Abstract
Interest in highly parallelized analysis of single molecules and single cells is growing rapidly. Here we develop hydrogel-based virtual microfluidics as a simple alternative to complex engineered microfluidic systems for the compartmentalization of nucleic acid amplification reactions. We applied digital multiple displacement amplification (dMDA) to purified DNA templates, cultured bacterial cells, and human microbiome samples in the virtual microfluidics system and demonstrated recovery and whole-genome sequencing of single-cell MDA products. Our results from control samples showed excellent coverage uniformity and markedly reduced chimerism compared with single-cell data obtained from conventional liquid MDA reactions. We also demonstrate the applicability of the hydrogel method for genomic studies of naturally occurring microbes in human microbiome samples. The virtual microfluidics approach is a simple and robust method that will enable many laboratories to perform single-cell genomic analyses.
Applications from de novo genome discovery to dissecting genomic diversity in biomedicine1–6 are driving tremendous interest in highly parallelized analysis of nucleic acids at the level of single molecules and single cells. These ‘digital’ single-molecule and single-cell assays require parallel clonal amplification of individual nucleic acid templates to generate a sufficient number of genomic replicates for detection and/or sequence library construction. High-throughput assays are needed for accurate copy number analysis with absolute quantification of DNA sequences7,8 and fragments9 in genomics and prenatal diagnostics. Such assays are also needed to overcome nonspecific background for the detection of rare sequence targets8 in microbial communities and blood plasma-based diagnostics6. In the burgeoning single-cell analysis field1, high-throughput and high-fidelity single-cell whole-genome10–12 and whole-transcriptome13 amplification (WGA and WTA) are needed for the discovery and validation of new genomes2–4,14–16 and the analysis of genomic5,12,14,17 and functional heterogeneity.
A wide variety of approaches have been explored for the microfluidic compartmentalization of single molecules and single cells across a large number of small discrete reactors, including SBS plates11, high-density microfluidic arrays18, engineered lab-on-chip systems19–22, and multi-phase micro-droplet systems12,23–26(Supplementary Table 1 and Fig. 1a). However, existing methods require complex instrumentation and microfabricated consumables that prevent the broadest deployment of digital assays. The characteristics desired in single-cell processing systems are resistance to extrinsic contamination and cross-compartment mixing27, low reaction volume/high-throughput, stability under temperature changes, and compartment access for the addition and removal of reagents and samples. Finally, sequence data quality is an overriding concern in single-cell genomics, as biases and artifacts such as chimeric fragments commonly formed in WGA and WTA processes can severely impact results.
Figure 1.
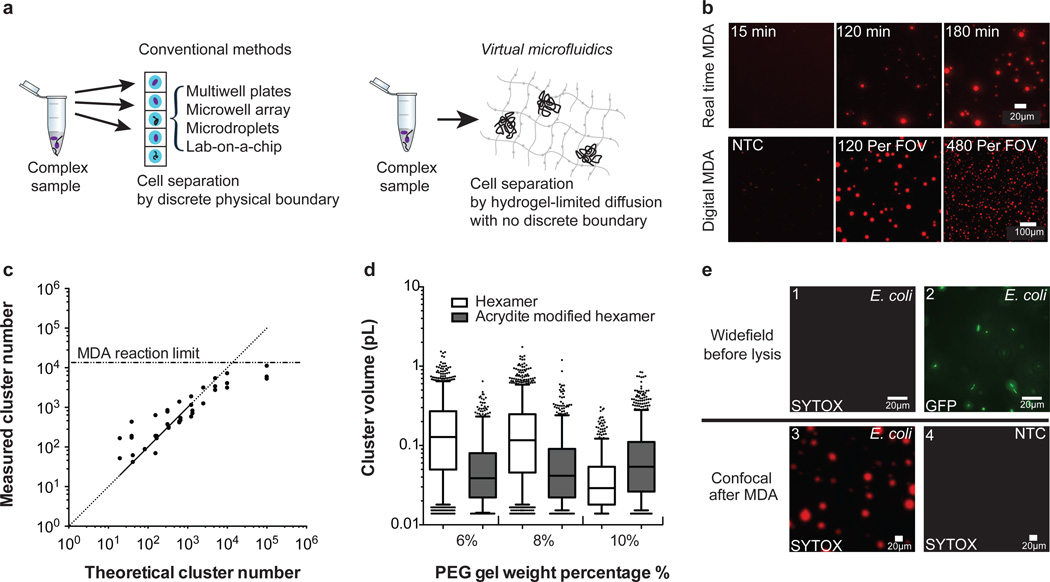
Application of Virtual Microfluidics for single molecule and single cell analysis. (a) Methods for single-cell analysis. Conventional approaches for single-cell analysis require discrete physical boundaries. Virtual microfluidics relies on hydrogel-limited diffusion to compartmentalize templates and reaction products. (b) Real time and digital MDA in PEG hydrogel. The top row shows time-lapse epifluorescence images (SYTOX Orange DNA stain) illustrating MDA cluster growth. Each template molecule gives rise to an amplified cluster of DNA in the gel. The bottom row demonstrates increasing DNA cluster number per 650 nm × 650 nm field of view (FOV) with increasing input template concentration. (c) Calibration curve illustrates linear relationship between template concentration and cluster number (n = 2 or 3 FOV at each concentration). (d) MDA cluster size is correlated with gel weight percentage and affected by acrydite modified hexamer anchorage. Data are shown as 5 %-95 % box plots with outliers scattered and center line for median. (n = 1334, 1587, 684, 704, 869, 1301). (e) Whole genome amplification of E. coli. 1) Before lysis, lab cultured E. coli are alive and are not detected by SYTOX Orange staining. 2) The same field of view shown where individual hydrogel-encapsulated E. coli are readily identifiable by GFP expression prior to MDA 3) After MDA, product clusters were readily identified by SYTOX Orange staining (different FOV than part 2). The high DNA content of clusters led to image saturation in this case. 4) No MDA product clusters were identified by SYTOX Orange staining after MDA in a negative control (no E. coli) sample.
