Structure-function analysis reveals that chitin binding by Pf-Avr4, a fungal Avr4 effector, is largely separable from recognition by the cognate Cf-4 receptor.
Abstract
Chitin is a key component of fungal cell walls and a potent inducer of innate immune responses. Consequently, fungi may secrete chitin-binding lectins, such as the Cf-Avr4 effector protein from the tomato pathogen Cladosporium fulvum, to shield chitin from host-derived chitinases during infection. Homologs of Cf-Avr4 are found throughout Dothideomycetes, and despite their modest primary sequence identity, many are perceived by the cognate tomato immune receptor Cf-4. Here, we determined the x-ray crystal structure of Pf-Avr4 from the tomato pathogen Pseudocercospora fuligena, thus providing a three-dimensional model of an Avr4 effector protein. In addition, we explored structural, biochemical, and functional aspects of Pf-Avr4 and Cf-Avr4 to further define the biology of core effector proteins and outline a conceptual framework for their pleiotropic recognition by single immune receptors. We show that Cf-Avr4 and Pf-Avr4 share functional specificity in binding (GlcNAc)6 and in providing protection against plant- and microbial-derived chitinases, suggesting a broader role beyond deregulation of host immunity. Furthermore, structure-guided site-directed mutagenesis indicated that residues in Pf-Avr4 important for binding chitin do not directly influence recognition by Cf-4 and further suggested that the property of recognition is structurally separated or does not fully overlap with the virulence function of the effector.
INTRODUCTION
Fungi employ a variety of mechanisms to infect and colonize the host tissue, thus causing disease. Pathogenesis on plants is a multilayered process consisting of several steps, including recognition of the host, penetration, and invasive growth. Among these, overcoming the host immune system is arguably the most crucial and determining step in the infection process. Unlike humans and other animals, plants possess two lines of defense responses against invading pathogens. The first line is a set of basal (innate) immune responses, mediated by transmembrane pattern recognition receptors (PRRs) that recognize conserved pathogen-associated molecular patterns (PAMPs) to activate PAMP-triggered immunity (PTI) (Jones and Dangl, 2006). PTI is an important barrier to microbial infections, and in response, pathogens have evolved an array of low molecular weight effector proteins that mask PAMPs and/or suppress PTI to enable disease. To overcome effectors, plants have acquired a second line of defense responses that rely on effector recognition by specialized immune receptors. Specifically, effector-triggered immunity utilizes intracellular or transmembrane resistance (R) proteins that, unlike PRRs, perceive cognate pathogen effectors with high specificity to induce abrupt defense responses in the form of a hypersensitive response (HR) (Jones and Dangl, 2006; Stergiopoulos and de Wit, 2009). Unlike PAMPs, most fungal effectors were assumed to be species- or perhaps lineage-specific, appearing later in the evolutionary history of pathogens to facilitate infection (Jones and Dangl, 2006; Stergiopoulos and de Wit, 2009). Indeed, Ecp6, a secreted effector from the tomato pathogen Cladosporium fulvum (synonym Passalora fulva), was until recently among the very few fungal effectors with homologs in numerous other fungi, mainly due to the presence of LysM domains in this protein, a motif that is widespread among microbes of diverse taxa and lifestyles (Bolton et al., 2008; de Jonge and Thomma, 2009). At the same time, we have also shown that homologs of the Avr4 and Ecp2 effectors from C. fulvum are present in some phylogenetically related species of Dothideomycetes and beyond (Stergiopoulos et al., 2010, 2012). Why some effectors are broadly conserved while others are not is poorly understood, but we have hypothesized that core fungal effectors may have conserved virulence functions that facilitate infections on a wide range of hosts. Alternatively, given their broad distribution in fungi with diverse lifestyles, including pathogens and nonpathogens, core effectors may also serve roles beyond deregulation of host immunity during infections, as for example interactions with other microbes in a pathogen’s environment (Stergiopoulos et al., 2010, 2012).
Cf-Avr4 (for C. fulvum Avr4) is a small, secreted effector that binds chitin present in fungal cell walls, thereby protecting them against host-derived chitinases during infection (Joosten et al., 1994; van den Burg et al., 2006). Disulfide bond assignment has shown that Cf-Avr4 contains a disulfide-bond pattern similar to carbohydrate-binding module family 14 (CBM14) lectins, suggesting that Cf-Avr4 utilizes a CBM14 fold to interact with chitin (van den Burg et al., 2003). CBM14s are modules of roughly 65 to 70 residues that bind specifically to chitin, a β(1-4) linked N-acetyl-d-glucosamine (GlcNAc) polysaccharide, which is a major structural constituent of the fungal cell wall and a potent inducer of PTI in plants (Suetake et al., 2000; Boraston et al., 2004; Chang and Stergiopoulos, 2015a). To date, a molecular-based mechanistic understanding of the CBM14-ligand interaction is lacking and tertiary information on CBM14 lectins is restricted to tachycitin, a small antimicrobial chitin-binding lectin that is found in horseshoe crab hemocytes and exhibits local sequence similarity and a similar disulfide-bond pattern as predicted for Cf-Avr4 (Kawabata et al., 1996). Specifically, the putative 73-residue structure of tachycitin was resolved using NMR spectroscopy and is thought to consist of a globular domain containing two β-sheets arranged in a distorted β-sandwich fold and stabilized by five disulfide bonds (Suetake et al., 2000). However, experimental validation by x-ray crystallography of the NMR-based structure of tachycitin is still needed.
To date, putative functional orthologs of Cf-Avr4 have been identified in different fungi of the Dothideomycete class, including the banana pathogen Mycosphaerella fijiensis (Mf-Avr4) (synonym Pseudocercospora fijiensis), the pine tree pathogen Dothistroma septosporum (Ds-Avr4), the poplar pathogen Septoria musiva (Sm-Avr4) (synonym Sphaerulina musiva), and several others (Stergiopoulos et al., 2010; de Wit et al., 2012). In tomato (Solanum lycopersicum), Cf-Avr4 is recognized by the cognate trans-membrane receptor-like protein Cf-4, eliciting an HR. Remarkably, despite their low sequence similarity, the majority of Avr4 homologs are still perceived by Cf-4 and elicit an HR in tomato (Stergiopoulos et al., 2010; de Wit et al., 2012). How plant immune receptors such as Cf-4 are able to perceive pathogen effectors that are so sequence diverse remains unknown. One possibility is that they may recognize core effectors indirectly via their virulence function. Alternatively, they may directly perceive a common structural fold shared among core effectors or specific amino acids that dictate their intrinsic function, and thus are not readily mutable. For example, the broad recognition of Avr4 effectors by Cf-4 may be due to the conserved chitin-binding domain (ChtBD) or specific residues that facilitate binding of Avr4 to chitin. Answers to such questions would require an understanding of the structural properties of Avr4 and the way by which it interacts with chitin (Wirthmueller et al., 2013).
Here, we functionally and structurally characterized Pf-Avr4, a member of the Avr4 effector family from the tomato pathogen Pseudocercospora fuligena. Furthermore, by comparing the activity of Pf-Avr4 with Cf-Avr4, we sought to provide answers to critical questions regarding the biology of core effector proteins in fungi, including (1) whether their biochemical and biological function as well as contribution to virulence are conserved among different fungal species, (2) whether they may serve a role beyond the realm of plant-microbe interactions, and (3) what is the molecular basis for their broad-recognition by cognate immune receptors. Through a combination of biochemical and biological assays, we show that Pf-Avr4 and Cf-Avr4 share a common specificity for (GlcNAc)6 and provide mycelial protection against both plant- and microbial-derived chitinases, suggesting that the function of these core effectors is not solely restricted to plant infections, but are likely involved in interactions with other microorganisms as well. In addition, by biochemical and structural analyses, we elucidate the molecular basis for chitin binding by Pf-Avr4, whereas by subsequent site-directed mutagenesis of residues implicated in ligand binding, we reveal that such residues are not individually targeted for recognition by Cf-4. Instead, our studies highlight the dependence of recognition on an ordered Pf-Avr4 structure, as partially unfolded proteins are unstable and susceptible to proteolytic cleavage, implying that specificity in recognition of this effector family by Cf-4 is mediated from the combined effect of multiple residues that define local tertiary folds or overall conformational properties of Avr4 to initiate immune responses. Overall, our studies significantly advance our understanding of core effector proteins in fungi and provide a conceptual framework on how these can be pleiotropically recognized by single cognate immune receptors.
RESULTS
The P. fuligena Avr4 Effector Protein (Pf-Avr4) Is a Member of the Avr4 Core Fungal Effector Family
We have recently determined the genome sequence of the plant pathogenic fungus P. fuligena, a hemibiotrophic fungus that causes black leaf mold in tomato and is phylogenetically related to the banana pathogen M. fijiensis and other Dothideomycete fungi, including C. fulvum and D. septosporum. Query of the P. fuligena genome sequence with the Avr4 effector protein from C. fulvum (Cf-Avr4) identified a cysteine-rich protein of 48% similarity and 38% identity at the amino acid level to Cf-Avr4 that was termed Pf-Avr4 (P. fuligena Avr4) (Supplemental Figure 1A). The 128-residue-long Pf-Avr4 is predicted to consist of a 23-residue N-terminal signal peptide and a 105-residue mature protein with eight cysteine residues. A putative CBM14 chitin-binding domain (Pfam01607) (Chang and Stergiopoulos, 2015b) is present in its sequence, spanning residues Cys-45 to Cys-103 (Supplemental Figure 1B). Global sequence alignments indicated that Pf-Avr4 shares a similar cysteine spacing pattern (C1-X9-C2-X5-C3-X7-C4-X5-C5-X15/16-C6-X12/14-C7-X7-C8) with Avr4-like effector proteins from other Dothideomycetes, suggesting that they all adopt an analogous disulfide-bond pattern. In addition, three highly conserved aromatic residues are present within the predicted CBM14 domains of the Avr4 effector family members (C6-X5-W-X6/8-WC7-X1-W/Y/T-X5-C8), which may be involved in saccharide binding (Boraston et al., 2004; Jiménez-Barbero et al., 2006) (Supplemental Figure 1B).
Pf-Avr4 and Cf-Avr4 Are Functional Orthologs That Bind Chitin and Protect Fungal Cell Walls against Chitinases of Plant and Microbial Origin
We have previously shown that members of the Avr4 effector family bind chitin to protect it against plant chitinases (Stergiopoulos et al., 2010). We thus first examined whether this biochemical and consequently biological function would extend to Pf-Avr4. The selective binding of Cf-Avr4 to chitin was previously established using a qualitative in vitro affinity precipitation assay, which tested binding of this lectin to the insoluble polysaccharides crab-shell chitin, chitosan, cellulose, xylan, curdlan (β-1,3-glucan), and lichenan (van den Burg et al., 2006; Stergiopoulos et al., 2010). Using this in vitro polysaccharide affinity precipitation assay, we examined the general binding specificity of Pf-Avr4 against the key structural polysaccharides of plant (cellulose and xylan) and fungal (chitin, chitosan, and curdlan) origin that the protein is likely to encounter during host infection (Figure 1A). For the ligand-binding assays, N-terminally 6xHis-tagged Pf-Avr4 was heterologously expressed and subsequently purified via affinity and size-exclusion chromatography from culture filtrates of the methylotrophic yeast Pichia pastoris. Under the assay conditions (pH 8.0, 150 mM sodium chloride, 25°C) (van den Burg et al., 2006), neither Pf-Avr4 nor Cf-Avr4 was found to bind cellulose, xylan, curdlan, or commercial chitosan that was fully deacetylated under strong basic conditions (60% sodium hydroxide) and high heat (110°C), as evidenced by the presence of protein predominantly in the supernatant fraction (unbound protein fraction), and not in the pelleted material (bound protein fraction). However, when binding was tested against shrimp-shell chitin, >60% of Pf-Avr4 and Cf-Avr4 was detected in the pelleted fraction, demonstrating clear binding to chitin (Figure 1A). Also, due to the qualitative nature of this assay, no discernable difference between Pf-Avr4 and Cf-Avr4 in binding to the shrimp-shell chitin could be observed.
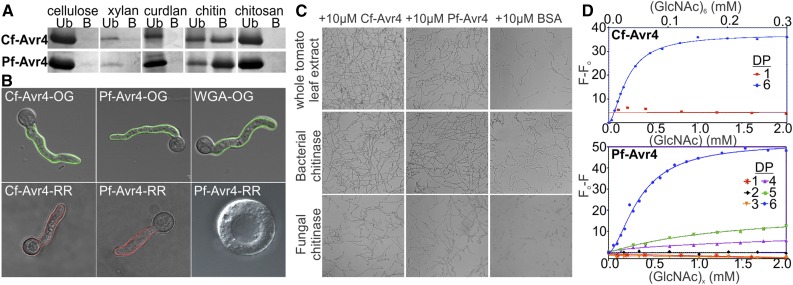
Figure 1.
Pf-Avr4 Is a Functional Ortholog of Cf-Avr4 That Binds Chitin and Protects against Chitinases.
(A) Affinity of Cf-Avr4 and Pf-Avr4 for several infection-related polysaccharides of fungal and plant origin was assayed using an in vitro polysaccharide precipitation assay (Ub, unbound fraction; B, bound fraction). Cf-Avr4 and Pf-Avr4 bind specifically to chitin and not to any of the other polysaccharides tested.
(B) Pf-Avr4-Oregon Green (OG) and Pf-Avr4-Rhodamine Red (RR) localizes to the fungal cell wall similarly to Cf-Avr4 and the WGA chitin-binding control. Pf-Avr4 associates specifically with the cell wall since Pf-Avr4-RR does not interact with the fungal protoplast.
(C) Pf-Avr4, like Cf-Avr4, is able to protect fungi from degradation by plant-, bacterial-, and fungal-derived chitinases, which is evidenced as fungal mycelial growth beyond that of the BSA controls. Cf-Avr4 offers slightly greater protection, as compared with Pf-Avr4, against tomato whole-leaf extracts with chitinase activity, while Cf-Avr4 and Pf-Avr4 provide equal protection against bacterial- and fungal-derived chitinases.
(D) Cf-Avr4 binds (GlcNAc)6 with a 10-fold greater affinity than does Pf-Avr4. Tryptophan fluorescence-based binding assays were used to quantify Cf-Avr4’s and Pf-Avr4’s affinity for chito-oligosaccharides since Pf-Avr4 was unamenable to ITC. Cf-Avr4 exhibited affinity for (GlcNAc)6 but not for GlcNAc, while Pf-Avr4 displayed measurable affinity for (GlcNAc)4-(GlcNAc)6.
The specific binding of Pf-Avr4 to chitin suggests that it has a similar biological function to Cf-Avr4, in protecting fungal cell walls against enzymatic degradation by chitinases (van den Burg et al., 2006). We examined this hypothesis by first observing the localization pattern of Pf-Avr4 on the cell walls of germinated conidia of Trichoderma viride using Pichia-produced protein conjugated to the fluorescent dye Oregon Green 488 (OG) or Rhodamine Red (RR) (Figure 1B). Wheat germ agglutinin protein (WGA) and Cf-Avr4 conjugated to OG or RR were used as positive controls for localization. Localization of the proteins was examined on germinated conidia of T. viride because the inner chitin layer in the cell walls of this fungus is only partially covered by the overlaying polysaccharide layers of β1,6-/β1,3-glucans and mannoproteins during the early stages of growth. Incubation of T. viride germlings with Pf-Avr4-OG or Pf-Avr4-RR showed a clear accumulation of the protein on the mycelial surface, while treatment with chitinases and glucanases to remove the entire cell wall abolished localization of Pf-Avr4 on the fungal surface (Figure 1B). Together, these results indicate that Pf-Avr4 specifically associates with the fungal cell wall through binding to chitin. As expected, a similar localization pattern was also observed for Cf-Avr4 and WGA (Figure 1B).
