but facing opposite to D49, L152 and M197. This reciprocal change in water accessibility on two opposite TM5 faces is consistent with a TM5 re-orientation in response to zinc binding. This result suggested that the transport site in apo-YiiP was constitutively open to the intracellular cavity, in agreement with a low-resolution cryo-EM structure of an apo-YiiP homolog. Millisecond time-resolved XF-MS was also used to monitor the time course of closing of the inter-cavity water pathway in response to rapid zinc binding to the detergent solubilized YiiP. The analysis showed the reciprocal pattern of steady-state responses for residues on opposite faces of TM5; the exponential rise in the accessibility of M197 and D49 mirrored the exponential decrease of the M151 and M159/M160 accessibilities at an average rate of 1.8 s−1 (Fig. 9b ▸). The rates of water accessibility changes for these four positions along TM5 were identical within experimental error, suggesting that TM5 underwent a rigid-body re-orientation upon zinc binding. XF-MS data suggested that the zinc binding energy in the hydrophobic core at the intra-cellular interface might be transformed into mechanical energy to reorient TM5 and close the L152 gate. This conformational change can alternatively expose the transport site to the intracellular and extracellular cavity. The difference in the pH at extra- and intra-cellular cavities can modulate the protonation state of His153, a critical residue for Zn2+ binding in the transport site, which in turn can regulate Zn2+ binding.
Figure 8.
XF-MS probes proton-coupled Zn2+ transfer mechanism in Zn transporter YiiP. (a) Bar plot showing radiometric water accessibility changes in response to Zn2+ binding measured by ratio (R) of  OH labeling rates for residues with increase (blue), decrease (red) and no change (grey) in accessibility after rapid Zn2+ exposure. (b) Time courses of water accessibility changes for the indicated residues. Solid lines represent single exponential fits, which provide rate constants of conformational changes associated with Zn2+ binding and translocation. (c) X-ray footprinting reveals a hydrophobic gate at residue L152, which controls the opening of the inner cavity water pathway for zinc-proton exchange in the YiiP transporter. The cross-sectional view shows the position of TM helices, which separate two cavities at the intra-cellular (IC) and extra-cellular (EC) sides. Residues are color coded as in (a). XF-MS results suggest the protein conformational change alternates the membrane-facing on–off mode of zinc coordination (in D49) and protonation–deprotonation (H153) of the transport site in a coordinated fashion. (d) The red arrow indicates the proposed water pathway, which connects EC with IC after excluding the residue L152 from the surface drawing of the TM helices. Results from Gupta et al. (2014 ▸).
OH labeling rates for residues with increase (blue), decrease (red) and no change (grey) in accessibility after rapid Zn2+ exposure. (b) Time courses of water accessibility changes for the indicated residues. Solid lines represent single exponential fits, which provide rate constants of conformational changes associated with Zn2+ binding and translocation. (c) X-ray footprinting reveals a hydrophobic gate at residue L152, which controls the opening of the inner cavity water pathway for zinc-proton exchange in the YiiP transporter. The cross-sectional view shows the position of TM helices, which separate two cavities at the intra-cellular (IC) and extra-cellular (EC) sides. Residues are color coded as in (a). XF-MS results suggest the protein conformational change alternates the membrane-facing on–off mode of zinc coordination (in D49) and protonation–deprotonation (H153) of the transport site in a coordinated fashion. (d) The red arrow indicates the proposed water pathway, which connects EC with IC after excluding the residue L152 from the surface drawing of the TM helices. Results from Gupta et al. (2014 ▸).
