SUMMARY
A major advance in the search for an HIV vaccine has been the development of a near-native Envelope trimer (BG505 SOSIP.664) that can induce robust autologous Tier 2 neutralization. Here, potently neutralizing monoclonal antibodies (nAbs) from rabbits immunized with BG505 SOSIP.664 are shown to recognize an immunodominant region of gp120 centered on residue 241. Residue 241 occupies a hole in the glycan defenses of the BG505 isolate with fewer than 3% of global isolates lacking a glycan site at this position. However, at least one conserved glycan site is missing in 89% of viruses, suggesting the presence of glycan holes in a majority of HIV isolates. Indeed, serum evidence is consistent with targeting of holes in natural infection. The immunogenic nature of breaches in the glycan shield has been under-appreciated in previous attempts to understand autologous neutralizing antibody responses and has important potential consequences for HIV vaccine design.
eTOC Blurb
This study describes monoclonal rabbit antibodies elicited by the HIV immunogen BG505 SOSIP.664. Previous reports showed high titer serum neutralization against BG505 virus but the epitope remained elusive. McCoy et al. show the neutralizing epitope is a breach in HIV’s glycan shield and suggest other HIV strains contain similar holes.
Introduction
The development of a protective vaccine remains the most attractive option for halting the global spread of HIV. Antibody-based HIV vaccine research has focused on the HIV envelope glycoprotein (Env), which is the only viral protein targeted by neutralizing antibodies (nAbs) during natural infection. Extensive progress has been made in identification and characterization of nAbs from HIV-infected individuals that are both potently neutralizing and active against a broad array of circulating HIV strains (reviewed in (Burton and Hangartner, 2016; Hraber et al., 2014; Moore et al., 2015; West et al., 2014)). Passive transfer of such broadly neutralizing Abs (bnAbs) into non-human primates protects against viral challenge even at low antibody concentrations (reviewed in (van Gils and Sanders, 2014)), suggesting that such bnAbs, if induced by vaccination, will prevent HIV infection. Many studies have investigated the coevolution of virus and nAbs. In HIV-infected patients who develop bnAbs, the early potent autologous neutralizing response broadens concomitantly with increased viral diversity as the virus escapes from the autologous nAbs. (Bhiman et al., 2015; Garces et al., 2015; Liao et al., 2013; Moore et al., 2009; Wei et al., 2003; Wibmer et al., 2013). Escape is frequently associated with the addition of glycans (Bunnik et al., 2008; Moore et al., 2009; Rong et al., 2009; van Gils et al., 2010).
In contrast to the plethora of bnAbs derived from HIV-positive donors, and the multiple epitopes and mechanisms by which they achieve breadth and potency, few immunization studies have induced even moderately potent autologous neutralization against typical resistant (Tier 2) primary viruses never mind against multiple heterologous viruses. Most Env subunit immunizations have yielded non-nAbs or Abs that only neutralize highly susceptible (Tier 1) viruses (Chen et al., 2013; Narayan et al., 2013; Qin et al., 2015; Qin et al., 2014; Schiffner et al., 2013; Seaman et al., 2010; Vaine et al., 2008; Zhang et al., 2015). One system in which strong autologous Tier 2 serum nAb responses have been observed following Env immunization is that of rabbits immunized with B41 and BG505 SOSIP.664 and SOSIP.v4 trimers (de Taeye et al., 2015; Sanders et al., 2013; Sanders et al., 2015). These soluble trimers closely mimic the native, membrane-embedded functional trimer on the surface of virions and have the potential to be key components of a nAb-eliciting vaccine (Burton and Mascola, 2015; Hraber et al., 2014; Lee et al., 2016; Moore et al., 2015; West et al., 2014). Despite these advances in the presentation of native-like HIV trimers and the reproducible induction of greater autologous neutralizing titers, little is understood about the epitopes targeted. In part, this is because the neutralization response to Env immunization in model systems has most commonly been assessed by serology alone (Narayan et al., 2013; Qin et al., 2014; Schiffner et al., 2013; Sundling et al., 2010; Zhang et al., 2015). Attempts to map serum neutralization following BG505 SOSIP.664 immunization have been limited to those epitopes easily dissected with the use of CD4-binding site mutants, peptides to linear epitopes, and alanine-scanning panels (Sanders et al., 2015). Some mAbs have been isolated from immunization studies with Env proteins that are not good native trimeric mimics (Chen et al., 2013; Qin et al., 2015), notably in non-human primates (NHPs) (Phad et al., 2015; Sundling et al., 2012; Sundling et al., 2014). Recently, V5-specific mAbs that neutralize the autologous transmitter/founder virus were isolated from immunized NHPs (Bradley et al., 2016). BnAbs have only been induced by immunization as rare clones in camelids (McCoy et al., 2012; McCoy et al., 2014), and in transgenic mice encoding mature bnAb heavy chains in response to BG505 SOSIP.664 immunization (Dosenovic et al., 2015).
In this study, we determined the specificities and molecular mechanisms underlying the potent autologous anti-HIV response induced by BG505 SOSIP.664 in rabbits by characterizing mAbs from three rabbits one week after their final immunization. Our findings reveal a neutralizing specificity that targets holes in the glycan shield characteristic of a given isolate and that conventional serological analysis has largely overlooked. This work highlights the potential challenges posed by viral glycans to any strategy to elicit bnAbs via first inducing autologous nAbs, a pattern that has been hypothesized to occur in natural HIV infection.
Results
BG505-specific mAbs isolated from three immunized rabbits
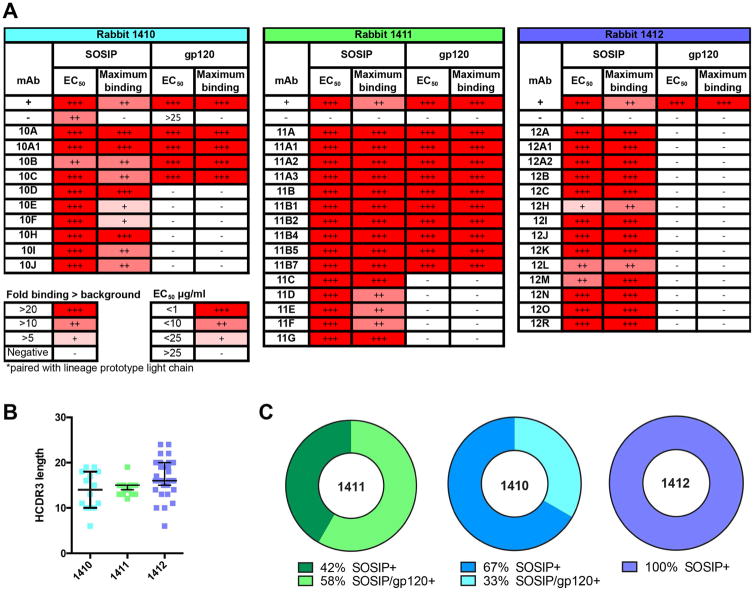
Following immunization with BG505 SOSIP.664, three rabbits were selected that exhibited strong serum neutralizing activity against the autologous BG505 virus (Sanders et al., 2015). The rabbits were designated 1410, 1411 and 1412. Note that autologous BG505 virus contains a T332N substitution to match the corresponding SOSIP.664 protein at that position (Sanders et al., 2015). Peripheral blood mononuclear cells (PBMCs), which were harvested seven days after the final immunization (week 25), and those which bound BG505 SOSIP.664 were isolated by single-cell sorting as described in experimental procedures (Table S1), enabling production of 39 mAbs from rabbits 1410–12 (Figure 1A). The mAbs exhibited a range of ELISA binding activities against the BG505 SOSIP.664 immunogen, with half maximal binding concentrations (EC50) ranging from 0.01 to 19.6 μg/ml (Figure 1A, Figure S1). Different levels of maximum binding at the highest concentration assayed (25 μg/ml) ranged from 5 to >20 fold above background, independent of the EC50 value (Figure 1A).