We next tested the ability of Pf-Avr4 to protect germlings of T. viride against hydrolysis by chitinases under in vitro conditions (Figure 1C). A similar assay has been used to demonstrate the protection properties of Cf-Avr4 (van den Burg et al., 2006). Addition of whole tomato leaf extract with chitinase activity (Supplemental Figure 2) on germlings of T. viride fully inhibited further growth of the fungus, while combined application of whole tomato leaf extract and either 10 μg of Pichia-produced Pf-Avr4 or Cf-Avr4 restored mycelial growth. Growth in the presence of Cf-Avr4 was higher compared with that of Pf-Avr4, suggesting that Cf-Avr4 binds to chitin and/or competes with plant-derived chitinases for the chitin substrate more efficiently than does Pf-Avr4. Notably, the protective function of Pf-Avr4 and Cf-Avr4 was also observed when germlings of T. viride were challenged with enzymatic solutions of bacterial (Streptomyces griseus) or fungal (T. viride) derived chitinases that were supplemented with basic β-1,3-glucanases, and in this case, both proteins provided equal levels of protection (Figure 1C). Thus, the two Avr4 effector proteins provide general protection against chitinases of either plant or microbial origin, suggesting that they may promote survival and fitness of the fungi on a plant host or their environment in general. For the above experiments, BSA was used as negative control and little to no mycelial growth was detectable after chitinase treatment for these samples (Figure 1C).
In Planta Assays Demonstrate That Pf-Avr4 Is Expressed and Localizes to the Fungal Cell Wall during Infection of the Host, Contributing to Fungal Virulence on Susceptible Plants
Cf-Avr4 is transcriptionally activated upon entry and intercellular growth of C. fulvum in tomato, whereas silencing of Cf-Avr4 reduces virulence of the fungus on its host (Joosten et al., 1994, 1997; van Esse et al., 2007). Since Cf-Avr4 and Pf-Avr4 share a common biochemical and biological function in vitro, we examined whether Pf-Avr4 would also be expressed and promote virulence of P. fuligena on tomato. Inoculation assays with P. fuligena were performed on the susceptible cultivar LA3940 as cv Moneymaker that was previously used in assays with C. fulvum has intermediate levels of susceptibility to this fungus (Zahn et al., 2011). As P. fuligena fails to sporulate under in vitro growth conditions, plant inoculations were performed by spraying leaves with mycelial fragments that were obtained by maceration of the fungal hyphae. Although this method enables inoculations with difficult fungi, it presents many challenges in standardizing the amount of viable inoculum sprayed on the plants and increases the experimental variation.
Expression analysis of Pf-Avr4 under in vitro growth conditions in liquid potato dextrose broth (PDB) media and during mycelial infection of tomato revealed that Pf-Avr4 is predominately expressed in planta, following a transient expression pattern in which transcription is steadily increased during the initial symptomless (biotrophic) phase of the infection, reaches its maximum at ∼9 d postinoculation (dpi) and decreases thereafter (Figure 2A). The expression profile of Pf-Avr4 is thus similar to that of Cf-Avr4, whose expression also gradually increases during the biotrophic phase and decreases during the necrotrophic stage of the infection (Joosten et al., 1994, 1997; Ökmen, 2013; Collemare et al., 2014).
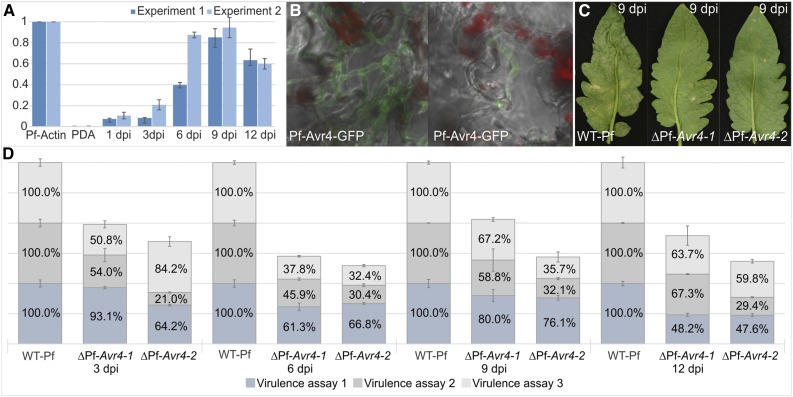
Figure 2.
Pf-Avr4 Is a Virulence Factor of P. fuligena.
(A) Expression of Pf-Avr4 relatively to fungal Actin under in vitro growth in liquid PDB media and during infection of the susceptible tomato cultivar LA3940 (compatible interaction). Tomato leaves were inoculated with mycelia of P. fuligena, and leaf samples were collected at 1, 3, 6, 9, and 12 dpi. Pf-Avr4 expression at each time point was calculated relative to P. fuligena actin that was set to 1.0 RQ. The mean RQ levels from two different biological experiments are shown. Standard deviations from four technical repetitions of the quantitative real-time PCR (qPCR) for each experiment are indicated with black bars for each time point.
(B) Pf-Avr4 localizes to the cell walls of P. fuligena during infection of the tomato host as determined by analysis of P. fuligena transformants expressing GFP-tagged Pf-Avr4 under the control of its native promoter. Growth of the fungus in the palisade mesophyll of the infected leaves is shown, as determined by Z-stack analysis of the images produced on a confocal microscope. Red dots are autofluorescence from the chloroplasts.
(C) Symptoms induced by the wild-type P. fuligena (WT-Pf) strain and two Pf-Avr4 deletion mutants (ΔPf-Avr4-1 and ΔPf-Avr4-2) on leaves of cv LA3940, as macroscopically seen at 9 dpi under the assay conditions.
(D) Quantification of the fungal biomass produced by the ΔPf-Avr4-1 and ΔPf-Avr4-2 mutants during infection of cv LA3940 relatively to the WT-Pf strain (set to 1.0 RQ). Virulence assays were performed three times and infected material was collected at 1, 3, 6, 9, 12, and 15 dpi from six to nine randomly selected leaves of two to three plants that were sprayed at the beginning of the experiment with the fungal mycelia. Subsequent fungal biomass quantification was done using qPCR and the corresponding macroscopic pictures of the disease symptoms for virulence assays 2 and 3 are shown in Supplemental Figure 4. Samples from 15 dpi were not analyzed, as the plant tissue was totally necrotic at this time point, thus yielding very low quality of RNA. Each stacked bar for each strain and time point is separated in three sections that represent the three virulence assays, respectively. sd from three technical repetitions of the qPCR is indicated for each section of the stacked bars. The analysis shows a slower buildup in fungal biomass by the two ΔPf-Avr4 mutants inside the host tissue as compared with the WT-Pf control strain. The large variation in the fungal biomass measured for each strain in different virulence assays or at different time points of the infection within the same virulence assay is largely attributed to the fact that inoculations were performed using fungal mycelia.
We next examined the localization pattern of Pf-Avr4 on the hyphae of P. fuligena during infection of tomato by analysis of P. fuligena transformants expressing GFP-tagged Pf-Avr4 under the control of its native promoter. Fluorescent imaging of mycelia growing in the palisade mesophyll of the infected leaves showed that the Pf-Avr4-GFP fusion protein aggregated at the fungal septa and peripheral cell wall, while the intensity of the fluorescence was higher at the hyphal apices where chitin is most accessible (Figure 2B). As expected, we did not observe any GFP fluorescence from mycelia growing on the surface of solid synthetic media such as potato dextrose agar (PDA). Thus, these studies further confirmed that Pf-Avr4 is produced during infection of the tomato host and localizes on the fungal hyphae to likely protect the exposed, during in planta growth, chitin against host chitinases.
Finally, we questioned whether Pf-Avr4 would also be required for full virulence of P. fuligena on its tomato host. Therefore, we generated two Pf-Avr4 deletion mutants (ΔPf-Avr4-1 and ΔPf-Avr4-2) of P. fuligena, in which Pf-Avr4 was replaced by a hygromycin resistance cassette and subsequently tested their virulence on cv LA3940. PCR-based analysis of the ΔPf-Avr4 mutants indicated a single integration event of the hygromycin resistance cassette in the Pf-Avr4 locus and the absence of any ectopic insertions in the genome of these transformants, thus validating their correctness (Supplemental Figure 3). Subsequent tomato leaf inoculations with the wild-type and ΔPf-Avr4 mutants showed a reduction in virulence of the mutants on cv LA3940, as determined by visual inspection of the inflicted symptoms on the leaves (Figure 2C; Supplemental Figure 4) and subsequent quantification of the fungal biomass at different stages of the infection process (Figure 2D). In detail, visual inspections of plants inoculated with the two ΔPf-Avr4 mutants showed that disease symptoms appeared on the infected leaves at almost the same time point (∼5 to 6 dpi) with symptoms inflicted by the wild-type strain. However, further progression of the disease was decelerated in case of the two ΔPf-Avr4 mutants, indicating that the deletion of Pf-Avr4 impairs, to some extent, fungal virulence (Supplemental Figure 4). These results were further corroborated by quantitative real-time PCR (qPCR), which indicated a slower buildup in fungal biomass by the two ΔPf-Avr4 mutants inside the host tissue as compared with the wild-type control strain (Figure 2D). Although the inoculation experiments were performed using mycelial fragments, which hindered a clear quantification of fungal biomass and resulted in a large variation from one experiment to another, infections with the deletion mutants of Pf-Avr4 did exhibit an overall decrease in fungal biomass and thus disease severity. For example, compared with the wild-type strain, at 6 dpi the fungal biomass of the ΔPf-Avr4-1 mutant was reduced by 38.7% in the first virulence assay, 54.1% in the second, and 62.2% in the third, whereas at 9 dpi the reduction in fungal biomass for this mutant was 20% in the first experiment, 41.2% in the second, and 32.8% in the third. Similar results were also obtained when analyzing the data for the ΔPf-Avr4-2 mutant (Figure 2D). Nonetheless, despite such large differences in absolute values of the measured biomass or the severity of the disease symptoms appearing on the plants (Figure 2D; Supplemental Figure 4), the general trend remained the same in these virulence assays, indicating that Pf-Avr4 contributes to virulence of P. fuligena on a susceptible tomato host, as does Cf-Avr4 for C. fulvum (van Esse et al., 2007).
Pf-Avr4 and Cf-Avr4 Share Binding Specificity, but Differ in Their Precise Affinity for (GlcNAc)6
Although both Pf-Avr4 and Cf-Avr4 bind to high molecular weight chitin (Figure 1A), it is unknown whether they also share specificity for the same length chito-oligosaccharides. Subtle differences in specificity could imply differences in the way by which they interact with chitin, whereas a conformity in specificity for the same length chito-oligosaccharides would suggest a similar binding-site topography and mechanism of ligand binding (Boraston et al., 2004). Conservation in binding-site topography in members of the Avr4 effector family is important because it could make this region a prime target for recognition by the cognate Cf-4 resistance protein. Therefore, we sought to determine Pf-Avr4’s and Cf-Avr4’s binding specificity to different length chito-oligosaccharides.
Target chito-oligosaccharides for Pf-Avr4 were expected to range between GlcNAc-(GlcNAc)6, since previous analyses using isothermal titration calorimetry (ITC) and tryptophan fluorescence-based binding assays showed that Cf-Avr4 interacts weakly with (GlcNAc)3 and binds more strongly to (GlcNAc)6 (van den Burg et al., 2004). As Pf-Avr4 proved unamenable to ITC (Supplemental Figure 5A), binding of the lectins to chito-oligosaccharides was tested by measuring the change in intrinsic tryptophan fluorescence of Pf-Avr4 and Cf-Avr4 upon titration with the chito-oligosaccharides. This method can be used as an indirect way to monitor ligand binding, as binding stimulates a change in the local tryptophan environment that can be measured as shifts in the emission wavelength and intensity of the intrinsic tryptophan fluorescence. Cf-Avr4 and Pf-Avr4 contain two (Trp-63 and Trp-71) and three (Trp-88, Trp-94, and Trp-97) tryptophans, respectively, within their putative CBM14 domains, and a change in the intrinsic tryptophan fluorescence of both proteins is detectable upon addition of chito-oligosaccharides (Figure 1D; Supplemental Figure 5B). Specifically, binding of Cf-Avr4 to (GlcNAc)6 was characterized by a decrease in tryptophan fluorescence, whereas binding of Pf-Avr4 to (GlcNAc)6 resulted in an increase in fluorescence. This quenching versus enhancing effect upon binding is largely determined by the differences in the local environment of their excited tryptophans, which can be influenced both by neighboring residues in the protein as well as by the composition and nature of the interacting ligand (Burstein et al., 1973).
The intrinsic fluorescence of Cf-Avr4 showed a peak emission at 347 nm, and upon binding to (GlcNAc)6 exhibited a blue shift to 339 nm at full saturation (Figure 1D; Supplemental Figure 5B). Saturation of Cf-Avr4 required ∼0.15 mM (GlcNAc)6 and was accompanied by quenching of the Cf-Avr4 fluorescence by 37.14% from the maximal fluorescence (ΔFmax = 37.14). Under the conditions of this experiment, Cf-Avr4 reached half saturation at a (GlcNAc)6 concentration of 31.37 μM (Figure 1D, Table 1). As a negative control, Cf-Avr4 was titrated with GlcNAc up to 2 mM, and no change in Cf-Avr4 emission fluorescence was detected, as indicated by a flat, linear tryptophan fluorescence profile (Figure 1D, Table 1).
Table 1. Tryptophan Fluorescence-Based Binding Data Detailing the Binding Affinity of Cf-Avr4, Pf-Avr4, and Pf-Avr4 ChtBD Mutants to Chito-Oligosaccharide Substrates.
| Tested Binding Partners | ΔFmaxa | Half ΔFmax (ΔFmax/2)b |
|---|---|---|
| Cf-Avr4c + GlcNAc | NB | NBd |
| Cf-Avr4 + (GlcNAc)6 | 37.14 (±0.91) | 0.03 (±0.002)e |
| Pf-Avr4c + GlcNAc | NB | NB |
| Pf-Avr4 + (GlcNAc)2 | NB | NB |
| Pf-Avr4 + (GlcNAc)3 | NB | NB |
| Pf-Avr4 + (GlcNAc)4 | WB | WBe |
| Pf-Avr4 + (GlcNAc)5 | 20.20 (±13.56) | 1.37 (±1.81) |
| Pf-Avr4 + (GlcNAc)6 | 51.88 (±2.18) | 0.35 (±0.03) |
| (GlcNAc)6f + Pf-Avr4WT g | 31.37 (±1.07) | 0.29 (±0.02) |
| (GlcNAc)6 + Pf-Avr4W88A | 10.50 (±1.55) | 0.58 (±0.10) |
| (GlcNAc)6 + Pf-Avr4N89A | NB | NB |
| (GlcNAc)6 + Pf-Avr4D90A | 15.19 (±1.54) | 0.89 (±0.07) |
| (GlcNAc)6 + Pf-Avr4N91A | 35.03 (±1.18) | 0.29 (±0.02) |
| (GlcNAc)6 + Pf-Avr4W94A | NB | NB |
| (GlcNAc)6 + Pf-Avr4D96A | 14.94 (±2.41) | 0.09 (±0.04) |
| (GlcNAc)6 + Pf-Avr4W97A | NB | NB |
Maximum change in fluorescence upon binding saturation (ΔFmax). Numbers in parenthesis represent the se of the mean calculated as follows: (sd of experimental data points) divided by (the square root of the number of data points).
Concentration (mM) of chito-oligosaccharide required to achieve half-saturation (ΔFmax/2). Numbers in parenthesis represent the se of the mean associated with the half-saturation values, calculated based on the half-saturation determined from the tryptophan fluorescence experiments collected in duplicate or triplicate.
Protein was produced in P. pastoris.
NB, no binding.
WB, weak binding.
