Abstract
Organs-on-a-chip has emerged as a powerful tool for pharmacological and physiological studies. A key part in the construction of such a model is the ability to pattern or culture cells in a biomimetic fashion. Most of the reported cells-on-a-chip models integrate cells on a flat surface, which does not accurately represent the extracellular matrix that they experience in vivo. Electrospinning, a technique used to generate sub-micron diameter polymer fibers, has been used as an in vitro cell culture substrate and for tissue engineering applications. Electrospinning of fibers directly into a fully sealed fluidic channel using a conventional setup has not been possible due to issues of confining the fibers into a discrete network. In this work, a dynamic focusing method was developed, with this approach enabling direct deposition of electrospun fibers into a fully sealed fluidic channel, to act as a matrix for cell culture and subsequent studies under continuous flowing conditions. Scanning electron microscopy of electrospun polycaprolactone fibers shows that this method enables the formation of fibrous layers on the inner wall of a 3D-printed fluidic device (mean fiber size = 1.6 ± 0.6 μm and average pore size = 113 ± 19 μm2). Cells were able to be cultured in this 3D scaffold without the addition of adhesion proteins. Media was pumped through the channel at high flow rates (up to 400 μL/min) during a dynamic cell culture process and both the fibers and the cells were found to be strongly adherent. A PDMS fluidic device was also prepared (from a 3D printed mold) and coated with polycaprolactone fibers. The PDMS device enables optical detection and confocal imaging of cultured cells on the fibers. Finally, macrophages were cultured in the devices to study how the fibrous scaffold can affect cell behavior. It was found that under lipopolysaccharide stimulation, macrophages cultured on PCL fibers inside of a channel secreted significantly more cytokines than those cultured on a thin layer of PCL in a channel or directly on the inner wall of a channel. Overall, this study represents a new approach and technique for in vitro cell studies, where electrospinning can be used to easily and quickly create 3D scaffolds that can improve the culture conditions in microfluidic devices.
Introduction
A significant research area in microfluidics is on-chip cell culture. With fluidic control, cell culture can be optimized and also integrated with on-chip analysis.1, 2 The concept of “Organs-on-a-Chip” represents a class of fluidic devices for in vitro cell culture that can mimic key structures and functions of in vivo tissues and organs.3, 4 Compared to static cell culture, using a fluidic approach provides not only continuous nutrient supply and waste removal but also gradient control, mimicking of in vitro physiological microenvironments (i.e., shear stress), the possibility of constructing a complete circulatory system, and the creation of in vitro organ models for pharmacology and physiology studies.2, 5 To this end, the Ingber group reported a poly(dimethylsiloxane) (PDMS)-based lung-on-a-chip model that contained multiple cell types to reconstitute the functional alveolar-capillary interface of the human lung.6 Due to the reusability, ruggedness, and integrative properties, 3D-printed fluidic devices have emerged recently as a platform for in vitro cell studies and a few 3D-printed cells-on-a-chip models have been successfully developed.7–9 For example, a fluidic device containing pancreatic β-cells, endothelial cells and erythrocytes was recently reported, which enabled the investigation of cell-cell interactions between the three cell types.10
To construct an on-chip organ that is functional and representative of in vivo conditions, an important technical issue that needs to be addressed is the way cells are cultured in the microchip-based fluidic network.2, 11, 12 However, most of the reported models so far culture the cells either on a bare polymer (i.e., PDMS) or in a microchannel coated with adhesion factors such as fibronectin and collagen.13, 14 It is readily apparent that flat, 2D surfaces do not represent the complex, three-dimensional extracellular matrix (ECM) that cells experience in vivo.5, 15, 16 However, limited research has been done to integrate ECM-resembling scaffolds within a fluidic device.
Electrospun scaffolds for in vitro cell culture have gained substantial academic interest.17 Some key features of electrospun fibers such as non-woven fibrous structure, high porosity, spatial interconnectivity and high surface area closely resemble the characteristics of the native ECM.18 Electrospinning is a technique that processes polymer solutions into fibers with diameters on the micrometer to nanometer scale. A standard electrospinning system consists of a syringe with a metal cannula, a syringe pump, a high-voltage power supply and a grounded collector electrode. When a polymer solution is drawn into a metal cannula and charged with a large potential, the electric field between the charged cannula and the grounded collector electrode overcomes the surface tension of the droplet and generates a charged Taylor cone, which can be elongated by the electrostatic force. This cone whips through the air towards the collector, creating dry fibers through evaporation of solvent.19 There have been many reports of applying electrospun fibers as in vitro cell culture matrices. For example, Mo and colleagues reported the co-culture of smooth muscle cells and endothelial cells on electrospun fibers. Their results showed the cells can proliferate on the fibers, exhibiting characteristic morphologies.18 Several other tissue specific cell types have also been successfully cultured on electrospun fibers, as are summarized in a recent review by Wang and colleagues.20
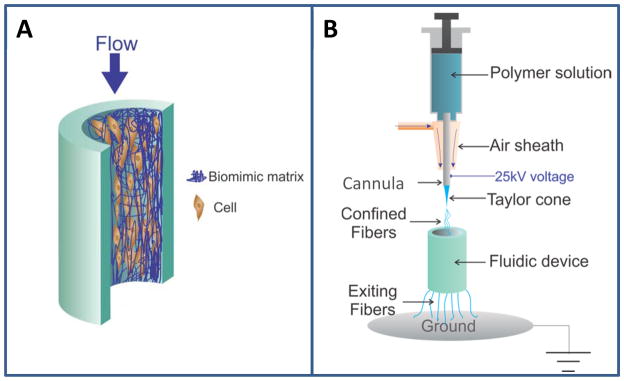
With regards to microchip-based approaches, if electrospun fibers can be integrated in a fluidic network, the device can be an ideal platform to study cells and tissues in vitro, with the resulting scaffold being similar to the native ECM and under concomitant flow conditions (see Figure 1A for a schematic of the concept). There are few reports showing the integration of electrospun fibers in a fluidic device, all of which are based upon sealing a layer of electrospun fibers between a substrate (on which the fibers are first sprayed) and a channel slab.21, 22 In addition, with these approaches the electrospun fibers only reside on one side of a square channel, which limits the area and capacity for cell culture, as well as the creation of a true 3D ECM scaffold. Moreover, some techniques used in these reports, such as nano-gold electrode array21 add to the cost and complexity of the device. Therefore, the goal of this work was to develop a simple yet effective method to directly electrospin polymer fibers into fully sealed fluidic channels to create a true 3D cell culture scaffold.
