Abstract
We have utilized a PCR-based analysis of single-strand conformation polymorphisms to identify polymorphisms that can be used for mapping cloned DNA sequences in the mouse. We have found that single-strand conformation polymorphism analysis of sequences that are potentially less subject to conservation (i.e., intron and 3' untranslated regions) is a relatively efficient means of detecting polymorphisms between inbred strains. Fifty percent of the tested primer pairs were polymorphic between inbred strains and 90% were polymorphic between mouse species, which is a frequency comparable to that found for microsatellite repeat sequences. We have found that this technique can be readily used to determine the strain distribution pattern in a recombinant inbred series and is a simple and rapid means to obtain a map position for cloned sequences. When this strategy was tested on a number of previously mapped cloned genes, the strain distribution patterns obtained were consistent with that to be expected on the basis of the known map position. We also tested the utility of this approach for characterizing genes that have not been previously mapped. Dvl, the mouse homolog of the putative Drosophila dishevelled gene, and Adfp, encoding an adipocyte differentiation-related protein, were found to map to chromosome 4. These results were confirmed using single-strand conformation polymorphism analysis of an interspecific backcross.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avner P., Amar L., Dandolo L., Guénet J. L. Genetic analysis of the mouse using interspecific crosses. Trends Genet. 1988 Jan;4(1):18–23. doi: 10.1016/0168-9525(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Buchberg A. M., Moskow J. J., Buckwalter M. S., Camper S. A. Mouse chromosome 11. Mamm Genome. 1991;1(Spec No):S158–S191. doi: 10.1007/BF00656492. [DOI] [PubMed] [Google Scholar]
- Campbell G. R., Zimmerman K., Blank R. D., Alt F. W., D'Eustachio P. Chromosomal location of N-myc and L-myc genes in the mouse. Oncogene Res. 1989;4(1):47–54. [PubMed] [Google Scholar]
- Copeland N. G., Jenkins N. A. Development and applications of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991 Apr;7(4):113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- Dietrich W., Katz H., Lincoln S. E., Shin H. S., Friedman J., Dracopoli N. C., Lander E. S. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992 Jun;131(2):423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearne C. M., McAleer M. A., Love J. M., Aitman T. J., Cornall R. J., Ghosh S., Knight A. M., Prins J. B., Todd J. A. Additional microsatellite markers for mouse genome mapping. Mamm Genome. 1991;1(4):273–282. doi: 10.1007/BF00352339. [DOI] [PubMed] [Google Scholar]
- Huppi K., Mock B. A., Schricker P., D'Hoostelaere L. A., Potter M. Organization of the distal end of mouse chromosome 4. Curr Top Microbiol Immunol. 1988;137:276–288. doi: 10.1007/978-3-642-50059-6_42. [DOI] [PubMed] [Google Scholar]
- Jackson I. J. A cDNA encoding tyrosinase-related protein maps to the brown locus in mouse. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4392–4396. doi: 10.1073/pnas.85.12.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. P., Serrero G. Isolation and characterization of a full-length cDNA coding for an adipose differentiation-related protein. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):7856–7860. doi: 10.1073/pnas.89.17.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt M., Luty J. A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet. 1989 Mar;44(3):397–401. [PMC free article] [PubMed] [Google Scholar]
- Love J. M., Knight A. M., McAleer M. A., Todd J. A. Towards construction of a high resolution map of the mouse genome using PCR-analysed microsatellites. Nucleic Acids Res. 1990 Jul 25;18(14):4123–4130. doi: 10.1093/nar/18.14.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly K. F., Elliott R. W. RI Manager, a microcomputer program for analysis of data from recombinant inbred strains. Mamm Genome. 1991;1(2):123–126. doi: 10.1007/BF02443789. [DOI] [PubMed] [Google Scholar]
- McMahon A. P., Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990 Sep 21;62(6):1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Mullins J. J., Zeng Q., Gross K. W. Mapping of the mouse atrial natriuretic factor gene. Evidence for tight linkage to the Fv-1 locus. Hypertension. 1987 May;9(5):518–521. doi: 10.1161/01.hyp.9.5.518. [DOI] [PubMed] [Google Scholar]
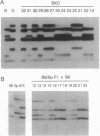
- Orita M., Suzuki Y., Sekiya T., Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989 Nov;5(4):874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Silver J., Buckler C. E. Statistical considerations for linkage analysis using recombinant inbred strains and backcrosses. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1423–1427. doi: 10.1073/pnas.83.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinardi L., Mazars R., Theillet C. Protocols for an improved detection of point mutations by SSCP. Nucleic Acids Res. 1991 Jul 25;19(14):4009–4009. doi: 10.1093/nar/19.14.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990 Aug 30;346(6287):847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- Todd J. A., Aitman T. J., Cornall R. J., Ghosh S., Hall J. R., Hearne C. M., Knight A. M., Love J. M., McAleer M. A., Prins J. B. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991 Jun 13;351(6327):542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]