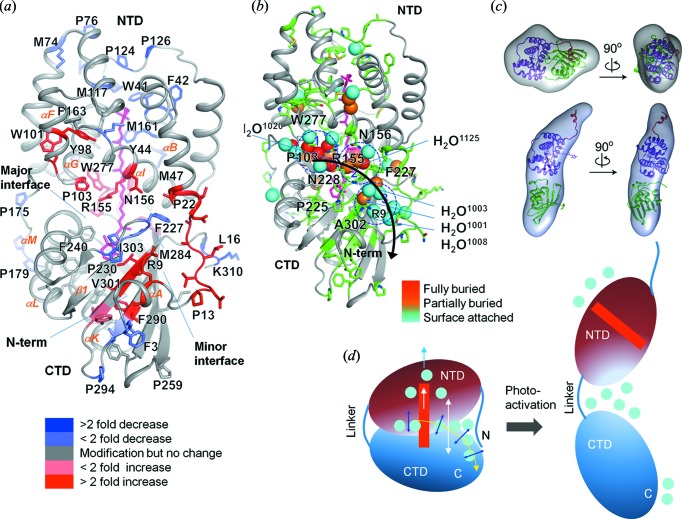
Figure 9.
XF-MS probes bound water mediated signal transfer pathway in OCP. (a) Solvent accessibility changes from dark adapted-OCPO to illuminated-OCPR are visualized on the structure of OCPO (3MG1). The modified residues are represented by sticks and the carotenoid is shown in pink. The color coding represents the ratio of rate constants between these two states. (c) The proposed signal propagation pathway from the carotenoid through the water side-chain hydrogen-bonding network to the protein surface that facilitates carotenoid shift, dissociation of NTD-CTD and detachment of the N-terminal helix from CTD. Conserved waters are shown in spheres and color codes indicate their depth from the surface of OCP. The modified residues are shown by green sticks. The results demonstrate disruption and reorganization of multiple close-packing interactions, mediated by both side chains and bound waters. (d) Ab initio bead reconstructions (gray volume) based on the SAXS results are shown for OCPO and OCPR. The subunit of OCPO from the crystal structure is docked into the volume envelope with the far N-terminal helix (red), NTD (purple) and CTD (green). The SAXS results show dissociation of NTD and CTD. (d) Schematic of the photoactivation of OCPO showing regions with the largest conformational rearrangement associated with changes in the hydrogen-bonding network and water rearrangements. Reproduced from Gupta et al. (2016 ▸).
6. Structural mechanism of the photoactivation of orange carotenoid protein
Photoprotective mechanisms are of fundamental importance for the survival of photosynthetic organisms. In cyanobacteria, the orange carotenoid protein (OCP), when activated by intense blue light, forms OCPR, which binds to the light harvesting antenna and triggers the dissipation of excess captured light energy. X-ray crystal structures of inactive OCP (OCPO) and the N-terminal part of OCPR (RCP) are available. A comparison of these two structures shows that photoactivation results in a 12 Å movement of the carotenoid inside the N-terminal half of OCP. Comparative XF-MS studies on OCPO and RCP showed that, in RCP, specific residues adjacent to the carotenoid binding region in the N-terminal domain (NTD) are protected. This result directly supported carotenoid translocation in solution similar to that in the crystal structure (Leverenz et al., 2015 ▸). However, RCP is only a substructure of the full-length OCPR. A hybrid approach comprising XF-MS, small-angle X-ray scattering (SAXS) and HDX-MS dispelled the uncertainty about the structural changes in the wild-type OCP and provided mechanistic details of the conformation changes for the photoactivation of full-length OCPO to OCPR (Gupta et al., 2015 ▸). While XF-MS revealed detailed residue-specific solvent accessibility changes that supported both domain dissociation and carotenoid translocation in the full-length protein, synergistic application of SAXS and HDX-MS methods complemented the XF-MS results by showing that photoactivation underwent a dissociation of the NTD and C-terminal domain (CTD) without substantially altering the secondary structure. XF-MS identified several functionally important residues involved in a hydrogen-bonding network with bound as well as conserved water molecules at the major and minor interfaces of the NTD and CTD (Fig. 9 ▸). The study provided a model for the photoactivation process in which the carotenoid translocation induces changes in the water–protein network connecting the carotenoid site with the protein surface. The hybrid approach was necessary to delineate the detailed molecular mechanism underlying photoprotection initiation in cyanobacteria. Further studies on OCP-phycobilisome and OCP-FRP interactions are in progress, which will contribute to a detailed understanding of the photoprotection cycle in photosynthetic systems.