Figure 1.
(A) Concept of the cells-on-a-chip module presented in this work. A fibrous matrix is coated inside of a channel network for 3D cell culture. Reagents (media or other solutions) are allowed to flow through the network of cells and matrix, providing continuous nutrient supply and waste removal. (B) Experimental setup of electrospinning directly into an assembled device with the aid of dynamic fiber focusing. A polymer solution is pushed by a syringe pump through a steel cannula (i.d.=300 μm, o.d.=550 μm), which is connected to a 25 kV voltage supply. The electric field force generates a Taylor cone at the end of the cannula, which then undergoes Rayleigh instability and eventual fiber formation. Without being focused, the fibers whip over a large area and thus cannot be confined into a channel device. An air sheath device was designed and fabricated in this work, which can be placed around the cannula. When air flow is introduced to the sheath from the side port, the air pressure surrounding the Taylor cone dynamically focused the fibers into the channel, with the extra fibers coming out of the other end of the device.
Due to the vigorous whipping movement of fibers generated by electrospinning, it is not possible to directly electrospin fibers into a small enclosed channel. Air flow has been used to focus electrospun fibers onto a flat target.23 In this work, we utilized 3D-printing to create a customized air sheath device to dynamically focus electrospun fibers into a fully closed fluidic channel. A 3D-printed fluidic device was fabricated and coated with electrospun fibers with this method, and we characterized the parameters that affect fiber formation within the device. SEM imaging indicates that a layer of fibrous scaffold was created on the inner wall of the fluidic channel. Fibroblasts were used to test cell compatibility of the scaffold, the results of which showed that viable cells with the expected morphology and size can be cultured directly on the spun fibers, without any collagen or fibronectin coating. Due to the high transparency of poly(dimethylsiloxane) (PDMS), which makes it ideal for optical detection/imaging of cells, a PDMS fluidic device was also fabricated using a 3D-printed mold. With the same air sheath method, a fibrous scaffold was coated on the inside of the PDMS channel. Subsequent cell culture and confocal microscopic imaging confirmed the biocompatibility of the scaffold in a PDMS device. To further investigate the potential effects of the in-channel fibrous scaffold on cell activity, macrophages were cultured on the 3D-printed microfluidic devices with coated PCL fibers. After stimulating the cells with lipopolysaccharides (LPS), a commonly used reagent to trigger immune response in macrophages,26,27 the cells cultured on fibers secreted significantly more cytokines (i.e., interleukin-6, IL-6, and vascular endothelial growth factor, VEGF) than those cultured on more traditionally used thin layer coatings (i.e., fibronectin). These results suggest that the air sheath method can be used to directly electrospin fibers into fully closed fluidic devices made of different materials, leading to an in-channel scaffold for 3D cell culture and subsequent cell studies under constant flow conditions. It is also clear that electrospinning can be used to easily and quickly create 3D scaffolds that can improve the culture conditions in microfluidic devices.
Experimental
Fabrication of the 3D-printed fluidic device and the 3D-printed air sheath device
The 3D-printed devices were designed using Autodesk Inventor Professional 2015 (San Rafael, CA, USA). The standard tessellation language file (.STL file) was used by the 3D-printer (Objet Eden 260 V, Stratasys, Ltd, Edina, MN, USA) to create the devices. The material used in this work was called Full Cure 720 (Stratasys, Ltd, Edina, MN, USA), the composition of which is propriety, but approximately containing 10–30% isobornyl acrylate, 10–30% acrylic monomer, 15–30% acrylate oligomer, 0.1–1% photo initiator, as is listed on the website of Stratasys. The devices were semi-transparent upon being printed. The assembly schematics of the devices, with design details and dimensions, can be found in Figure S1 of the supplementary information.
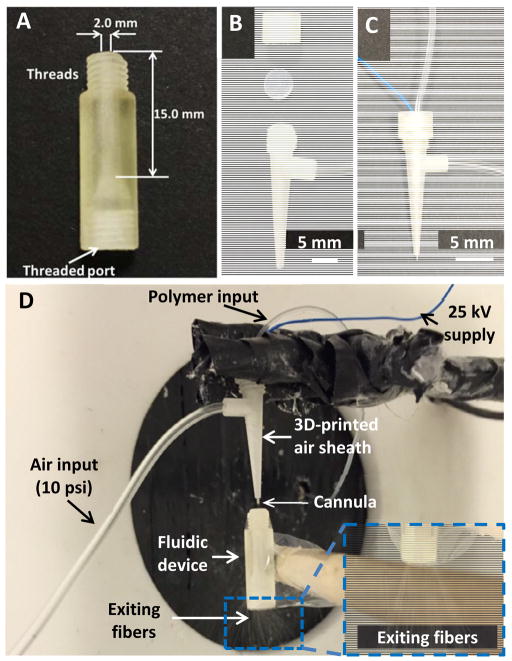
A male threaded part and a female threaded port (which fits commercial finger tight adapters) were designed on the fluidic device for easy connection to other devices/instruments (Figure 2A). The air sheath consists of a cone with a side connection port to air tanks, and a ring lid (open on the top). Threads were printed on both parts so that they can be simply integrated with each other, with septa in between. A metal cannula (300 μm i.d.× 550 μm o.d., New England Small Tube Company, NH, USA) connected to a piece of Tygon tubing (0.02″ i.d.× 0.06″ o.d., Cole-Parmer, IL, USA) can be placed through the air sheath, together with a piece of aluminum wire that can be connected to a high voltage supply (Figure 2B). The design details of the sheath device can be found in Figure S2 in the supplementary information.
Figure 2.
(A) Image of the 3D-printed fluidic device. A cylinder channel that is 2 mm diameter and 15 mm in length was fabricated; fibers were then directly electrospun into the inside of the channel to create a 3D cell culture matrix. A male threaded part and a threaded port (which fits commercial finger tight adapters) were designed at the ends of the device for easy connection to other devices and instruments; (B) Image of the 3D-printed air sheath device that aids electrospinning directly into channels. It consists of a cone with a side connection port and a cap. Threads were printed on both parts so that they can be integrated easily, with a septa being placed between the parts; (C) A steel cannula (connected to Tygon tubing) was placed through the air sheath device, together with an aluminum wire (coiled on the cannula) that can be connected to a high voltage supply; (D) Picture of the setup for electrospinning fibers into the 3D-printed fluidic device. A polymer solution was pumped through the steel cannula (to which 25 kV was applied) that was surrounded by the air sheath device. The side port of the air sheath device was connected to a compressed air line that provided an air flow at 10 psi. The 3D-printed fluidic device was then placed under the cannula to collect fibers. It can be observed that fibers were exiting the other end of the device, which indicated successful coating of fibers on the inside of the channel.