Figure 1. Binding properties of BG505 SOSIP.664 trimer-elicited rabbit mAbs.
(A) Binding of rabbit mAbs derived from the animal indicated above each table to BG505 SOSIP.664 and gp120 was assayed by streptavidin capture ELISA. The EC50 values were calculated using Prism and the maximum binding at 25 μg/ml shown as fold increase relative to background with a non-HIV specific rabbit mAb. (B) The HCDR3 lengths of all rabbit heavy chains are shown for each individual animal. The median is represented by the horizontal line and the interquartile range by the vertical line. (C) The percentage of mAbs specific for either BG505 SOSIP.664 or for both BG505 SOSIP.664 and gp120 are shown below the pie charts for the three animals indicated. See also Figures S1.
The heavy-chain CDR3s (HCDR3s) of the mAbs have median lengths of 14, 15 and 16 for rabbits 1410, 1411 and 1412, respectively, which are similar to those previously reported for rabbits (Lavinder et al., 2014), with individual HCDR3 sequences up to 24 amino acids (Figure 1B). The median lengths and presence of longer HCDR3s are similar to observations made on human Abs (Lavinder et al., 2014). Sequence analysis of the mAb heavy chains revealed multiple separate clonal lineages from each rabbit as expected for a polyclonal response. However, a number of clonally related sequences were also recognized. Each mAb was named with the rabbit identifier (10, 11 or 12) and then a unique alphabetical lineage identifier (e.g. A, B etc). This naming was sufficient to identify the prototype Abs: 10A, 10B, 11A, 11B etc. Additional lineage members were then assigned a further number: 10A1, 11A1, 11A2 etc. Of the multi-member lineages, all mAbs in the 10A, 11A and 11B families bound to both BG505 gp120 and SOSIP.664 proteins as did mAbs 10B and 10C.
Potent autologous neutralizing activities of rabbit mAbs
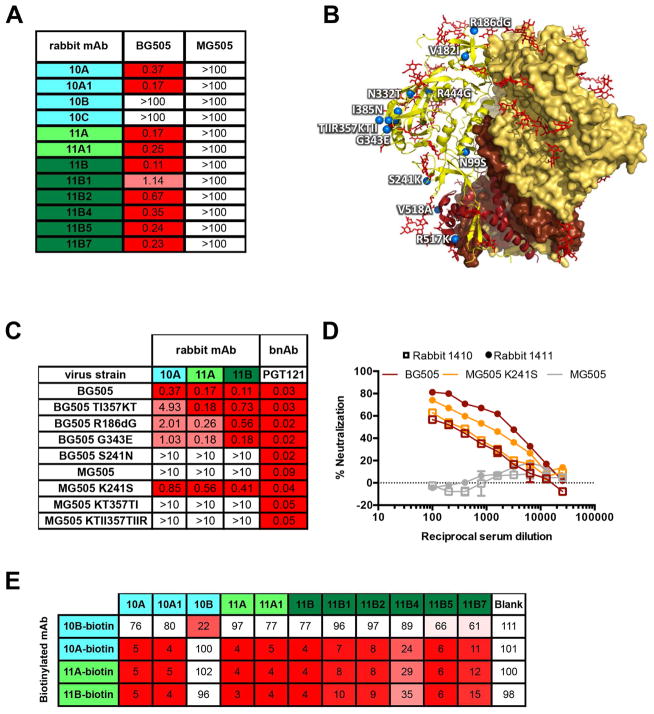
The rabbit mAbs were evaluated for their ability to neutralize BG505, the related maternal parent virus MG505.A2 (Wu et al., 2006), and a small panel of heterologous viruses. The mAbs belonging to the 10A, 11A and 11B lineages potently neutralized BG505 with IC50 values ranging from 0.1–1.1 μg/ml, but did not neutralize MG505.A2 (Figure 2A) or any heterologous viruses (Table S2). mAbs 10B and 10C were not able to neutralize the Tier 2 BG505 virus, but were able to neutralize heterologous Tier 1 viruses (Table S2), and as such represent off-target responses (de Taeye et al., 2015; Sanders et al., 2015).
Figure 2. Potently neutralizing mAb families converge on a common binding site on Env trimer.
(A) The rabbit mAbs were titrated against the indicated pseudoviruses in the TZM-bl luciferase reporter assay and the IC50 values shown were calculated using Prism. (B) Crystal structure of the BG505 SOSIP.664 trimer (PDB: 4NC0). The gp120 subunit is represented in yellow and the gp41 subunit in brown. N-linked glycans are highlighted in red sticks. Residues at which the MG505.A2 strain differs from the BG505 immunogen are highlighted as blue spheres and indicated with white labels. (C) The rabbit mAbs were titrated against the pseudovirus mutants indicated in the left-hand column in the TZM-bl luciferase reporter assay and the IC50 values were calculated using Prism. (D) The sera of rabbits 1410 and 1411 were titrated against the BG505, MG505 and MG505 K241S pseudoviruses in the TZM-bl luciferase reporter assay. (E) The percent binding of biotinylated rabbit mAbs was tested in the presence of the indicated non-biotinylated competitors, where 100% was the absorbance measured in the absence of competitor. See also Figure S2 and Tables S2, S4 and S5.