7. Future directions
For the past decade, XF-MS has been integrated effectively with many other structural and biochemical techniques to provide a comprehensive picture of macromolecular complexes and their functional states. Examples highlighted in this review included hybrid methods combining information on local changes in solvent accessibility deduced from the profiles of hydroxyl radical reactivity with static atomic structures obtained from X-ray crystallography, global structures obtained from SAXS and cryo-EM, and backbone accessibility measurements obtained from HDX-MS. Crystallographic structure information, in particular, is often used as a reference point to enrich footprinting interpretations by providing details at atomic resolution for individual components or complexes in native or non-functional states (Kamal et al., 2007 ▸; Takamoto et al., 2007 ▸; Gupta et al., 2004 ▸). In this case, XF-MS analysis is used to interpret protein conformational changes associated with the dynamics of proximally bound water, which are determined from the available crystal structures of the native state. XF-MS is now established as a direct method for pinpointing both positional and dynamical aspects of water–protein interactions. However, there are only a limited number of studies which correlate the location of water molecules in protein crystal structures with their role in the protein function. In one such study, crystal structures and database analysis were used to identify protein functional surfaces and cavities and assess conformational changes for bound and unbound states (Tseng et al., 2010 ▸). Another study investigated the existence of conserved cavities and the intriguing role of bound water in these cavities in gating of ion channels (Ostmeyer et al., 2013 ▸). These studies strongly put forward the critical question of how the functional cooperativity between bound water in conserved cavities and ligand binding in distal domains can control the overall function of large protein complexes. We anticipate that theoretical predictions of conserved cavities by bioinformatics can be combined with the experimentally determined functional sites obtained by XF-MS (which includes information about the sites of bound water and proximal cavities). This approach could provide a strategy to classify and model proteins according to their conserved surface or cavity topography and correlate the results with their functional divergence. In addition, more recent studies have shown the importance of bound water networks in ligand binding and the complicated role of water in the overall thermodynamics of binding (Breiten et al., 2013 ▸; Beuming et al., 2012 ▸). The XF-MS method offers a new tool for optimizing ligand design by mapping the locations and thermodynamic properties of water molecules at protein binding sites and, at the same time, XF-MS data can be used to develop, validate and refine molecular dynamics approaches that determine water–protein side-chain interactions.
Another exciting future application is the use of X-ray footprinting for protein crystallography, which could yield valuable information in several important areas. First, footprinting data on proteins in crystal form could be used to delineate protein–protein contacts in very small and weakly diffracting crystals. By comparing solvent accessibility of proteins in crystals to solvent accessibility in solution, protections due to crystal contacts could be identified and used to engineer more ordered, and therefore better diffracting, crystals. Second, XF-MS applied to proteins in solution as a function of denaturant concentration would yield information on regions of disorder by allowing calculation of free energies of unfolding on a residue-by-residue basis, which could be used to engineer optimized protein crystals. Third, XF-MS could be used to investigate radiation damage in protein crystals, yielding information on the different structural changes at various residues as a result of radiation exposure. XF-MS is ideally suited to this endeavor; XF-MS is able to detect both oxidation addition reactions (i.e. addition of O atoms and/or OH groups) and subtraction reactions, such as decarboxylation of aspartate and glutamate. Indeed, the response of each amino acid to hydroxyl radicals is unique, can be detected post-irradiation with LC-MS/MS, and can be investigated as a function of dose. For example, in a series of preliminary experiments, crystals were irradiated at the ALS footprinting beamline 3.2.1. The crystals were then divided into two batches: one batch was digested and analyzed for residue specific changes as in a standard XF-MS experiment, and the other batch was run on ALS macromolecular crystallography beamline 5.0.1. These experiments were repeated with different exposure times, correlating the global damage as measured by loss of diffraction, with specific damage as measured by changes to residues in the crystals (Ralston, unpublished data).
Currently, dedicated XF-MS beamlines are under development at both the ALS and NSLS II. These beamlines are designed for fully automated sample handling, microsecond irradiation timescales and time-resolved experiments, and will open the door to even further advances in the technique of XF-MS.