Fabrication of the PDMS fluidic device
Figure S3 in the supplementary information shows the process to fabricate a PDMS device. Briefly, a mold was 3D-printed in separate parts. Upon using, the parts were simply assembled and PDMS (prepolymer: curing reagent = 10:1) was poured into the mold. After being incubated at 75 °C for 30 min, the parts of the mold were separated off the cured PDMS device.
Electrospinning into a fluidic device
The polycaprolactone (PCL) polymer (M.W.~80,000, Sigma-Aldrich, MO, USA) was dissolved in 1,1,1,3,3,3-Hexafluoro-2-propanol (HFP) at room temperature. The concentration of PCL used in this study was 15% (w/v). After the polymer solution was homogenized on a shaker, it was loaded in a 5 mL syringe fitted with a piece of Tygon tubing (0.02′ i.d.× 0.06′ o.d., Cole-Parmer, IL, USA) via commercial adapters (IDEX, CA, USA). A steel cannula (300 μm i.d.×550 μm o.d., New England Small Tube Company, NH, USA) was connected at the end of the Tygon tubing and was placed through the air sheath. A piece of aluminum wire was coiled around the pin and connected to a 25 kV supply. The side port of the air sheath was connected to an air tank and 10 psi of air flow was applied to dynamically focus fibers coming out of the cannula. Once started, the polymer solution was driven by a syringe pump at a flow rate of 80 μL/min. A 3D-printed fluidic device or a PDMS-based device was placed 2 mm below the cannula to collect fibers. The device was coated 5 times, with each coating lasting 10 sec. After each coating compressed air was blown through the channel for 10 sec to help dry the coated fibers. Figure 2D shows the process to deposit fibers on the inside of a fluidic device.
SEM Characterization of coated devices
The coated fluidic device was split in half along the channel using a blade, which was then sputter coated with gold at 30 mA for 40 sec (Denton Vacuum LLC, NJ, USA). The sputter coated piece was then examined by a scanning electron microscope (SEM, Inspect F50 model, FEI, OR, USA) at 10 kV acceleration voltage. The fiber and pore sizes were analyzed using the ImageJ software. For fiber size determination, at least 50 fibers were measured on each SEM image using ImageJ. The pore size of the fibrous scaffold was determined by the area measurement tool29, 30 in ImageJ and at least 20 pores were measured on each SEM image.
To observe the thickness of the scaffold on the inner wall of a fluidic channel, the 3D-printed fluidic device was transected using a blade to show the cross section of the channel, followed by air focused electrospinning as mentioned above. After being coated, the cross section of the device was sputter coated and the SEM image acquired as described above.
Culture of human dermal fibroblasts (HDFs) on fiber coated devices
In 3D-printed devices
Three fiber coated devices were connected end to end by the threads (the male part connected to the threaded port of another device) and then soaked in isopropanol (IPA) for 12 hours. The devices were then taken out of IPA and placed in UV light in a cell hood for 24 hours for sterilization. A 15 mL plastic test tube was processed as a container for cell culture on the fiber coated devices. A 0.5 cm diameter hole was drilled through the cap of the test tube, and a piece of 0.4 μm pore size polycarbonate membrane (Sigma-Aldrich, MO, USA) was sealed between the tube and the opened cap. This container was also sterilized by soaking it in IPA (12 hours) and drying it in UV light for 24 hours ).
Primary human dermal fibroblasts (HDF, normal, human, adult, ATCC, VA, USA) with passage numbers of 1 to 4 were used in this study. When the cells were confluent in a T-75 flask, the DMEM/F-12 media buffered with 10% fetal bovine serum and 1% antibiotic-antimycotic (ThermoFisher Scientific, MO, USA) was removed and 7 mL trypsin/EDTA solution (ThermoFisher Scientific, MO, USA) was added. After 5 min in an incubator (37 °C, 5% CO2), the resulting cell suspension was transferred into a 15 mL plastic test tube, which was then centrifuged at 1500 g for 5 min to pellet the cells. After removing the supernatant, the cells were re-suspended in 2 mL fresh media. An aliquot of 100 μL of the suspension was pipetted into a 700 μL centrifuge tube, followed by adding 100 μL buffered trypan blue solution (ThermoFisher, MO, USA). The amount of viable cells was then determined using a hemocytometer. After this measurement the 2 mL cell suspension was further diluted to a density of 1 million viable cells per mL. There was usually 5 to 6 mL diluted cell suspension acquired after these steps.
The diluted cell suspension was then transferred into the sterilized and dried test tube, followed by soaking the 3D-printed devices in the suspension. The test tube was recapped with a piece of porous membrane in between, which facilitates gas exchange between the media and the incubator atmosphere. The test tube was then placed horizontally in an incubator (37 °C, 5% CO2) for 12 hours (Figure S4A), during which, the test tube was periodically rotated every 1 hour to make sure cells adhered homogeneously around the channel, instead of only on one side. After the 12-hour static culture, a dynamic cell culture step was performed to determine fiber and cell adherence to the inner wall of the fluidic device. As shown in Figure S4B in the supplementary information, the end of the top device was connected via a commercial finger tight fitting to a piece of Tygon tubing (0.02′ i.d. × 0.06″ o.d., Cole-Parmer, IL, USA), which connected to a peristaltic pump (Cole-Parmer, IL, USA). The other end of the Tygon tubing was immersed in the media in the vial. The peristaltic pump circulated fresh media through the devices for another 24 hours in the incubator.
In PDMS devices
After the devices were sterilized by the same method described above, the HDF suspension was gently pipetted through the channels. The devices were then placed in a 50 mm petri dish, with the addition of fresh media to immerse the devices. The petri dish was then placed in a 37 °C incubator. The HDFs were cultured in this way for 2 days on the PDMS devices.
