SUMMARY
All eukaryotic cells prepare for cell division by forming a “mitotic spindle”—a bipolar machine made from microtubules (MTs) and many associated proteins. This device organizes the already duplicated DNA so one copy of each chromosome attaches to each end of the spindle. Both formation and function of the spindle require controlled MT dynamics, as well as the actions of multiple motor enzymes. Spindle-driven motions separate the duplicated chromosomes into two distinct sets that are then moved toward opposite ends of the cell. The two cells that subsequently form by cytokinesis, therefore, contain all the genes needed to grow and divide again.
The mitotic spindle is a cytoskeletal machine that organizes duplicated DNA during cell division. Each step of the mitotic process is illuminated by discoveries in a range of research fields and organisms.
1. INTRODUCTION
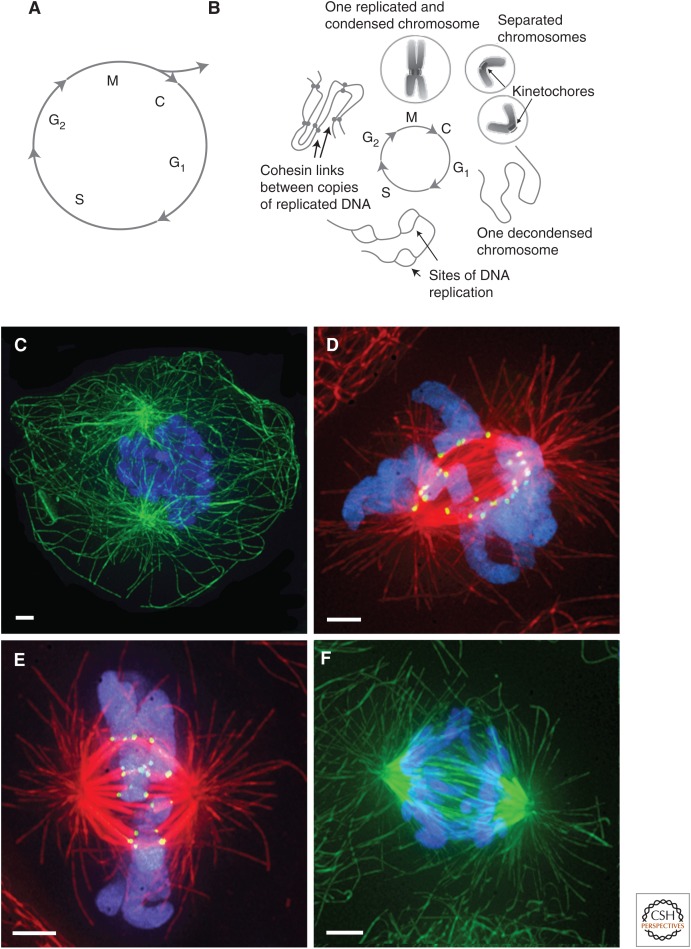
All cellular cycles of growth and division include a time of synthesis called “interphase” during which DNA is replicated, and all other cellular constituents are made in sufficient quantities to supply the needs of two cells. During mitosis, these materials are rearranged, so after “cytokinesis” each daughter cell will be fully endowed to grow and divide again (Fig. 1A,B).
Figure 1.
The cellular growth and division cycle. (A) Cartoon of the main segments of the cell cycle. During interphase (G1, S, G2), the cell accomplishes sufficient biosynthesis to become two. In mitosis (M), cell parts are reorganized so the mitotic spindle can achieve the equipartition of the chromosomes and centrosomes, leaving the distribution of more numerous components, such as ribosomes, to the laws relating to large numbers and the process of cytokinesis (C), in which the cell itself divides into two daughters. In tissues, cells can continue further rounds of division or can exit the cell cycle. (B) Chromosome behavior during the cell cycle. During the first gap phase, G1, cells have only the chromosome they were given at the previous cell division; each decondensed chromosome is a single DNA duplex. This phase is followed by a period of chromosome replication in S phase, and then a further gap phase, G2, in which the newly replicated sister chromatids are held together by cohesins. During mitosis, the condensed sister chromatids are separated in a process highly dependent on interactions between microtubules (MTs) and chromosomal kinetochores. (C–F) Immunofluorescence images of mitotic cells, sourced from the mammalian rat kangaroo PtK1 strain. In prophase (C), the chromosomes (blue) condense inside a still-patent nucleus while MTs (green) organize in the cytoplasm. In prometaphase (D), the spindle MTs (red) gain access to the chromosomes (blue) and attach to the kinetochores (yellow) that will subsequently govern most chromosome motions. By metaphase (E), the chromosomes are quite accurately aligned on the spindle midplane. During anaphase (F), they segregate, moving toward opposite poles of the spindle, and the spindle itself elongates. Scale bars, 2 µm. (A,B, Reprinted, with permission, from McIntosh et al. 2012; C–F, previously unpublished micrographs, kindly provided by Jennifer DeLuca, Department of Biochemistry and Molecular Biology, Colorado State University.)
Accurate segregation of the cell’s already duplicated DNA is the largest task faced by a dividing cell because the DNA of any complete genome is considerably longer than the diameter of the cell. For example, human DNA sums to almost 2 m, whereas most of our cells are only less than 1/100,000th of that size. Segregating these long strands with high fidelity is the job of the mitotic spindle. Success in this task is essential for the success of any organism.
During its replication, each DNA duplex becomes tied to its sister by “cohesins” (Peters et al. 2008). For mitosis, these linked DNA duplexes, called “chromatids,” become compacted into objects that are small relative to the diameter of the cell. The result is the compacted chromosomes that were initially described in the 19th century, and they are familiar from classic pictures of cell division. Chromosome condensation occurs during the mitotic period called “prophase” (Fig. 1B,C); its events are essential for successful cell division. When the spindle forms during prometaphase (Fig. 1D), it attaches to the chromosomes and organizes them into the twofold-symmetric structure of “metaphase” (Fig. 1E). The successful segregation of chromosomes during “anaphase” (Fig. 1F) is enhanced by the twofold symmetry achieved at metaphase. Once the metaphase chromatids have separated far enough, two fully functional nuclei can form in distant parts of the cell during “telophase.” Cytokinesis will then produce two cells that can repeat these events at the end of the next interphase (see Glotzer 2016).