Acknowledgments
We thank Rich Celestre for help with the studies performed at the Advanced Light Source. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231. The National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. The Center for Synchrotron Biosciences at the National Synchrotron Light Sources is supported by NIBIB under P30-EB0966. Studies on potassium channel work are supported by the Welcome Trust. Studies on YiiP are supported by NIH and DOE. Study on OCP is supported by NIH and DOE. This research used resources of the Joint BioEnergy Institute supported by the Office of Science, Office of Biological and Environmental Research, US DOE under contract DE-AC02-05CH11231.
References
- Angel, T. E., Chance, M. R. & Palczewski, K. (2009). Proc. Natl Acad. Sci. USA, 106, 8555–8560. [DOI] [PMC free article] [PubMed]
- Angel, T. E., Gupta, S., Jastrzebska, B., Palczewski, K. & Chance, M. R. (2009). Proc. Natl Acad. Sci. USA, 106, 14367–14372. [DOI] [PMC free article] [PubMed]
- Aryal, P., Sansom, M. S. & Tucker, S. J. (2015). J. Mol. Biol. 427, 121–130. [DOI] [PMC free article] [PubMed]
- Aye, T. T., Low, T. Y. & Sze, S. K. (2005). Anal. Chem. 77, 5814–5822. [DOI] [PubMed]
- Ball, P. (2008). Chem. Rev. 108, 74–108. [DOI] [PubMed]
- Bavro, V. N., De Zorzi, R., Schmidt, M. R., Muniz, J. R., Zubcevic, L., Sansom, M. S., Vénien-Bryan, C. & Tucker, S. J. (2012). Nat. Struct. Mol. Biol. 19, 158–163. [DOI] [PMC free article] [PubMed]
- Bertini, I., Hajieva, P., Luchinat, C. & Nerinovski, K. (2001). J. Am. Chem. Soc. 123, 12925–12926. [DOI] [PubMed]
- Bertini, I., Huber, J. G., Luchinat, C. & Piccioli, M. (2000). J. Magn. Reson. 147, 1–8. [DOI] [PubMed]
- Beuming, T., Che, Y., Abel, R., Kim, B., Shanmugasundaram, V. & Sherman, W. (2012). Proteins, 80, 871–883. [DOI] [PubMed]
- Bhat, T. N., Bentley, G. A., Boulot, G., Greene, M. I., Tello, D., Dall’Acqua, W., Souchon, H., Schwarz, F. P., Mariuzza, R. A. & Poljak, R. J. (1994). Proc. Natl Acad. Sci. USA, 91, 1089–1093. [DOI] [PMC free article] [PubMed]
- Bohon, J., D’Mello, R., Ralston, C., Gupta, S. & Chance, M. R. (2014). J. Synchrotron Rad. 21, 24–31. [DOI] [PMC free article] [PubMed]
- Bohon, J., Jennings, L. D., Phillips, C. M., Licht, S. & Chance, M. R. (2008). Structure, 16, 1157–1165. [DOI] [PMC free article] [PubMed]
- Braden, B. C., Fields, B. A. & Poljak, R. J. (1995). J. Mol. Recognit. 8, 317–325. [DOI] [PubMed]