Inspired by earlier work culturing microbes in hydrogels28, polymerase cloning in polyacrylamide gels using PCR29 and in agarose gels using MDA in conjunction with flow cytometry30, we developed and tested bulk poly(ethylene glycol) (PEG) hydrogels as a general and facile platform for compartmentalizing single molecules and single cells without discrete partitions. This approach, which we call virtual microfluidics (Fig. 1a), enables massively parallel single-molecule and single-cell amplification in virtual sub-divisions without the need for engineered micro-devices, multi-phase liquid systems, or instrumentation for cell sorting or microfluidics control. We selected hydrolytically degradable PEG hydrogels that covalently crosslink under mild conditions31. The chemically selective crosslinking reaction used in our method does not damage templates or inhibit subsequent reactions, and forms gels that are stable to high temperatures. The mesh size of PEG gels formed using 4-arm PEG crosslinkers is about 25 nm at 10 % (w/v)31, which allows diffusion of small molecules, oligonucleotides, and enzymes but immobilizes cells and high-molecular weight nucleic acids32. If desired, PEG gels can be functionalized33 to selectively immobilize low molecular weight species by attachment to the gel matrix.
Virtual microfluidics enables high throughput whole genome amplification and serial reagent exchange in an easy-to-use, benchtop format that requires no special equipment or environmental control. Here we first test the performance of in-gel amplification as an analytical method for molecular counting assays. Then we show preparative amplification and recovery of single bacterial genomes for ex-situ analysis of lab-cultured control cells and human gut microbes by next-generation sequencing (NGS).
MDA34 is a well-characterized WGA method commonly used to enable single-cell genome sequencing10,14–16,35. To evaluate the virtual microfluidics concept for WGA, we tested dMDA9,26 of purified Lambda phage DNA in the hydrogel format (Fig. 1bcd and Supplementary Fig. 1). Our estimate of 10 pg DNA content per cluster (Supplementary Fig. 2) suggests that the total MDA product concentration approached endpoint product concentrations typical of conventional liquid MDA reactions (~800 ng/µL)27. In addition to its use in preparative amplification, hydrogel microfluidics is also applicable as a simple, microfabrication-free platform for popular digital quantification assays such as digital PCR (Supplementary Fig. 3). We varied parameters in the virtual microfluidics system to test how in situ single-molecule MDA reactions can be controlled (Fig. 1d, Supplementary Fig. 4 and 5), observing that the smaller pore sizes in higher density gels limit the spread of DNA products.
We then applied digital MDA at the single-cell level using the virtual microfluidics system. Figure 1e shows the identification of individual log-phase E. coli cells in the hydrogel by fluorescence microscopy. We lysed the embedded cells by heat treatment and carried out MDA on the denatured genomic DNA, observing the appearance of MDA clusters at the reaction endpoint (Fig. 1e.3).
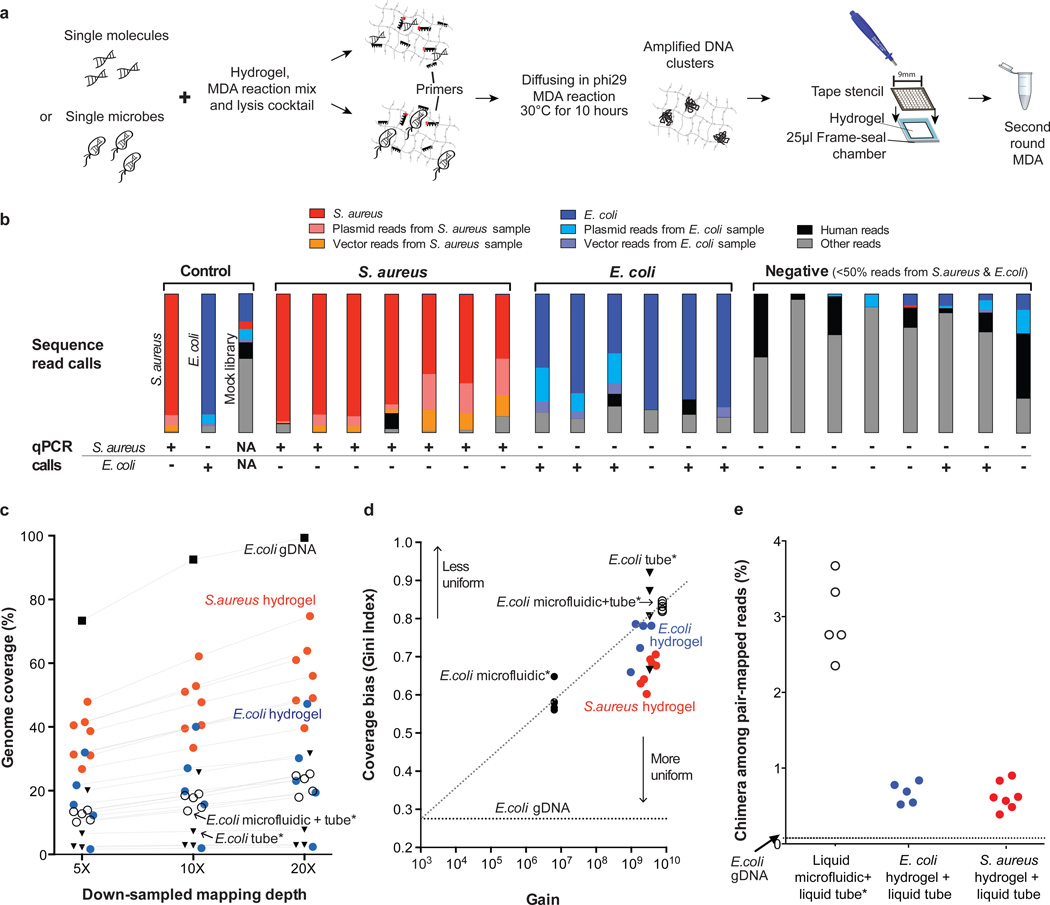
Next, we tested the potential of virtual microfluidics to support single-cell shotgun genome sequencing (Fig. 2). We mixed log-phase Escherichia coli (BL21) and Staphylococcus aureus subsp. aureus (RN6390/8325) strains at about 200,000 cells/mL and embedded the cells in a 300 micron thick PEG hydrogel. We used a mixed-input approach to ensure sensitive identification of any cross-contamination among single-cell samples and any contamination of single cell samples from other sources (including E. coli DNA contamination). The embedded cells were lysed by enzymatic and heat treatment, and MDA reagents were introduced by diffusion into the gel. 80 sub-samples from the gel (of 60 nL each) were recovered manually in a grid pattern as indicated in Figure 2a. Each punch sample was re-amplified to 109 – 1010 overall fold-amplification in a second-round 20 µL liquid MDA reaction. Real-time PCR (QPCR) assays for E. coli and S. aureus genome sequences were applied to diluted aliquots from each sample (Supplementary Table 2, Table 3). The QPCR results were well-approximated by a random cell dispersion model (Supplementary Note).
Figure 2.