(GlcNAc)6 was used at a concentration of 2 mM.
Wild-type Pf-Avr4 and ChtBD mutants were produced in E. coli.
In comparison, the intrinsic fluorescence of Pf-Avr4 displayed a peak fluorescence emission at 341 nm, while binding events did not alter Pf-Avr4’s intrinsic fluorescence emission wavelength, which remained stable at 341 nm (Figure 1D; Supplemental Figure 5B). Pf-Avr4 did not display measurable affinity for GlcNAc-(GlcNAc)3, which is evident from the flat, linear tryptophan fluorescence profiles produced upon titration of these chito-oligosaccharides up to 2 mM (Figure 1D, Table 1). Pf-Avr4 exhibited slight affinity for (GlcNAc)4 as this titration produced a measurable change in tryptophan fluorescence (ΔFmax = 6.0 at 2 mM) compared with titration with (GlcNAc)-(GlcNAc)3 (ΔFmax = 0 at 2 mM) (Figure 1D). The magnitude of Pf-Avr4’s affinity for (GlcNAc)4 falls just above the detection threshold of the assay but is too low to discern half-saturation binding parameters. Pf-Avr4 displayed higher affinity for (GlcNAc)5, as a larger change in tryptophan fluorescence (ΔFmax = 20.2) was detected and half-saturation was achieved with 1.37 mM (GlcNAc)5 (Figure 1D, Table 1). Furthermore, Pf-Avr4 binds with significantly greater affinity to (GlcNAc)6, as an 51.88% increase in tryptophan fluorescence was observed (ΔFmax = 51.88), and half-saturation was reached at 0.35 mM (GlcNAc)6 (Figure 1D, Table 1). Taken together, Pf-Avr4, like Cf-Avr4, displays measurable affinity for (GlcNAc)4-(GlcNAc)6; however, the two proteins differ in their precise ligand affinity, as Cf-Avr4 binds (GlcNAc)6 with 10-fold greater affinity than does Pf-Avr4. Such differences in binding affinities are frequently present among members of the same CBM family and are mainly the result of slight differences in the amino acid composition of their substrate binding sites (Christiansen et al., 2009).
The 1.7-Å X-Ray Crystal Structure of Pf-Avr4 Reveals a Compact, Globular Protein Stabilized by a Network of Intramolecular Interactions
To understand how Pf-Avr4 functions on a molecular level, we attempted to cocrystallize Pf-Avr4 with its chitin ligand; however, despite intensive screening, only crystals of ligand-free Pf-Avr4 were obtained. For crystallization, Pichia-produced Pf-Avr4 was readily crystallized by sitting-drop vapor diffusion in 2.0 M ammonium sulfate and 0.1 M HEPES, pH 7.5, at room temperature. Experimental phases were determined by single-wavelength anomalous dispersion for sulfur, using Cu-Kα radiation (λ = 1.54 Å) from a rotating anode x-ray source. Pf-Avr4 has a high sulfur content, which is primarily due to the abundance of cysteine residues within its sequence, thus making phase determination via this method possible. The resulting structure (Figure 3A; Supplemental Movie 1) was refined using high-resolution synchrotron data to an atomic resolution of 1.7 Å in the space group P3221 with two Pf-Avr4 molecules per asymmetric unit (Table 2).
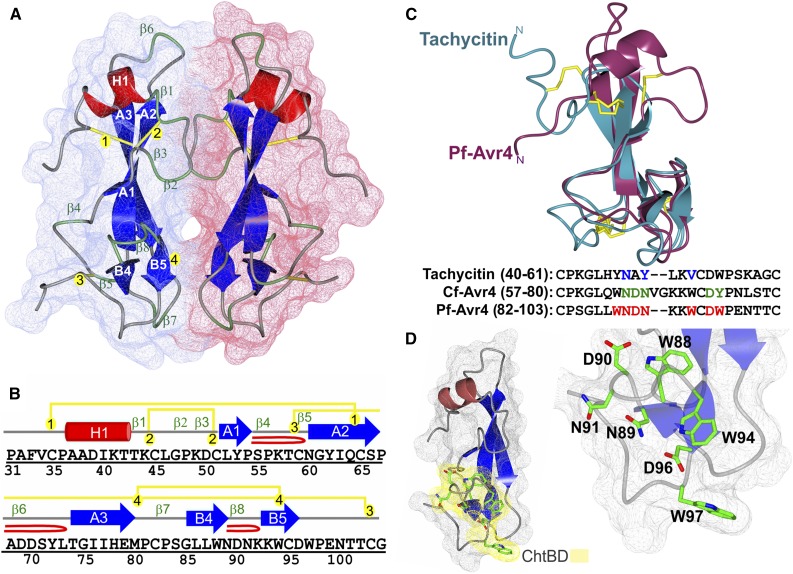
Figure 3.
The X-Ray Structure of Pf-Avr4 Solved to 1.7-Å Resolution.
(A) The overall fold and surface filling of Pf-Avr4, spanning residues Pro-31 to Gly-105. Pf-Avr4 was crystallized with two molecules per asymmetric unit (chain A, red; B, blue), which are held together by many indirect water-mediated contacts (not shown) and three hydrogen-bond direct protein-protein contacts (not shown). The structure of Pf-Avr4 reveals a compact, globular protein stabilized by four disulfide bonds and an extensive network of intramolecular forces. Pf-Avr4 consists of a single N-terminal helix (red, H1) followed by a distorted β-sandwich fold, comprised of two β-sheets, one three-stranded, and the other two-stranded. The five β-strands are depicted in blue, labeled by their sheet (A and B) and placement within the structure (1 to 5). β-Turns are labeled in green (β1-8), and disulfide bonds are drawn in yellow and labeled by order in the structure (1 to 4).
(B) The secondary structure sequence alignment for Pf-Avr4, residues Pro-31 to Gly-105. Secondary structure elements are color-coded and labeled as depicted in (A). β-Hairpins are depicted by a red loop.
(C) Pf-Avr4 (maroon) aligns to CBM14 family member tachycitin (cyan) (RMSD 1.98 Å, for 52 aligned α-carbons), encompassing the distorted β-sandwich motif and putative ChtBD of tachycitin. Sequence alignment of tachycitin, Cf-Avr4, and Pf-Avr4 through their respective putative ChtBDs enabled the identification of seven residues (red, bold) on Pf-Avr4 that may have a key role in Pf-Avr4-chitin binding. Putative functional residues are shown for tachycitin (blue) and Cf-Avr4 (green).
(D) Pf-Avr4’s ChtBD (yellow) is located at the C terminus, and individual ChtBD residues (green) are illustrated and labeled on the ChtBD-magnified structure on the right.
Table 2. Data Collection and Refinement Statistics for Pf-Avr4 (PDB Code: 4Z4A).
| Cu Anode | SSRL 7-1 | |
|---|---|---|
| X-Ray Source | ||
| Wavelength (Å) | 1.5418 | 1.12709 |
| Temperature (K) | 85 | 100 |
| Space group | P3221 | P3221 |
| Unit-cell parameters (Å, o) | a = b = 57.07, c = 126.63 | a = b = 57.1, c = 126.98 |
| α = β = 90, γ = 120 | α = β = 90, γ = 120 | |
| Resolution (Å) | 1.90 (2.0–1.9) | 42.35–1.70(1.74–1.70) |
| Rmergea (%) | 5.8 (53.9) | 5.2 (67.1) |
| CC1/2 | 99.9 (84.7) | |
| <I/σ(I)> | 53.4 (4.19) | 17.94 (2.61) |
| No. of reflections | 1,146,434 (61,399) | 181,689 (11,050) |
| No. of unique reflections | 19,624 (2,762) | 26,958 (1,966) |
| Completeness (%) | 99.9 (99.7) | 99.0 (99.6) |
| Redundancy | 58.4 (22.2) | 6.74 (5.62) |
| Refinement Statistics | ||
| Resolution (Å) | 35.0–1.70 (1.74–1.70) | |
| No. of reflections (F>0) used in refinement | 25,549 (1,962) | |
| Rfactorb (%) | 18.3 (26.0) | |
| Rfreec (%) | 21.0 (28.9) | |
| RMS bond length (Å) | 0.016 | |
| RMS bond angle (°) | 1.709 | |
| Overall B Value (Å2) | 24.7 | |
| Wilson Plot B Value (Å2) | 24.8 | |
| Ramachandran plot statisticsd | ||
| Residues | 147 | |
| Most favored region | 143/147 | |
| Allowed region | 147/147 | |
| Disallowed | 0/147 |
Rmerge = [∑h∑i|Ih – Ihi|/∑h∑iIhi], where Ih is the mean of Ihi observations of reflection h. Numbers in parenthesis represent highest resolution shell.
Rfactor, ∑||Fobs| − |Fcalc|| / ∑|Fobs| × 100 for 95% of recorded data.
Rfree = ∑||Fobs| − |Fcalc|| / ∑|Fobs| × 100 for 5%.
From MolProbity (Chen et al., 2010).
The crystal structure of Pf-Avr4 spans residues Pro-31 to Gly-105, with the N terminus (residues Thr-24 to Thr-30) and C terminus (residues Val-106 to Gly-128) exhibiting a high degree of disorder and, therefore, not included in the final model (Figure 3B). Overall, it reveals a globular protein composed of a single N-terminal α-helix (H1) followed by a distorted β-sandwich fold, a common fold among CBMs (Boraston et al., 2004), which is formed by a central β-sheet, composed of three antiparallel β-strands (A1, A2, and A3), and a C-terminal β-sheet, comprised of two antiparallel β-strands (B4 and B5). A four-residue-long type I β-turn (β8) links B4 and B5. While roughly 30% of Pf-Avr4 folds into organized α/β secondary structure, 70% of the structure is comprised of highly ordered connecting loops. In total, there are eight β-turns (β1-β8) (types I, II, and IV) present in each Pf-Avr4 chain, all of which are four residues in length, and three β-hairpin loops, ranging in length from three to seven residues. Due to the ordered nature of these β-turns, which include extensive interactions between side chain and main chain nitrogen and oxygen atoms, they were well resolved in the structure. Analysis of Pf-Avr4’s dimeric crystal packing revealed that the two monomer chains (A and B) are held together by many water-mediated interactions and three interchain hydrogen bonds. One hydrogen-bond pair is located between Tyr72/A and Tyr72/B (2.7-Å bond distance), while the second and third hydrogen-bond pairs are situated in the C-terminal domain between the backbone oxygen of Ser84/A and the side-chain nitrogen of Trp97/B (2.9 Å). An equivalent bond is present between Ser84/B and Trp97/A. The dimeric assembly of Pf-Avr4 observed in the crystal is consistent with SEC-MALS data, which indicate that Pf-Avr4 behaves as a dimeric species in solution (Supplemental Figure 6). As expected, all eight cysteines present in Pf-Avr4 participate in disulfide bonds, matching those predicted for Cf-Avr4 (van den Burg et al., 2003) and forming the following disulfide pairs: Cys35-Cys65, Cys45-Cys51, Cys59-Cys103, and Cys82-Cys95 (Figures 3A and 3B). Collectively, these disulfide bonds and tight loop regions greatly enhance the overall stability of the Pf-Avr4 structure.
Structure-Based Analysis Positions Pf-Avr4’s Chitin-Binding Site in the C-Terminal β-Sheet
As the first structural representative of the Avr4 core effector family, the structure of Pf-Avr4 provides novel molecular insight into the substrate-binding architecture of these family members and of the putative contacting residues between the ligand and the protein. An understanding of these two properties is key to further examination of whether a casual relation exists between Pf-Avr4 ligand-binding function and the property of recognition by Cf-4. Thus, we used the tertiary structure of Pf-Avr4 along with information from other members of the CBM14 family and knowledge of conserved ligand-binding features in CBM families (Boraston et al., 2004) in order to determine Pf-Avr4’s chitin-binding site and identify crucial residues required for binding chitin.
A structural homology search through DaliLite v.3 (Dietmann et al., 2001; Holm et al., 2008) identified tachycitin (PDB: 1DQC), a small chitin-binding lectin and member of the CBM14 family from the horseshoe crab Tachypleus tridentatus (Kawabata et al., 1996; Suetake et al., 2000), as the top structural homolog for Pf-Avr4 (Figure 3C). Overall, tachycitin’s NMR solution structure corresponds very well with that of Pf-Avr4, aligning with an RMSD of 1.98 Å over 52 residues, encompassing ∼50% of each structure. Of marked importance, the proposed chitin-binding domain (ChtBD) architecture in tachycitin is also conserved in Pf-Avr4 and consists of two β-strands connected by a β-hairpin loop, together forming a single β-sheet (Suetake et al., 2000). Key residues implicated in tachycitin’s interaction with chitin are Asn-47, Tyr-49, and Val-52, though experimental evidence supporting the role of these residues in ligand binding is lacking (Suetake et al., 2000). When the proposed ChtBD of tachycitin is structurally aligned to Pf-Avr4, the corresponding residues in Pf-Avr4 are Asn-89, Asn-91, and Trp-94, suggesting that these residues may be involved in the Pf-Avr4-chitin interaction (Figure 3C).
Additional putative binding residues can be identified from the sequence alignment of the presumed ChtBD of Cf-Avr4 with Pf-Avr4. The ChtBD of Cf-Avr4 was delineated through NMR titration experiments, which suggested Asn-64, Asp-65, Asn-66, Asp-73, and Tyr-74 as prominent residues in Cf-Avr4’s interaction with chitin (van den Burg et al., 2004). These residues are conserved in Pf-Avr4, with the exception of Tyr-74, which in Pf-Avr4 is replaced with a tryptophan (Trp-97) (Figure 3C), a similar but slightly bulkier aromatic residue. Aromatic residues are fundamental components of the protein-carbohydrate binding mechanism, as they are involved in hydrophobic stacking interactions and offer many coordination points for contact with the sugar ligand (Boraston et al., 2004; Chen et al., 2013; Hudson et al., 2015). In this respect, Pf-Avr4 has a third tryptophan (Trp-88), in addition to Trp-94 and Trp-97, located in the putative ChtBD that could potentially participate in the Pf-Avr4-chitin interaction.
Taken together, Pf-Avr4’s ChtBD is thought to contain Asn-89, Asp-90, Asn-91, Asp-96, Trp-94, and Trp-97, with potential contribution from neighboring Trp-88 (Figure 3D). When mapped on the tertiary structure of Pf-Avr4, the ChtBD resides in the C-terminal region of the protein and is composed of a four-residue β-hairpin loop flanked by β-strands B4 and B5 (Figure 3D). Additionally, the connecting loop contains an antiparallel G1 β-bulge (Chan et al., 1993), formed by hydrogen-bonding between Asn89(O)-Lys93(N) (2.9 Å) and Asn89(O)-Trp94(N) (3.2 Å). This ordered motif likely contributes to and maintains the local structure of the ChtBD, dictating the arrangement of B5. Moreover, Trp-88, Trp-94, and Trp-97 coordinate many contacts between solvent molecules and neighboring residues, thus providing a strong support for the overall ChtBD fold. Trp-88 sits on the C-terminal β-sheet B with its side chain pointing toward the N terminus and packed against β-sheet A, while Trp-94 and Trp-97, which are also positioned on β-sheet B, are solvent exposed and face in the opposing direction (Figure 3D), suggesting that they may interact directly with chitin, while Trp-88 has an indirect role in ligand binding.