An important attribute of mitosis is the accuracy with which chromosome segregation is accomplished. Budding yeasts make only one mistake in about 100,000 divisions (Hartwell and Smith 1985), although cultured mammalian cells are considerably less accurate (Bakhoum et al. 2014). Accurate chromosome segregation is obviously important for biological success, but mitosis is impressive in a less obvious way. The mechanical tools of a cell are nanomachines: the subunits of cytoskeletal fibers and the proteins that interact with them as controllers, links, and motors. Most chromosomes are three orders of magnitude bigger than a typical cellular motor, and the extents of mitotic movement are approximately 10-fold larger still. Thus, the spindle must use minute components to accomplish a huge and important job with stringent fidelity. On top of that, the spindle is responsible for guiding the segregation of some nonchromosomal cell parts: centrosomes, which include the centrioles that can serve as basal bodies for growing flagella, the Golgi complex, and, in cells in which mitochondria or chloroplasts are few in number, these large organelles as well. Thus, the spindle is an important machine that is well worth our best efforts to understand.
Cell division is requisite for all forms of life, but its importance is emphasized by the distress caused by its failure. Chromosome loss in unicellular organisms commonly leads to death, a significant negative selection! In multicellular organisms, inaccurate mitosis leads to aneuploidy, an aspect of cancerous progression. Excess cell division is an obvious component of cancer, and insufficient division can promote conditions such as anemia. It is no wonder that many scientists have pursued this subject over many years. The resulting knowledge is impressive but certainly not complete, as the text below will show. The following focuses on eukaryotic cells, but even with this limitation there is sufficient variation that many interesting specifics will be glossed. For more detailed descriptions, see Dumont and Mitchison (2012) and McIntosh et al. (2012). Moreover, chromosome segregation in prokaryotes is now understood well enough to show intriguing similarities and informative differences from mitosis (Erickson et al. 2010).
2. THE EVENTS AND MECHANISMS OF MITOSIS
2.1. During Prophase, Cells Get Ready for Division
2.1.1. Prophase in the Nucleus
The onset of visible chromosome condensation defines the beginning of prophase. This compaction acts on strands of “chromatin”—that is, the fibers that DNA forms as it wraps around histone octamers. Prophase compaction decreases the length and increases the thickness of each chromosome, individualizing the strands of DNA into visible units (Figs. 1B,C and 2A,B). As compaction occurs, transcription shuts down, in part because transcription factors become displaced (Martinez-Balbas et al. 1995). The mechanisms for prophase compaction have been debated for years, but progress has been confounded by our limited understanding of chromosome structure. Chromatin is made from charged polymers whose organization changes with conditions; what you see depends on how you look, a situation that has led to controversy. Electron tomography of frozen-hydrated chromatin, a reliable approach, shows a matrix of crisscrossed strands interlinked by fibers (Konig et al. 2007), at least some of which include “condensins,” a family of proteins able to encircle one or more pieces of double-stranded DNA and affect its folding (Thadani et al. 2012). Previous models for chromosome structure posited roles for supercoiling and scaffolds, but the existence of these structures has not been supported by the recent evidence (Uhlmann 2014). Posttranslational modifications of chromatin, such as phosphorylation and acetylation of histones and other chromatin proteins, are very likely to be involved in compaction, given the need to balance charges as this polyelectrolyte condenses. However, the data now available are not sufficient to know how chromosome structure changes during compaction.
As chromatin condenses, many additional events help the cell to prepare for division. The nucleolus disperses, sometimes as fragments but generally by releasing many nucleoprotein particles into the nucleoplasm, where they look like ribosomes. This event is probably the result of condensing the DNA that had served as the nucleolar organizer. Components of both the nuclear pore complexes and the lamin fibers that line the inner surface of the nuclear envelope in many eukaryotic cells become hyperphosphorylated, probably by the serine/threonine-specific cyclin-dependent kinase 1 (CDK1). These fibers disperse as soluble proteins, although lamin-B remains associated with the nuclear envelope membranes. Disappearance of the lamin network weakens the envelope, so other factors can now disrupt it in the cells of most animals and all higher plants, where nucleoplasm and cytoplasm mix for mitosis.
The dispersal of the envelope is a controlled event. Microtubules (MTs) of the growing spindle have been seen to push on the envelope, but these jabs are not essential for envelope dispersal because the process will occur in the presence of drugs that block MT polymerization. Transient increases in cytoplasmic Ca2+ concentration and associated increases in the activity of protein kinase C have been linked with envelope dispersal (Steinhardt and Alderton 1988), but the impact of this activity is not yet understood. As it disperses, the envelope forms vesicles and cisternae that mingle with the endoplasmic reticulum (ER), and so integral envelope proteins, too, become dispersed (Yang et al. 1997).
In many microscopic eukaryotes, the nuclear envelope does not break down at mitosis—the spindle forms inside the nucleus, leading to a “closed mitosis.” This strategy requires that the proteins of the mitotic spindle enter the nucleoplasm, presumably using cell cycle–regulated nuclear localization signals. A closed mitosis is found in Giardia, an organism from near the root of the eukaryotic tree, but it is also found in ciliates, amoebae, and some fungi, which are our much closer relatives. Between completely closed and fully open mitoses, there are many gradations, as in the green alga, Chlamydomonas, or in the nuclei of Drosophila embryos at syncytial blastoderm, in which mitotic envelopes appear intact except at their polar regions, where centrosomes are localized in the cytoplasm. MTs pass through these “windows” to enter the nucleoplasm and interact with the chromosomes (O’Toole et al. 2003). The reasons for a given spindle organization in a particular cell type are still matters for speculation, but some interesting hypotheses have been advanced (Sazer et al. 2014).