Culture of macrophages in 3D printed devices
Microfluidic devices with electrospun fibers on the inner wall were prepared as described above. In addition, devices with a thin layer of PCL coated on the inside were prepared by pumping 15% PCL solution through a channel and subsequent drying in a fume hood for 24 hours. The PCL thin layer coated channels were then filled with 0.5 mg/mL fibronectin solution and allowed to dry in a cell hood for 24 hours. As a control, a microfluidic device without any PCL coating was also prepared with the same fibronectin treatment method (coated with fibronectin only).
RAW 264.7 mouse macrophages purchased from ATCC (VA, USA) were subcultured following the manufacturer’s instructions. When the cells were confluent in a flask, they were detached by using a scraper and the suspension centrifuged. The cell pellet was resuspended in 1 mL DMEM media (with 10% FBS and 1% pen-strep), followed by cell counting on a hemacytometer. The 1 mL cell suspension was then diluted with the media to a final concentration of 2 million cells/mL. The microfluidic devices were then filled with the diluted cell suspension, placed in a petri dish, and the device was placed in an incubator (37 °C, 5% CO2) for three days before use.
Cell Evaluation
MTS Cell Proliferation Assay with the 3D-printed fluidic device
A CellTiter96 MTS assay kit (Promega, WI, USA) was used to determine the number of viable cells in proliferation cultured on the 3D-printed fluidic device per manufacturer protocol. After the cell culture process was finished, the devices were detached and each device was placed immediately in a glass vial containing 2 mL warmed fresh media. An aliquot of 500 μL assay solution was then added to each vial, followed by a thorough mixing. The vials were placed in an incubator (37 °C, 5% CO2) for 1 hour. The caps of the glass vials were loosened during the incubation to facilitate O2 and CO2 exchange between the media and the incubator atmosphere.
Four standards were prepared to quantify viable cells in proliferation. HDFs cultured in a flask were trypsinized and resuspended as described above. Then different amounts of cell suspension was added into four glass vials, followed by adding fresh media to make a total volume of 2 mL in each vial. Four vials containing 0, 0.40 × 105, 1.56 × 105, 4.76 × 105 cells were prepared. Four bare fluidic devices were placed in the vials, to compensate for possible absorption/adsorption of the MTS reagents onto the devices (as well as any possible light scattering). An aliquot of 500 μL of assay solution was then added to each vial, followed by a thorough mixing. The vials were incubated in the same way as described above.
After incubation, an aliquot of 50 μL of solution was sampled from each vial and loaded in a 96-well plate, followed by an absorption measurement at 490 nm using a plate reader (Molecular Devices LLC, CA, US). The absorbance values of the four standards were plotted versus cell amount, which was used as a calibration curve to quantify the amount of viable cells cultured on a fluidic device.
SEM Imaging of cells cultured in the 3D-printed fluidic device
After the cell culture period, the fluidic device was split in half along the channel direction using a blade. The split pieces were fixed in 10 % formalin (buffered in phosphate buffer solution) for 30 min at room temperature. The pieces then underwent sequential ethanol dehydration with 50%, 70%, 80%, and 95% ethanol for 10 min each, followed by two 10 min rinses in 100 % ethanol (Sigma-Aldrich, MO, US). After the pieces were air dried, they were sputter coated with gold at 30 mA for 40 sec, followed by SEM imaging.
Confocal imaging of the cells cultured in the PDMS device
The HDFs cultured on the fibers coated in a PDMS channel were rinsed by warm (37 °C) Hanks Balanced Salt Solutions (HBSS, Sigma-Aldrich, MO, USA), after which, 4% formaldehyde solution (in PBS) was pipetted into the channel to fix the cells for 10 min at 37 °C. After rinsing off the remaining formaldehyde, the Alexa Actin 555 reagent (LifeTechnology, WA, USA) was used to stain actin of the cells for 20 min at 37 °C. 4′,6-diamidino-2-phenylindole solution (DAPI, LifeTechnology, WA, USA) was used to soak the cells during the imaging process using a confocal microscope (Leica, Germany). A device coated with fibers (without cells) was also stained and imaged with the same method to see if the polymer fibers interfere with cell imaging under the microscope.
Macrophage stimulation and multiplex detection of cytokines
A LPS solution (1 mg/L) was made by dissolving the proper amount of LPS powder (Sigma-Aldrich, MO, USA) in DMEM media. The LPS containing media was then circulated through the microfluidic channels using a setup similar to Figure S4B in the supplementary information. After stimulating the cells with LPS for 12 hours, 500 μL of the circulating solution was collected for cytokine detection. As a control, DMEM media without LPS was also used to circulate through the devices for 12 hours, and the cytokines from these samples were also measured.
A mouse cytokine/chemokine magnetic bead panel multiplex kit (Millipore, MA, USA) was used to detect the cytokines secreted from the macrophages stimulated by LPS solution. A 96-well plate was prepared following the manufacturer’s instructions, and a Luminex MAGPIX instrument (Luminex, TX, USA) was used for fluorescence measurement of the cytokines. In a well, a 25 μL solution of microbeads coated with specific antibodies against cytokines (from the commercially available kit) was mixed with 25 μL of the collected circulating solution, as well as the provided assay buffer (25 μL) and matrix solution (25 μL).. After cytokine capture onto the beads during an overnight incubation at 4 °C, the detection antibody (tagged with a fluorophore) was added. Following a 1 hour incubation period, the beads were introduced into the Luminex MAGPIX system, where they were detected by laser induced fluorescence. Standards were prepared by diluting the stock solution (from the commercial kit) and detected in the same way as the sample solutions. Based on the fluorescence intensity of the standards and the samples, the concentration of cytokines was calculated by the built-in software.
Results and Discussion
There have been numerous examples of using microfluidic devices and liquid manipulation to culture cells under flow conditions. As aforementioned, compared to static cell culture, flow-based cell culture can be a step forward to better represent in vivo microenvironments (i.e., shear stress) and conditions (i.e., continuous nutrient supply and waste removal).11 The term “Organs-on-a-Chip” has been proposed recently to recapture the main functions of certain organs on a fluidic device, which can be potentially applied for drug toxicity assessment, drug screening, and fundamental physiological studies.4, 15, 28 Although 2D cell culture matrices such as collagen and fibronectin layers have been widely applied in culture flasks and microfluidic devices, little research has been done to incorporate in vivo representative ECM analogue fibers in a fluidic device.22 Cells and tissues are embedded within 3D, fibrous ECM in vivo, which have proven to be able to regulate cellular activities.29 In other words, even under flow conditions, if cells are not cultured on an ECM analogue scaffold, they may not be able to fully mimic in vivo conditions.