Single-cell whole genome sequencing from E. coli and S. aureus hydrogel WGA samples. (a) Virtual microfluidics WGA workflow. (b) Sequence read classification using BLAST against the corresponding databases (Supplementary Note). The samples were ordered based on the ratio of S. aureus to E. coli reads from shotgun sequencing and the fraction of reads not from S. aureus or E. coli. Two negative samples are classified as false positive PCR calls. Positive S. aureus and E. coli samples with matching PCR calls are included in downstream analyses. (c) Genome coverage in S. aureus and E. coli hydrogel punch samples compared with published single-cell E coli data produced using conventional liquid MDA reactions*. One E. coli outlier library showed extremely poor genome coverage. This library had low complexity (37 % duplicate reads), which points to poor library quality rather than MDA as the cause for low genome coverage. (all samples were randomly down-sampled based on mapped reads and bootstrapped 10 times; error in all cases was smaller than the symbols plotted). (d) Coverage distribution bias. Gini Index (derived from Lorenz curve) reports the genome coverage bias of single-cell E. coli and S. aureus punches compared to the same published liquid-MDA single-cell E. coli data as a function of amplification gain. (e) Chimera frequency in the virtual microfluidics samples is significantly reduced versus published E. coli data produced using standard liquid MDA reactions.
* Indicates liquid MDA data from de Bourcy et al 2014.
We sequenced lllumina short-insert libraries produced from randomly selected punch samples and positive-control gDNA samples (MiSeq v2 500 cycles). Quality-filtered reads were then mapped to de novo assemblies of the positive-control gDNA datasets and sequence databases (Supplementary Table 4, Table 5, and Supplementary Fig. 6). The positive punch samples showed strong enrichment (Fig. 2b) for reads mapping to the expected reference genome (Supplementary Figure 7) while the negative punch samples showed enrichment for human reads, reads with poor mapping quality, and E. coli (possibly contaminants from the reagents and/or laboratory environment), and were similar to the results from a mock library (Supplementary Table 6). The lack of E. coli and S. aureus cross-contamination in the positive punch samples indicates that virtual microfluidics can resolve single-cell amplification products.
At 20× mean coverage (Supplementary Table 7), approximately 30 % of the E. coli genome and about 60 % of the S. aureus genome were covered in each single-cell sample (Fig. 2c). The coverage values for E. coli are in-line with typical single microbe genome sequencing at similar sequencing effort21,36 (Supplementary Note). The superior coverage performance in S. aureus may be attributable to the lower GC content of S. aureus (33 %) compared with E. coli (51 %), better accessibility (deproteination) of the genome after lysis, and/or higher average genome equivalents per cell in S. aureus resulting from cell cycle dynamics.
To rigorously evaluate sequence coverage distribution, we calculated the Gini Index (a measure of inequity ranging from 0 to 1) for each of our single-cell datasets and previously published single-cell E. coli liquid MDA datasets for which raw read data were available and fold-amplification was known21 (Fig. 2d). The coverage uniformity in our single-cell punch samples compares favorably with published single-cell datasets at similar amplification gain.
We then analyzed the occurrence of chimeric reads (Supplementary Note), which are known to occur with high frequency in MDA by a cross-priming mechanism37. Chimeric reads directly confound de novo assembly, analysis of rearrangements, and mapped read counting. Our single-cell datasets contained about 0.5 % chimeric reads, approximately five-fold lower than previously published short-read datasets produced using liquid single-cell MDA samples (Fig. 2e and Supplementary Table 4). The occurrence of chimeric reads spanning more than 10 kb of the template is even lower (about 0.1 %, Supplementary Fig. 8), raising the possibility of extracting long-range information from single-cell MDA samples using long-read sequencing and other technologies. This dramatic reduction in the occurrence of chimeric reads can be understood by restricted diffusion of the MDA intermediates that prevents cross-priming by isolating each portion of the product mixture. It may be the case that substantially all of the chimeras we observed in the punch samples were generated during the liquid-phase secondary amplification reactions. Based on these results, it is likely beneficial to run MDA in PEG hydrogels for all applications at all scales.