Many prophase events in addition to lamin dispersal are controlled by the protein kinase CDK1, but additional mitotic kinases have been identified. Polo kinase and its relatives localize to the centrosomes and are essential for the increase in size and activity of this organelle at mitosis (Lane and Nigg 1996). The serine/threonine kinase Aurora A is essential for the prophase maturation of centrosomes in nematodes (Hannak et al. 2001) and fruit flies (Giet et al. 2002). Modifications by these enzymes enable the accumulation of γ-tubulin ring complexes, which initiates more centrosome-associated MTs. Other kinases accumulate on chromosomes, particularly at the chromosomal primary constriction called the “centromere,” where they become involved in mitotic quality control. Identifying the substrates and interactions for each of these regulatory factors is a major theme in current mitosis research.
2.1.2. Prophase Changes in the Cytoplasm
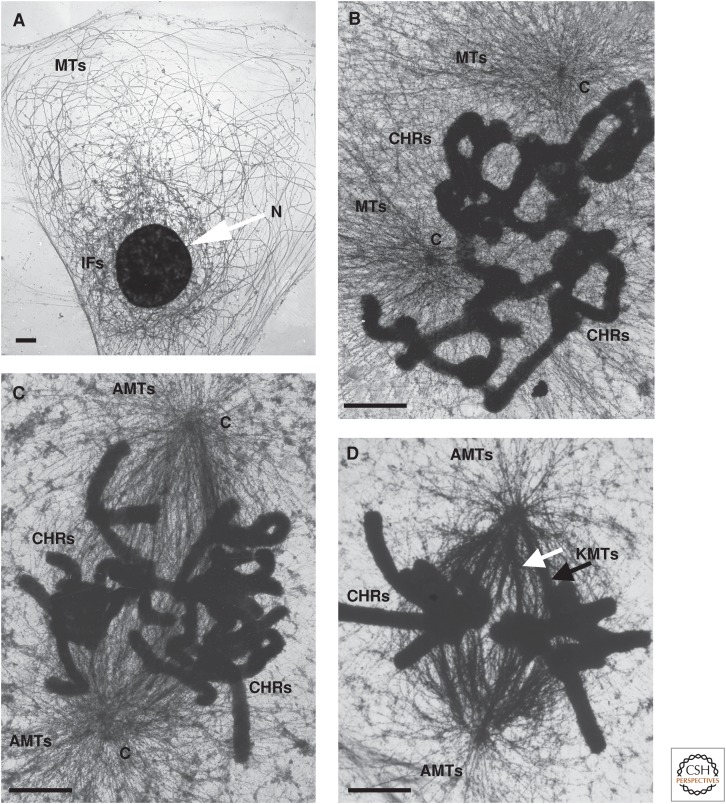
Protein synthesis slows during prophase because ribosomes transit messenger RNA (mRNA) more slowly, leading to an accumulation of polysomes. The mechanism for this change is not known, but it has been suggested that it serves both to protect mRNA during mitosis and to allow a rapid restart of protein synthesis on the completion of division (Sivan et al. 2007). Both MTs and microfilaments lose their interphase stability and largely dissolve as their components respond to increased protein phosphorylation (Maller 1986; Vandre et al. 1986). Although stress fibers disappear, some actin remains fibrous at the cell cortex to be used for cytokinesis; additional actin assemblies can form and rotate about the cell in anticipation of contractile ring formation (Mitsushima et al. 2010). As the interphase MTs disappear, new ones form (Fig. 2A,B). In organisms with centrosomes, these structures initiate the MTs that will become the spindle. The resulting MTs are more labile than their interphase counterparts, largely as a result of phosphorylation of MT-associated factors, such as “tog” domain proteins (Ma and Poon 2011), which catalyze tubulin polymerization, and stathmin, a small protein that binds tubulin and blocks its assembly (Gadea and Ruderman 2006). Intermediate filaments made from vimentin also disperse for mitosis following phosphorylation by CDK1 (Chou et al. 1990), but cytokeratins persist and collapse onto the nucleus as their associated MTs disappear. They form a cage that surrounds the nucleoplasm even after the nuclear envelope has dispersed (Zieve et al. 1980).
Figure 2.
Electron micrographs of cells as they form a mitotic spindle. Rat kangaroo PtK1 cells were cultured on gold grids coated with a thin layer of plastic and carbon, lysed with 0.2% Triton X100 and 1 mm MgCl2, in a PIPES-HEPES buffer, pH 7.2, and then fixed with 2% paraformaldehyde and 0.1% glutaraldehyde. These samples were quenched in 0.2 mg/mL NaBH4 in 1:1 ethanol:phosphate-buffered saline, then stained with a monoclonal antibody against tubulin, followed by a rabbit antimouse IgG bound to 10-nm colloidal gold, followed by fixation in osmium tetroxide, and then by drying with the critical-point method. Microtubules (MTs) in the interphase and mitotic cells are nicely contrasted, and the chromosomes are stained by osmium. (A) Interphase: The nucleus (N) contains decondensed chromatin. Intermediate filaments (IFs) surround the nucleus. (B) Early prometaphase: CHRs, chromosomes; C, centrosome. (C) Late prometaphase: AMTs, astral microtubules. (D) Metaphase: KMTs, kinetochore microtubules. Scale bars, 1 µm. (Images kindly provided by Mary Morphew, University of Colorado, Boulder.)
Cytoskeletons of multicellular plants display an additional prophase modification that is significant for cytokinesis—they form a band of actin and MTs that girds the cell just inside its plasma membrane. This “preprophase band” is transient, but it marks the cell cortex and establishes the place where cytokinesis will occur as anaphase ends (Zhang et al. 1990; Lipka et al. 2014). The molecular mechanisms by which these marks define the position of cytokinesis are still under investigation.