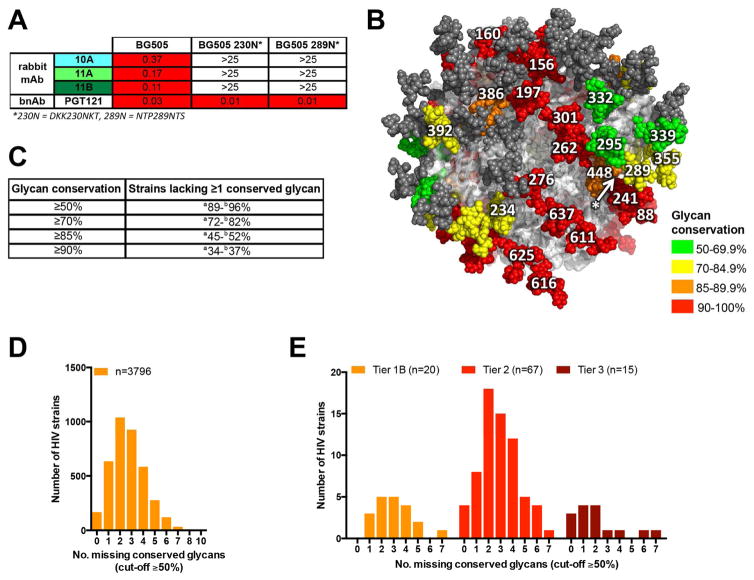
The MG505.A2 virus differs in amino-acid sequence from BG505 at 13 residues (Figure 2B). Thus, one or more of these 13 changes is crucial for loss of neutralization activity by nAbs 10A, 11A and 11B. Strikingly, these three nAbs were able to neutralize a point variant of MG505 that restored the BG505 serine at position 241, denoted MG505 K241S, with IC50 values close to those seen for BG505 and thus identifying the 241 position as crucial for nAb recognition (Figure 2C). Both BG505 and MG505 viral sequences are atypical at the 241 position, which normally encodes a potential N-linked glycosylation site (PNGS) in 97% of HIV strains (http://www.hiv.lanl.gov/) and neutralization of BG505 S241N was not observed (Figure 2C). Importantly, the 241-dependent nAbs comprise a major proportion of the BG505 serum neutralizing activity of rabbits 1410 and 1411 as the MG505 K241S virus is neutralized by the sera at titers almost as high as the autologous virus BG505 (Figure 2D). Furthermore, an additional 7 of 9 serum samples from BG505 SOSIP.664-immunized rabbits (Sanders et al., 2015) exhibited the same phenotype (Figure S2). In a separate study, immunization with BG505 SOSIP.664 incorporated onto liposomes followed by soluble BG505 SOSIP.664 boosting resulted in the same 241-dependent activity in 6 of 8 animals with autologous neutralizing titers (Figure S2). Therefore, it appears that sensitivity to position 241 is a major feature of autologous nAbs induced in rabbits by BG505 SOSIP.664 immunization.
Rabbit nAbs target a common Env region not previously characterized
Given the observations above, we reasoned that it was likely that the isolated mAbs recognize overlapping epitopes. Competitive BG505 SOSIP.664 binding studies tested the ability of each mAb to prevent binding of the biotinylated prototype lineage members 10A, 11A and 11B. As expected, within each lineage, the members showed mutual inhibition of binding (Figure 2E). In addition, the members of all three potently neutralizing lineages competed to varying degrees with the biotinylated prototypes of both other lineages as compared to a non-neutralizing mAb 10B. This suggests that all three sets of nAbs converge on a common overlapping binding site (Figure 2E) despite having arisen in two different animals and belonging to three distinct clonal families.
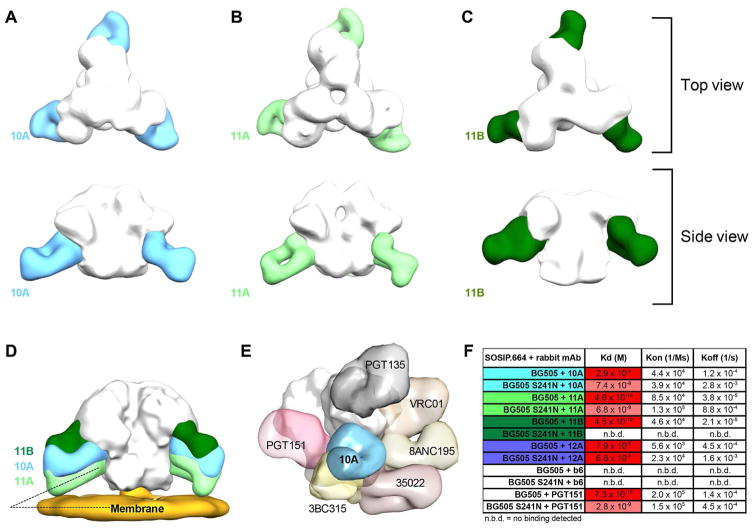
As an alternate approach to investigate the region of Env bound by the nAbs, we carried out negative-stain electron microscopy (NS-EM) analysis of BG505 SOSIP.664 trimers in complex with rabbit mAb fragment antigen binding (Fab) of 10A, 11A, and 11B. The results indicate a common epitope for nAbs 10A, 11A, and 11B situated on gp120 between the V4 loop and glycan N88 (Figure 3A–C; Figure S3). All three Abs bind with a stoichiometry of three Fabs per trimer. Remarkably, the three Fabs bind the trimers at an angle of approach from the membrane-facing portion of the Env ectodomain (Figure 3D). This angle is different from all characterized bnAbs isolated from human donors to date that either approach the trimer from the apex or bind parallel to the membrane. The lack of constraints in the solubilized membrane-free immunogen likely makes induction of nAbs with this approach angle possible. However, the rabbit mAbs neutralize BG505 pseudovirus, in which Env is of course anchored to the membrane, and therefore shows that this constraint does not appear to strongly interfere with Ab binding.
Figure 3. Negative-stain electron microscopy shows a common site of Ab binding to the BG505 SOSIP trimer.
3D reconstructions of (A) nAb 10A, (B) nAb 11A and (C) nAb 11B each in complex with BG505 SOSIP.664. (D) nAbs 10A, 11A,11B share a steep angle of approach from the C-terminal end of the trimer. (E) Comparison of nAb 10A to previously identified bnAbs. (F) Kd values and kinetic parameters for mAb/SOSIP binding studies using an Octet RED96 BLI system. Papain-digested mAb Fabs were used in all experiments above. See also Figure S3.
When the 3D EM reconstructions are compared to known bnAb sites, they reveal a region of the Env surface not previously described as being a recognition site for bnAbs (Figure 3E). Centrally placed in this region is residue 241, which, as described above, is a serine in BG505 and not a PNGS as commonly found in other strains. The region thus constitutes a “hole” in the glycan shield. Interestingly, introduction of the S241N mutation into BG505.664 does not completely abolish binding of nAbs 10A or 11A to the trimer when assayed by ELISA or biolayer interferometry (BLI) (Figure 3F; Figure S4). However, the off rates for both nAbs are dramatically increased in the presence of N241, illustrating that the glycan alters the epitope in some manner. The increased off rates result in ~10-fold decrease in measured binding affinities of the two Fabs to BG505 SOSIP.664 (Figure 3F). Rabbit nAb 11B did not bind to the S241N mutant of BG505 SOSIP.664, suggesting that its epitope is slightly different from those of 10A and 11A. When viewed from the side, the three Abs bind at different vertical levels of Env, with 11B binding slightly closer to the apex, followed by 10A, and 11A binding closer to the membrane (Figure 3D). This difference is further corroborated by competition assays with human gp120-gp41 interface mAbs (Figure 4A). Of interest, earlier studies on a panel of BG505 alanine-scanning mutant viruses (Sanders et al., 2015) did not identify residue 241 as critical for neutralization; S241A mutant and wild-type BG505 viruses were equally neutralized by immune sera. However, this may be due to the similarity of S and A. We re-examined the serum alanine-scanning data from rabbits 1410 and 1411 and compared that data with similar alanine-scanning data for mAbs 10A and 11A (Table S3). This showed that sera and mAbs have a shared sensitivity to certain viral mutants, which do not overlap with their epitopes, raising the possibility that the effects of the substitutions are allosteric in nature.