The residues predicted to participate in binding to chitin also interact extensively with surrounding residues in the structure. Asn-89, which sits at the end of B4, interacts with the side chain of Asp-96 [Asn89(ND2)↔Asp96(OD2)], as well as with the backbone nitrogen of Asn-91 [Asn89(OD1)↔Asn91(N)] and Lys-92 [Asn89(OD1)↔Lys92(N)]. Asn-89 is central to maintaining the local structure of the ChtBD, as it plays a pivotal role in dictating the β-bulge motif. Asp-90, the first residue residing on the β-hairpin loop connecting B4 and B5, interacts with the side chains of Trp-88 [Asp90(OD1)↔Trp88(NE1)] and Lys-93 [Asp90(OD1)↔Lys93(NZ)]; similarly, Asp-96, located at the end of B5, coordinates with the side chain of Trp-94 [Asp96(OD2)↔Trp94(NE1)], main chain nitrogen of Trp-97 [Asp96(OD1)↔Trp97(N)], and side chain of Asn-100 [Asp96(OD2)↔Asn100(ND2)]. The side chains of Asn-91 and Trp-97 do not interact with neighboring residues, although Trp-97 is involved in a hydrogen bond with Ser-84 of the neighboring protein chain. The side chains of binding residues Asn-89, Asn-91, Trp-94, Asp-96, and Trp-97 all point in the same direction, toward the C terminus of the protein, with the exception of Asp-90, which may implicate a more supplementary role for this residue in binding chitin (Figure 3D). Measured end to end, Asn-91 to Trp-97 form a shallow binding cleft of 19.3 Å in length.
Site-Directed Mutagenesis Confirms the Location of the Chitin-Binding Site in Pf-Avr4 and the Importance of Aromatic and Adjacent Polar Residues for (GlcNAc)6 Binding
In order to define the involvement and degree of contribution that each predicted binding residue has on Pf-Avr4 (GlcNAc)6-binding function, alanine point mutations at residues Trp-88 (W88A), Asn-89 (N89A), Asp-90 (D90A), Asn-91 (N91A), Trp-94 (W94A), Asp-96 (D96A), and Trp-97 (W97A) were produced in the Rosetta-gami-B Escherichia coli strain, and their affinity for (GlcNAc)6 was determined using our tryptophan fluorescence-based binding assay and compared with the wild-type Pf-Avr4 (Pf-Avr4WT) protein produced with the same bacterial expression system. Binding titrations and data analyses were performed as described above for the Pichia-produced Pf-Avr4, using a maximum of 2 mM (GlcNAc)6 for each titration.
When assaying the interaction between (GlcNAc)6 and the E. coli-produced Pf-Avr4WT and ChtBD mutants, Pf-Avr4WT achieved half saturation at a concentration of 0.29 mM (GlcNAc)6 with a ΔFmax of 31.37 (Supplemental Figure 7A; Table 1). The E. coli-produced Pf-Avr4WT (GlcNAc)6-binding behavior was commensurate with the Pichia-produced Pf-Avr4.
Of the seven Pf-Avr4 ChtBD mutants assayed, N91A was the only mutation that had no measurable effect on the interaction between Pf-Avr4 and (GlcNAc)6, as Pf-Avr4N91A reached half saturation at 0.29 mM (GlcNAc)6 (ΔFmax=35.03) nearly identical to that of Pf-Avr4WT (Supplemental Figure 7A; Table 1). In contrast, Pf-Avr4’s binding affinity for (GlcNAc)6 was significantly reduced, abolished, or driven below the assay detection limit by the N89A, W94A, and W97A mutations, as evidenced by the flat, linear tryptophan fluorescence profiles of the corresponding Pf-Avr4 protein mutants (Supplemental Figure 7A; Table 1). In comparison, alanine mutations at Trp-88 and Asp-90 led to a reduction in Pf-Avr4’s affinity for (GlcNAc)6. Specifically, Pf-Avr4W88A’s affinity for (GlcNAc)6 was reduced 2-fold, requiring the addition of 0.58 mM (GlcNAc)6 to reach half saturation (ΔFmax = 10.50), while Pf-Avr4D90A exhibited a 3-fold reduction in affinity and achieved half saturation at 0.89 mM (GlcNAc)6 (ΔFmax = 15.19).
Unlike the other ChtBD mutants, Pf-Avr4D96A required less (GlcNAc)6 to reach half-saturation (0.09 mM) but also exhibited a markedly different fluorescence profile compared with the other mutants. As the concentration of (GlcNAc)6 increased, Pf-Avr4D96A displayed an initial, rapid change in tryptophan fluorescence until ∼1 mM (GlcNAc)6, achieving a ΔFmax of 14.94, and from 1 mM to 2 mM (GlcNAc)6 the magnitude of fluorescence change greatly decreased (Supplemental Figure 7A; Table 1). However, based on structural analysis, it is not surprising that a mutation to D96 strongly alters Pf-Avr4’s tryptophan fluorescence profile. D96 is the only ChtBD residue that interacts directly with a tryptophan predicted to be directly involved in binding chitin, forming a hydrogen bond to the indole nitrogen of W97. This interaction is predicted to greatly influence and limit the flexibility of solvent-exposed W97, thus strongly contributing to the overall fluorescence profile of Pf-Avr4. Due to the anomalous effect that the D96A mutation has on tryptophan fluorescence, measurements of fluorescence changes are likely less reliable for this mutant, as is reflected in the relatively large se of the mean associated with, and unique to, the concentration of (GlcNAc)6 required for half saturation and ΔFmax.
To further examine the differences in chitin-binding ability among the Pf-Avr4 ChtBD mutants, and to do so in a more biologically meaningful context, we performed protection assays of T. viride germlings against chitinases. In these assays, T. viride germlings were treated individually with either Pf-Avr4WT (positive control), BSA (negative control), or one of the Pf-Avr4 ChtBD mutants. When challenged with whole tomato leaf extract, fungal germlings treated with BSA displayed minimal growth, while those treated with Pf-Avr4WT exhibited strong growth that was relatively comparable to germling growth in the absence of chitinases (i.e., water-challenged germlings) (Supplemental Figure 7B). In agreement with the biochemical tryptophan fluorescence assays, germlings treated with Pf-Avr4D90A and Pf-Avr4N91A grew similarly to those treated with Pf-Avr4WT when challenged with whole tomato leaf extract (Supplemental Figure 7B), suggesting that these mutants provided equal protection against the plant-derived chitinases. This assay also enabled a clearer understanding of Pf-Avr4D96A-chitin interaction behavior, as this mutant provided equal to Pf-Avr4WT level of protection against plant-derived chitinases, indicating that the D96A mutation does not largely alter the affinity (increase or decrease) of Pf-Avr4 for its chitin substrate. Pf-Avr4 ChtBD mutants that did not display a measurable affinity for (GlcNAc)6 did not also offer significant protection against chitinases: Pf-Avr4W94A displayed low to mild protection, whereas Pf-Avr4W97A and Pf-Avr4N89A afforded the fungi no to minimal protection (Supplemental Figure 7B). Due to a substantial reduction in affinity for chitin, ChtBD mutants Pf-Avr4W94A, Pf-Avr4W97A, and Pf-Avr4N89A provided greatly reduced protection against chitinases likely because they are unable to bind cell wall chitin as efficiently as Pf-Avr4WT and the other ChtBD mutants. Pf-Avr4W88A, which also provided no to minimal protection, exhibited reduced affinity for (GlcNAc)6, but is also the most sensitive ChtBD mutant to protease cleavage (see next section). Therefore, under the apoplastic conditions assayed here (treatment with whole tomato leaf extract), Pf-Avr4W88A does not provide protection against chitinases as it is likely proteolyzed faster than it can bind the fungal cell wall. Collectively, these biological assays corroborated the biochemical results, as the differences in binding affinity among the ChtBD mutants also translated into differences in their ability to protect T. viride germlings against plant-derived chitinases.
Point Mutations in Residues Required for (GlcNAc)6 Binding Do Not Fully Abolish Recognition by Cf-4
We previously determined that many Avr4 effector family members are perceived by Cf-4, eliciting an HR in tomato (Stergiopoulos et al., 2010; de Wit et al., 2012). In determining the structure of Pf-Avr4, we sought to examine whether Pf-Avr4 is recognized by Cf-4 and, if so, decipher whether the property of recognition by Cf-4 overlaps with its chitin-binding function. We hypothesized that since Avr4 homologs share highest sequence identity within their predicted ChtBDs, it is possible that the indispensability of the ChtBD makes it a prime target for recognition by Cf-4, either through overall ChtBD folding properties or specific key binding residues. In this respect, our ChtBD mutants provided an ideal framework with which to examine the importance of the overall ChtBD topology as well as of specific (GlcNAc)6-binding residues to Cf-4-mediated recognition of Pf-Avr4.
To assay for recognition, the E. coli-produced Pf-Avr4WT and ChtBD mutants were infiltrated into tomato leaves of cv Purdue 135 (+Cf-4) and cv Moneymaker (MM) (-Cf-4) at 1, 5, and 10 μg/mL. E. coli-produced Cf-Avr4 was used as a positive control and the presence of HR on the infiltrated leaf sectors was assessed at 5 d postinfiltration. Infiltration of Pf-Avr4WT into leaves of cv Purdue 135 produced a strong HR over the entire infiltrated sectors at all tested concentrations, whereas infiltrations into MM leaves did not elicit HR (Figure 4A; Supplemental Figure 8A). Notably, infiltrations with Pf-Avr4WT triggered a stronger HR response than Cf-Avr4, as Pf-Avr4WT was able to elicit a strong HR at 1 μg/mL, while Cf-Avr4 did not at this concentration. The above results were reproducible irrespectively of whether protein infiltrations were done with the E. coli- or Pichia-produced Pf-Avr4WT and Cf-Avr4 proteins and suggest that Pf-Avr4WT recognition by Cf-4 is more robust compared with Cf-Avr4, thus further indicating that chitin-binding affinity does not correlate with Cf-4 recognition.
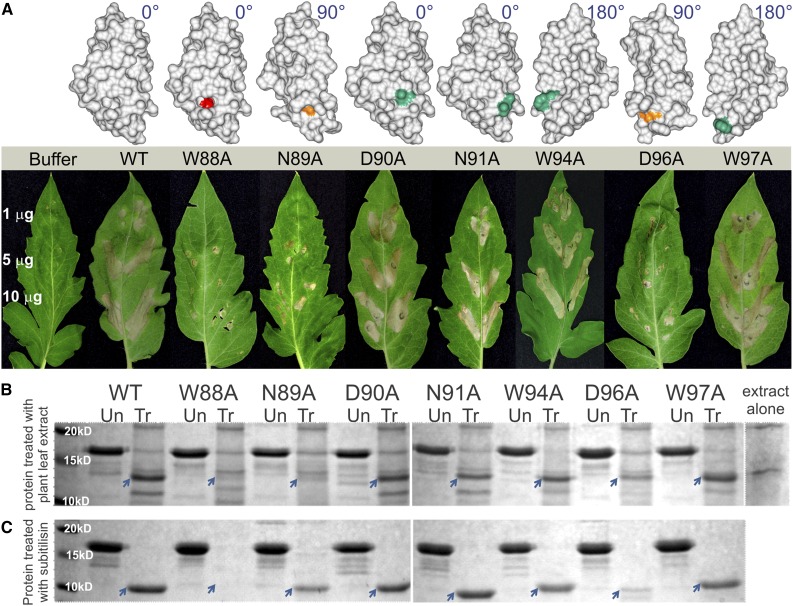
Figure 4.
Mutations of Pf-Avr4 That Destabilize the Architecture of ChtBD and Are Not Recognized by Cf-4 Are More Susceptible to Proteolysis under Apoplastic Conditions.
(A) ChtBD mutants were infiltrated into tomato leaves of cultivar Purdue 135 (+ Cf-4) and cv Moneymaker (MM) (− Cf-4) at 1, 5, and 10 μg/mL to determine their ability to be recognized by Cf-4 and elicit an HR. The location of each ChtBD mutation is highlighted on the surface-modeled structure of Pf-Avr4 (gray) with structural orientation indicated by degrees rotated from the wild-type structure. Mutations that elicited an HR equivalent to Pf-Avr4WT are depicted in green, mutations that displayed reduced HR are shown in orange, and the one with abolished HR is colored in red.
(B) To assess the proteolytic vulnerability of the ChtBD mutants under apoplastic conditions, Pf-Avr4WT and ChtBD mutants were treated with whole tomato leaf protein extract (Un, untreated protein; Tr, protein treated with plant leaf extract). The 12-kD Pf-Avr4WT cleavage product (indicated with a blue arrow) corresponds to the full-length protein minus the N-terminal tag, as confirmed by Edman degradation sequencing.
(C) Pf-Avr4WT and ChtBD mutants were treated with subtilisin, a nonspecific protease (Un, untreated protein; Tr, subtilisin-treated) to further assess proteolytic sensitivity. Subtilisin digested Pf-Avr4WT from a 17-kD protein to a 10-kD product, corresponding to the full-length, mature protein sequence. Of the mutants, only Pf-Avr4W88A, Pf-Avr4N89A, and Pf-Avr4D96A show significant susceptibility.
When assaying the ChtBD mutants, Pf-Avr4 mutations at Asp-90, Asn-91, Trp-94, and Trp-97 to alanine produced an HR of the same intensity as the Pf-Avr4WT at all concentrations tested (Figure 4A; Supplemental Figure 8A). In contrast, mutations N89A and D96A produced an intermediate HR phenotype at protein concentrations >5 μg/mL, as evidenced by the reduced necrotic lesions in the infiltrated leaf sectors. Also, mutation W88A resulted in the complete loss of HR at protein concentrations of 1 and 5 μg/mL, and in minimal or absent HR at a protein concentration of 10 μg/mL. In all cases, infiltrations in tomato leaves of MM did not elicit an HR. Taken together, these results suggest that the Pf-Avr4 ChtBD mutants are still recognized by Cf-4, although to a varying degree as infiltrations with the same protein concentrations can result in necrotic lesions of different intensities.
To further examine the specificity of the interaction between Cf-4 and Pf-Avr4WT or the ChtBD mutants, we performed agroinfiltration experiments, where we transiently coexpressed Cf-4 with each protein in Nicotiana benthamiana leaves, using the Agrobacterium tumefaciens transient transformation assay (Van der Hoorn et al., 2000). Coinfiltrations at 1:2 (A6000.5:A6001.0), 1:1 (A6000.5:A6000.5), and 1:0.5 (A6000.5:A6000.25) ratios of Cf-4:Pf-Avr4WT induced a strong HR in the infiltrated leaf sectors within 5 d postinfiltration (Supplemental Figure 8B). In addition, there was no discernable difference in the intensity of the induced HR when Pf-Avr4WT was coinfiltrated with Cf-4 (A6000.5) at cell suspension densities of A6001.0 and A6000.5, while coinfiltrations with effector cell densities of A6000.25 resulted in slightly weaker and patchy HR in the infiltrated sectors. When assaying the ChtBD mutants, all mutants affected an HR pattern similar to Pf-Avr4WT with the exception of Pf-Avr4W88A, which elicited a slightly weaker HR when infiltrated at cell densities of A6000.25 and A6000.5 (Supplemental Figure 8B). These results substantiate the protein infiltration assays and indicate that point mutations in residues critical for (GlcNAc)6 binding result in avirulent forms of Pf-Avr4 that are perceived by Cf-4 when effector and receptor are present in sufficient protein amounts.
The Ability of the Pf-Avr4 ChtBD Mutants to Elicit an HR in Tomato Is Inversely Correlated to Their Vulnerability to Proteolytic Cleavage in the Protease-Rich Tomato Leaf Apoplast
Although all of the Pf-Avr4 ChtBD mutants are recognized by Cf-4, the temporal differences among some mutants (i.e., W88A, N89A, and D96A) to elicit an HR of the same intensity when infiltrated at equal protein concentrations, poses the question of whether this is due to destabilized interaction with the Cf-4 resistance protein or due to the conformational stability of these mutants in the protease-rich tomato leaf apoplast. In this respect, it has been previously reported that loss of Cf-4 recognition by natural isoforms of Cf-Avr4 is due to the instability and rapid degradation of these proteins in the tomato leaf apoplast (van den Burg et al., 2003). Therefore, we examined whether a causal relation exists between the ability of the ChtBD mutants to elicit an HR in tomato and their vulnerability to proteolytic cleavage.