2.1.3. Prophase Restructuring Is Important
Mitotic changes in cytoskeletal organization are significant for four reasons. First, many of the molecules that make the spindle and the cytokinetic machinery are the very molecules that built the interphase cytoskeleton—a nice example of cellular economy. Second, the disappearance of the interphase cytoskeleton leads animal cells to round up, giving them a symmetry that encourages a uniform distribution of their contents, aiding the equipartition of organelles at cytokinesis. Third, the solubilization of interphase polymers decreases cytoplasmic viscosity, facilitating the diffusive randomization of medium-sized cytoplasmic objects (e.g., multienzyme complexes, ribosomes, and small vesicles). Diffusive motions are also important in plant mitosis because cytoplasmic streaming ceases at this cell cycle stage, removing the customary engine of the plant cell for rapid cytoplasmic mixing. When cytokinesis divides a cell in two, diffusion has already achieved the approximate equipartition of many cellular constituents without the action of a special machine such as the spindle. This process helps to ensure the capacity of daughter cells to grow and divide again. Finally, a rounded cell facilitates reorientation of the spindle in response to external cues (Fink et al. 2011)—this allows cytokinesis to place daughter cells correctly. Spindle orientation is particularly important in multicellular plants, in which cells, once born by cytokinesis, do not move relative to one another.
Analogous prophase dispersal occurs with some cytomembranes. During prophase, the Golgi complex of a HeLa cell fragments into numerous vesicles and tubules (Barr 2004). By metaphase, there are thousands of such structures distributed quite evenly through the cytoplasm (Shima et al. 1997). In some animal cells, however, Golgi vesicles cluster around the spindle poles. In yeasts, in contrast, the Golgi appears to be segregated actively by the actin cytoskeleton (Barr 2002). The ER in many cells behaves differently—it moves to the cell periphery, lying just under the plasma membrane (West et al. 2011). The mechanism for this repositioning is not known, but it does not have the properties of diffusion. Nonetheless, it accomplishes the same goal of fostering organelle equipartition by subsequent cell cleavage.
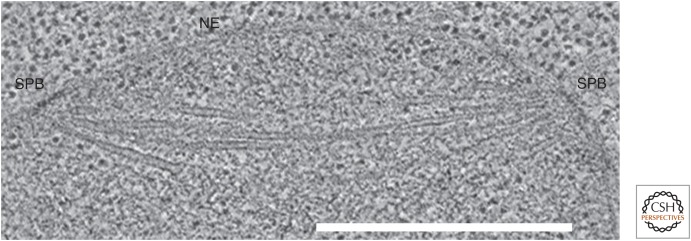
Prophase is said to end when the nuclear envelope disperses, but the spindle commonly begins to form before then. In cells with centrosomes, it first appears as highly dynamic MTs growing radially from these well-defined objects (Fig. 1C). Sometimes the two centrosomes of a cell are still close as MT growth begins, but they separate as the spindle forms; in other cases, they are already separated and lie on opposite sides of the nucleus, and so MTs form as two distinct asters (Fig. 2B). Both pathways can be seen in the same type of cultured cell, and both lead to a normal metaphase spindle, but segregation errors are fewer if the centrosomes separate early (Silkworth et al. 2011). In a closed mitosis, the end of prophase is hard to define; spindle formation simply starts when chromosome condensation is sufficient. Budding yeasts are unusual, however, in forming a spindle as the chromosomes begin to replicate; a bipolar spindle is already present in G2 (Fig. 3). Clearly, the metaphase structure can be reached by many routes.
Figure 3.
Formation of a closed mitotic spindle occurs within the nucleus. Slice from an electron tomogram of a dividing budding yeast cell during prometaphase. The nuclear envelope (NE), the spindle pole bodies (SPBs), and the microtubules are clear, but, in this cell type, chromosome condensation is not sufficient to make the chromatin obvious. Scale bar, 1 µm. (Image kindly provided by Eileen O’Toole, University of Colorado, Boulder; reprinted, with permission, from McIntosh et al. 2012.)
The early stages of spindle formation in higher plants are intriguing because these cells lack centrosomes. In the endosperm of the African blood lily, a sheath of MTs forms around the prophase nucleus (De Mey et al. 1982). The axis of this sheath “anticipates” the axis of the mitotic spindle that will form a little later. As the sheath disperses, regions at both ends of the sheath, lying just outside the nuclear envelope, become the sites from which many MTs form and enter the nucleus as the envelope disperses. Thus, although these cells lack centrosomes, the path to metaphase seems quite similar to that in most animal cells.
2.2. During Prometaphase the Chromosomes Engage with the Spindle and Become Organized
2.2.1. Chromosomes Form Specializations for Spindle Attachment
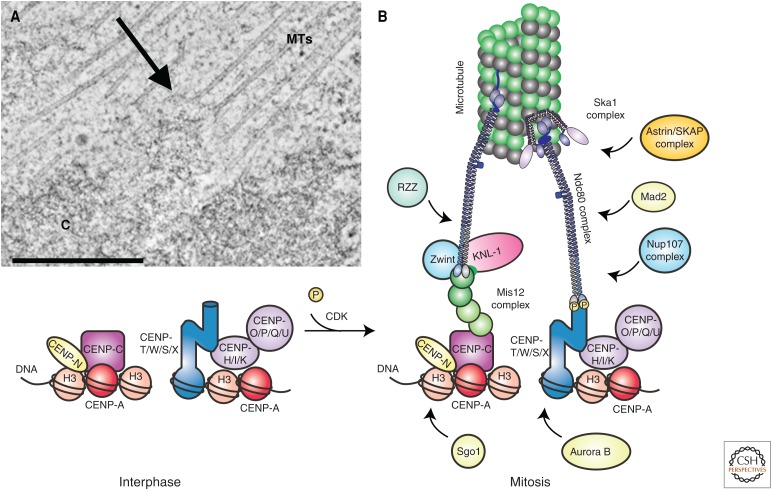
The most important parts of a chromosome for spindle attachment are its structures known as kinetochores, one on each chromatid (Fig. 4A). These structures are built from many proteins. In human cells, approximately 100 different polypeptides assemble into multiple protein complexes that, in turn, assemble on specialized chromatin (Fig. 4B) (Cheeseman 2014). Kinetochores include several fibrous proteins that bind to MT walls—the Ndc80 complex and KNL1, which are almost ubiquitous, and proteins such as CLASP1 and CENP-F, which are found in vertebrates and have putative analogs in additional organisms. Kinetochores also include motor enzymes that bind MTs and generate forces to influence both chromosome position and MT dynamics. The identities of kinetochore motors are not conserved, but their functions are. Although the data are incomplete, probably all kinetochores bind a minus end–directed motor. In animal cells this motor is dynein, whereas in yeasts it is kinesin-14. Kinetochores also bind at least one plus end–directed motor. In almost all cells, this includes one or more kinesin-8 proteins and, in animal cells, a dimer of kinesin-7 (CENP-E) as well. Kinetochores also bind a disassembly-promoting kinesin. In yeasts, this is one or more kinesin-8 proteins, which can induce MT shortening either directly or by promoting catastrophes. In animal cells, kinetochores also bind kinesin-13, a catalyst of MT depolymerization that lacks motility. In summary, MT-binding proteins, motors with both directionalities, and activities that regulate MT dynamics are universal parts of the mechanisms by which kinetochores grasp spindle MTs. It must be added, however, that recent work on kinetochores in trypanosomes has shown a considerable divergence from the norm (Akiyoshi and Gull 2014). As the study of mitosis expands to organisms with greater phylogenetic spread, more exceptions than are currently anticipated may be identified.