Due to the high surface area to volume ratio, porosity and biocompatibility, electrospun fibers have become an optimal scaffold for ECM mimicking in many studies.18, 19 Even though there are some reports trying to integrate electrospun fibers in a fluidic device with intricate techniques and procedures, there lacks a simple and direct way to combine such fibers within a fluidic channel.21, 22 Our results here provide a new but simple method to directly coat electrospun fibers inside a fully sealed fluidic channel. Using traditional electrospinning techniques, where the electric field is the sole driving force, fiber deposition tends to be widespread with large amounts of overspray, which cannot be confined into a small, closed fluidic channel. In this work, we developed a dynamic air focusing method to help focus electrospun fibers, the concept which is demonstrated in Figure 1B. Compared with a classical electrospinning setup, an air sheath was utilized so that air flow can be applied to focus the fibers into a fluidic device. As shown in Figure 2B, the air sheath device was fabricated by 3D-printing, consisting of two parts: a cap and a cone with a side port. The two parts can be joined by the printed threads, with a septa placed in between. The top of the cap is open so that a steel cannula at the end of a piece of Tygon tubing, as well as an aluminum wire (coiled on the cannula) can be placed through the air sheath (Figure 2C). The side port acts as the sheath air inlet after being connected to a compressed air line. It was observed that an optimal sheath air flow velocity is desired to dynamically focus the fibers. As shown in Figure S5 in the supplementary information, with an increase in the sheath air pressure, the fiber size tends to decrease (Figure S5A), while the pore size tends to become larger (Figure S5B). Figure S5C shows the SEM images of the fibers formed under different air pressures. When using a sheath air pressure below 5 psi, the cone is not effectively focused into the fluidic channel, while use of a pressure higher than 15 psi leads to drying of the polymer solution at the cannula tip, causing clogging issues. It can also be seen that at a 5 psi sheath air pressure, the fibers are chaotically distributed and beaded, with large deviation of fiber sizes. At 7.5 psi, however, the fibers are more orientated along the channel and the fiber size is more uniform. At 10 and 15 psi, the fibers are even smaller. Therefore, considering the fiber and pore sizes as well as fiber uniformity, the optimized sheath air pressures were determined to between 10 and 15 psi (2.9 and 3.4 m/s). The size of the tip of the air sheath device was also optimized. The o.d. of the steel cannula was 550 μm (300 μm i.d.), and the optimized air sheath tip size was determined to be 800 μm in diameter. A smaller tip size led to limited space between the air sheath and the cannula, causing air flow problems out of the sheath, while air flow out of a larger tip size could not focus the fibers effectively.
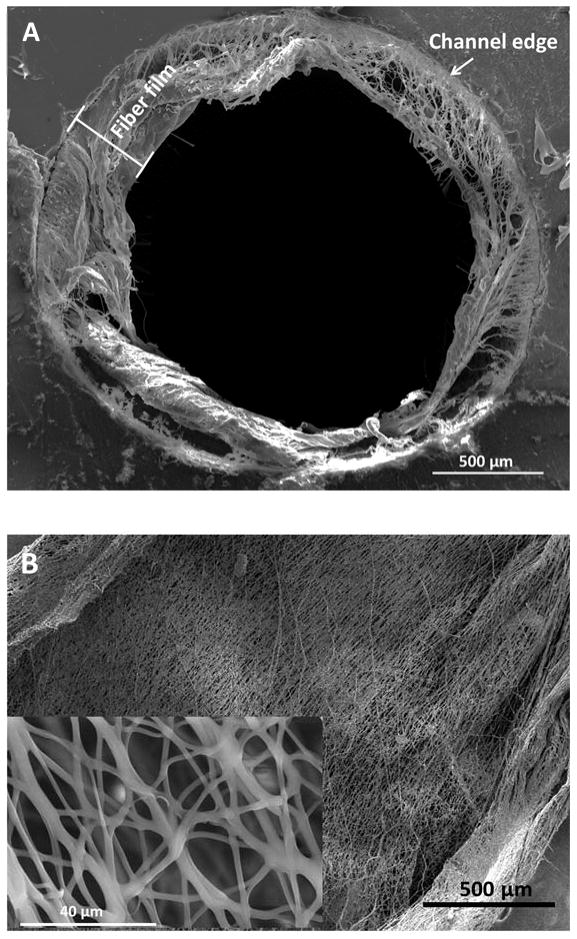
As shown in Figure 2D, a 3D-printed fluidic channel was utilized for coating with electrospun fibers. 3D-printing enabled the integration of a threaded port and a male threaded part on the device (Figure 2A), which makes it easy to be connected with other devices and instruments. After fibers were formed at the tip of the cannula, the sheath air focused the fibers going through the fluidic device. The exiting fibers from the other end of the fluidic device indicated successful coating of fibers on the inner wall of the channel. As shown in Figure S6 in the supplementary information, without the sheath of air, the polymer fibers tended to overspray around the fluidic device, with none being directed into the channel. To obtain sufficient fibers on the inside of a channel, a device was coated 5 times, with each coating step lasting 10 sec. Due to the limited space in a closed channel, the solvent (HFP) utilized in these PCL solutions may not be able to evaporate efficiently, resulting in welded fibers that can reduce the porosity and surface area of the scaffold (which may adversely affect subsequent cell culture). Therefore, a drying step in between each coating was applied by blowing compressed air through the channel for 5 sec. The coated fibers were examined using SEM. Figure 3A shows the SEM image of the cross section of a fiber coated channel. It can be seen that the 3D scaffold was coated on the inner wall of the channel, the thickness of which was measured to be 379 ± 15 μm (mean ± standard deviation). The structure of the coated substrate in the channel was further detected using SEM in a view along the channel axis. As shown in Figure 3B, the electrospun substrate on the inner wall of the channel is highly fibrous and porous. The inset demonstrates the substrate on a smaller scale, which clearly suggests that fine fibers and pores were fabricated. ImageJ analyses on the SEM images showed that the mean fiber size was 1.6 ± 0.6 μm and the average pore size was 113 ± 19 μm2 (mean of 4 devices ± standard deviation). For comparison, the SEM image of a bare channel (Figure S7A in the supplemental information) only shows a ridged surface in the channel area, which results from the resolution of the 3D-printer. A thin-layer coated device was also prepared by pumping 15% PCL solution through the channel, followed by drying in a fume hood at room temperature for 24 hours. As shown in Figure S7B in the supplemental information, the ridges of the 3D-printed part were covered by a smooth layer of PCL, but without porous or fibrous structures.