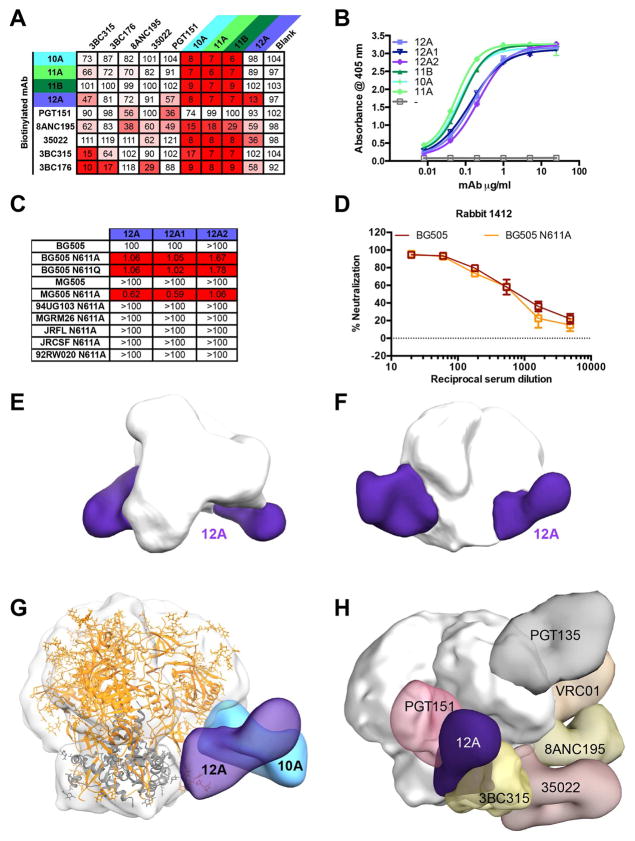
Figure 4. Glycan-sensitive potent neutralization by rabbit mAbs.
(A) The competitor non-biotinylated mAbs listed in the top row were pre-incubated with Avi-tagged BG505 SOSIP.664 protein. Binding of the biotinylated mAbs listed in the first column is expressed as percentage binding where 100% was the absorbance measured in the absence of competitor. (B) Binding of neutralizing rabbit mAbs indicated in the legend was assayed by ELISA using streptavidin-coated plates to capture Avi-tagged biotinylated BG505 SOSIP.664. (C) The rabbit mAbs were titrated against the pseudovirus mutants indicated in the top row in the TZM-bl luciferase reporter assay and the IC50 values were calculated using Prism. (D) The sera of rabbit 1412 were titrated against the BG505 and BG505 N611A pseudoviruses in the TZM-bl luciferase reporter assay. (E, F) Negative-stain EM 3D reconstructions of Ab 12A in complex with BG505 SOSIP.664. (G) nAb 12A shows a different angle of approach from nAbs isolated from rabbits 1410 and 1411. (H) The binding site of nAb 12A is shown relative to select bnAbs and exhibits similarities to gp120-gp41 interface bnAb PGT151. See also Figures S3–4 and Tables S3–5.
The non-Tier 2-nAbs, 10B and 10C did not compete significantly for the shared site bound by the nAbs (Table S5). Epitope mapping using a D368R mutant BG505 gp120 and a V3 peptide-Fc fusion construct indicated that 10B binds to V3 and 10C to the CD4bs of gp120, in addition, one non-nAb from rabbit 1410 bound to post-fusion MN gp41 soluble protein (data not shown). A proportion of non-nAbs were found to compete with human bnAbs 8ANC195, 35022, 3BC315 and 3BC176, which bind the gp120-gp41 interface (Table S4). This is similar to the results described in (Hu et al., 2015) following BG505 SOSIP.664 immunization of mice. IgG was purified from non-neutralizing mouse serum and found to compete most strongly with 3BC315 alone (62%) or in combination with 35022 (74%).
A second weak autologous nAb specificity is identified that is glycan sensitive
The third rabbit from which mAbs were isolated, 1412, also yielded a neutralizing lineage, designated 12A, comprising 3 members, all of which bind BG505 SOSIP.664 but not gp120 (Figure 1A, Figure 4B). The activity of the 12A prototype was identified in the original neutralization screen (Table S2). However, its neutralization potency was very weak with IC50 values approaching 100 μg/ml, and lower affinity for SOSIP trimers relative to 10A, 11A, and 11B (Figure 3F, 4B). As 12A required gp41 or a trimer configuration for binding, it was tested against gp41 mutant pseudoviruses previously generated to characterize the human bnAb PGT151 (Blattner et al., 2014; Falkowska et al., 2014). Of these, an N611A BG505 variant was much more sensitive (~100-fold) to 12A family neutralization than the wild-type BG505 virus (Figure 4C). However, nAb 12A did not show any major increase in breadth in terms of neutralization (Figure 4C) regardless of the absence of N611.
In contrast to the 241-dependent response described above, the N611-dependent response is unlikely to be a major component of the neutralization activity of the donor rabbit (1412) as the serum neutralized wild type and N611A variant BG505 isolates similarly (Figure 4D). However, competition studies indicate that the N611-dependent response overlaps, in terms of Env footprint, to a degree with the 10A, 11A and 11B nAbs and many non-nAbs isolated (Figure 4A, Table S5). Notwithstanding, EM analysis illustrates that the binding site of rabbit nAb 12A is indeed different from that of 10A, 11A, and 11B, and the antibody exhibits a steeper angle of approach to Env, a feature that is more in common for human bnAbs. In fact, 12A binds an epitope closer to PGT151 and does not always bind with a stoichiometry of 3 Fabs per trimer (Figure 4E–F). Overall, it is noteworthy that another nAb response has been identified to target an epitope in the vicinity of that targeted by the 241-centered Abs and that potent neutralization is again dependent on the absence of glycosylation at a key position.