Figure 4.
Structure of the kinetochore. (A) Slice from an electron tomogram of a rat kangaroo PtK1 cell in prometaphase. A chromosome (C) and the associated microtubules (MTs) are easy to distinguish in the tomogram; their point of connection is the kinetochore (arrow). (B) Diagram of kinetochore composition and structure. At mitotic entry, phosphorylation (P) by activated CDK–cyclin-B promotes assembly of the outer kinetochore on a platform of constitutive kinetochore proteins. For information about the kinetochore proteins shown here, see Cheeseman 2014. CDK, cyclin-dependent kinase. (Reprinted, with permission, from Cheeseman 2014, © Cold Spring Harbor Laboratory Press.)
2.2.2. Spindles Are Well Designed to Form Useful Attachments to Chromosomes
Spindle MTs display dynamic instability (see Goodson and Jonasson 2016) with rates of catastrophe that exceed those of interphase by approximately 10-fold (Belmont et al. 1990). Thus, growing MTs frequently transition to shrinking. They can then either experience a rescue to start growing again or shorten to nothing, freeing their γ-tubulin ring complex to initiate another MT. Both in this way and through diffusive rotations (Kalinina et al. 2013), MTs probe the region in which chromosomes reside, repeatedly renewing their chance to encounter a kinetochore. When they do, they are stabilized by kinetochore binding and removed from the dynamic pool. This “search and capture” scenario was first proposed in an insightful model for chromosome–spindle attachment (Kirschner and Mitchison 1986). Mathematical formulations of the concept are quite successful in accounting for the ability of the spindle to make certain that MTs attach to all kinetochores in a timely manner (Magidson et al. 2015).
In an open mitosis, the centrosome-initiated MTs grow into the nuclear space as soon as the nuclear envelope disperses. If they emanate from already separated centrosomes, they invade the chromosome mass from two sides, presenting MTs to the chromosomes in a roughly twofold-symmetric way. In both animals and plants, the number of MTs that participate in this invasion is often greater than that which the spindle poles themselves can initiate. This increase in number is accomplished by “augmin,” a complex that binds to the walls of existing MTs and initiates a new polymer (Kamasaki et al. 2013; Petry et al. 2013), increasing the barrage of MTs that peppers the chromosomes, and thereby increasing the chances of their binding to the spindle. In cells whose centrosomes had not separated before envelope dispersal, the spindle grows as the centrosomes separate. Chromosomes then attach all over this elongating bipolar array, commonly binding to MT walls. As their ultimate position will be at MT plus ends, considerable rearrangement is then required (see below). Closed mitoses usually begin chromosome attachment when the poles are still close together, so they too must have mechanisms to rearrange their initial attachments and achieve the architecture characteristic of metaphase.
Because metaphase chromosomes all include two chromatids, each with its own kinetochore, some chromosomes wind up by chance with their “sister” kinetochores associated with MTs that grew from each of the two spindle poles—in which case, no subsequent rearrangement is required. However, the chances of this scenario are not sufficient for accurate mitosis. For example, sister kinetochores will sometimes attach to MTs growing from only one pole. Spindles correct this and other mistakes with impressive reliability. A myriad of details are available from several extensive reviews on how spindles correct faulty chromosome attachments (Nicklas 1997; Bakhoum et al. 2014). This article will focus on the basic principles and the mechanisms that underlie them.
2.2.3. When Improper Attachments Form, the Spindle Can Often Correct Them
Sometimes kinetochores interact with an MT end that by chance grew right into them, but initial kinetochore binding is more common with an MT wall (Magidson et al. 2011). This is not surprising as there are so many more tubulins in MT walls than at their ends. The problem a cell must solve is how to rearrange things so each kinetochore binds to the ends of MTs that are coming from only one spindle pole (Fig. 4A). Failure in this process leads to a single kinetochore attaching to both spindle poles, known as a “merotelic” connection, which is counterproductive for accurate chromosome segregation. There are several mechanisms for either avoiding such connections or correcting them. The facts that each copy of the replicated chromosome contains a kinetochore and that two spindle poles initiate MTs that come into the nucleoplasm from opposite directions give a good start on getting attachments right. More important, though, is that kinetochores are actively motile on the MTs to which they bind. The minus end–directed activity of dynein is initially dominant, pulling each kinetochore toward the pole from which that MT came. This attachment is, however, not stable—it stabilizes only when under tension, as shown by elegant experiments with chromosome micromanipulation in living cells (Nicklas et al. 1998). When sister kinetochores are attached to sister poles, that chromosome is being pulled in opposite directions, putting its centromere under the tension that promotes attachment stability. Most other arrangements of spindle attachment lack this feature and are therefore unstable, so they tend to dissolve, whereas proper attachments persist. This is the primary mechanism by which proper spindle attachments are achieved, but MTs can also grow out from the kinetochore region and interact with pole-initiated MTs, enhancing attachment (Kitamura et al. 2010).