Next, we tested the potential of virtual microfluidics for single-cell genome sequencing using samples from the Fiji Community Microbiome project (FijiCOMP). The FijiCOMP samples contain a vast uncharacterized diversity of microbial species that differ from those found in the microbiome of Western subjects. The procedure for processing these human stool samples was similar to those described above for lab-cultured E. coli and S. aureus, with modifications for initial sample processing and lysis (online methods).
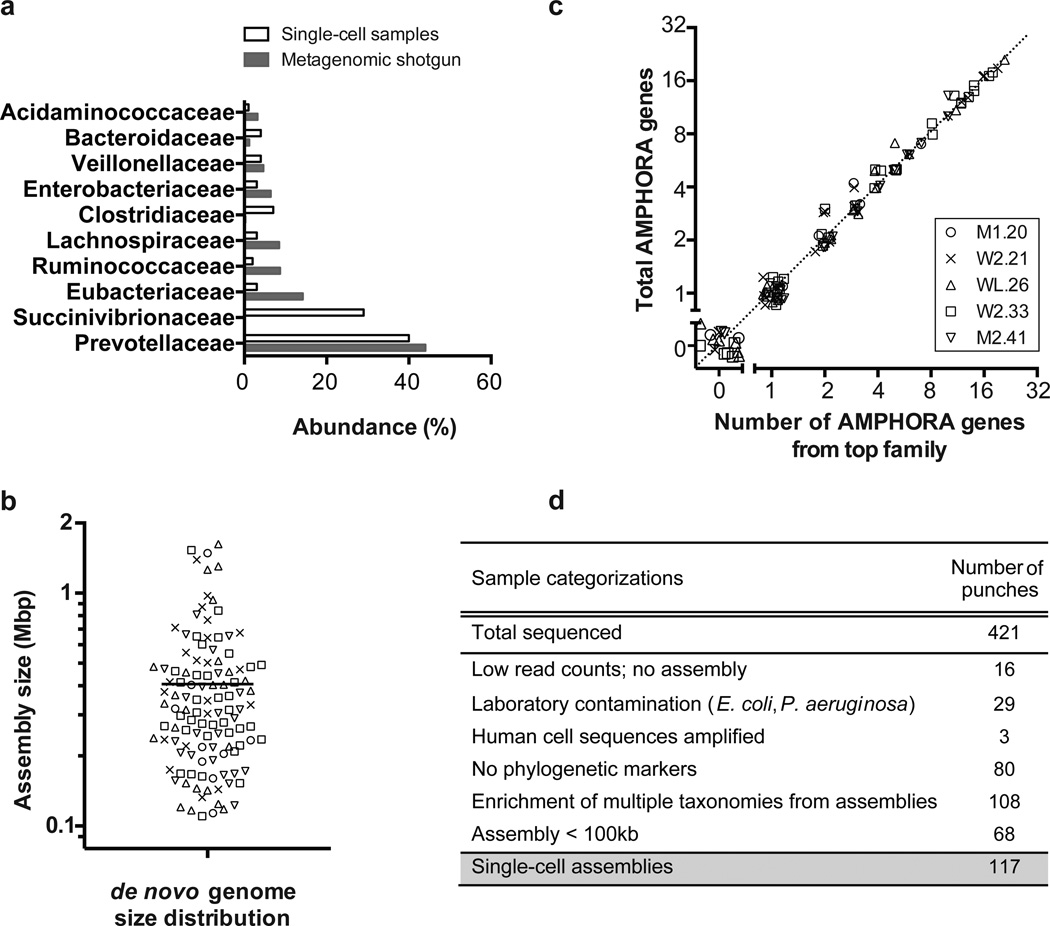
We processed a total of 421 hydrogel punch samples and compared the distribution of organisms detected in our hydrogel samples with the distribution observed from shotgun metagenomic profiling, which showed that the same top microbial families were observed using both approaches (Fig. 3a and Supplementary Table 8). Interestingly, the second most abundant microbial family found in the single-cell dataset, the Succinivibrionaceae, was not initially detected (but later confirmed) in the shotgun metagenomic data using standard methods for taxonomic assignment such as MetaPhlAn38. This discrepancy highlights the importance of unbiased approaches like single-cell analysis for organisms that are less well represented in reference databases.
Figure 3.
Fiji microbiome project single-cell whole genome sequencing (n = 117 single-cell datasets). (a). Single-cell and metagenome shotgun taxonomies. Here we compared the distributions of taxonomic families from single-cell assemblies that originated from 5 different donor individuals, with the distributions of microbial families determined from metagenome shotgun sequencing (Samples were weighted according to the number of single cells analyzed from each stool sample, see Supplementary Table 8). We were able to capture single-cell datasets from at least 9 of the top 10 bacterial families observed by shotgun metagenomic analysis of the Fijian samples. (b) de novo assembly size ranging from 100 kbp to 2 Mbp were obtained from single-cell sequencing data. The line indicates the mean assembly size. (c) AMPHORA gene analysis. We used BLAST to identify phylogenetically conserved single copy marker genes (AMPHORA genes) in the single-cell samples. Here we show the total number of AMPHORA genes and the number of AMPHORA genes from the top phylogenetic family for each sample. That these values are approximately equal (dotted line) supports the assertion that each dataset arises from an individual bacterial cell. A Gaussian-distributed random jitter (µ = 0, σ2 = 0.1) was added to enhance the visualization of these results. (d) Overview of FijiCOMP single-cell hydrogel samples.