The incidence of missing glycan sites in HIV Env trimer generally
The 241 glycan site is conserved in 97% of HIV-1 isolates. The wild-type BG505 virus lacks not only this site but also the conserved glycan sites at 289 and 332, which are 72% and 67% conserved, respectively, in other isolates (Figure S5). However, the BG505 immunogen and viruses used in this study were engineered to contain N332 (Sanders et al., 2015). The 289 glycan site is in proximity to the 241 site and an absence of a glycan at this position could contribute to a larger glycan hole. To assess this we generated a 289 glycan site knock in virus, and also a 230 glycan site knock in virus; this second glycan is absent in BG505 but not conserved in >50% of strains. The reintroduction of either glycan was able to ablate the neutralization activity of all three nAbs. However, as neither glycan is present in MG505, they do not prevent neutralization of the resistant MG505 pseudovirus, which is rendered sensitive to neutralization by the substitution of lysine for serine at position 241 (Figure 2C). BG505 is not unusual in lacking highly conserved glycan sites. Analysis of 3792 unique sequences (www.lanl.gov) shows that 96% of strains are missing one or more conserved (defined as ≥50% incidence) glycan sites (Table 1). This does not necessarily mean that 96% of isolates have a glycan hole. Nearby glycan sites might provide a compensatory glycan that could, in effect, substitute for the missing glycan (Sok et al., 2013). Considering N332 and N334 glycan sites (which are mutually exclusive), approximately 33% of isolates lack the conserved N332 glycan site, but, of this subset, 27% of isolates have the N334 site. Thus 94% of isolates have a glycan site at either N332 or N334 and the presence of a glycan hole in this region may be as low as 6%. Different approaches are possible for estimating the probability of nearby glycans compensating for a given glycan and leading to reduced likelihood of a glycan hole. Using one approach based on potential glycan sites within 4 residues of a missing glycan compensating for that glycan, the figure of 96% above is reduced to 89% of strains (Table 1). However, it should be noted that this number is based on one approach to calculation of potential glycan substitution and further detailed experimental studies are warranted to explore the phenomenon of glycan substitution and its potential role in glycan holes. Alternatively, if missing glycan sites in a given isolate are in close spatial proximity they may create a larger hole, as is suggested by the ablation of neutralization activity by the introduction of a glycan at 289 or 230 in the case of BG505. The individual glycan sites that are absent in viruses that lack multiple conserved glycan sites vary between isolates. In some cases, they are in close proximity and may contribute to a larger glycan hole, such as that seen in BG505, while others are well separated and these strains are predicted to have multiple discrete glycan holes. Across 3792 unique Env sequences, no individual pair of glycan sites was found to be missing more often than would be predicted by their individual conservation level (Figure S5). Of the 3407 unique strains that lack two or more conserved glycan sites, 33 strains are missing glycan sites at 241 and 289 (including BG505), but almost one third (1030 strains) are missing 289. Across these 1030 strains, the other most commonly missing glycan sites are 234, 295, 339, 355 and 625 (Figure S5). Of these, 295, 339 and 355 are within 22Å of the 289 glycan site in Env (Figure 5B), so their combined absence with 289 could produce a larger glycan hole similar to that found in BG505 in approximately 20% of strains, when considering just this one 72% conserved glycan site as the focal point. Thus the formation of a larger hole in the glycan shield in strains lacking multiple conserved glycan sites is not unlikely. However, how many missing glycan sites are needed to provide an immunodominant target for neutralizing activity remains to be determined experimentally from further immunization studies.
Table 1. Summary of glycan conservation across HIV viruses.
A) Different definitions of “conserved” (≥ 50 or ≥ 70 or ≥ 85 or ≥ 90%) listed in the first column, glycan sites in variable loops were not included in this analysis due to difficulties in aligning the variable loops of Env, given the high levels of diversity in both loop length and sequence. The percentage of strains lacking at least one conserved glycan site is shown in the second column according to alignment to HXB2 PNGS. The third column shows the same figure if allowance is made for nearby glycan sites that could in principle substitute for a given glycan (see Experimental Procedures).
| Glycan conservation | % HIV strains missing a conserved glycan site | % HIV strains missing a conserved glycan site when proximal glycan sites are considered to substitute for one another | Conserved glycan site residue numbers (HXB2) |
|---|---|---|---|
| ≥50 | 96% | 89% | 88, 156, 160, 197, 234, 241, 262, 276, 289, 295, 301, 332, 339, 355, 386, 392, 448, 611, 616, 625, 637 |
| ≥70 | 82% | 72% | 88, 156, 160, 197, 234, 241, 262, 276, 289, 301, 339, 355, 386, 392, 448, 611, 616, 625, 637 |
| ≥85 | 52% | 45% | 88, 156, 160, 197, 241, 262, 276, 301, 386, 448, 611, 616, 625, 637 |
| ≥90 | 37% | 34% | 88, 156, 160, 197, 241, 262, 276, 301, 611, 616, 625, 637 |
Figure 5. Distribution and prevalence of HIV Env glycan holes.
(A) Rabbit mAbs were titrated against the pseudovirus mutants indicated in the left-hand column in the TZM-bl luciferase reporter assay and the IC50 values were calculated using Prism. No neutralization activity is indicated by an IC50 of >25. The mAbs are no longer able to neutralize viruses in which either the N230 or N289 glycan sites are restored. (B) Model of a fully glycosylated BG505 SOSIP.664 trimer constructed using PDB: 5ACO with glycans 184, 465, 332, 289 and 241 (which are absent from the PDB file) modeled onto the structure. The glycan site found at 230 in <50% of strains, not including BG505, is indicated by * and an arrow points to the space that would be occupied by the 230 glycan as residue 230 lies behind the 448 glycan between 241, 295 and 289. Glycan sites depicted on all three protomers of the trimer are color-coded by their level of conservation across 3792 unique Env sequences aligned to HXB2 PNGS (glycan site residue numbers are shown on only one protomer). (C) Percentage of strains lacking at least one conserved glycan according to different definitions of “conserved” (≥ 50 or ≥ 70 or ≥ 85 or ≥ 90%). For each definition, two percentage values are given: (a) is the lower estimate for strains lacking at least one conserved glycan, calculated by counting conserved glycan sites aligned to HXB2 PNGS for each strain and also counting nearby glycan sites that could in principle provide a compensating glycan as explained in the text. (b) Is the upper estimate for strains lacking at least one conserved glycan, calculated by only counting conserved glycan sites aligned to HXB2 PNGS for each strain (see Table 1 for additional information). (D) The number of HIV strains lacking between 0 and 7 PNGS at Env positions where a PNGS is conserved at the ≥50% glycan site cut-off level. (E) The number of missing glycan sites, at positions where a PNGS is conserved by 50% or more, among a subset of viruses (n=102) viruses described in (Seaman et al., 2010) stratified by neutralization resistance tier. See also Figure S5.
Discussion
This study has identified a highly immunogenic neutralizing site on the HIV Env BG505 SOSIP.664 trimer, centered on residue 241 of gp120, that is recognized by three separate potent nAb lineages (10A, 11A and 11B) derived from two immunized rabbits. Viral mutagenesis and mapping studies show that the potent autologous serum neutralizing activity in the two donor animals can be largely assigned to the 241-dependent specificity. In addition, serum studies on a larger number of animals shows the prevalence of this neutralizing specificity in 67–75% of rabbits that were capable of neutralizing BG505 (including animals in separate immunization studies). Therefore, immunization of rabbits with BG505 SOSIP.664 produces a reproducible immunodominant autologous Tier 2 neutralizing response directed to a common epitope region. Of note, another nAb lineage was identified in a third rabbit that recognized a different, but overlapping epitope at the gp120-gp41 interface.