Additional mechanisms for getting proper chromosome attachment are numerous (Khodjakov and Rieder 2009; Foley and Kapoor 2012). If kinetochore dynein pulls a kinetochore poleward, bringing that chromosome into the vicinity of a spindle pole, the connected kinetochore is now peppered with MTs, providing such a density of ingrowing MTs that any MT from the opposite pole is likely to be competed away. Meanwhile, the sister kinetochore is turned to face MTs growing from the opposite pole; this and other mechanical strategies certainly contribute to mitotic fidelity. In addition, however, kinetochores include Aurora B, a protein kinase that can phosphorylate several kinetochore components involved in binding MTs—for example, the NDC80 complex (DeLuca et al. 2011). This modification reduces MT affinity, promoting detachment. Thus, MT–kinetochore interactions are subject to enzymatic “softening” until Aurora B is turned off. Tension at the kinetochore achieves this goal, probably through pulling the MT end away from the opposite kinetochore, out of the region where Aurora B can interact with its substrates and weaken the connections (Dewar et al. 2004; Lampson and Cheeseman 2010).
2.2.4. Prometaphase Is Prolonged by Improperly Attached Kinetochores through the Action of the Spindle Assembly Checkpoint (SAC)
Kinetochores possess additional kinase activities (e.g., MPS1) whose activity controls the localization of additional kinetochore components (London and Biggins 2014), probably in response to MT attachment or forces generated in the spindle (Joglekar and Aravamudhan 2016). These kinases are parts of a remarkable “checkpoint” that inhibits the onset of anaphase until all chromosomes are properly attached to the spindle (McIntosh 1991; Roberts et al. 1994; Hardwick and Murray 1995). This checkpoint is based on a biochemical cascade, initiated at unattached kinetochores, that inhibits a polyubiquitin ligase, the “cyclosome” or anaphase-promoting complex (APC/C) (Hoyt 2001). When the last chromosome is properly attached, this cascade is shut off, probably as a result of both tension at the kinetochore and MT binding. Now the APC/C can polyubiquitinate key proteins, leading to their degradation by the proteasome, which in turn activates a different protease called “separase.” As explained below, this enzyme degrades connections between sister kinetochores, allowing anaphase to start. Thus, kinetochores are cleverly designed to achieve proper spindle attachment, and the routes toward this goal are several, perhaps even more numerous than we now recognize.
Kinetochore chemistry and physiology have become growth industries. Thanks to modern methods for molecular genetics, biochemistry, and biophysics, much has now been learned about the stages in the assembly, maturation, and function of kinetochores. We also know the spatial arrangement of kinetochore proteins along the spindle axis at metaphase (Wan et al. 2009), how kinetochores bind MTs through multiple fibrous proteins (Cheeseman 2014), and how they can move on MTs to enhance their chances of accurate segregation (Kapoor et al. 2006). We even know that a kinetochore can retain a firm hold on a spindle MT while the MT depolymerizes at the kinetochore (Coue et al. 1991). For yeasts, this mechanism is now quite well understood. It relies on a protein complex called Dam1, comprising 10 different polypeptides, which form a prolate protein complex that assembles into a ring around each kinetochore-associated MT (KMT) (Westermann et al. 2006). These rings are bound to kinetochores, probably by the NDC80 complex (Tien et al. 2010); this supercomplex provides a robust and reliable connection between a chromosome and a spindle MT, an issue of great importance when a chromosome associates with only one or a very few MTs (Volkov et al. 2013). The Dam1 complex is not found outside the fungi, but comparable mechanisms using different proteins to bind kinetochores at the ends of dynamic MTs are now subjects of intense investigation in other groups of organisms (Schmidt et al. 2012; Volkov et al. 2015).
2.3. Alternative Pathways for Forming a Spindle
The pathways for spindle formation described above are not universal. Some spindles that perform meiosis are formed largely through MT initiation in the vicinity of the chromosomes, thanks to a chromosome-generated gradient in the concentration of the small GTPase, Ran, in its GTP-associated form (Kalab and Heald 2008). This gradient affects several aspects of MT initiation and interaction, leading to a thicket of MTs immediately around the chromosomes. The resulting fibrous assemblage sorts out into a bipolar array, thanks to the action of kinesin-5 and then dynein and/or other minus end–directed motors that help to cluster MT minus ends (Heald et al. 1996). Also involved is the fibrous protein “nuclear mitotic apparatus protein 1” (NUMA1), which binds dynein and interacts with MTs, especially their ends, helping MTs to motor over their neighbors and make the bipolar array found at metaphase (Chakravarty et al. 2004; Elting et al. 2014; Sikirzhytski et al. 2014). When this pathway was first discovered, it seemed to be an alternative to the centrosome-mediated processes described above, but further research has shown that both paths can coexist in a single mitotic cell (Kalab and Heald 2008). Experiments knocking down the cellular machinery that makes the Ran–GTP gradients suggest, however, that this mechanism is not essential in mitotic cell division (Goshima et al. 2007). One must add, however, that centrosomes too seem to be dispensable; certainly centrioles are not needed, as mutant Drosophila that cannot make centrioles grow to adulthood (Basto et al. 2006). In experiments in which centrosomes are damaged by laser irradiation, the spindles will still form (Khodjakov et al. 2000). Thus, multiple spindle-forming mechanisms coexist—it seems that cells have several strategies for spindle formation, thereby maximizing the chances that this essential process occurs without flaws.
Understanding the essential features of mitosis is in some ways facilitated by studies of biological diversity. For example, the distinction between open and closed mitosis becomes blurred when we look at mitosis in a group of protozoans called hypermastigote flagellates. In Barbulanumpha, the nuclear envelope does not break down, making it a closed mitosis. However, the spindle forms in the cytoplasm and interacts with the chromosomes through the nuclear envelope. Each kinetochore attaches to the inner surface of the envelope and forms an MT attachment site on the cytoplasmic face of the envelope, allowing each chromosome to experience forces generated in the cytoplasm (Ritter et al. 1978). Thus, nuclear or cytoplasmic localizations of spindle parts are not of great significance. The important thing is that sister kinetochores interact with sister spindle poles. Some older studies of dinoflagellates suggested that existing organisms include variations in spindle design that might help us understand the way mitosis evolved when eukaryotes first formed (Kubai 1975). However, the cells used in these studies have not yet been reexamined with modern methods that might give more reliable descriptions of their conserved and divergent features.