Figure 3.
(A) An SEM view of the cross section of a channel coated with electron fibers. A layer of fibers were coated on the inner wall of the device. (B) An SEM image of fibers in a view along the channel. The inset is a zoomed in view of the fibers. The results show that the electrospun scaffold on the inside of a channel is porous and fibrous. The fiber width was measured to be 1.6 ± 0.6 μm, while the average pore size being 113 ± 19 μm2.
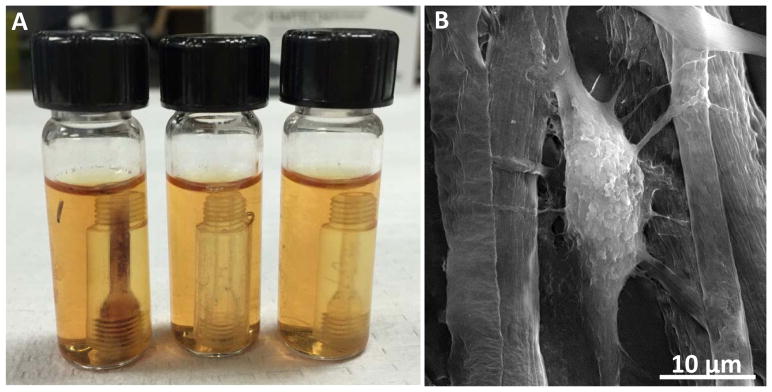
HDFs were used to test the biocompatibility of the scaffold by a static seeding and dynamic culturing process. Three fluidic devices were connected end-to-end by the printed threads to enhance throughput. As shown in Figure S2A, the three devices were placed in a test tube containing HDF suspension for static seeding. After incubation for 12 hours, the devices were taken out and connected via a commercial finger tight adapter to a peristaltic pump for dynamic HDF culturing (Figure S2B). Hollow fiber chambers (HFCs), which are opaque and large volume (hundreds of mL) perfusion-based cell culture platforms, are commonly used in pharmaceutical studies and a MTS assay is commonly used to evaluate cell proliferation in a HFCs.30 MTS assay changes its color form light orange to purple with viable cells, with the purple color intensity being proportional to the amount of viable cells.31 As shown in Figure 4A, even though the 3D printed devices were semi-transparent, a layer of dark purple could be observed with MTS assay on the inside of the left device, which was fiber coated on the inside. This result indicates that proliferative HDFs were successfully cultured on the fibrous scaffold. In contrast, HDFs cannot be cultured on a bare fluidic device (the middle vial in Figure 4A) or a thin-layer coated device (the right device in Figure 4A). The purple solution from the first vial in Figure 4A was then pipetted out for absorption measurement to quantify the amount of viable cells. It was determined that 4.1 (± 0.8) × 105 viable cells can be cultured on such a scaffold on the inside of a device (mean of 5 devices ± standard deviation). The morphology of the cells was examined by SEM. As shown in Figure 4B, the spindle morphology and the size of the cell are consistent with in vivo HDFs.32 Some pseudopodia were also formed by the cell to attach to surrounding fibers, which further confirmed that the cells can adhere onto the fibrous scaffold, even under 400 μL/min flowing condition during the dynamic culture process.
Figure 4.
(A) MTS assay to quantify viable HDFs on different devices. The three devices from left to right are: a PCL fiber coated device, a bare device (no fibers coated), and a PCL layer coated device. The same HDF seeding and culture process was conducted on all three devices. A layer of dark purple was observed on the inside of the channel of the left device, which indicated viable cells were adhering on the fibers coated on the channel. The solution from the vials was pipetted into a 96-well plate for absorption measurements, which revealed that 4.13 × 105 (± 0.76 × 105) cells were cultured on such a device. The middle and the right devices, however, did not show color change in the channels, indicating no cells were adhered onto the inner walls of the devices. (B) SEM image of an HDF cultured on the coated fibers in a fluidic device. Some pseudopodia were formed by the cell to attach surrounding fibers, which further confirmed that HDFs can adhere on the fibers
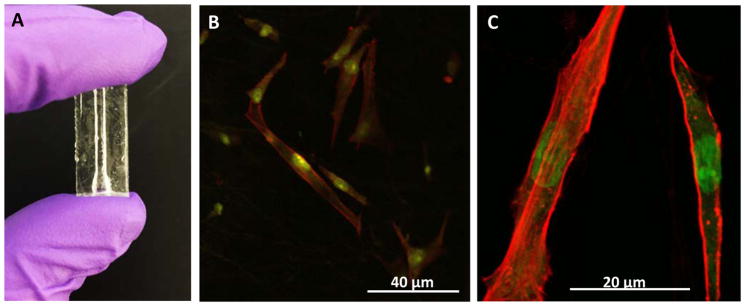
These results suggest that viable cells can be cultured on the in-channel scaffold in a dynamic way. 3D-printing enables the device to be rugged and robust, which allows for media circulation for a long time without structural or functional impairments (Figure S4B). No fiber detachment was observed during the flowing experiments, which indicates strong adherence of the electrospun fibers on the inner wall of the fluidic device. In order to enable imaging of cells cultured on such a scaffold on the inside of a fluidic channel, a PDMS version of the 3D-printed fluidic device was fabricated with a molding method (Figure S3). As shown in Figure 5A, the PDMS device is transparent. PCL fibers were also coated on the inner wall of the PDMS channel using the air focused electrospinning method. HDFs were cultured with the same method in these PDMS devices. The cells were then stained by Alexa Actin 555 (for actin) and DAPI (for nuclei) for confocal imaging. Figures 5B and 5C are confocal images of HDFs cultured on the fibers deposited on the inside of a PDMS channel, which showed the feasibility of optical observation and imaging of cells cultured on fibrous scaffolds inside of a fluidic device. A PDMS device with coated PCL fibers only (no cells) was also stained and imaged in the same manner. It was found that the polymer fibers are not stained by the reagents, and thus the fibers do not interfere with cell imaging.
Figure 5.