We carried out de novo assembly of the single-cell datasets and assigned taxonomy to ribosomal gene sequences and 31 “single copy” bacterial marker genes (at the family level)39(Supplementary Table 9). This analysis enabled us to make crude assessments of genome coverage and sample purity in the FijiCOMP single-cell datasets (for which we lack strain-specific bona fide reference sequence). Of the 293 assemblies (up to 12 Mbp), we classified 117 as single amplified genomes with assembly size greater than 100 kb and strong enrichment of sequences from a single taxonomy (Fig. 3bc and see Fig. 3d for the fate of all samples). Overall, the data quality observed from these human microbiome bacteria was consistent with the results of our studies with lab-cultured Gram-negative and Gram-positive samples and demonstrates the applicability of the hydrogel method to real-world samples, including lysis and amplification of a variety of naturally occurring microbes.
Virtual Microfluidics offers a broadly applicable method for single-cell analysis that does not requires special equipment or environmental controls to produce useful single-cell genome sequence data. For sensitive applications, extra measures against contamination (such as use of a laminar flow cabinet, separate pre/post amplification laboratories and equipment) can further reduce contamination.
Virtual microfluidics enables high-throughput digital assays and preparative single-cell whole-genome amplification without microfabricated consumables or expensive instrumentation (instrumentation for sequence data generation is still required). The primary throughput limitation in this initial demonstration is the volume sub-sampled (60 nL) when product clusters are retrieved, which limits the number of sub-samples that can be retrieved from a single hydrogel. Throughput could be increased by using a thinner gel with more surface area and/or reducing the punch size from the 500 µm and 1 mm diameters we employed here. The fraction of retrieved samples containing a single-cell WGA reaction product can be improved by using image data to guide product retrieval. The thin hydrogel format affords excellent physical access for imaging, equipment, and reagents, which enables an assortment of sub-sampling approaches including punch/pickers, localized hydrogel dissolution, and localized affinity tagging or barcoding. Barcoding approaches could conceivably enable retrieval of all amplified products en masse while allowing in silico demultiplexing to sort sequence reads according to cell of origin.
Virtual microfluidics establishes a new paradigm in single-molecule and single-cell analysis with dramatically different characteristics than established microfluidic approaches. Besides reducing the production of chimeras in MDA, the unique physical characteristics of the engineered hydrogel environment may provide a means for enhancing coverage extent and uniformity from WGA and WTA samples through the self-limiting reactivity within each virtual compartment, similar to a recently reported emulsion approach12. In addition, the straightforward addition and removal of reagents to/from product clusters en mass and excellent optical access ideally suit the virtual microfluidics system for rare-cell assays incorporating in situ labeling of cells or product clusters (Supplementary Note). We expect that virtual microfluidics will find application as a low-cost digital assay platform and as a high-throughput platform for single-cell sample preparation.
ONLINE MATERIALS AND METHODS
Cross-linked PEG hydrogel formation
Hydrogel components, including 4-arm PEG acrylate (MW 10,000) and HS-PEG-SH (MW 3,400), were obtained from Laysan Bio. For every 25 µL of 10 % (wt/v) cross-linked hydrogel, 1.6 mg of 4-arm PEG acrylate and 1.1 mg of HS-PEG-SH were dissolved in pH 7.4 PBS (Invitrogen). It was briefly vortexed and centrifuged to ensure mixing and it was allowed to sit on bench for 10 min while the hydrogel components cross-linked through the reaction between the thiol and acrylate groups.
In-gel PCR
The primers (Supplementary Table 3) used for PCR on purified λ DNA (48 kbp, NEB) were ordered through IDT. A 25 µL hydrogel PCR reaction consisted of 2 U of VentR(exo-) polymerase (NEB), 1× ThermoPol Reaction Buffer (NEB), 0.4 mM dNTP (NEB), 1 µM Primers, 5 % DMSO (Sigma), 0.5 mg/mL BSA (NEB), 1.6 mg 4-arm PEG acrylate in PBS, 1.1 mg HS-PEG-SH in PBS, and λ DNA template (NEB) of various concentrations. The 25 µL above components were loaded in a 9 mm by 9 mm frame-seal chamber (Bio-rad). The following thermal protocol was ran on an MJ Research PTC-100 twin tower thermal cycler: 30 °C for 30 min (gel polymerization), 98 °C for 3 min; 98 °C for 30 sec, 57 °C for 30 sec, 72 °C for 1 min for 40 to 60 cycles; 72 °C for 5 min and hold at 4 °C. The gel was stained with 500 nM SYTOX Orange nucleic acid dye (Invitrogen).
In-gel MDA
A 25 µL hydrogel MDA reaction consisted of 0.5 µL of REPLI-g sc Polymerase (Qiagen), 1× phi 29 buffer (NEB), 50 µM Random Hexamers (IDT; including two phosphorothioate bonds at 3’ terminus), 2.5 % DMSO, 0.4 mM dNTP, 0.5 mg/mL BSA, 500 nM SYTOX Orange (Invitrogen) and denatured λ DNA. Λ DNA was denatured (using alkaline buffer “D1”, Qiagen) and neutralized (buffer “N1”, Qiagen) according to Qiagen REPLI-G sc kit protocol prior to hydrogel encapsulation. All MDA and gel components, except polymerase and SYTOX Orange dye, were UV treated for 30 min using the Stratalinker UV crosslinking instrument (Stratagene) to render contaminating background DNA incompetent for MDA. The 25 µL reaction mixture was loaded in a 9 mm by 9 mm frame-seal chamber (Bio-rad, about 300 µm in height). The gel was sealed in the chamber with a plastic cover and maintained at 30 °C for 8 hours or longer in the MJ Research PTC-100 twin tower thermal cycler. After the reaction, we imaged the gel using Nikon ECLIPSE Ti inverted microscope or Nikon ultra-fast laser scanning confocal microscope (MIT Koch Institute Microscopy Core Facility).