2.4. Following Spindle Attachment, the Chromosomes Commonly Migrate to the Spindle Equator to Form the “Metaphase Plate”
Chromosome movement to the equatorial plane of the spindle is a common, although not universal, aspect of mitosis. Its value comes from its putting the chromosomes on the same “starting line” before the onset of their separation at anaphase—this arrangement (Fig. 2D) should minimize the chances of a single chromosome being left behind as the chromosomes segregate. The mechanisms for “congression” to the spindle equator are still under investigation, but several experimental results illuminate the pathways. If prometaphase chromosomes are severed by irradiation with a microbeam, the fragment that lacks kinetochores is pushed away from the nearby spindle pole (Rieder and Salmon 1994). Likewise, chromosome arms in animal cells are pushed toward the spindle equator and then outward from the spindle axis, as if they were being pushed away from both poles at once. This action has been called the “polar ejection force,” and it probably contributes to the gradual motion of prometaphase chromosomes to the spindle equator. One model for this force is the pressure exerted on chromosome arms as dynamically unstable MTs grow into condensed chromatin, encountering resistance as the MTs try to elongate. Another model is based on kinesins-4 and -10, which bind to chromatin and interact weakly with MT walls, walking toward their plus ends (Brouhard and Hunt 2005). Knocking down either of these motors by RNA interference does not prevent formation of the metaphase plate, but, in HeLa cells, the double knockdown does block it (Wandke et al. 2012), suggesting that these motors represent an important mechanism for chromosome motion to the spindle equator.
A different mechanism for congression is provided by the plus end–directed kinesin CENP-E. This kinetochore-associated motor can interact with the walls MTs whose ends are not bound to its kinetochore. The motor drives toward MT plus ends and shifts that kinetochore away from the nearby pole (Kapoor et al. 2006). Still other mechanisms are suggested by experiments based on aberrant meiotic chromosomes with three kinetochores. At metaphase, two kinetochores on these chromosomes associate with one pole and one with the other. These chromosomes adopt a position off the spindle equator, positioned so the sum of the two short kinetochore-associated fibers equals the length of the one longer fiber (Hays et al. 1982; Hays and Salmon 1990). This observation implies that spindles can exert pole-directed forces on KMTs that are proportional to their number and length—properties that would help to assure the metaphase configuration. The mechanism that generates this force is not yet known, but it might depend on a motor such as kinesin-12, which is concentrated in bundles of KMTs (Sturgill et al. 2014). It seems probable that, as with spindle formation, there are multiple mechanisms working to achieve congression. Probably different cells rely more heavily on one mechanism than another, but all the mechanisms can combine to achieve the same arrangement, another example of the willingness of cells to use multiple mechanisms to achieve an important goal.
2.5. The Metaphase Spindle Is a Complex Steady State
During prometaphase, and even after chromosome congression is complete, spindle MTs are remarkably dynamic. MTs not bound to kinetochores turn over with a half-time of ∼30 sec (Saxton et al. 1984). The total amount of polymer is approximately constant, as measured by MT fluorescence, but it is not in equilibrium; the dynamic instability of MTs requires the continuous hydrolysis of considerable amounts of GTP to maintain a steady-state amount of polymer. Moreover, the more stable spindle MTs (e.g., KMTs) show a “flux” toward the spindle poles (Mitchison 1989). This slow motion has been seen best with speckle imaging (Waterman-Storer and Salmon 1997); its rate is almost as fast as the subsequent motions of chromosomes in anaphase. It requires kinesin-13 at the spindle poles to catalyze depolymerization of MTs at their minus ends and tubulin addition at kinetochores or other sites of plus end localization (Zhang et al. 2007). In some cells, this flux also requires the action of kinesin-5 to slide interdigitating MTs away from the spindle equator (Brust-Mascher et al. 2009), consuming additional chemical energy to maintain the apparently stable metaphase state. The reasons for metaphase MT flux are not obvious, although clever models have been proposed (Matos et al. 2009). However, flux is not essential for mitosis because yeast spindles do not do it (Maddox et al. 2000).
2.6. The Beginning of Metaphase Is Ill-Defined, but Its Termination Is One of the Most Important Transitions in the Whole Cell Cycle
The onset of metaphase is hard to specify because most chromosomes do not stay at the equator—they oscillate along the spindle axis with an amplitude that varies from chromosome to chromosome and from cell to cell. It is therefore hard to say when congression has really been achieved, although some cells, such as human epithelial cells in culture, show a “sharpening” of the metaphase plate just before anaphase begins. The so-called “metaphase arrest,” achieved by drugs that prevent MT formation, is actually a prometaphase arrest, because congression does not occur. The end of metaphase, however, is obvious; it is the time when sister chromatids begin to separate. The transition from metaphase to anaphase is significant because it is a process with no return. The proper attachment of all chromosomes initiates the silencing of the SAC, which activates protein polyubiquitination and subsequent proteolysis of numerous regulatory proteins—securin, as mentioned above, but also cyclin B, which leads to the inactivation of CDK1. Now phosphatases can undo the phosphorylations that were characteristic of mitotic entry. Thus, proper attachment of all the chromosomes to the spindle leads to a major cell cycle transition. In cultured mammalian cells, there is an interval of ∼20 min between the last chromosome attachment and the onset of anaphase (Rieder et al. 1995).