To investigate whether the ChtBD mutants were more susceptible to proteolysis, we assayed their ability to withstand proteolytic cleavage by treating them with whole leaf plant extracts from tomato. In this assay, Pf-Avr4WT was digested from a 17-kD protein band to a 12-kD band, which based on N-terminal Edman degradation sequencing was determined to correspond to full-length Pf-Avr4WT minus the N-terminal tag sequence (Figure 4B). Of the ChtBD mutants, the ones with substitutions D90A, N91A, W94A, and W97A were proteolyzed to the same 12-kD product as observed for Pf-Avr4WT. By contrast, ChtBD mutants with substitutions W88A and N89A were significantly degraded by the proteases present in the whole tomato leaf extract, as very faint cleavage products are seen at 12 kD (Figure 4B). This result supports the hypothesis that alanine point mutations at W88 and N89 alter the stability or folding properties of the ChtBD, potentially by increasing the flexibility of the ChtBD loop and subsequently increasing Pf-Avr4’s susceptibility to proteolytic digestion. Similarly, Pf-Avr4D96A shows only a slight cleavage product band at 12 kD, and while the intensity of this band is greater than those of Pf-Avr4W88A and Pf-Avr4N89A, it is still far less intense compared with Pf-Avr4WT (Figure 4B). This suggests that the point mutation D96A also increases the sensitivity of Pf-Avr4 to proteolysis, though to a lesser extent than alanine mutations at W88 and N89. Based on the Pf-Avr4 structure, D96 interacts with three residues in the ChtBD region (N89, N91, and K92), and through these interactions has an indirect, though not negligible, contribution to the overall structural integrity of the ChtBD.
Finally, to further confirm that degradation of the ChtBD mutants Pf-Avr4W88A, Pf-Avr4N89A, and Pf-Avr4D96A was due to proteolysis, E. coli-produced Pf-Avr4WT and ChtBD mutants were treated with subtilisin, a broadly specific serine protease that cleaves flexible, accessible protein sequence, favoring an uncharged residue in the P1 position (Philipp and Bender, 1983). Subtilisin processed Pf-Avr4WT to its 10-kD mature product, as evidenced by the appearance of a 10-kD cleavage band for Pf-Avr4WT and the ChtBD mutants bearing substitutions N89A, D90A, N91A, W94A, D96A, and W97A (Figure 4C). Pichia-produced Pf-Avr4 was used as a negative control, whereas disulfide-disrupted Pichia-produced Pf-Avr4, which is predicted to have a more flexible structure due to loss of one or more disulfide bonds, was used as a positive control. Correspondingly, subtilisin processed the Pichia-produced Pf-Avr4 to its 10-kD mature form and completely proteolyzed the disulfide-disrupted Pf-Avr4, as no full-length protein or cleavage products were detected by SDS-PAGE (Figure 4C). The results of the subtilisin treatment are very similar to those of the treatment with tomato whole leaf extract. Pf-Avr4W88A is completely proteolyzed, indicating a high degree of structural flexibility that is uncharacteristic of Pf-Avr4WT, while Pf-Avr4D96A is only partially degraded, which is evidenced by the presence of a very faint 10-kD cleavage product. Interestingly, subtilisin treatment did not fully degrade Pf-Avr4N89A, as a 10-kD product, although reduced by ∼80% compared with Pf-Avr4WT, is still present. It is possible that more complete proteolysis of Pf-Avr4N89A, which is achieved with the tomato whole leaf extract, requires the action of additional proteases that are present in the whole tomato leaf extract. These results confirm that alanine point mutations at residues W88, N89, and D96 result in conformationally unstable and sensitive to proteolytic degradation proteins.
DISCUSSION
In this work, we succeeded in determining the x-ray crystal structure of Pf-Avr4, a member of the Avr4 fungal effector family from the tomato pathogen P. fuligena. In addition, through a combination of structure-guided mutagenesis, biochemical, and biological assays with Pf-Avr4 and Cf-Avr4, we offer new insight into the biology of core effector proteins in fungi and discuss a conceptual framework for their pleiotropic recognition by single cognate immune receptors.
Functional Conservation within the Avr4 Effector Family Suggests Evolutionary Constraints
Our studies show, that despite the diversity in their amino acid sequences, functional and ligand specificity is retained between Pf-Avr4 and Cf-Avr4, as both proteins bind chitin present in fungal cell walls and provide mycelial protection against enzymatic degradation by chitinases (van den Burg et al., 2006). Furthermore, although binding to high molecular weight chitin is shown to be a conserved feature among members of the Avr4 effector family (Stergiopoulos et al., 2010; de Wit et al., 2012; Mesarich et al., 2016), by detailed ligand-binding analysis, we demonstrate that the specificity between Pf-Avr4 and Cf-Avr4 extends further to the length and mode of interaction with their common (GlcNAc)6 ligand oligosaccharide. This suggests that the Avr4 family, and perhaps other core effector families as well, evolves under functional and structural constraints, and such features could be potentially explored in the designing and genetic engineering of immune receptors with pleiotropic recognition specificities (Vleeshouwers and Oliver, 2014). However, Cf-Avr4 binds (GlcNAc)6 with 10-fold greater affinity than does Pf-Avr4, but such differences in binding affinities are common among members of the same CBM family and can be largely attributed to slight differences in the amino acid composition of their respective ligand-binding domains (Christiansen et al., 2009; Hudson et al., 2015). For instance, Cf-Avr4 and Pf-Avr4 share 67% sequence identity through their respective ChtBDs, and it is plausible that this disparity could account for such differences in affinity for (GlcNAc)6. Regardless, Pf-Avr4 is still able to protect against plant chitinases and thereby contribute to fungal virulence, as shown by the specific expression and localization of this effector on the fungal cell walls of P. fuligena during tomato infection, and the reduced virulence of the ΔPf-Avr4 mutants on a susceptible cultivar. However, as fungi use multiple layers of defense against host chitinases, including masking chitin under a layer of β-glucans and other polysaccharides (Bowman and Free, 2006), converting chitin into chitosan (El Gueddari et al., 2002), secreting chitinase inhibitors (Lange et al., 1996), and possibly other chitin-binding effectors with a role in protection (for example, Ecp6 effectors are shown to partially protect against chitinases in some fungal species) (Marshall et al., 2011), a complete loss of pathogenicity in the ΔPf-Avr4 mutants was not observed and might also explain the differences in contribution to virulence between Pf-Avr4 and Cf-Avr4. Overall, our data indicate that Pf-Avr4 and Cf-Avr4 are true functional orthologs and physiologically relevant effectors for fungal infections in plants.
Avr4 Proteins May Facilitate General Fungal Fitness and Survival
Intriguingly, next to protecting against plant chitinases during tomato infection, Cf-Avr4 and Pf-Avr4 also provide protection against bacterial- and fungal-derived chitinases, suggesting that they could have a dual role in plant pathogenesis and antimicrobial protection. Thus, perhaps at odds with traditional beliefs that effectors have evolved to facilitate host infections, our data imply a broader role for Avr4 effectors beyond deregulation of host immunity, in ecological competence of the producing organism. In support of this hypothesis, we have recently shown that, as with other core fungal effector proteins (Stergiopoulos et al., 2012), putative Avr4 homologs are found in 51 fungal species with diverse lifestyles, including saprophytes and plant, animal, and human pathogens, indicating that pathogenic lifestyle does not determine the patchy phylogenetic distribution of the Avr4 effector family in diverse fungal lineages (Chang and Stergiopoulos, 2015b). Such a dynamic birth-and-death mode of evolution is frequently observed in stress-response related genes involved in interactions with other organisms, such as adaptive immunity or pathoadaptation to new hosts (Ota and Nei, 1994; Nei and Rooney, 2005). Finally, studies in human pathogens have also clearly shown that many virulence traits, including effectors secreted by the Type VI secretion system (T6SS), can have dual roles in both parasitic and environmental fitness (Jani and Cotter, 2010). Thus, it is plausible that some core effector families serve general ecological fitness and survival of the producing organism and not just parasitic infections of the host (Stergiopoulos et al., 2012).
Structural Determination of the Avr4 Effector Family to Atomic Resolution
Obtaining accurate and high-resolution protein structures is a major challenge in structural biology and key to further biological studies. The 1.7-Å x-ray structure of Pf-Avr4 presented here represents the first three-dimensional structure of a member of the Avr4 effector family and CBM14 lectins in general. Previous attempts to structurally characterize members of this family were unsuccessful as, although Cf-Avr4 could be partially characterized using NMR techniques (van den Burg et al., 2004), the presence of numerous prolines and resonance peak overlaps prevented its 3D structure solution. The NMR-derived data for Cf-Avr4 were, nonetheless, useful to map secondary structure elements of the protein as well as to detect residues whose environment changes upon interaction with (GlcNAc)3, suggesting that they could be involved in ligand binding. However, an experimental validation of the 3D structural properties of Avr4 as well as of the involvement of specific residues in binding to chitin was so far missing. In this respect, the x-ray structure of Pf-Avr4 presented here reveals that the N-terminal α-helix of the protein does not contain any cysteine residues involved in disulfide bonds, in contrast to the NMR-based secondary structure assignments of Cf-Avr4, which suggested that the N-terminal helix is longer and contains the second conserved cysteine involved in a disulfide bond (van den Burg et al., 2004). Likewise, NMR titration experiments with Cf-Avr4 indicated Asn-64, Asp-65, Asn-66, Asp-73, and Tyr-74 as the prominent residues involved in ligand binding. Our functional profiling confirmed that residues in Pf-Avr4 equivalent to Cf-Avr4-Asn64 and Cf-Avr4-Tyr74 (i.e., Asn-89 and Trp-97) contribute to (GlcNAc)6 binding (van den Burg et al., 2004), but also showed that the equivalent of Cf-Avr4-Asn66 residue in Pf-Avr4 (i.e., Asn-91) had no effect on (GlcNAc)6 binding, and in addition identified Trp-94, a residue that is conserved between Pf-Avr4 and Cf-Avr4 but was not predicted by the NMR data, as key to binding chitin. Next to Cf-Avr4, a few discrepancies were also identified between Pf-Avr4 and the NMR-derived structure of tachycitin (Suetake et al., 2000), a member of the CBM14 lectin family that shares a similar overall fold with Avr4 (van den Burg et al., 2003). Specifically, superimposing of the two structures reveals that the core of the proteins contains two β-sheets, an N-terminal three-stranded antiparallel β-sheet and a C-terminal two-stranded antiparallel β-sheet. Both structures are also stabilized by four disulfide bonds between eight conserved cysteine residues. However, the two proteins differ in that Pf-Avr4 contains an N-terminal seven-residue α-helix (Ala-37 to Thr-43), whereas in tachycitin, this helix is replaced by a short one-turn C-terminal α-helix that is not found in Pf-Avr4. Taken together, the determination of Pf-Avr4’s solution structure by x-ray diffraction enabled us to expand the repertoire of the Avr4 and CBM14 families.
Elucidating the Molecular Basis for Chitin Binding in the Avr4 Effector Family
Our structure-function analyses further allowed us to specify more precisely the location and composition of the chitin-binding site as well as to portray a mechanistic model of Pf-Avr4’s interaction with chitin. CBMs generally do not undergo conformational changes when binding to their ligands but rather the tertiary structure employed by these modules provides a platform for substrate binding (Boraston et al., 2004; Hashimoto, 2006). Pf-Avr4’s ChtBD is located in the C terminus of the protein, consisting of residues positioned on β-strands B4 and B5 and their connecting β-hairpin loop. At the molecular and mechanistic level, the aromatic amino acids Trp-88, Trp-94, and Trp-97 and their adjacent polar residues Asn-89, Asp-90, and Asp-96 partake, directly or indirectly, in the Pf-Avr4-(GlcNAc)6 interaction, with Asn-89, Trp-94, and Trp-97 being critical to (GlcNAc)6 binding. In particular, Asn-89 largely determines the local structure of the connecting loop between B4 and B5, whereas Trp-94 and Trp-97 are poised to directly interact with the substrate through their aromatic side chains that can form hydrophobic stacking interactions with the nonpolar face of the GlcNAc ring, thus allowing many contact points with (GlcNAc)6. In contrast, Trp-88 and Asp-90, whose side chains interact with each other and face opposite the binding surface, have an indirect role in (GlcNAc)6 binding and likely promote proper orientation of binding residues and protein stability. Trp-88, in particular, interacts with many neighboring residues and is sandwiched between the two β-sheets in the protein core, thus functioning as a molecular linchpin that stabilizes the structure. Likewise, Asp-96, which faces toward the C terminus of Pf-Avr4, interacts with three other residues implicated in binding (GlcNAc)6, i.e., it is forming van der Waals contacts with Trp-97 and is hydrogen-bonded to the indole nitrogen of Trp-94 and side chain of Asn-89, thus being critical to maintaining the local structure of the ChtBD. Together, these studies offer a comprehensive characterization of the molecular basis for functional specificity in the Avr4 effector family and portray a mechanistic picture of the underlying molecular mechanism for binding chitin that was so far absent.
Individual Chitin-Binding Residues Do Not Have a Direct Effect on Pf-Avr4's Interaction with Cf-4
Despite their diversity in primary structures, most of the Avr4 proteins examined so far, including Pf-Avr4, have the ability to trigger a Cf-4-mediated HR (Stergiopoulos et al., 2010; de Wit et al., 2012; Mesarich et al., 2016). However, the molecular basis for the pleiotropic recognition of effectors by single cognate immune receptors is still poorly understood. Knowledge on the features that underlie pleiotropy in recognition of effectors could be key to engineering immune receptors for broad-spectrum resistance (Vleeshouwers and Oliver, 2014). With a more accurate structural definition of Pf-Avr4’s ChtBD location and composition, we also examined whether the avirulence and virulence functions of the protein can be attributed to the ChtBD region. The underlying hypothesis was that given the indispensability of the ChtBD for the virulence function of Avr4 and the conservation of the ChtBD architecture among members of this effector family, it is possible that the broad recognition of Avr4 effectors by Cf-4 is conferred by the local fold of this region or specific binding residues. In addition, since Avr4 may facilitate general fitness and protection of the fungus in its environment, and not just during infection of the plant host, mutating residues that disturb the virulence function of this effector will entail a higher fitness cost to overcome resistance. Thus, residues that are central to chitin binding or dictate the architecture of the ChtBD are likely to be under structural and functional constraints; consequently, they may have been exploited by Cf-4 as prominent targets for recognition.