In-gel real-time dMDA
MDA hydrogel reactions were set up as described above and conducted at room temperature for 6 hours on a Nikon ECLIPSE Ti Epi-Fuorescence Microscope excited with a Lumencor Spectra X light engine (Lumencor) with fluorescent emissions filtered through a SpGold filter (Semrock). MATLAB was used to capture time-lapse image stacks through a Nikon 20×/0.4 NA objective and Hamamatsu C11440 camera with 15 min intervals, 100 ms exposure time, and 10 % Lumencor excitation power. All samples were stained with 500 nM SYTOX Orange. Each E. coli MDA cluster or mammalian cell image stack was cropped and processed as described below.
In-gel single-microbe MDA - cultured E. coli and S. aureus
Antibiotic resistant Staphylococcus aureus subsp. aureus (GFP) NCTC 8325 and Escherichia coli (RFP) BL21 strains were obtained as cryogenic stocks. For each culture, the frozen stock was inoculated in 5 mL LB broth and cultured at 37 °C overnight. 10 µL of 25 mg/mL Chloramphenicol was added to S. aureus culture and 5 µL of 50 mg/mL Ampicillin was added to the E. coli culture. 50 µL and 20 µL of each overnight culture were added to fresh 5 mL LB broth with the respective antibiotic concentration. After two hours incubation (to achieve exponential growth phase), 1 mL of each culture (O.D. 600 nm = 0.2) were centrifuged for 2 min at >10 krpm and the pellet was washed with 500 µL PBST (1% Tween-20) twice. The equal ratio mixture of microbes were diluted to 206,000 cells/mL and 1 µL of each was encapsulated in the same hydrogel sample to produce an average of less than 1 microbe per 500 µm diameter view. In addition to hydrogel MDA reaction mix described above, lysozyme (Sigma, final concentration 2.5 mg/mL) and lysostaphin (Sigma, final concentration 0.1 mg/mL) were added to the mix. The hydrogel was left at RT to let crosslink for 20 mins and cross-linked hydrogels were incubated at 37 °C for 1 hour for microbe lysis and heated to 95 °C for 5 min to denature genomic DNA before rapid quenching on ice. 1 µL of REPLI-g sc Polymerase (Qiagen) diluted in 2 µL water was then added on top of the hydrogel and allowed to diffuse into the gel. Next, the gel chamber is resealed and MDA was conducted for 10 hours. After the MDA reaction, the sample was heated to 65 °C for 5 min to deactivate phi29 polymerase.
In-gel single-microbe MDA - human gut microbiome samples
We received ethics approvals for human subjects research from the Columbia University IRB, Massachusetts Institute IRB, Broad Institute IRB, and two research ethics committees in Fiji: HRERC at CMNHS, FNU and FNHRERC at MoHFiji Ministry of Health. Fiji Community Microbiome Project (FijiCOMP) study participants from 5 agrarian villages within the Fiji Islands provided stool samples stored in 20% glycerol within 30 minutes of voiding and were frozen at −80 °C (Five participants: M1.20, W2.21, WL.26, W2.33, M2.41) (Supplementary Table 8). 10 µL of thawed cells were resuspended in 500 µL PBST (0.1 %). Samples were sonicated for 20 seconds and filtered through 35 µm Nylon mesh and 5 µm membrane (Pall Corp.) to collect filtrate with a 500 µL PBST wash. Samples were further diluted 1 to 500 ~ 1 to 2000 fold in PBST to reach the final concentration of ~30 cells/µL. The diluted cell samples (2 µl) then underwent alkaline lysis (1.5 µL D2 buffer) for 15 minutes at room temperature, after which the solution was neutralized (1.5 µL Stop solution). Hydrogel monomer mix (1.3 mg 4-Arm PEG Acrylate and 0.9 mg SH-PEG-SH) and MDA master mix were pipetted gently down the wall of each sample tube. MDA master mix includes 1× phi 29 buffer (NEB), 50 µM Random Hexamers with two phosphorothioate bonds at 3’ terminus, 2.5 % DMSO, 0.4 mM dNTP, 0.5 mg/mL BSA, 500 nM SYTOX Orange (Invitrogen) and 1 µL REPLI-g SC Polymerase (Qiagen). Only gentle tapping was used to ensure reagent mixing, in order not to disrupt the lysed and denatured microbial genomes. 25 µL of each microbial suspension was added into a frame-seal chamber, the sealed chamber was incubated at 30 °C for 12 hours, followed by 65 °C for 5 mins.
Image acquisition and analysis
Z stack images were taken by Nikon ultra-fast laser scanning confocal microscope with pinhole = 1.2, HV = 112, offset = 0, laser wavelength = 561 nm, laser power = 1.3 to 1.5, using a 20× objective on Galvano mode. Acquisition speed was 1 frame/sec and z step size was 0.95 µm. On the inverted microscope, z stack images were taken with the exposure time 100 ms, Lumencor excitation power 10 %, binning size 2 and z step size 10 µm. Both z Stacks were first processed into max intensity projections in FIJI(FIJI is just image J). Max projection tif files were then loaded into MATLAB. Background was obtained by applying a Gaussian filter of hsize 200 and sigma 50. All max projections were background-subtracted and thresholded at 2× – 2.5× standard deviations above the mean intensity. Cluster count, cluster area (radius), and cluster mean intensity were obtained with the bwconncomp and regionprops functions.