- Breiten, B., Lockett, M. R., Sherman, W., Fujita, S., Al-Sayah, M., Lange, H., Bowers, C. M., Heroux, A., Krilov, G. & Whitesides, G. M. (2013). J. Am. Chem. Soc. 135, 15579–15584. [DOI] [PubMed]
- Brunne, R. M., Liepinsh, E., Otting, G., Wüthrich, K. & van Gunsteren, W. F. (1993). J. Mol. Biol. 231, 1040–1048. [DOI] [PubMed]
- Bushnell, G. W., Louie, G. V. & Brayer, G. D. (1990). J. Mol. Biol. 214, 585–595. [DOI] [PubMed]
- Carugo, O. & Bordo, D. (1999). Acta Cryst. D55, 479–483. [DOI] [PubMed]
- Chao, Y. & Fu, D. (2004). J. Biol. Chem. 279, 12043–12050. [DOI] [PubMed]
- Chaplin, M. (2006). Nat. Rev. 7, 861–866. [DOI] [PubMed]
- Chaudhuri, B. N., Gupta, S., Urban, V. S., Chance, M. R., D’Mello, R., Smith, L., Lyons, K. & Gee, J. (2011). Biochemistry, 50, 1799–1807. [DOI] [PMC free article] [PubMed]
- Conti Nibali, V. & Havenith, M. (2014). J. Am. Chem. Soc. 136, 12800–12807. [DOI] [PubMed]
- Cui, P. X., Wang, Y., Chu, W. S., Guo, X. Y., Yang, F. F., Yu, M. J., Zhao, H. F., Dong, Y. H., Xie, Y. N., Gong, W. M. & Wu, Z. Y. (2014). Sci. Rep. 4, 7453. [DOI] [PMC free article] [PubMed]
- Denisov, V. P. & Halle, B. (1996). Faraday Discuss. 103, 227–244. [DOI] [PubMed]
- Denisov, V. P., Peters, J., Hörlein, H. D. & Halle, B. (1996). Nat. Struct. Biol. 3, 505–509. [DOI] [PubMed]
- Downard, K. M., Maleknia, S. D. & Akashi, S. (2012). Rapid Commun. Mass Spectrom. 26, 226–230. [DOI] [PubMed]
- Frankel, L. K., Sallans, L., Bellamy, H., Goettert, J. S., Limbach, P. A. & Bricker, T. M. (2013). J. Biol. Chem. 288, 23565–23572. [DOI] [PMC free article] [PubMed]
- Frölich, A., Gabel, F., Jasnin, M., Lehnert, U., Oesterhelt, D., Stadler, A. M., Tehei, M., Weik, M., Wood, K. & Zaccai, G. (2009). Faraday Discuss. 141, 117–130. [DOI] [PubMed]
- Furutani, Y., Shichida, Y. & Kandori, H. (2003). Biochemistry, 42, 9619–9625. [DOI] [PubMed]
- Garczarek, F. & Gerwert, K. (2006). Nature (London), 439, 109–112. [DOI] [PubMed]
- González Fernández-Niño, S. M., Smith-Moritz, A. M., Chan, L. J., Adams, P. D., Heazlewood, J. L. & Petzold, C. J. (2015). Front. Bioeng. Biotechnol. 3, 44. [DOI] [PMC free article] [PubMed]
- Gowen, A. A., Amigo, J. M. & Tsenkova, R. (2013). Anal. Chim. Acta, 759, 8–20. [DOI] [PubMed]
- Gupta, S., Bavro, V. N., D’Mello, R., Tucker, S. J., Vénien-Bryan, C. & Chance, M. R. (2010). Structure, 18, 839–846. [DOI] [PMC free article] [PubMed]
- Gupta, S., Celestre, R., Feng, J. & Ralston, C. (2016). Synchrotron Radiat. News, 29, 39–44.