Despite the rationality of this hypothesis, single alanine substitutions of residues in Pf-Avr4 that are critical to (GlcNAc)6 binding still yielded avirulent forms of the protein that could trigger a Cf-4 mediated HR, indicating that these ChtBD residues are not directly recognized by Cf-4. Although point mutations in residues W88, N89, and D96 can partially disrupt recognition, the produced isoforms are vulnerable to proteolytic degradation within the protease-rich apoplastic environment of tomato and elicit an HR on a protein concentration-dependent manner, as seen by transient expression in N. benthamiana. Combined, the two features indicate, on one hand, that these binding-site residues make important contributions to the structure of the ChtBD domain and subsequently to the conformational stability of the protein, but, on the other, that they likely do not mediate a direct interaction with Cf-4. Thus, the overall analysis of the ChtBD mutants illustrates that disruption of recognition by introduced mutations in the ChtBD is mediated indirectly, through reduced stability and increased proteolytic sensitivity of the partially unfolded protein, rather than through direct residue or fold recognition in solely this region. It should be noted here that, although not yet experimentally proven, the interaction between Avr4 and Cf-4 is presumed to occur through direct physical binding of the two proteins rather than through indirect recognition mechanisms (van den Burg et al., 2006; van Esse et al., 2007). Recent studies by Mesarich et al., 2016 also indicated that the C-terminal region of Cf-Avr4 between Cys-57 and Cys-82, a region that largely overlaps with the ChtBD and contains most of the residues implicated in ligand binding, is critical for recognition of Cf-Avr4 by Cf-4. The same study also identified Pro-58 (or Pro-87 if residue count includes the signal peptide) in Cf-Avr4 as key to recognition of Avr4 proteins by Cf-4 (Mesarich et al., 2016). This residue is also conserved in Pf-Avr4 (Pro-83) and is located in a relatively neutral loop region that is N-terminal to the ChtBD domain. However, as prolines are structurally important residues with a pivotal role in directing protein folding (MacArthur and Thornton, 1991; Deber et al., 2010), it is plausible that mutating this residue would alter the structure of the β-hairpin loop that encompasses the ChtBD domain, thereby rendering the protein sensitive to proteolytic degradation (Mesarich et al., 2016). Indeed, an alanine substitution of Pro-83 in Pf-Avr4 resulted in a partially unstable protein that triggers a Cf-4-mediated HR, as seen by treatment of the protein with subtilisin and infiltrations in cv Purdue 135 (+Cf-4) (Supplemental Figure 9). In this later case, infiltrations of the Pf-Avr4WT at the concentrations of 1, 5, and 10 μg/mL resulted in a strong HR for all three concentrations, whereas infiltrations of the Pf-Avr4P83A mutant triggered an HR only at the concentrations of 5 and 10 μg/mL but not at 1 μg/mL or lower. These results indicate that this residue is not directly recognized by Cf-4 and most likely mediates avirulence by influencing protein stability.
Taken together, our studies indicate that individual amino acids that are pivotal for binding chitin are not likely the ones targeted for recognition by Cf-4, suggesting that the Pf-Avr4 ligand-binding function is structurally distinct or does not fully overlap with the property of recognition. Instead, the studies highlight a direct link between recognition of Pf-Avr4 and conformational stability, a property that is further closely linked to proteolytic susceptibility. As the protein needs to function in a protease-rich environment of the apoplast, it is perhaps not surprising that Pf-Avr4 and other Avr4 effector proteins generally adopt a well ordered and rigid globular conformational structure that is stabilized by a network of intramolecular forces and does not allow for excessive fluctuation. It is plausible that this feature has consequently been exploited by Cf-4, in evolving the ability to recognize the overall conformational properties of Avr4 or multiple ordered local tertiary folds to initiate immune responses, further suggesting that Cf-4 is functioning as a PRR rather than a typical R protein, although it elicits an HR. For example, analogous to the models proposed for recognition of diverse peptides by the major histocompatibility complex (MHC) molecules or of MHC-peptide complexes by T cell receptors, loss of antigenicity in one epitope of Avr4 could shift recognition to other epitopes of the protein and/or Cf-4 may undergo “induced fit” conformational changes upon an initial loose contact with different isoforms of Avr4 to allow the formation of a stable complex (Nowak, 1996; Wu et al., 2002; Armstrong et al., 2008). Such flexibility in receptor-ligand interaction might also explain why natural allelic variants of Cf-Avr4 that escape recognition by Cf-4 have mutations in cysteine residues that disturb protein folding, thereby rendering the protein sensitive to proteolytic degradation (van den Burg et al., 2003) or why Cf-4 can recognize highly polymorphic Avr4 effector proteins that nonetheless share a conserved cysteine-bond pattern and predicted secondary structure (Stergiopoulos et al., 2010). Although still a hypothesis, if true, it would be interesting to examine whether other surface immune receptors that perceive core apoplastic effectors have also exploited the link between an ordered 3D protein structure needed for conformational stability and proteolytic resistance to recognize overall effector conformational properties or multiple folds. Overall, maintaining an ability to recognize effector folding patterns rather than specific residues would present with the evolutionary advantage of expending the recognition specificity of immune receptors, such as Cf-4, to multiple isoforms of core effectors or diverse effectors that do not share similarity in their primary structures (Cesari et al., 2013).
METHODS
Cloning and Protein Expression
Pseudocercospora fuligena Avr4 (Pf-Avr4), residues 24 to 128, was cloned into the pPic9 vector (Invitrogen) between the SmaI and NotI restriction sites, resulting in an N-terminal 6xHistidine-FLAG tag, and was integrated into the genomic DNA of Pichia pastoris strain GS115. To produce recombinant Pf-Avr4, P. pastoris was cultured in a fermentor and protein expression was induced with continuous methanol addition over 72 h at 30°C. Cultures were centrifuged to remove cell debris and the culture supernatant was concentrated and extensively dialyzed against 50 mM Tris-HCl, pH 8.0, 50 mM sodium chloride, and 5 mM imidazole. The resulting material was applied to a NiNTA column (Qiagen), and protein was eluted in 50 mM Tris-HCl, pH 8.0, 50 mM sodium chloride, and 250 mM imidazole. The final purification step was done using a Superdex 75, 120 mL column (GE Healthcare) into 25 mM Tris-HCl, pH 8.0, and 50 mM sodium chloride. After the final purification step, Pf-Avr4 was highly pure (>90%) as determined by SDS-PAGE and was further analyzed by SEC-MALS analysis on a 1260 Infinity purifier (Agilent Technologies; Wyatt Technology) using a Superdex 200 10/300 GL, 25 mL, column (GE Healthcare). Column output was analyzed by a MiniDAWN TREOS MALS detector (Wyatt Technology) with a laser source at 658.8 nm, followed by an Optilab T-rEX refractometer (Wyatt Technology) with a 658.0-nm light source. Analysis was performed under the tryptophan binding assay conditions (50 mM potassium phosphate, pH 7.0, and 150 mM sodium chloride) at 25°C. Cladosporium fulvum Avr4 (Cf-Avr4) was cloned into the pPic9 vector (Invitrogen) and purified and analyzed as described for Pf-Avr4. Cf-Avr4 was highly pure after the final chromatography step (>99%) and was used for subsequent experiments. Protein was stored at −80°C.
Wild-type Pf-Avr4 and ChtBD mutants (W88A, N89A, D90A, N91A, W94A, D96A, and W97A) were cloned between the SalI and NotI restriction sites of a modified pCDF-Duet-1 vector (Novagen), which contains two 6xHistidine tags and a Rhinovirus 3C protease cleavage site N-terminal to the MCS. Protein expression (Pf-Avr4WT and ChtBD mutants) was induced in Rosetta-gami-B(D3) Escherichia coli cells with 1 mM IPTG for 16 h at 18°C. Harvested cells were pelleted, resuspended in 50 mM Tris-HCl, pH 8.0, 250 mM sodium chloride, and 5 mM imidazole, and lysed by microfluidization. Cleared lysate was loaded onto Ni-NTA resin (Qiagen), the column was washed in 50 mM Tris-HCl, pH 8.0, 250 mM sodium chloride, and 30 mM imidazole, and protein was eluted in 50 mM Tris-HCl, pH 8.0, 250 mM sodium chloride, and 500 mM imidazole. Protein was then run over a Superdex 75 column with a 120 mL bed volume (GE Healthcare) into 25 mM Tris-HCl, pH 8.0, and 50 mM sodium chloride. This expression system provided a high-throughput method of protein production, in which most mutants could be produced with a yield of roughly 10 mg of protein per liter of cell culture in half the time it would take to produce the protein through Pichia fermentation. E. coli-produced Pf-Avr4WT eluted from a size exclusion column and SEC-MALS system with a very similar elution profile to and solution behavior as Pichia-produced Pf-Avr4, and the ChtBD mutants show a similar size exclusion elution profile to Pf-Avr4WT. No significant difference in activity was observed between protein produced in E. coli versus that produced in Pichia, as determined using the in vitro saccharide precipitation assay and tryptophan fluorescence binding assay to (GlcNAc)6.
Plant Materials and Growth Conditions
Three tomato (Solanum lycopersicum) cultivars were used in this study, namely, cv LA3940, which is known to be susceptible to P. fuligena, cv Moneymaker (MM) that has intermediate levels of susceptibility to P. fuligena and does not carry any functional Cf resistance genes (also known as MM-Cf-0), and cv Purdue 135, which is known to carry a functional Cf-4 resistance gene. Plants were grown in a growth chamber with 16 h of artificial light and 70% humidity at 27°C for 6 weeks. All three tomato lines were obtained from the Tomato Genetic Resource Center at UC Davis. The Nicotiana benthamiana plants used in our transient expression assays were grown under similar conditions but at a temperature of 25°C.
Quantification of Pf-Avr4 Expression Levels
Infections of the susceptible tomato cv LA3940 with P. fuligena were conducted in a temperature, humidity, and light controlled growth chamber as described previously (Zahn et al., 2011), with some modifications. Specifically, P. fuligena was initially grown on half PDA medium in the dark at 25°C for 1 week. Because the P. fuligena isolate used for the experiments is unable to sporulate under in vitro culture conditions, the inoculation experiments were performed using fungal mycelia instead of spores. For this purpose, the mycelia from the initial culture of the fungus on half PDA were harvested and subcultured in 100 mL liquid PDB medium for 4 d in order to increase fungal biomass. The suspension was subsequently filtered through two layers of cheesecloth and the mycelial concentration was calculated with a hemocytometer. Mycelium fragments (105 per milliliter) with an average mycelial size ranging from 50 to 150 μm were prepared in 10% PDB inoculation solution. The mycelium fragments were sprayed on the lower part of leaves using a spray bottle (Delta Industries). After spraying, the inoculated plants were kept in the dark at 95% humidity and 25°C for 2 d. Six inoculated leaves from two plants were collected at 1, 3, 6, 9, and 12 dpi and were immediately frozen in liquid nitrogen and stored at −80°C until RNA extractions. Total RNA was isolated from inoculated leaves with TRIzol reagent (Life Technologies) following the manufacturer’s instructions. cDNA was generated by using gene-specific primers and Maxima reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer’s protocol. Quantitative real-time PCR was conducted with SsoFast EvaGreen Supermix (Bio-Rad Laboratories) with P. fuligena actin primers Pf-Actin-F/Pf-Actin-R and Pf-Avr4 qPCR-specific primers Pf-Avr4_qPCR-F/Pf-Avr4_qPCR-R (Supplemental Table 1), in a CFX96 Touch Real-Time PCR detection system (Bio-Rad Laboratories). Real-time PCR conditions included an initial 95°C denaturation step for 10 min followed by denaturation for 15 s at 95°C, annealing for 30 s at 60°C, and extension for 30 s at 72°C for a total of 40 cycles. The Ct (threshold cycle) values from the CFX96 system were used for determining the relative amount of RNA. P. fuligena actin expression was used as an endogenous control for calculating the relative expression level of Pf-Avr4 during infection. The raw expression values for Pf-Avr4 was determined using the equation: 2−ΔCt(Pf-Avr4-Pf-Actin), following the method of Livak and Schmittgen (2001). Raw expression values were then normalized based on the expression of Actin, which was considered to be 1. The final expression value was the mean of four qPCR repetitions from two individual inoculation experiments. The expression of Pf-Avr4 in vitro was determined under growth in liquid PDB medium at 25°C, with shaking at 200 rpm for 4 d in the dark.
In Vitro Polysaccharide Precipitation Assays
Binding of Pf-Avr4 and Cf-Avr4 to insoluble polysaccharides was tested by mixing 15 μg of lectin with 20 mg of each polysaccharide in 50 mM Tris-HCl, pH 8.0, and 150 mM sodium chloride binding buffer solution with a reaction volume of 700 μL for 2 h at 25°C under constant rotation (van den Burg et al., 2004, 2006). The polysaccharides tested included cellulose, xylan, shrimp shell chitin, chitosan, deacetylated chitosan, and curdlan AL (all Sigma-Aldrich, except curdlan AL, which was obtained from InvivoGen). To separate bound from unbound protein, the reactions were centrifuged at 14,000 rpm for 10 min, and the supernatant (unbound protein) was collected and stored at 4°C. The pelleted saccharide/bound protein (bound protein) was washed three times with 50 mM Tris-HCl, pH 8.0, and 150 mM sodium chloride. After the final wash step, 1% SDS was added to the pellet fraction and the samples were incubated at 100°C for 10 min to elute the bound protein. Bound protein was recovered by centrifuging the samples at 14,000 rpm for 10 min and collecting the supernatant. TCA/acetone precipitation was performed on the unbound and bound fraction samples by adding cold 20% (w/v) TCA to each sample, vortexing briefly, and incubating the samples at 4°C overnight. The samples were then centrifuged at 14,000 rpm for 25 min at 4°C. The supernatant was aspired off and the pelleted protein was washed with ice-cold 100% acetone and brief vortex, and centrifuged at 14,000 rpm for 20 min at 4°C. The acetone was aspirated and the resulting protein samples were allowed to dry at room temperature for 30 min before analysis by SDS-PAGE and Coomassie Brilliant Blue stain. In order to produce fully deacetylated chitosan, commercial chitosan (Sigma-Aldrich), which had a degree of deacetylation of ≥75%, was treated with an excess of 60% (w/v) sodium hydroxide and incubated at 100°C for 1 h (Chang et al., 1997). The chitosan was then washed with water until the supernatant pH was neutral and dried at 40°C to complete dryness. The binding reactions were incubated at room temperature with continuous rotation for 2 h.
Localization of Cf-Avr4 and Pf-Avr4 on Fungal Mycelia
Localization studies of Cf-Avr4 and Pf-Avr4 on germlings of Trichoderma viride were done according to van den Burg et al. (2006) with slight modifications. Cf-Avr4 and Pf-Avr4 were labeled via their N-terminal free amine groups with Oregon green 488-X, succinimidyl (OG) (excitation/emission wave 496/524 nm; Life Technologies). Pf-Avr4 was also labeled separately with the amine-reactive Rhodamine Red-X, succinimidyl Ester (RR) (excitation/emission wavelength: 560/580 nm; Life Technologies) following the manufacturer’s recommendations. Nonreacted dye was separated from labeled protein by size-exclusion chromatography and protein was eluted in 25 mM Tris-HCl, pH 8.0, and 50 mM sodium chloride. T. viride spores were germinated overnight in rich liquid medium consisting of yeast extract (10 g/L) and glucose (30 g/L). Germlings with no more than two hyphae cells were stained with 4 μM Cf-Avr4-OG, Pf-Avr4-OG, and Pf-Avr4-RR or 4 μM Wheat Germ Agglutinin-OG (Life Technologies) in PBS for 1 h at room temperature. After staining, the germinated fungi were washed three times with PBS buffer and transferred on microscope slides. The fluorescent signal was detected with a Zeiss LSM710 confocal microscope at 40× magnification using an excitation wavelength of 488 nm for OG and 532 nm for RR. Protoplasts of T. viride were generated in 0.6 M potassium chloride by overnight treatment of 1-week-old fungal mycelium with a mixture of 15 mg/mL of lysing enzymes from Trichoderma harzianum (Sigma-Aldrich; cat. no. L1412) that contained β-glucanase, cellulase, protease, and chitinase activities, and 81.25 units/mL of Lyticase from Arthrobacter luteus (Sigma-Aldrich; cat. no. L2524).