For whole gel (25 µL, 9mm by 9mm) microbe density approximation, we imaged the gel with a 4× objective in a 5 × 5 grid with a 31 % overlap. The 25 images were stitched using the FIJI stitching function. Fluorescent DNA clusters were counted and only gels with the appropriate clusters’ range and dispersion (60~80 per gel) were selected for hydrogel cluster retrieval. Images of the sampled locations were acquired but not used to guide sampling, sample preparation, or data analysis in this case.
MDA product cluster retrieval
In order to identify and retrieve a regular array of punches (not guided by cluster image data), we produced a tape stencil to guide the punch tool (Adhesive Applications High Tack Silicone Film Tape). We laser cut the double-sided tape with a 9 × 9 array of 500 µm diameter circles that has a center-to-center distance of 947 µm (Full Spectrum Laser LLC MLE-40). The tape stencil was applied on top of the frame seal plastic cover. The gel was peeled off the glass slide by allowing it to adhere to the plastic cover. The gel is then punched with a 1 mm diameter steel punch (Militek) and the micro-samples collected in a 96 well lobind twinteck plate (Eppendorf). The steel punch was cleaned with bleach and 70 % Ethanol after each use.
Secondary liquid MDA and PCR screening - cultured E. coli and S. aureus
The retrieved hydrogel punch (approximately 0.06 µL of hydrogel and 10 pg of DNA if a cluster was captured) was dissolved and denatured in 1 µL of 1 M KOH with 0.1 mM EDTA and 0.1 M DTT at 72 °C for 10 min before neutralization in 1 µL stop solution (Qiagen REPLI-g single cell kit). The neutralized product was added to 12.5 µL REPLI-g sc reaction mix with 1 µL of phi29 polymerase. The secondary MDA reaction was incubated for 10 hours before polymerase deactivation at 65 °C for 5 min. The DNA products from MDA were cleaned by the SPRI procedure in 1.8:1 beads to DNA volume (Beckman Coulter). Each sample was analyzed for the presence of S. aureus and E. coli marker loci by four sets of primers in standard qPCR reactions with Jumpstart Taq 2× ready mix (Sigma Aldrich), 1× Evagreen (Biotium), 1× ROX (Invitrogen) and 1 µM primers in Stratagene M3005. Both melting curve analysis and agarose gel electrophoresis (not shown) were used to support the QPCR results.
Secondary In-gel MDA - human gut microbiome samples
Hydrogel punches (approximately 0.24 µL of hydrogel and 10 pg of DNA if a cluster was captured) were dissolved and denatured in 1 µL of 400 mM KOH with 0.1 mM EDTA and 0.1 M DTT at 72 °C for 10 min before neutralization in 1 µL stop solution (Qiagen REPLI-g single cell kit). The neutralized product was added to 8 µL hydrogel and MDA master mix to reach a final volume of 10 µL for second round MDA reaction in hydrogel. The MDA reaction was incubated for 10 hours at 30 °C before polymerase deactivation at 65 °C for 5 min. The 10 µL gel was dissolved with 10 µL 400 mM KOH for 5mins at 72°C, and then neutralized with 6.6 µL 2.5% acetic acid.
WGS library construction & sequencing
We quantified the purified MDA products using the Qubit/Quant-IT HS assay (Thermo Fisher Scientific) and normalized samples to 5 ng/µL. All SPRI procedures were conducted on the Bravo robotic system (Agilent Technologies). 5 ng of purified DNA was then added to 1 µL of 5× tagmentation DNA buffer, 2 µL H2O and 1 µL Nextera Tagmentation DNA enzyme (Illumina). The mixture was first incubated at 58 °C for 10 min. With the addition of 0.5 µL of 1 % SDS, it was then incubated at 68 °C for 10 min, 4 °C for 3 min and 25 °C for 3 min to stop the tagmentation reaction. Another SPRI clean-up was carried out, followed by PCR library barcoding using Index primer N7 and S5 (Illumina) with the thermal protocol: 72 °C for 3 min, 98 °C for 30 sec, 12 cycles of 98 °C for 10 sec, 60 °C for 30 sec, 72 °C for 30 sec and a 5 min final extension at 72 °C. Samples were barcoded uniquely in the PCR step using standardized custom sample barcodes (Broad Institute Genomics platform). The PCR products were purified with SPRI twice with 1:1 beads to DNA volume and library quantification was carried out with the Quant-It assay (Thermo Fisher Scientific) and the KAPA library quantification kit (KAPA Biosystems). Library normalization and pooling were conducted on the Janus Mini Varispan workstation (PerkinElmer). For E. coli and S. aureus samples, an average of 0.7 million paired-end reads were allocated for each sample in a MiSeq 500 cycle v2 run (Illumina). For stool samples, about 1 M reads (> 50×) were allocated to each sample on HiSeq 2500 2×101/125 runs (Illumina). See Supplementary Note and Figures for all details regarding sequencing analysis.
Supplementary Material
Table 1.
overview of FijiCOMP single-cell hydrogel samples
| Sample categorizations | Number of punches |
|---|---|
| Total sequenced | 421 |
| Low read counts; no assembly | 16 |
| Laboratory contamination (E. coli, P. aeruginosa) | 29 |
| Human cell sequences amplified | 3 |
| No phylogenetic markers | 80 |
| Enrichment of multiple taxonomies from assemblies | 108 |
| Assembly < 100kb | 68 |
| Single-cell assemblies | 117 |
Acknowledgments
The authors thank S. Kim, D. Feldman and A. Kulesa for advice on bioinformatics, microscopy and image analysis, N. Ranu and the Hung lab (Broad Institute) for bacterial samples, L. Griffith, G. Lagoudas, J. Borrajo and L. Morinishi for helpful discussions, and members of Griffith lab (MIT) especially C. Chopko, J. Valdez and H. Lee for hydrogel expertise. This work was supported in part by a Lawrence Summers Fellowship from the Broad Institute (L.X.), a Career Award at the Scientific Interface from the Burroughs Welcome Fund (P.C.B.), and grants from: the Center for Microbiome Informatics and Therapeutics at MIT, a National Human Genome Research Institute, grant number U54HG003067 to the Broad Institute, the Center for Environmental Health Sciences at MIT, and the Fijian Ministry of Health.