- Gupta, S., Celestre, R., Petzold, C. J., Chance, M. R. & Ralston, C. (2014). J. Synchrotron Rad. 21, 690–699. [DOI] [PMC free article] [PubMed]
- Gupta, S., Chai, J., Cheng, J., D’Mello, R., Chance, M. R. & Fu, D. (2014). Nature (London), 512, 101–104. [DOI] [PMC free article] [PubMed]
- Gupta, S., D’Mello, R. & Chance, M. R. (2012). Proc. Natl Acad. Sci. USA, 109, 14882–14887. [DOI] [PMC free article] [PubMed]
- Gupta, S., Guttman, M., Leverenz, R. L., Zhumadilova, K., Pawlowski, E. G., Petzold, C. J., Lee, K. K., Ralston, C. Y. & Kerfeld, C. A. (2015). Proc. Natl Acad. Sci. USA, 112, E5567–E5574. [DOI] [PMC free article] [PubMed]
- Gupta, S., Mangel, W. F., McGrath, W. J., Perek, J. L., Lee, D. W., Takamoto, K. & Chance, M. R. (2004). Mol. Cell. Proteom. 3, 950–959. [DOI] [PubMed]
- Gupta, S., Sullivan, M., Toomey, J., Kiselar, J. & Chance, M. R. (2007). J. Synchrotron Rad. 14, 233–243. [DOI] [PubMed]
- Halle, B. (2004). Philos. Trans. R. Soc. B, 359, 1207–1223. [DOI] [PMC free article] [PubMed]
- Hambly, D. M. & Gross, M. L. (2005). J. Am. Soc. Mass Spectrom. 16, 2057–2063. [DOI] [PubMed]
- Hare, S., Vos, A. M., Clayton, R. F., Thuring, J. W., Cummings, M. D. & Cherepanov, P. (2010). Proc. Natl Acad. Sci. USA, 107, 20057–20062. [DOI] [PMC free article] [PubMed]
- Huang, W., Ravikumar, K. M., Chance, M. R. & Yang, S. (2015). Biophys. J. 108, 107–115. [DOI] [PMC free article] [PubMed]
- Jiang, J. S. & Brünger, A. T. (1994). J. Mol. Biol. 243, 100–115. [DOI] [PubMed]
- Kamal, J. K., Benchaar, S. A., Takamoto, K., Reisler, E. & Chance, M. R. (2007). Proc. Natl Acad. Sci. USA, 104, 7910–7915. [DOI] [PMC free article] [PubMed]
- Kamal, J. K. & Chance, M. R. (2008). Protein Sci. 17, 79–94. [DOI] [PMC free article] [PubMed]
- Kandori, H. (2000). Biochim. Biophys. Acta, 1460, 177–191. [DOI] [PubMed]
- Kaur, P., Kiselar, J. G. & Chance, M. R. (2009). Anal. Chem. 81, 8141–8149. [DOI] [PMC free article] [PubMed]
- Kaur, P., Kiselar, J., Yang, S. & Chance, M. R. (2015). Mol. Cell. Proteom. 14, 1159–1168. [DOI] [PMC free article] [PubMed]
- Kerfeld, C. A., Sawaya, M. R., Brahmandam, V., Cascio, D., Ho, K. K., Trevithick-Sutton, C. C., Krogmann, D. W. & Yeates, T. O. (2003). Structure, 11, 55–65. [DOI] [PubMed]
- Kiselar, J. G. & Chance, M. R. (2010). J. Mass Spectrom. 45, 1373–1382. [DOI] [PMC free article] [PubMed]
- Kiselar, J. G., Janmey, P. A., Almo, S. C. & Chance, M. R. (2003). Proc. Natl Acad. Sci. USA, 100, 3942–3947. [DOI] [PMC free article] [PubMed]
- Klinger, A. L., Kiselar, J., Ilchenko, S., Komatsu, H., Chance, M. R. & Axelsen, P. H. (2014). Biochemistry, 53, 7724–7734. [DOI] [PMC free article] [PubMed]
- Konermann, L., Ahadi, E., Rodriguez, A. D. & Vahidi, S. (2013). Anal. Chem. 85, 2–9. [DOI] [PubMed]
- Kuhn, L. A., Siani, M. A., Pique, M. E., Fisher, C. L., Getzoff, E. D. & Tainer, J. A. (1992). J. Mol. Biol. 228, 13–22. [DOI] [PubMed]
- Kuntz, I. D. Jr, Brassfield, T. S., Law, G. D. & Purcell, G. V. (1969). Science, 163, 1329–1331. [DOI] [PubMed]
- Leverenz, R. L., Sutter, M., Wilson, A., Gupta, S., Thurotte, A., Bourcier de Carbon, C., Petzold, C. J., Ralston, C., Perreau, F., Kirilovsky, D. & Kerfeld, C. A. (2015). Science, 348, 1463–1466. [DOI] [PubMed]
- Levinson, N. M. & Boxer, S. G. (2014). Nat. Chem. Biol. 10, 127–132. [DOI] [PMC free article] [PubMed]
- Liljenzin, J. (2002). In Radiochemistry and Nuclear Chemistry. Oxford: Butterworth-Heinemann.