The in vivo cellular localization pattern of Pf-Avr4 in its native fungal species P. fuligena was examined by analysis of chimeric Pf-Avr4 fused at its C terminus with GFP. For this purpose, a 6x-glycine linker and Superfolder GFP (sGFP) were fused to the 3′ coding region of Pf-Avr4 by PCR and the gene fusion was cloned into the pBHt2 binary vector under the control of the Pf-Avr4 native promoter and terminator (Mullins et al., 2001). The pBHt2 binary vector was modified such that the hygromycin B phosphotransferase gene (HYG), which is under the control of the Aspergillus nidulans trpC promoter, was replaced by the neomycin resistance gene (neo), which confers resistance to Geneticin (G418). pBHt2-Pf-Avr4-GFP was next transformed into Agrobacterium tumefaciens (AGL1) by electroporation. Agrobacterium-mediated fungal transformation was performed by mixing pBHt2-Pf-Avr4-GFP transformed agrobacteria (A600 0.5) with mycelia fragments of P. fuligena in 1:1 ratio and spread over a nitrocellulose filter membrane (Sigma-Aldrich) placed on induction medium (AIM agar: 2.05 g/L dibasic potassium phosphate, 1.45 g/L monobasic potassium phosphate, 0.15 g/L sodium chloride, 0.5 g/L magnesium sulfate, 0.1 g/L calcium chloride, 0.0025 g/L iron sulfate, 0.5 g/L ammonium sulfate, 1.8 g/L glucose, 40 mM MES, pH 5.3, and 1.5% agar) amended with 200 μM of acetosyringone for 3 d at 25°C. A second round of selection of transformants was performed by transferring the nitrocellulose filter membranes once colonies had appeared onto half PDA medium (18.5 g/L) amended with 150 μg/mL Geneticin (Life Technologies; cat. no. 10131035). Finally, the presence of the transgene in all transformants was verified by PCR, and GFP expression and the Pf-Avr4 cellular localization pattern in the transformants were examined by epifluorescence microscopy in which actively growing hyphae were observed under a Leica LAS-AF SPE confocal microscope (Leica Camera) at 40× magnification and an excitation wavelength of 488 nm.
Cf-Avr4- and Pf-Avr4-Mediated Protection of T. viride Germlings against Chitinase Activity
The protection ability of Cf-Avr4, Pf-Avr4WT, and Pf-Avr4 ChtBD mutants was challenged by different chitinases as described previously with several modifications (van Esse et al., 2007). T. viride spores (104 spores) were pregerminated in half PDB medium (12 g/L) for 15 h at 25°C. Fifty microliters of germinated spores were mixed with 50 μL 0.2 μg/μL whole tomato leaf protein extract and 10 μg Avr4 protein in a total reaction volume of 150 μL, and the reaction was incubated at 25°C for 8 to 10 h. To detect the ability of the Avr4 proteins to protect T. viride germlings against bacterial chitinase (chitinase from Streptomyces griseus; Sigma-Aldrich; cat. no. C6137) and fungal chitinase (chitinase from T.viride; Sigma-Aldrich; cat. no. C8241) activity, T. viride germlings were treated with 5 units of Zymolyase (Zymo Research; cat. no. E1004), 0.2 units of either bacterial chitinase or fungal chitinase, and 10 μg Avr4 protein in a total reaction volume of 150 μL. Zymolase contains β-1,3 glucanase and β-1,3-glucan laminaripentao-hydrolase activity and was used to remove the surface glucan layer on the fungal cell wall, exposing the underlying chitin. The reactions were incubated at 25°C for 8 to 10 h, after which time images were taken with a Nikon Diaphot inverted tissue culture microscope.
Targeted Gene Replacement of Pf-Avr4 by the Hygromycin Resistance Cassette
Targeted gene replacement of Pf-Avr4 in the genome of P. fuligena by the hygromycin resistance cassette was performed by a double crossover approach via an Agrobacterium-mediated transformation, as described by Frandsen et al. (2008). Briefly, pBht2KOI binary plasmid was modified from the original pBht2 plasmid (Mullins et al., 2001) by inserting ApaI, SpeI, and AvrII restriction enzyme sites at the 3′ end of hygromycin B phosphotransferase resistance cassette (HYG). A 400-bp DNA fragment of upstream sequences from the Pf-Avr4 open reading frame as well as a 1-kb DNA fragment of downstream sequences were amplified from genomic DNA of P. fuligena using primer pairs SalI-Pf-Avr4-F/Pf-Avr4-HindIII-R and SpeI-Pf-Avr4-F/Pf-Avr4-ApaI-R, respectively (Supplemental Figure 3 and Supplemental Table 1). The two amplified PCR products were subsequently double-digested with SalI/HindIII (the 400-bp fragment) and SpeI/ApaI (the 1-kb fragment) and cloned into pBht2KOI vector. Agrobacterium-mediated fungal transformation was conducted as described above in Methods (see section referring to the localization of Cf-Avr4 and Pf-Avr4 on fungal mycelia). Putative transformants were selected on half-PDA medium amended with 100 µg/mL of hygromycin B, whereas the integration of the HYG-cassette in the Pf-Avr4 locus was subsequently verified by PCR using the primer pair Pf-Avr4_F/Pf-Avr4_R (Supplemental Table 1). This primer pair amplifies in case of a successful targeted Pf-Avr4 replacement event a 2640-bp fragment, whereas in the absence of a homologous recombination event in this gene locus it amplifies a 792-bp fragment (Supplemental Figure 3). To further confirm the absence of any ectopic integrations of the HYG cassette in the background of a ΔPf-Avr4 deletion mutant, we followed a genome walking approach according to the manufacturer's instruction (Clontech Laboratories). Briefly, genomic DNA of the wild-type Pf-Avr4 strain and two ΔPf-Avr4 deletion mutants (ΔPf-Avr4-1 and ΔPf-Avr4-2) was digested with restriction enzymes EcoRV or PvuII and adaptors were ligated to the end of the digested DNA fragments. Primer pairs AP1/Pf-Avr4_GW-5′ and AP1/Pf-Avr4_GW-3′ (Supplemental Table 1) were used for amplifying DNA fragments upstream and downstream of HYG-cassette, respectively. Using these primers, only a single PCR fragment of the same size was obtained for the Pf-Avr4WT and the two ΔPf-Avr4 deletion mutants, indicating a single integration event. Further DNA sequencing of obtained PCR fragments verified the presence of a single integration event of the HYG cassette in the Pf-Avr4 locus and the absence of any additional ectopic integration events.
Virulence Assays on the Susceptible Tomato Cultivar LA3940
Inoculations on the susceptible tomato cv LA3940 with the wild-type P. fuligena strain (WT-Pf) as well as two ΔPf-Avr4 deletion mutants (ΔPf-Avr4-1 and ΔPf-Avr4-2) were performed with mycelial fragments, as described above in Methods (see section referring to the quantification of Pf-Avr4 expression levels). Briefly, mycelial fragments (105 fragments per mL) of each strain were sprayed on the leaves of three 6-week-old tomato plants, respectively. Three leaves from each inoculated plant were collected at 1, 3, 6, 9, 12, and 15 dpi and were immediately frozen in liquid nitrogen and then stored in −80°C until RNA extractions. Total RNA was isolated from inoculated leaves using the TRIzol reagent (Life Technologies) according to the manufacturer’s instructions and was subsequently treated with RNase-free DNase (ThermoFisher Scientific) in order to remove any residual contaminations by genomic DNA. cDNA was synthesized according to the protocol of the iScript cDNA synthesis kit (Bio-Rad Laboratories). Quantitative real-time PCR (qPCR) was conducted using SsoFast EvaGreen Supermix (Bio-Rad Laboratories), according to the manufacturer’s instructions, on a CFX96 Touch Real-Time PCR detection system (Bio-Rad Laboratories). First, expression of the fungal actin (Actin) under infection with the Pf-Avr4WT strain was quantified by qPCR, using primers Pf-Actin-F and Pf-Actin-R (Supplemental Table 1). Expression of the tomato RPL2 (ribosomal protein large subunit 2: PL2T) gene was used as an endogenous control. RPL2 was amplified with primers RPL2-F and RPL2-R (Supplemental Table 1). Real-time PCR conditions included an initial 95°C denaturation step for 10 min, followed by denaturation for 15 s at 95°C, annealing for 30 s at 60°C, and extension for 30 s at 72°C for a total of 40 cycles. For relative quantification of the fungal biomass produced by the ΔPf-Avr4-1 and ΔPf-Avr4-2 mutants compared with the WT-Pf strain, the expression of Pf-Actin in leaves inoculated with the WT-Pf strain was set to 100% and the expression of Pf-Actin in leaves inoculated with the ΔPf-Avr4-1 and ΔPf-Avr4-2 mutants was calculated relative to the WT-Pf. Virulence assays were repeated three times and each qPCR was repeated in triplicate as well.
Tryptophan Fluorescence-Based Binding Assays
Fluorescence measurements were performed on an Aminco Bowman Series 2 luminescence spectrometer (SIM-AMINCO Spectronic Instruments). Tryptophan residues were excited at 295 nm, and emission spectra were collected from 300 to 420 nm using an excitation and emission slit width of 4 nm. Fluorescence binding reactions were performed in a 10-mm path length quartz cuvette with a total reaction volume of 2.4 mL. Each binding reaction contained 4 μM protein (Cf-Avr4 or Pf-Avr4) in 50 mM potassium phosphate, pH 7.0, and 150 mM NaCl buffer. The temperature was controlled to 25°C ± 1°C with a Fisher Scientific Isotemp Refrigerated Circulator Model 900 circulating water bath (ThermoFisher Scientific). Initial intrinsic fluorescence measurements of unbound Pf-Avr4 and Cf-Avr4 were taken prior to the start of each titration experiment. Chito-oligosaccharides were then titrated into the cuvette by hand and allowed to equilibrate for 3 min, under constant stirring by a magnetic stir bar. An emission spectrum was collected upon each addition of chito-oligosaccharide. An equilibration time of 3 min between titration points was found to be sufficient, as the lectin-chito-oligosaccharide solution had achieved maximal fluorescence within this time. Chito-oligosaccharides were titrated to a maximum concentration of 2 mM because it was assumed that 2 mM, at 500× molar excess, was more than sufficient to detect binding, if binding was occurring. Chito-oligosaccharides tested were as follows: GlcNAc (Sigma-Aldrich), (GlcNAc)2-(GlcNAc)6 (Megazyme). Fluorescence emission spectra were corrected for background fluorescence due to lectin dilution, buffer, and chito-oligosaccharide concentration. The inner filter effect was determined to be negligible since absorbance readings taken at 295 nm of the titration end-point solutions were found to be <<0.05 in all cases (Ward, 1985). The fluorescence measurements were translated into corresponding saturation binding curves with the concentration (in mM) of chito-oligosaccharide on the x axis and change in fluorescence (ΔF) on the y axis. These plots were fit with a nonlinear least-squares regression using GraphPad Prism 6 software (GraphPad Software), enabling determination of the total change in fluorescence (ΔFmax) upon binding saturation and the concentration of chito-oligosaccharide required to achieve half-saturation (ΔFmax/2) of the lectin. The tryptophan fluorescence binding profiles shown in Supplemental Figure 7A are the mean titration values, which is the average of experiments collected in duplicate or triplicate. The se of the mean is also shown and was calculated as follows: (sd of experimental data points) divided by (the square root of the number of data points).
Crystallization, Structure Determination, and Homology Identification for Pf-Avr4
The sitting-drop vapor diffusion method was used to set up crystallization trials in 96-well plates. Three commercially available screens, JCSG (Qiagen), SaltRX, and Index (Hampton Research), were used for the initial screening. For each screen, 1.5 μL of 15 mg/mL of protein was mixed with the reservoir solution in a 1:1 ratio to form a 3-μL drop and the plates were sealed with tape. Initial crystallization hits were further optimized using 24-well sitting drop method. The best diffracting crystals were grown in 0.1 M HEPES, pH 7.5, and a range of ammonium sulfate from 1.6 to 2.0 M. All of the crystallization experiments were performed at room temperature and were kept and monitored in a temperature-controlled room at 21°C. Crystals belong the space group P3221 with cell parameters: a = 57.1Å, c = 126.6 Å, and contain two monomers per asymmetric unit with a Matthews coefficient of 2.19 Å3/D (Matthews, 1968).
Crystals were briefly soaked in a cryoprotectant composed of 30% (v/v) ethylene glycol in mother liquor then flash-cooled in liquid nitrogen. Experimental phases were determined using sulfur single-wavelength anomalous dispersion. A highly redundant data set was collected on a rotating copper anode x-ray source using Cu-Kα radiation (λ = 1.54 Å) using a single crystal (Table 2). Diffraction data were processed with the Proteum software package (Bruker AXS). The phases were determined and a partial model built using the PHENIX software package (Adams et al., 2010). The model building was completed manually using the program COOT (Emsley et al., 2010).
High-resolution diffraction data were collected at a wavelength of 1.12709 Å on the ADSC Quantum 315r detector at Beamline 7-1 of the Stanford Synchrotron Radiation Lightsource with a 100-μm focused beam. Images were recorded with a 0.3° oscillation and an exposure time of 10 s. Diffraction data were integrated and scaled with XDS and XSCALE (Kabsch, 2010b, 2010a). The structure from the rotating anode x-ray diffraction data was refined against the high synchrotron data using the program REFMAC5 (Murshudov et al., 2011). Data collection and refinement statistics are listed in Table 2. Structure images were generated in CCP4mg (McNicholas et al., 2011), and the secondary structure sequence alignment was generated in PDBsum (de Beer et al., 2014). Identification of PfAvr4 structural homologs was performed with the DaliLite v.3s server (Dietmann et al., 2001; Holm and Rosenström, 2010), using Chain A of the PfAvr4 structure as the query structure on July 6, 2015. This search identified two structural homologs, tachycitin (PDB: 1DQC) with a Z-score of 4.7 and Blo 1 12 (PDB: 2MFK) with a Z-score of 4.1. Z-scores above 2.0 are considered significant and usually indicate the presence of a similar fold (Holm et al., 2008).
Isolation of Intracellular Chitinases and Proteases from Tomato Whole Leaf Extracts
Whole tomato leaf protein extracts were prepared from 100 g of fresh cv Moneymaker leaves. Leaves were homogenized in water, and protein was purified using ammonium sulfate precipitation at 4°C. The resulting protein extract was dialyzed against water. The protein extract was filtered and concentrated prior to use in assays. For the protection assay, the protein extract was further purified using a G-50 column (Roche). Total protein was quantified by taking an A280 measurement with an absorbance of 1 equal to 1 mg/mL.
Colorimetric Determination of Whole Tomato Leaf Extract Chitinolytic Activity
To measure the chitinase activity of the whole tomato leaf extract, 7 mg of chitin azure (Sigma-Aldrich) was mixed with ∼400 μg of whole tomato leaf extract (total protein) in 100 mM sodium acetate pH 5.0 buffer. The total reaction volume was 100 μL, and 100 μg of bacterial chitinase (Sigma-Aldrich) was used as a positive control. Reactions were incubated at 25°C for 8.5 h shaking at 1200 rpm, and the chitin azure was pelleted at 14,000 rpm for 10 min prior to taking absorbance readings. An absorbance reading (A560) was taken on the supernatant fraction and samples were blanked against a chitin azure buffer only sample. Experiments were performed in duplicate.
Agrobacterium-Mediated Transient Coexpression of Avr4 Effectors and Cf-4
Agroinfiltration-based transient expression in N. benthamiana was performed as described previously (Ma et al., 2012). Binary expression vectors of pMOG800 (Van der Hoorn et al., 2000) with Cf-Avr4, Pf-Avr4WT, or Pf-Avr4 ChtBD mutants were constructed by cloning the Avr4 cDNA encoding for the mature proteins downstream of the PR1A signal sequence of Nicotiana tabacum for secretion into the apoplast and under the control of the CaMV 35S promoter and NOS terminator. Plasmids were transformed into Agrobacterium (GV3101) by electroporation. Agrobacteria containing binary vectors were grown in 3 mL LB medium with 50 μg/mL kanamycin and 25 μg/mL rifampicin at 28°C overnight. Ten microliters of initial culture was inoculated into 25 mL LB-mannitol medium (LB medium with 10 g/L mannitol) containing 50 μg/mL kanamycin and 25 μg/mL rifampicin under same growth condition for 2 d, and cultures were grown to an optical density (A600) of 0.8 to 2.0. After centrifuging agrobacteria at 2800g for 15 min, the pellet was resuspended in MMAi medium (5 g/L Murashige and Skoog basal salts, 20 g/L sucrose, 10 mM MES, and 200 μM acetosyringone) and incubated for 2 h at room temperature. Agrobacteria culture containing binary vectors of Cf-4 was mixed with agrobacteria containing binary vectors of Cf-Avr4, Pf-Avr4WT, or Pf-Avr4 ChtBD mutants at 1:2 (A6000.5:A6001.0), 1:1 (A6000.5:A6000.5), and 1:0.5 (A6000.5:A6000.25) ratios of Cf-4:Avr4. The mixtures of agrobacteria were infiltrated into the leaves of N. benthamiana plants using a 1-mL syringe. All HR images were taken after 6 dpi.