Tier 2 autologous nAb responses have been described previously through immunization with DNA prime-Env boost regimes (Crooks et al., 2015; Narayan et al., 2013), with a DNA/VLP-Env regime (Crooks et al., 2015) and with Env gp140 (Bradley et al., 2016), but such responses, although encouraging, were sporadic, occurring in few animals. The latter identified an autologous nAb lineage, from 1 of 6 NHPs immunized with transmitter/founder Env. The nAbs bind a V5 epitope that can be blocked by the introduction of a glycan at position 463 (<50% conserved), which was observed in the donor virus 6 months after transmission. In the DNA/VLP-Env study, sera from a small number of rabbits immunized with VLP-associated JR-FL Env potently neutralized the JR-FL isolate, which lacks a glycan site at position 197, which is also an unusual feature compared to other isolates (Crooks et al., 2015). Furthermore, resistant viruses derived from several Tier 2 isolates, when engineered to eliminate the glycan site at N197, were also neutralized by the sera. The data were interpreted to suggest a hole at position 197 in the glycan shield that was being targeted by the immune sera. Importantly, in these studies neutralization was only observed in a small fraction of immunized animals, in contrast to the study described herein. In another study, Tier 1 nAbs induced during infection of NHP with a virus lacking the N160 glycan site have been shown to bind a V2/V3 epitope that is occluded by the re-introduction of the N160 glycan (Krachmarov et al., 2011). Thus the principle of targeting a hole in the glycan shield following Env immunization is clearly supported. The extent to which glycan holes contribute to antibody response to BG505 SOSIP.664 and additional Env trimers derived from other HIV strains are currently under investigation in multiple species. Retrospectively, it is interesting that the absence of this highly conserved glycan was not predicted to be a target of autologous serum neutralization. It was, however, noted in structural studies that the 241 glycan was missing in BG505, necessitating this glycan to be modeled into a recent high-resolution structure of glycosylated Env trimer to more fully represent circulating strains (Pancera et al., 2014).
A number of studies suggest that holes in the glycan shield can play an important role in neutralizing antibody responses in humans. First, many studies of natural infection have described autologous neutralizing responses from which viruses escape by addition of a glycan site that may correspond to the filling of holes (Bunnik et al., 2008; Moore et al., 2009; Rong et al., 2009; van Gils et al., 2010; Wei et al., 2003). As a single example, Wei et al (Wei et al., 2003) showed that viruses from one donor (WEAU) lacked the highly conserved glycan at position 234 until day 212 when escape from the autologous plasma was observed. Notably, removal of the glycan site from the day 212 escape virus resulted in a 20-fold increase in contemporaneous plasma neutralization (Wei et al., 2003). Second, study of inferred precursors of the PGT121 family of bnAbs suggests that the early response is directed to an Env lacking certain glycan sites (Sok et al., 2014) and later matures to the more fully glycosylated Env (Garces et al., 2015). Third, the lack of a glycan at position 276 is suggested as important for the initiation of VRC01-class antibody lineages targeting the CD4 binding site and designed Env immunogens lacking the glycan N276 are efficient in binding and activation of VRC01-class precursors (Jardine et al., 2013; Jardine et al., 2015; Lynch et al., 2015). Fourth, two donors infected with viruses lacking the N332 glycan site developed autologous nAbs that were postulated to target the 332 region (Moore et al., 2012). Subsequently, escape viruses from the same donors acquired the N332 site and developed bnAb lineages targeting the N332 glycan, although whether the bnAb and autologous nAb lineages are related is not known (Moore et al., 2012). Indeed, whether autologous nAbs can be the precursors of bnAb lineages is unclear. Data from natural infection suggest that autologous nAbs may pressure the patient virus to expose immunotypes which trigger the induction of bnAbs (Moody et al., 2015; Moore et al., 2015).
How does antibody targeting of breaches in the glycan defenses of Env affect HIV vaccine development that seeks to elicit bnAbs? We have shown that the majority of Tier 2 isolates lack at least one conserved glycan site, with the absence of two conserved sites being the most common occurrence. For the existing Tier 2 Env immunogen BG505 SOSIP.664, one approach to increase the breadth of the response is to fill in the isolate-specific glycan holes in the hope that this will re-direct the Ab response to more cross-reactive epitopes that have already been defined by known bnAbs (Burton and Hangartner, 2016; Burton and Mascola, 2015; West et al., 2014). This strategy would effectively mimic the post-autologous nAb viral escape stage of natural infection, where autologous nAbs have selected for viruses with fewer immunogenic glycan holes, potentially resulting in greater bnAb epitope exposure. To this end, it would be invaluable to establish whether any bnAb precursors are present even at low to very low levels in the responses to BG505 SOSIP.664 immunization. A second approach is to design immunogens to broaden the Tier 2 isolate-specific responses described. For example, one might seek to introduce a glycan, perhaps of a shorter oligosaccharide chain, at position 241 as a boosting antigen. In this context, it is noteworthy that both 10A and 11A bind to BG505 S241N SOSIP.664 protein, albeit at a lower level than wild type. Thus, the results suggest that these potent nAbs can accommodate, to some degree, a glycan at position 241, unlike 11B. For BG505-derived immunogens both of these approaches could be facilitated by higher resolution structures of complexes of additional immunization-induced nAbs and trimers. Furthermore, our findings provide a rationale for the design of immunogens based on other HIV strains in terms of examining which conserved glycan sites are absent and filling these potential holes.
In summary, we have uncovered a paradigm of vulnerability on Env immunogens; glycan holes in Tier 2 stabilized Env trimers are immunogenic and can be a dominant target of autologous neutralization. The role of a glycan hole in autologous rabbit serum BG505 neutralization described in this study agrees with the established role of glycan-mediated escape from autologous neutralization during natural infection. Therefore, it is now critical to consider the glycan holes present in individual HIV isolates when designing immunogens and evaluating post-immunization neutralizing antibodies.
Experimental Procedures
FACS
Cryopreserved PBMCs were thawed, resuspended in 10 ml of RPMI 10% FCS, and collected by centrifugation at 600 × g for 5 min. Cells were washed with PBS and resuspended in 10 ml of PBS and collected by a second centrifugation step. Cells were resuspended in 100 μl of FWB (2% FCS PBS) with anti-rabbit IgM FITC (1:1000) and a streptavidin-APC tetramer of biotinylated anti-rabbit IgG. After 1 h on ice, cells were washed once with 10 ml of PBS, collected by centrifugation, and resuspended in 100 μl of FWB with 1 μl of a streptavidin-PE tetramer of biotinylated BG505 SOSIP.664 and 1 μl of a mixed streptavidin-BV786 tetramer of B41 and ZM197 SOSIP.664. After a further 1 h on ice, cells were washed once with 10 ml of PBS, collected by centrifugation and resuspended in 500 μl of FWB for sorting on a BD FACS Aria III. IgM-IgG+BG505+B41+ZM197+ lymphocytes were collected at 1 cell per well into Superscript III Reverse Transcriptase lysis buffer (Invitrogen) as previously described and immediately stored at −80°C prior to cDNA generation and single cell PCR (see Table S1).