- Lin, J., Balabin, I. A. & Beratan, D. N. (2005). Science, 310, 1311–1313. [DOI] [PMC free article] [PubMed]
- Ling, J., Cho, C., Guo, L. T., Aerni, H. R., Rinehart, J. & Söll, D. (2012). Mol. Cell, 48, 713–722. [DOI] [PMC free article] [PubMed]
- Lu, M., Chai, J. & Fu, D. (2009). Nat. Struct. Mol. Biol. 16, 1063–1067. [DOI] [PMC free article] [PubMed]
- Lu, M. & Fu, D. (2007). Science, 317, 1746–1748. [DOI] [PubMed]
- Maleknia, S. D. & Downard, K. M. (2012). Rapid Commun. Mass Spectrom. 26, 2311–2318. [DOI] [PubMed]
- Maréchal, A. & Rich, P. R. (2011). Proc. Natl Acad. Sci. USA, 108, 8634–8638. [DOI] [PMC free article] [PubMed]
- Muroda, K., Nakashima, K., Shibata, M., Demura, M. & Kandori, H. (2012). Biochemistry, 51, 4677–4684. [DOI] [PubMed]
- Nickels, J. D., O’Neill, H., Hong, L., Tyagi, M., Ehlers, G., Weiss, K. L., Zhang, Q., Yi, Z., Mamontov, E., Smith, J. C. & Sokolov, A. P. (2012). Biophys. J. 103, 1566–1575. [DOI] [PMC free article] [PubMed]
- Nucci, N. V., Pometun, M. S. & Wand, A. J. (2011). Nat. Struct. Mol. Biol. 18, 245–249. [DOI] [PMC free article] [PubMed]
- Orban, T., Jastrzebska, B., Gupta, S., Wang, B., Miyagi, M., Chance, M. R. & Palczewski, K. (2012). Structure, 20, 826–840. [DOI] [PMC free article] [PubMed]
- Ostmeyer, J., Chakrapani, S., Pan, A. C., Perozo, E. & Roux, B. (2013). Nature (London), 501, 121–124. [DOI] [PMC free article] [PubMed]
- Otting, G., Liepinsh, E. & Wüthrich, K. (1991). Science, 254, 974–980. [DOI] [PubMed]
- Padayatti, P. S., Wang, L., Gupta, S., Orban, T., Sun, W., Salom, D., Jordan, S. R., Palczewski, K. & Chance, M. R. (2013). Mol. Cell. Proteom. 12, 1259–1271. [DOI] [PMC free article] [PubMed]
- Papoian, G. A., Ulander, J. & Wolynes, P. G. (2003). J. Am. Chem. Soc. 125, 9170–9178. [DOI] [PubMed]
- Pearson, R. G. & Williams, E. L. (1987). J. Polym. Sci. A, 25, 565–573.