In Vitro Protease Sensitivity Determination of the Pf-Avr4 ChtBD Mutants
For treatment with whole leaf extract, 4 μg E. coli-produced Pf-Avr4WT and Pf-Avr4 ChtBD mutants (W88A, N89A, D90A, N91A, W94A, D96A, and W97A) were mixed with ∼90 μg of whole leaf plant extract in water and the reaction was incubated at room temperature for 2 h. The reaction was stopped by the addition of SDS-PAGE loading buffer and visualized by SDS-PAGE and Coomassie Brilliant Blue stain. For subtilisin treatment, 8 μg of E. coli-produced Pf-Avr4WT and Pf-Avr4 ChtBD mutants (W88A, N89A, D90A, N91A, W94A, D96A, and W97A) were mixed with 10 mM calcium chloride and 0.6% subtilisin protease (Sigma-Aldrich). Pichia-produced Pf-Avr4 and a disulfide-disrupted Pichia-produced Pf-Avr4 were used as negative and positive controls, respectively. Samples were incubated for 30 min at room temperature, and the digestion reactions were stopped with the addition of 10 mM PMSF and SDS-PAGE loading buffer. Samples were visualized by SDS-PAGE and Coomassie blue stain.
Isothermal Titration Calorimetry with Pf-Avr4 and Cf-Avr4
ITC experiments were performed on a NanoITC low volume calorimeter (TA Instruments) using a cell volume of 170 μL and stir rate of 270 rpm. Chito-oligosaccharide titrations were done with a 2.5 μL titration volume with a 3-min equilibration time between each titration point. The initial titration point was done with a 1 μL titrant volume, and this point was disregarded for model fitting. For all experiments, Pichia-produced Pf-Avr4 and Cf-Avr4 were dialyzed extensively against 50 mM potassium phosphate, pH 7.0, and 150 mM sodium chloride, and the chito-oligosaccharides were reconstituted in the same buffer to minimize mixing heats. For the (GlcNAc)3 titrations, the cell contained either Pichia-produced Pf-Avr4 or Cf-Avr4 at a concentration of 0.35 mM and the syringe contained 23 mM (GlcNAc)3 (Sigma-Aldrich). For the (GlcNAc)6 titrations, the cell contained either 0.1 mM Pichia-produced Pf-Avr4 or Cf-Avr4 and the syringe contained 2 mM (GlcNAc)6 (Megazyme). Data were corrected with a chito-oligosaccharide blank titration, which consisted of a titration of the respective chito-oligosaccharide into buffer at equal volumes and concentrations used for the experimental titrations, and was fit with an independent binding model using TA Instruments ITC analysis software. Experiments were performed in duplicate.
Accession Numbers
The nucleotide and corresponding amino acid sequence of Pf-Avr4 has been deposited in the NCBI GenBank (accession number KR108309). The Pf-Avr4 protein structure has been deposited in the protein data bank (PDB code: 4Z4A).
Supplemental Data
Supplemental Figure 1. The Pseudocercospora fuligena Avr4 effector protein (Pf-Avr4) is a member of the Avr4 core fungal effector family.
Supplemental Figure 2. Chitinolytic activity of the whole tomato leaf extract and of the bacterial chitinase, as measured by treatment of chitin azure at 25°C for 7.5 h.
Supplemental Figure 3. Targeted gene replacement of Pf-Avr4 by the hygromycin resistance cassette in the genome of Pseudocercospora fuligena.
Supplemental Figure 4. Virulence of the wild-type Pseudocercospora fuligena strain (WT-Pf) and two ΔPf-Avr4 deletion mutants (ΔPf-Avr4-1 and ΔPf-Avr4-2) on tomato cultivar LA3940.
Supplemental Figure 5. The ITC and raw tryptophan fluorescence binding profiles for Cf-Avr4 and Pf-Avr4.
Supplemental Figure 6. The solution behavior of Cf-Avr4 and Pf-Avr4 as determined by SEC-MALS analysis.
Supplemental Figure 7. Affinity of wild-type Pf-Avr4 and chitin-binding domain mutants for (GlcNAc)6 and their ability to provide protection against plant-derived chitinases.
Supplemental Figure 8. Pf-Avr4 chitin-binding domain mutants trigger a Cf-4 mediated hypersensitive response, although some at a protein concentration-dependent manner.
Supplemental Figure 9. The Pf-Avr4P83A mutant is partially susceptible to proteolysis by subtilisin and is recognized by Cf-4.
Supplemental Figure 10. The fluorescence behavior of Pf-Avr4D90A is unaffected by titration of sugars to which Pf-Avr4 does not display affinity.
Supplemental Table 1. Primers used for Pf-Avr4 gene expression analysis and targeted gene replacement.
Supplemental Movie 1. The x-ray structure of Pf-Avr4 rotating at a 360° angle on a vertical and horizontal axis.
Supplementary Material
Acknowledgments
This work was supported by the Research Investments in Science and Engineering (RISE) program of UC Davis (Award RI-091). B.S. was supported by a Human Frontier Science Program long-term postdoctoral fellowship (LT000674/2012). N.H. was supported by NIH training grant T32 GM007377. Portions of this research were carried out at the Stanford Synchrotron Radiation Light source, which is supported by the U.S. DOE under Contract DE-AC02-76SF00515. The SSRL-SMBP is supported by the DOE-BER and by the NIH, NIGMS (P41GM103393). The contents of this publication do not necessarily represent the official views of NIGMS or NIH. We acknowledge Jim Lincoln and Stephen Bolus for assistance during the early stages of this work. George Bruening, Savithramma Dinesh-Kumar, Gitta Coaker, and Dave Wilson are acknowledged for critical suggestions, comments, and reading of the manuscript.
AUTHOR CONTRIBUTIONS
A.C.K. and I.S. designed research, performed research, analyzed data, and wrote the article. L.-H.C. and N.H. performed research and analyzed data. A.S. and B.S. performed research. A.J.F. analyzed data and wrote the article.
References
- Adams P.D., et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong K.M., Piepenbrink K.H., Baker B.M. (2008). Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem. J. 415: 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M.D., et al. (2008). The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69: 119–136. [DOI] [PubMed] [Google Scholar]
- Boraston A.B., Bolam D.N., Gilbert H.J., Davies G.J. (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.M., Free S.J. (2006). The structure and synthesis of the fungal cell wall. BioEssays 28: 799–808. [DOI] [PubMed] [Google Scholar]
- Burstein E.A., Vedenkina N.S., Ivkova M.N. (1973). Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 18: 263–279. [DOI] [PubMed] [Google Scholar]
- Cesari S., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25: 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.W., Hutchinson E.G., Harris D., Thornton J.M. (1993). Identification, classification, and analysis of beta-bulges in proteins. Protein Sci. 2: 1574–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.L.B., Tsai G., Lee J., Fu W.R. (1997). Heterogeneous N-deacetylation of chitin in alkaline solution. Carbohydr. Res. 303: 327–332. [Google Scholar]
- Chang T.C., Stergiopoulos I. (2015a). Evolutionary analysis of the global landscape of protein domain types and domain architectures associated with family 14 carbohydrate-binding modules. FEBS Lett. 589: 1813–1818. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Stergiopoulos I. (2015b). Inter- and intra-domain horizontal gene transfer, gain-loss asymmetry and positive selection mark the evolutionary history of the CBM14 family. FEBS J. 282: 2014–2028. [DOI] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Enck S., Price J.L., Powers D.L., Powers E.T., Wong C.H., Dyson H.J., Kelly J.W. (2013). Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins. J. Am. Chem. Soc. 135: 9877–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen C., Abou Hachem M., Janecek S., Viksø-Nielsen A., Blennow A., Svensson B. (2009). The carbohydrate-binding module family 20--diversity, structure, and function. FEBS J. 276: 5006–5029. [DOI] [PubMed] [Google Scholar]
- Collemare J., Griffiths S., Iida Y., Karimi Jashni M., Battaglia E., Cox R.J., de Wit P.J.G.M. (2014). Secondary metabolism and biotrophic lifestyle in the tomato pathogen Cladosporium fulvum. PLoS One 9: e85877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T.A., Berka K., Thornton J.M., Laskowski R.A. (2014). PDBsum additions. Nucleic Acids Res. 42: D292–D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R., Thomma B.P.H.J. (2009). Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 17: 151–157. [DOI] [PubMed] [Google Scholar]
- de Wit P.J.G.M., et al. (2012). The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 8: e1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deber C.M., Brodsky B. Rath A. (2010). Proline residues in proteins. eLS, http://dx.doi.org/10.1002/9780470015902.a0003014.pub2. [Google Scholar]
- Dietmann S., Park J., Notredame C., Heger A., Lappe M., Holm L. (2001). A fully automatic evolutionary classification of protein folds: Dali Domain Dictionary version 3. Nucleic Acids Res. 29: 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Gueddari N.E., Rauchhaus U., Moerschbacher B.M., Deising H.B. (2002). Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol. 156: 103–112. [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen R.J.N., Andersson J.A., Kristensen M.B., Giese H. (2008). Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutelle S., Maurin M., Rougier F., Barbaut X., Bourguignon L., Ducher M., Maire P. (2008). The Hill equation: a review of its capabilities in pharmacological modelling. Fundam. Clin. Pharmacol. 22: 633–648. [DOI] [PubMed] [Google Scholar]
- Hashimoto H. (2006). Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 63: 2954–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Rosenström P. (2010). Dali server: conservation mapping in 3D. Nucleic Acids Res. 38: W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008). Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson K.L., Bartlett G.J., Diehl R.C., Agirre J., Gallagher T., Kiessling L.L., Woolfson D.N. (2015). Carbohydrate-aromatic interactions in proteins. J. Am. Chem. Soc. 137: 15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani A.J., Cotter P.A. (2010). Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8: 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Barbero J., Javier Cañada F., Asensio J.L., Aboitiz N., Vidal P., Canales A., Groves P., Gabius H.J., Siebert H.C. (2006). Hevein domains: an attractive model to study carbohydrate-protein interactions at atomic resolution. Adv. Carbohydr. Chem. Biochem. 60: 303–354. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Joosten M.H., Cozijnsen T.J., De Wit P.J. (1994). Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367: 384–386. [DOI] [PubMed] [Google Scholar]
- Joosten M.H.A.J., Vogelsang R., Cozijnsen T.J., Verberne M.C., De Wit P.J.G.M. (1997). The biotrophic fungus Cladosporium fulvum circumvents Cf-4-mediated resistance by producing unstable AVR4 elicitors. Plant Cell 9: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (2010a). Xds. Acta Crystallogr. D Biol. Crystallogr. 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (2010b). Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata S., Nagayama R., Hirata M., Shigenaga T., Agarwala K.L., Saito T., Cho J., Nakajima H., Takagi T., Iwanaga S. (1996). Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J. Biochem. 120: 1253–1260. [DOI] [PubMed] [Google Scholar]
- Lange J., Mohr U., Wiemken A., Boller T., Vögeli-Lange R. (1996). Proteolytic processing of class IV chitinase in the compatible interaction of bean roots with Fusarium solani. Plant Physiol. 111: 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Ma L., Lukasik E., Gawehns F., Takken F.L. (2012). The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol. Biol. 835: 61–74. [DOI] [PubMed] [Google Scholar]
- MacArthur M.W., Thornton J.M. (1991). Influence of proline residues on protein conformation. J. Mol. Biol. 218: 397–412. [DOI] [PubMed] [Google Scholar]
- Marshall R., Kombrink A., Motteram J., Loza-Reyes E., Lucas J., Hammond-Kosack K.E., Thomma B.P.H.J., Rudd J.J. (2011). Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156: 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B.W. (1968). Solvent content of protein crystals. J. Mol. Biol. 33: 491–497. [DOI] [PubMed] [Google Scholar]
- McNicholas S., Potterton E., Wilson K.S., Noble M.E. (2011). Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67: 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesarich C.H., Stergiopoulos I., Beenen H.G., Cordovez V., Guo Y., Karimi Jashni M., Bradshaw R.E., de Wit P.J. (2016). A conserved proline residue in Dothideomycete Avr4 effector proteins is required to trigger a Cf-4-dependent hypersensitive response. Mol. Plant Pathol. 17: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins E.D., Chen X., Romaine P., Raina R., Geiser D.M., Kang S. (2001). Agrobacterium-mediated transformation of Fusarium oxysporum: An efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91: 173–180. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Rooney A.P. (2005). Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39: 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A. (1996). Immune responses against multiple epitopes: A theory for immunodominance and antigenic variation. Semin. Virol. 7: 83–92. [Google Scholar]
- Ökmen B. (2013). Identification and Characterization of Novel Effectors of Cladosporium fulvum. PhD dissertation (Wageningen, The Netherlands: Wageningen University; ). [Google Scholar]
- Ota T., Nei M. (1994). Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol. Biol. Evol. 11: 469–482. [DOI] [PubMed] [Google Scholar]
- Philipp M., Bender M.L. (1983). Kinetics of subtilisin and thiolsubtilisin. Mol. Cell. Biochem. 51: 5–32. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., de Wit P.J. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47: 233–263. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., Kourmpetis Y.A., Slot J.C., Bakker F.T., De Wit P.J., Rokas A. (2012). In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins. Mol. Biol. Evol. 29: 3371–3384. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., van den Burg H.A., Okmen B., Beenen H.G., van Liere S., Kema G.H.J., de Wit P.J.G.M. (2010). Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA 107: 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetake T., Tsuda S., Kawabata S., Miura K., Iwanaga S., Hikichi K., Nitta K., Kawano K. (2000). Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif. J. Biol. Chem. 275: 17929–17932. [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Harrison S.J., Joosten M.H., Vervoort J., de Wit P.J. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19: 1420–1430. [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Spronk C.A., Boeren S., Kennedy M.A., Vissers J.P., Vuister G.W., de Wit P.J., Vervoort J. (2004). Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein-protein interactions: the chitin-binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin-binding domain. J. Biol. Chem. 279: 16786–16796. [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Westerink N., Francoijs K.J., Roth R., Woestenenk E., Boeren S., de Wit P.J., Joosten M.H., Vervoort J. (2003). Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278: 27340–27346. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn R.A.L., Laurent F., Roth R., De Wit P.J.G.M. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13: 439–446. [DOI] [PubMed] [Google Scholar]
- van Esse H.P., Bolton M.D., Stergiopoulos I., de Wit P.J., Thomma B.P. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20: 1092–1101. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V.G.A.A., Oliver R.P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 27: 196–206. [DOI] [PubMed] [Google Scholar]
- Ward L.D. (1985). Measurement of ligand binding to proteins by fluorescence spectroscopy. Methods Enzymol. 117: 400–414. [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Maqbool A., Banfield M.J. (2013). On the front line: structural insights into plant-pathogen interactions. Nat. Rev. Microbiol. 11: 761–776. [DOI] [PubMed] [Google Scholar]
- Wu L.C., Tuot D.S., Lyons D.S., Garcia K.C., Davis M.M. (2002). Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature 418: 552–556. [DOI] [PubMed] [Google Scholar]
- Zahn M., Teuber F., Bollig K., Horst W.J. (2011). Quantification of Pseudocercospora fuligena in tomato lines carrying introgressions from Solanum habrochaites using a qPCR assay. Plant Dis. 95: 394–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.