(A) PDMS fluidic device fabricated using a 3D-printed mold. (B and C) Confocal images of HDFs cultured on the fibers deposited inside of a PDMS device. Red represents stained actin by Alexa Actin 555 and green shows the nuclei stained by DAPI. A device with only PCL fibers (no cells) was also stained with the same method and observed under the microscope. It was found that the polymer fibers were not stained by the reagents, and thus did not interfere with cell imaging.
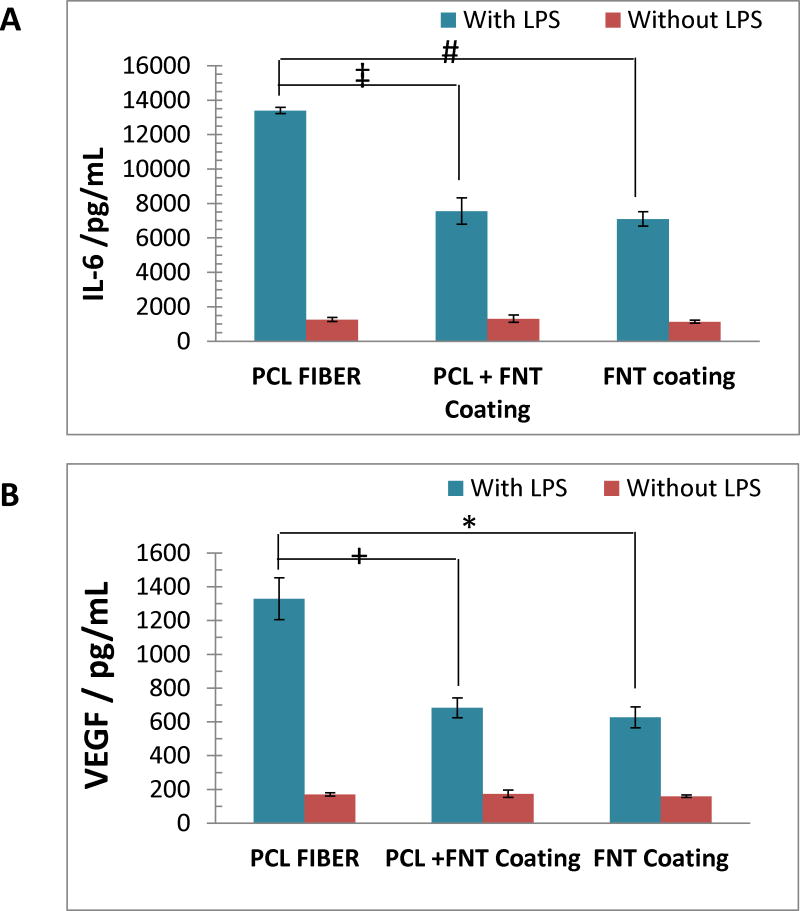
To investigate how the fibrous scaffold in a microfluidic channel can affect cell activity, macrophages-were utilized. The macrophages were cultured on the devices in a static fashion for 24 hours, after which time, LPS containing media was circulated through the devices for another 12 hours to stimulate cytokine production. Three different 3D-printed microfluidic devices were used for these studies: 1) a device with PCL fibers coated on the inner wall of the channel (3D scaffold); 2) a device with a thin PCL layer coated on the inside of the channel, followed by a thin layer of fibronectin coated on the PCL (2D coating); and 3) a device with fibronectin coated on the channel inner wall (2D coating). Macrophages are a type of immune cells that can engulf foreign agents entering the body, which include microbes and other particular matters. In addition, these cells can scavenge apoptotic cells and digest waste products from tissues.24 Macrophages play an important role in the inflammatory response by secreting cytokines such as IL-6 and VEGF that can serve as signaling molecules with other cells.24, 25 Therefore, the LPS stimulated cytokine secretion activity of macrophages cultured on the three devices was studied. Figure 6 shows the results of IL-6 and VEGF secretion profile. For both graphs, with LPS stimulation (blue bars), the macrophages secreted significantly more cytokines than unstimulated cells (red bars). There is no statistical difference at the 95% confidence level between the red bars (unstimulated secretion from macrophages), which indicates the cell counts on the three devices are the same. For macrophages cultured on the fibrous 3D scaffold (PCL FIBERS), significantly more IL-6 and VEGF were secreted than those cultured on flat substrates (PCL + FNT Coating and FNT Coating). IL-6 is a pro-inflammatory cytokine and VEGF is a healing cytokine, but the presence of both in elevated levels at a short time point would indicate an enhanced activation of the macrophages over controls.27 These results suggest that the macrophages cultured on the fibers, which mimic the 3D architecture of the native ECM, are more responsive to external stimulation (i.e., LPS) triggering macrophage activation and ultimately inflammation and healing. This also suggests that the 3D fibrous scaffold improves the culture environment for the macrophages studied here.
Figure 6.
Cytokine secretion from macrophages cultured on different substrates. (A) is the IL-6 secretion profile and (B) shows the secretion of VEGF. For both graphs, PCL FIBER represents a microfluidic channel with electrospun PCL fibers (a 3D cell culture substrate); PCL + FNT Coating means a microfluidic channel coated with a layer of PCL, which was then coated with fibronectin (a 2D cell culture substrate); FNT coating corresponds to a microfluidic channel that is coated with fibronectin (a 2D cell culture substrate). There is no significant difference at 95% confidence level between the red bars (natural secretions without LPS stimulation) that can indicate the cell counts on the three substrates are equal. By comparing the blue bars (secretion with LPS stimulation), it can be seen that in both graphs, cells cultured on the fibrous 3D scaffold secreted more cytokines than those cultured on 2D substrates (*, +, #, and ‡, all statistically different, p<0.001). These results suggest that the fibrous scaffold can be an important factor that affects cell physiology. (n=3, error bar=standard error of mean)
Conclusions
This work developed a new type of “Cells-on-a-Chip” module, which enables cell culture on an ECM analogue scaffold and cell studies under flow conditions. To the best of our knowledge, this is the first report of directly electrospinning fibers into a fully closed fluidic device. The application of the air sheath helped to eliminate overspray issues and enabled the direct focusing of fibers into a sealed fluidic channel. SEM imaging and subsequent image analyses indicated that fine fibers and porous structures can be constructed on the inside of a fluidic channel to act as a cell culture matrix. HDFs were seeded and cultured on fiber coated devices, the results of which indicate that the electrospun fibers enable cell adhesion and proliferation in their physiological morphology. Moreover, to make microscopic imaging of cells more feasible, a transparent PDMS device was fabricated using 3D-printed molds, with confocal imaging of the 3D scaffold being possible. Macrophages were then cultured on the fibrous scaffold and the secretion of cytokines was measured. It was found that macrophages cultured on the 3D fiber scaffold secrete more cytokines upon LPS stimulation, showing that the fibrous scaffold does positively affect the culturing of cells in devices. Overall, this work provides an in vivo representative protocol for cell culture on a fluidic device, which can be a step forward to develop better “Organs-on-a-Chip” models in the future. Future work will focus on evaluating other materials and cell types for use in these types of devices as well as integration of other functions such as detection modules so that high throughput 3D cell culture and analysis is possible.