- Persson, E. & Halle, B. (2008). J. Am. Chem. Soc. 130, 1774–1787. [DOI] [PubMed]
- Pryor, W. A. (1986). Annu. Rev. Physiol. 48, 657–667. [DOI] [PubMed]
- Ralston, C. Y., Sclavi, B., Sullivan, M., Deras, M. L., Woodson, S. A., Chance, M. R. & Brenowitz, M. (2000). Methods Enzymol. 317, 353–368. [DOI] [PubMed]
- Renthal, R. (2008). Protein Sci. 17, 293–298. [DOI] [PMC free article] [PubMed]
- Rich, P. R. & Maréchal, A. (2013). J. R. Soc. Interface, 10, 20130183. [DOI] [PMC free article] [PubMed]
- Rispens, T., Davies, A. M., Ooijevaar-de Heer, P., Absalah, S., Bende, O., Sutton, B. J., Vidarsson, G. & Aalberse, R. C. (2014). J. Biol. Chem. 289, 6098–6109. [DOI] [PMC free article] [PubMed]
- Royer, W. E. Jr, Pardanani, A., Gibson, Q. H., Peterson, E. S. & Friedman, J. M. (1996). Proc. Natl Acad. Sci. USA, 93, 14526–14531. [DOI] [PMC free article] [PubMed]
- Salanga, C. L., Dyer, D. P., Kiselar, J. G., Gupta, S., Chance, M. R. & Handel, T. M. (2014). J. Biol. Chem. 289, 14896–14912. [DOI] [PMC free article] [PubMed]
- Schilling, B., Rardin, M. J., MacLean, B. X., Zawadzka, A. M., Frewen, B. E., Cusack, M. P., Sorensen, D. J., Bereman, M. S., Jing, E., Wu, C. C., Verdin, E., Kahn, C. R., Maccoss, M. J. & Gibson, B. W. (2012). Mol. Cell. Proteom. 11, 202–214. [DOI] [PMC free article] [PubMed]
- Shacter, E. (2000). Drug Metab. Rev. 32, 307–326. [DOI] [PubMed]
- Sullivan, M. R., Rekhi, S., Bohon, J., Gupta, S., Abel, D., Toomey, J. & Chance, M. R. (2008). Rev. Sci. Instrum. 79, 025101. [DOI] [PubMed]
- Swartz, K. J. (2004). Nat. Rev. Neurosci. 5, 905–916. [DOI] [PubMed]
- Takamoto, K. & Chance, M. R. (2006). Annu. Rev. Biophys. Biomol. Struct. 35, 251–276. [DOI] [PubMed]
- Takamoto, K., Kamal, J. K. & Chance, M. R. (2007). Structure, 15, 39–51. [DOI] [PubMed]
- Tao, X., Avalos, J. L., Chen, J. & MacKinnon, R. (2009). Science, 326, 1668–1674. [DOI] [PMC free article] [PubMed]
- Tarek, M. & Tobias, D. J. (2000). Biophys. J. 79, 3244–3257. [DOI] [PMC free article] [PubMed]
- Tompa, K., Bánki, P., Bokor, M., Kamasa, P., Lasanda, G. & Tompa, P. (2009). Biophys. J. 96, 2789–2798. [DOI] [PMC free article] [PubMed]
- Tran, Q. T., Williams, S., Farid, R., Erdemli, G. & Pearlstein, R. (2013). Proteins, 81, 291–299. [DOI] [PubMed]
- Tseng, Y. Y., Chen, Z. J. & Li, W. H. (2010). Nucleic Acids Res. 38, D288–D295. [DOI] [PMC free article] [PubMed]
- Tsenkova, R. (2009). J. Near Infrared Spectrosc. 17, 303–313.
- Uysal, S., Vásquez, V., Tereshko, V., Esaki, K., Fellouse, F. A., Sidhu, S. S., Koide, S., Perozo, E. & Kossiakoff, A. (2009). Proc. Natl Acad. Sci. USA, 106, 6644–6649. [DOI] [PMC free article] [PubMed]
- Watson, C., Janik, I., Zhuang, T., Charvátová, O., Woods, R. J. & Sharp, J. S. (2009). Anal. Chem. 81, 2496–2505. [DOI] [PMC free article] [PubMed]
- Williams, M. A., Goodfellow, J. M. & Thornton, J. M. (1994). Protein Sci. 3, 1224–1235. [DOI] [PMC free article] [PubMed]
- Wong, J. W., Maleknia, S. D. & Downard, K. M. (2005). J. Am. Soc. Mass Spectrom. 16, 225–233. [DOI] [PubMed]
- Xu, G. & Chance, M. R. (2007). Chem. Rev. 107, 3514–3543. [DOI] [PubMed]
- Yoon, J., Oh, B., Kim, K., Park, J., Han, D., Kim, K. K., Cha, S. S., Lee, D. & Kim, Y. (2004). J. Biol. Chem. 279, 341–347. [DOI] [PubMed]
- Yuan, S., Filipek, S., Palczewski, K. & Vogel, H. (2014). Nat. Commun. 5, 4733. [DOI] [PubMed]
- Zimmerberg, J. & Parsegian, V. A. (1986). Nature (London), 323, 36–39. [DOI] [PubMed]