Antibodies and Fab generation
Rabbit Ab variable regions (Genbank accession numbers: KX571250-1324) were cloned into an expression plasmid adapted from the pFUSE-rIgG-Fc and pFUSE2-CLIg-rK2 vectors (Invivogen). Human and rabbit Abs were transiently expressed with the FreeStyle 293 Expression System (Invitrogen). Abs were purified using affinity chromatography (Protein A Sepharose Fast Flow, GE Healthcare) and the purity and integrity were checked by SDS–PAGE. To generate Fabs, rabbit IgG was digested with 2% papain (Sigma P3125) in digestion buffer (10 mM L-cysteine, 100 mM Na acetate pH 5.6, 0.3 mM EDTA) for 6 h then quenched with 30 mM iodoacetamide. Undigested IgG and Fc fragments were removed by affinity chromatography and the Fab-containing flow through was collected. Size-exclusion chromatography was performed using Superdex 200 10/300 resin (GE Healthcare) to remove papain and digestion byproducts.
Controls
For all neutralization, ELISA and flow cytometry assays, where indicated, the (+) positive control was the anti-V3 mAb R56 (PDB: 4JO1) and the (-) negative control was a hybrid using the heavy chain of R56 and light chain of R20 which cannot bind HIV Env (PDB: 4JO3).
Neutralization assays
Pseudovirus neutralization assays using TZM-bl target cells were carried out as previously described (Li et al., 2005).
Enzyme-linked immunosorbent assay (ELISA)
ELISA assays were performed as described in (Sanders et al., 2015) and (Derking et al., 2015).
Statistical methods
Statistical analyses were performed using Graph Pad Prism 6 for Windows, Graph Pad Software.
Site-directed mutagenesis
Env and mAb mutants were generated using the QuikChange site-directed mutagenesis kit (Agilent) according to the manufacturer’s instructions.
Biolayer interferometry
His-tagged BG505 SOSIP.664 trimers were loaded onto Ni-NTA biosensors and dipped into varying concentrations of rabbit Fab (1000, 500, 250, 125, 62.5, 32.25, 16 nM) using an Octet Red96 instrument (ForteBio). All samples were previously diluted in 1 X kinetics buffer (phosphate-buffered saline pH 7.2, 0.01% [w/v] BSA, 0.002% [v/v] Tween-20). Association was measured for 300 s followed by dissociation for 1200 s in 1 X kinetics buffer. Anti-HIV Fabs b6 and PGT151 were used as negative and positive controls, respectively, to ensure trimer integrity. A reference well containing 1 X kinetics buffer was subtracted from each data set and curves were aligned on the Y-axis using the baseline step and an interstep correction was applied between the association and dissociation curves. A 1:1 binding model, which assumes first-order kinetics and that Fabs bind to all available sites on the trimer at an equal rate, was fit to the data (see Figure S4).
Negative-stain electron microscopy
BG505 SOSIP.664 was incubated with a 6-molar excess of rabbit Fabs 10A, 11A, 11B, or 12A overnight at room temperature. Grid preparation, data collection, and data processing followed the same protocol described elsewhere (de Taeye et al., 2015) using Leginon and Appion software suites (Lander et al., 2009; Suloway et al., 2005), see Figure S3 for additional information. EM reconstructions have been deposited to the Electron Microscopy Data Bank under the accession numbers EMD-8309, EMD-8310, EM-8311, and EMD-8312.
Immunizations
The immunization of animals 1254,1256–7, 1274, 1278, 1279,1283–5, 1410, 1411 and 1412 was previously described (Sanders et al., 2015). In addition, 3 groups of New Zealand White rabbits (3409–3420) were immunized twice with either sham liposomes or liposomes embedded with post fusion gp41 or BG505 SOSIP.664. Proteins were conjugated to pegylated distearoylphosethanolamine (PEG-DSPE) using maleimide chemistry (see Figure S2). All animals then received three soluble BG505 SOSIP.664 protein boosts as detailed in Figure S2. The TSRI IACUC approved all experimental procedures involving rabbits 3409–3420.
Bioinformatics analysis
The analysis of glycan site conservation was based on sequence datasets from LANL (http://www.hiv.lanl.gov/). 3792 unique sequences were selected according to the PatientCode field. The PNGS sequon N-X-S/T, where X is any amino acid except proline, was used to search the alignment and tabulate the number of PNGS at each position. In addition, the number of PNGS at each position was also calculated to allow for nearby glycan substitution by discounting those strains where a glycan site is found up to 4 residues away from each conserved (≥50%) HXB2 PNGS site. Note that large sequence alignments containing both closely and distantly related sequences of varying length can contain local regions with gaps and may result in variation of assigned positions between different alignment algorithms. Regions with many gaps, such as the variable loops, were excluded from this analysis and only regions with no or minimal gaps were included in order to minimize PNGS counting errors. For the glycan modeling GlycanRelax (Pancera et al., 2010) was employed to generate conformations of Man8 at all predicted N-linked glycosylation motifs in PDB:4TVP and at positions 184, 241, 289, 332 and 465, which are naturally missing in BG505. A single representative model was chosen for each figure.
Supplementary Material
Highlights.
HIV neutralizing antibodies were isolated from rabbits immunized with BG505 SOSIP.664.
These antibodies target a hole in the glycan shield of the BG505.
Serum neutralization specificity maps to the same immunodominant glycan hole.
Most HIV strains lack a conserved glycan site that could be a neutralization target.
Acknowledgments
This work was supported by the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Grant UM1AI100663 (to D.R.B, I.A.W, A.B.W, W.R.S.), NIH HIVRAD Grant P01 AI110657 (to R.W.S., I.A.W, A.B.W) and the International AIDS Vaccine Initiative Neutralizing Antibody Consortium through the Collaboration for AIDS Vaccine Discovery grants OPP1084519 and OPP1115782. Additionally, this study was financially supported by a Marie-Curie Fellowship (FP7-PEOPLE-2013-IOF) to (L.E.M). M.J.vG received funding from the Aids fonds Netherlands (grant #2012041) and EMBO (grant #ASTF260-2013). R.W.S is a recipient of a Vidi grant from the Netherlands Organization for Scientific Research (grant #917.11.314) and a Starting Investigator Grant from the European Research Council (ERC-StG-2011-280829-SHEV). This work was partially funded by IAVI with the generous support of USAID and the Bill & Melinda Gates Foundation; a full list of IAVI donors is available at www.iavi.org. The contents of this manuscript are the responsibility of the authors and do not necessarily reflect the views of USAID or the US Government or the other funding bodies. The authors would like to thank James C Paulson for his support and Christina Corbaci for assistance with figure preparation.