Supplementary Material
Acknowledgments
Support from the National Institute of General Medical Sciences (Award Number R15GM084470-04) is acknowledged.
References
- 1.Chen J, Li J, Sun Y. Lab Chip. 2012;12:1753–1767. doi: 10.1039/c2lc21273k. [DOI] [PubMed] [Google Scholar]
- 2.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 3.Cooke BM, Usami S, Perry I, Nash GB. Microvasc Res. 1993;45:33–45. doi: 10.1006/mvre.1993.1004. [DOI] [PubMed] [Google Scholar]
- 4.Selimovic S, Dokmeci MR, Khademhosseini A. Curr Opin Pharmacol. 2013;13:829–833. doi: 10.1016/j.coph.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Healy KE. J Tissue Eng Regen M. 2014;8:13–13. [Google Scholar]
- 6.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonyar A, Santha H, Ring B, Varga M, Kovacs JG, Harsanyi G. Procedia Engineer. 2010;5:291–294. [Google Scholar]
- 8.Chen CP, Wang YM, Lockwood SY, Spence DM. Analyst. 2014;139:3219–3226. doi: 10.1039/c3an02357e. [DOI] [PubMed] [Google Scholar]
- 9.Gross BC, Erkal JL, Lockwood SY, Chen CP, Spence DM. Anal Chem. 2014;86:3240–3253. doi: 10.1021/ac403397r. [DOI] [PubMed] [Google Scholar]
- 10.Liu YL, Chen CP, Summers S, Medawala W, Spence DM. Integr Biol. 2015;7:534–543. doi: 10.1039/c4ib00243a. [DOI] [PubMed] [Google Scholar]
- 11.Meyvantsson I, Beebe DJ. Annu Rev Anal Chem. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- 12.Ozturk SS, Hu W-S. Cell culture technology for pharmaceutical and cell-based therapies. Taylor & Francis; Boca Raton: 2005. [Google Scholar]
- 13.Tourovskaia A, Figueroa-Masot X, Folch A. Lab Chip. 2005;5:14–19. doi: 10.1039/b405719h. [DOI] [PubMed] [Google Scholar]
- 14.Gomes ME, Sikavitsas VI, Behravesh E, Reis RL, Mikos AG. J Biomed Mater Res A. 2003;67A:87–95. doi: 10.1002/jbm.a.10075. [DOI] [PubMed] [Google Scholar]
- 15.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Ann Biomed Eng. 2012;40:1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 16.Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR, Khademhosseini A. Expert Opin Drug Dis. 2014;9:335–352. doi: 10.1517/17460441.2014.886562. [DOI] [PubMed] [Google Scholar]
- 17.Doshi J, Reneker DH. Ias. 1993;93(Pts 1–3):1698–1703. [Google Scholar]
- 18.Mo XM, Xu CY, Kotaki M, Ramakrishna S. Biomaterials. 2004;25:1883–1890. doi: 10.1016/j.biomaterials.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 19.Doshi J, Reneker DH. J Electrostat. 1995;35:151–160. [Google Scholar]
- 20.Wang XF, Ding B, Li BY. Mater Today. 2013;16:229–241. doi: 10.1016/j.mattod.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho DW, Matlock-Colangelo L, Xiang CH, Asiello PJ, Baeumner AJ, Frey MW. Polymer. 2011;52:3413–3421. [Google Scholar]
- 22.Lee KH, Kwon GH, Shin SJ, Baek JY, Han DK, Park Y, Lee SH. J Biomed Mater Res A. 2009;90A:619–628. doi: 10.1002/jbm.a.32059. [DOI] [PubMed] [Google Scholar]
- 23.McClure MJ, Wolfe PS, Simpson DG, Sell SA, Bowlin GL. Biomaterials. 2012;33:771–779. doi: 10.1016/j.biomaterials.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Sieweke MH, Allen JE. Science. 2013;342:946–953. doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 25.Duque GA, Descoteaux A. Front Immunol. 2014;5:1–12. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilcek J, Feldmann M. Trends Pharmacol Sci. 2004;25:201–209. doi: 10.1016/j.tips.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Meng FY, Lowell CA. J Exp Med. 1997;185:1661–1670. doi: 10.1084/jem.185.9.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moyer MW. Sci Am. 2011;304:19–19. doi: 10.1038/scientificamerican0311-19a. [DOI] [PubMed] [Google Scholar]
- 29.Alepee N, Bahinski A, Daneshian M, De Weyer B, Fritsche E, Goldberg A, Hansmann J, Hartung T, Haycock J, Hogberg HT, Hoelting L, Kelm JM, Kadereit S, Mcvey E, Landsiedel R, Leist M, Luebberstedt M, Noor F, Pellevoisin C, Petersohn D, Pfannenbecker U, Reisinger K, Ramirez T, Rothen-Rutishauser B, Schaefer-Korting M, Zeilinger K, Zurich MG. Altex-Altern Anim Ex. 2014;31:441–477. doi: 10.14573/altex1406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall LM, Krauthauser CM, Wexler RS, Hollingshead MG, Slee AM, Kerr JS. Anticancer Res. 2000;20:903–911. [PubMed] [Google Scholar]
- 31.Malich G, Markovic B, Winder C. Toxicology. 1997;124:179–192. doi: 10.1016/s0300-483x(97)00151-0. [DOI] [PubMed] [Google Scholar]
- 32.Scharffetter K, Wlaschek M, Hogg A, Bolsen K, Schothorst A, Goerz G, Krieg T, Plewig G. Arch Dermatol Res. 1991;283:506–511. doi: 10.1007/BF00371923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.