Footnotes
ACCESSION CODES
Raw sequencing data on E. coli and S. aureus are accessible at the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA279815 with BioSample accession numbers SAMN03451478-SAMN03451501. FijiCOMP metagenomic reads can be found under BioProject accession number PRJNA217052 with the accession numbers: SRX345831, SRX344363, SRX344765, SRX343094, SRX344442, SRX346405, SRX343839, SRX343780, SRX345901, SRX344600, SRX343866, SRX343411, SRX344189, SRX344380, SRX346966, SRX345329, SRX343800, and SRX344616. FijiCOMP virtual microfluidics 117 single cells are accessible with the BioSample accession numbers SAMN04461233-SAMN04461349.
AUTHOR CONTRIBUTIONS
P.C.B. and L.X. conceived the concept for this study. L.X. designed and implemented experiments and conducted data analysis. I.L.B. and E.J.A. provided Fiji microbiome samples. I.L.B. conducted analysis of gut microbe data. L.X., P.C.B. and I.L.B. wrote and all authors approved the manuscript.
DISCLOSURE DECLARATION (including any conflicts of interest)
The Broad Institute and MIT may seek to commercialize aspects of this work and related applications for intellectual property have been filed.
CODE AVAILABILITY
All custom MATLAB functions, python code, and shell scripts are included in the supplementary software zip file.
REFERENCES
- 1.Blainey PC, Quake SR. Dissecting genomic diversity, one cell at a time : Nature Methods : Nature Publishing Group. Nat Meth. 2013;11:19–21. doi: 10.1038/nmeth.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall IPG, Blainey PC, Spormann AM, Quake SR. A Single-cell genome for Thiovulum sp. Appl. Environ. Microbiol. 2012;78:8555–8563. doi: 10.1128/AEM.02314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin. Chem. 2015;61:79–88. doi: 10.1373/clinchem.2014.221366. [DOI] [PubMed] [Google Scholar]
- 7.Sykes PJ, et al. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 8.Vogelstain B, Kinzler KW. Digital Pcr. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blainey PC, Quake SR. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Research. 2011;39:e19–e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghunathan A, et al. Genomic DNA Amplification from a Single Bacterium. Appl. Environ. Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y, et al. Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proceedings of the National Academy of Sciences. 2015;112:11923–11928. doi: 10.1073/pnas.1513988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Gelder RN, et al. Amplified RNA Synthesized from Limited Quantities of Heterogeneous cDNA. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodsworth JA, et al. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat Comms. 2013;4:1854. doi: 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess M, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 17.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love KR, Bagh S, Choi J, Love JC. Microtools for single-cell analysis in biopharmaceutical development and manufacturing. Trends in Biotechnology. 2013;31:280–286. doi: 10.1016/j.tibtech.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 20.Landry ZC, Giovanonni SJ, Quake SR, Blainey PC. Methods in Enzymology. Vol. 531. Elsevier; 2013. pp. 61–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bourcy CFA, et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS ONE. 2014;9:e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsen T, Roberts RW, Arnold FH, Quake SR. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 24.Mazutis L, et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc. 2013;8:870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hindson CM, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Meth. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morinishi LS, Blainey P. Simple Bulk Readout of Digital Nucleic Acid Quantification Assays. JoVE. 2015:e52925–e52925. doi: 10.3791/52925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blainey PC. The future is now: single-cell genomics of bacteria and archaea - Blainey - FEMS Microbiology Reviews - Wiley Online Library. FEMS Microbiol. Rev. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podar M, Keller M, Hugenholtz P. Uncultivated Microorganisms. Vol. 10. Berlin Heidelberg: Springer; 2009. in; pp. 241–256. [Google Scholar]
- 29.Mitra RD, Church GM. In situ localized amplification and contact replication of many individual DNA molecules. Nucleic Acids Research. 1999;27:e34–e39. doi: 10.1093/nar/27.24.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen LZ, et al. Single virus genomics: a new tool for virus discovery. PLoS ONE. 2011;6:e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raeber GP, Lutolf MP, Hubbell JA. Molecularly Engineered PEG Hydrogels: A Novel Model System for Proteolytically Mediated Cell Migration. Biophysical Journal. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, Joseph S, Aluru NR. Effect of cross-linking on the diffusion of water, ions, and small molecules in hydrogels. J. Phys. Chem. B. 2009;113:3512–3520. doi: 10.1021/jp808145x. [DOI] [PubMed] [Google Scholar]
- 33.Phelps EA, et al. Maleimide Cross-Linked Bioactive PEG Hydrogel Exhibits Improved Reaction Kinetics and Cross-Linking for Cell Encapsulation and In Situ Delivery. Adv. Mater. 2011;24:64–70. doi: 10.1002/adma.201103574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. pnas.org. 2002 doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcy Y, et al. Dissecting biological ‘dark matter’ with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woyke T, et al. Decontamination of MDA Reagents for Single Cell Whole Genome Amplification. PLoS ONE. 2011;6:e26161. doi: 10.1371/journal.pone.0026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasken RS, Stockwell TB. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol. 2007;7:19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segata N, Waldron L, Ballarini A, Narasimhan V. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature. 2012 doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.