Footnotes
AUTHOR CONTRIBUTIONS
L.E.M, M.J.vG, G.O, M.S.M, A.B.W, R.W.S. and D.R.B designed the experiments. L.E.M, M.J.vG, G.O, T.M, M.S.M, J.L.T, M.P. performed the experiments. J.E.V, D.S, J.H, and R.W.S contributed critical reagents. L.E.M, G.O and D.R.B wrote the paper. L.E.M, M.J.vG, G.O, B.B, J.E.V, D.W.K, S.M, C.A.C, W.R.S, I.A.W, A.B.W, R.W.S and D.R.B. analyzed the data and edited the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, Kitchin D, Botha G, Gorman J, Garrett NJ, et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nature medicine. 2015;21:1332–1336. doi: 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley T, Fera D, Bhiman J, Eslamizar L, Lu X, Anasti K, Zhang R, Sutherland LL, Scearce RM, Bowman CM, et al. Structural Constraints of Vaccine-Induced Tier-2 Autologous HIV Neutralizing Antibodies Targeting the Receptor-Binding Site. Cell reports. 2016;14:43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. Journal of virology. 2008;82:7932–7941. doi: 10.1128/JVI.00757-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annual review of immunology. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nature immunology. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Vaine M, Wallace A, Han D, Wan S, Seaman MS, Montefiori D, Wang S, Lu S. A novel rabbit monoclonal antibody platform to dissect the diverse repertoire of antibody epitopes for HIV-1 Env immunogen design. Journal of virology. 2013;87:10232–10243. doi: 10.1128/JVI.00837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, et al. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS pathogens. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, et al. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS pathogens. 2015;11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, Torrents de la Pena A, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. Aids. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, et al. Murine Antibody Responses to Cleaved Soluble HIV-1 Envelope Trimers Are Highly Restricted in Specificity. Journal of virology. 2015;89:10383–10398. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov C, Lai Z, Honnen WJ, Salomon A, Gorny MK, Zolla-Pazner S, Robinson J, Pinter A. Characterization of structural features and diversity of variable-region determinants of related quaternary epitopes recognized by human and rhesus macaque monoclonal antibodies possessing unusually potent neutralizing activities. Journal of virology. 2011;85:10730–10740. doi: 10.1128/JVI.00365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. Journal of structural biology. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PloS one. 2014;9:e101322. doi: 10.1371/journal.pone.0101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. Journal of virology. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch RM, Wong P, Tran L, O’Dell S, Nason MC, Li Y, Wu X, Mascola JR. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. Journal of virology. 2015;89:4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LE, Quigley AF, Strokappe NM, Bulmer-Thomas B, Seaman MS, Mortier D, Rutten L, Chander N, Edwards CJ, Ketteler R, et al. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. The Journal of experimental medicine. 2012;209:1091–1103. doi: 10.1084/jem.20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy LE, Rutten L, Frampton D, Anderson I, Granger L, Bashford-Rogers R, Dekkers G, Strokappe NM, Seaman MS, Koh W, et al. Molecular evolution of broadly neutralizing Llama antibodies to the CD4-binding site of HIV-1. PLoS pathogens. 2014;10:e1004552. doi: 10.1371/journal.ppat.1004552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody MA, Gao F, Gurley TC, Amos JD, Kumar A, Hora B, Marshall DJ, Whitesides JF, Xia SM, Parks R, et al. Strain-Specific V3 and CD4 Binding Site Autologous HIV-1 Neutralizing Antibodies Select Neutralization-Resistant Viruses. Cell host & microbe. 2015;18:354–362. doi: 10.1016/j.chom.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nature medicine. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Ranchobe N, Lambson BE, Gray ES, Cave E, Abrahams MR, Bandawe G, Mlisana K, Abdool Karim SS, Williamson C, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS pathogens. 2009;5:e1000598. doi: 10.1371/journal.ppat.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Williamson C, Morris L. Virological features associated with the development of broadly neutralizing antibodies to HIV-1. Trends in microbiology. 2015;23:204–211. doi: 10.1016/j.tim.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan KM, Agrawal N, Du SX, Muranaka JE, Bauer K, Leaman DP, Phung P, Limoli K, Chen H, Boenig RI, et al. Prime-boost immunization of rabbits with HIV-1 gp120 elicits potent neutralization activity against a primary viral isolate. PloS one. 2013;8:e52732. doi: 10.1371/journal.pone.0052732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phad GE, Vazquez Bernat N, Feng Y, Ingale J, Martinez Murillo PA, O’Dell S, Li Y, Mascola JR, Sundling C, Wyatt RT, et al. Diverse antibody genetic and recognition properties revealed following HIV-1 envelope glycoprotein immunization. Journal of immunology. 2015;194:5903–5914. doi: 10.4049/jimmunol.1500122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Banerjee S, Agrawal A, Shi H, Banasik M, Lin F, Rohl K, LaBranche C, Montefiori DC, Cho MW. Characterization of a Large Panel of Rabbit Monoclonal Antibodies against HIV-1 gp120 and Isolation of Novel Neutralizing Antibodies against the V3 Loop. PloS one. 2015;10:e0128823. doi: 10.1371/journal.pone.0128823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Shi H, Banerjee S, Agrawal A, Banasik M, Cho MW. Detailed characterization of antibody responses against HIV-1 group M consensus gp120 in rabbits. Retrovirology. 2014;11:125. doi: 10.1186/s12977-014-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, Allen SA, Pinter A, Shaw GM, Hunter E, et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS pathogens. 2009;5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffner T, Kong L, Duncan CJ, Back JW, Benschop JJ, Shen X, Huang PS, Stewart-Jones GB, DeStefano J, Seaman MS, et al. Immune focusing and enhanced neutralization induced by HIV-1 gp140 chemical cross-linking. Journal of virology. 2013;87:10163–10172. doi: 10.1128/JVI.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. Journal of virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien JP, Menis S, Wickramasinghe L, et al. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Science translational medicine. 2014;6:236ra263. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS pathogens. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sundling C, Forsell MN, O’Dell S, Feng Y, Chakrabarti B, Rao SS, Lore K, Mascola JR, Wyatt RT, Douagi I, et al. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. The Journal of experimental medicine. 2010;207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O’Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Science translational medicine. 2012;4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Zhang Z, Phad GE, Sheng Z, Wang Y, Mascola JR, Li Y, Wyatt RT, Shapiro L, Karlsson Hedestam GB. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. Journal of immunology. 2014;192:3637–3644. doi: 10.4049/jimmunol.1303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaine M, Wang S, Crooks ET, Jiang P, Montefiori DC, Binley J, Lu S. Improved induction of antibodies against key neutralizing epitopes by human immunodeficiency virus type 1 gp120 DNA prime-protein boost vaccination compared to gp120 protein-only vaccination. Journal of virology. 2008;82:7369–7378. doi: 10.1128/JVI.00562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gils MJ, Bunnik EM, Burger JA, Jacob Y, Schweighardt B, Wrin T, Schuitemaker H. Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors. Journal of virology. 2010;84:3576–3585. doi: 10.1128/JVI.02622-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gils MJ, Sanders RW. In vivo protection by broadly neutralizing HIV antibodies. Trends in microbiology. 2014;22:550–551. doi: 10.1016/j.tim.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS pathogens. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SM, Overbaugh J. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. Journal of virology. 2006;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Fu H, Luallen RJ, Liu B, Lee FH, Doms RW, Geng Y. Antibodies elicited by yeast glycoproteins recognize HIV-1 virions and potently neutralize virions with high mannose N-glycans. Vaccine. 2015;33:5140–5147. doi: 10.1016/j.vaccine.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.