2.7. Anaphase Chromosome Segregation includes Two Components, Each of Which Is Mechanistically Complex
Each of the two chromatids in a metaphase chromosome becomes a chromosome of anaphase, identifiable as soon as separase has cleaved the cohesions that have held the sister strands together (Uhlmann 2001). The ensuing motions include a decrease in the distance of each chromosome from the pole it faces (anaphase A) (Fig. 5) and an increase in the separation between the spindle poles (anaphase B) (Fig. 6). Commonly, these motions occur in the sequence A and then B, but some cells do both at once, and others do one or the other. Each of these motions involves changes in the lengths of MTs and motions of MTs relative to one another, so both polymer dynamics and motor enzymes are involved. Exactly which mechanism contributes the most to each process differs from one cell type to another.
Figure 5.
Rat kangaroo PtK1 cell with separating chromosomes. Cultured cells were lysed and fixed as for Figure 2 and then embedded, sectioned, and imaged on a high-voltage electron microscope. Arrows labeled 1 and 2 indicate initial and secondary sites, respectively, of kinetochore microtubule (KMT) depolymerization during anaphase A. CHRs, chromosomes. Scale bar, 1 µm.
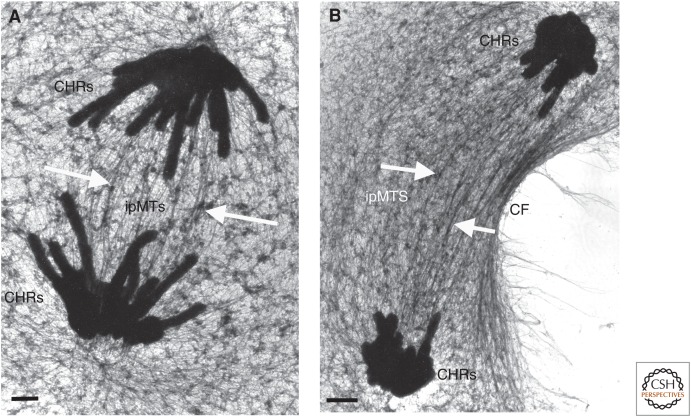
Figure 6.
Anaphase B. Rat kangaroo PtK1 cells fixed at mid (A) and late (B) stages of spindle elongation and prepared as in Figure 2. ipMTs are the interpolar microtubules, which interdigitate at the spindle midplane. These elongate by sliding and the addition of tubulin at the sites indicated by white arrows. CF, cleavage furrow; CHRs, chromosomes. Scale bars, 1 µm. (Images kindly provided by Mary Morphew, University of Colorado, Boulder.)
To describe the motions of spindle MTs that accompany chromosome segregation, MTs have been marked by several methods—photobleaching or photoactivation of tubulin fluorescence and speckle imaging with fluorescent tubulin. All these approaches have contributed to our understanding of spindle MT motions. Anaphase A commonly begins with a shortening of KMTs by loss of tubulin subunits at the kinetochores (Mitchison et al. 1986). In yeasts, this is the only component of anaphase A, but in many other cells there is also a loss of subunits at the poles (Maddox et al. 2002). Sometimes these losses are sequential, but, in insect spermatocytes, all the tubulin depolymerization is at the pole; in fact, kinetochores continue to add subunits throughout anaphase (LaFountain et al. 2004). Once again, we see variation in the details of mitotic processes. Probably this variation is due to differences in the activities or positions of specific spindle components, such as a kinase or a motor. Although potentially informative, such differences should not distract us from developing a sense of mitosis as a whole.
Dynein (Sharp et al. 2000; Yang et al. 2007) or kinesin-14 (Tanaka et al. 2007) at kinetochores may play roles in pulling the chromosomes poleward as KMTs depolymerize. In cultured animal cells, dynein can also pull the minus ends of KMTs poleward, thanks to its binding to NuMA1, which interacts with MT ends (Elting et al. 2014). This motor-dependent scenario has, however, been put in a different context by the discovery that, in both fission and budding yeasts, all minus end–directed motors are dispensable for minus end–directed chromosome motion (Grishchuk and McIntosh 2006; Tanaka et al. 2007). Indeed, an anaphase-A-like motion has been reconstituted in vitro by binding isolated chromosomes to MTs grown from a coverslip-associated polymerization initiator and then simply reducing the concentration of tubulin; soluble ATP and/or GTP are not required for chromosome motion at physiological speeds in vitro (Coue et al. 1991; Lombillo et al. 1995). These observations suggest that MT dynamics contribute to the forces acting on chromosomes. Polymer dynamics also seem to be the mechanism for chromosome segregation in eubacteria (Moller-Jensen et al. 2003), so perhaps a mechanism based on protein dynamics was the first machinery for chromosome segregation, even as prokaryotes and eukaryotes evolved from a common ancestor. In eukaryotes, that mechanism has since been overlaid with motor-dependent pathways that have probably added to its reliability, providing a selective advantage (McIntosh et al. 2010).
Studies based on the positions of MT ends (McIntosh et al. 1979) or on speckle imaging (Maddox et al. 2002) have shown that anaphase B is accompanied by the relative sliding of the overlapping non-KMTs that form the interpolar spindle (Figs. 5 and 6). Kinesin-5 effects this sliding in fly spindles, whereas other motors and MT cross-linkers antagonize it (Fig. 6) (Brust-Mascher and Scholey 2011). Interestingly, however, experiments in fungi and nematode embryos have shown that MT sliding near the spindle midplane actually acts as a brake on the rate of spindle elongation. The force for sliding elongation comes from dynein bound to the cell cortex, which interacts with MTs that grow from the spindle poles but project away from the body of the spindle—so-called “astral MTs” (Figs. 1D and 2D). These interactions drag the spindle poles toward the cell surface and contribute to spindle elongation (Aist et al. 1991; Fink et al. 2006; Civelekoglu-Scholey and Scholey 2007). Thus, anaphase B can be the result of either a “rear-wheel” or a “front-wheel” drive. In many cells, the extent of anaphase B is so great that there is an obvious elongation of the MTs involved. Direct observation with fluorescent tubulin has shown that this polymerization is at the MT plus ends where they overlap near the spindle midplane. Such polymerization allows further MT sliding to extend the elongation process (Saxton and McIntosh 1987), and assures that the two sets of chromosomes get sufficiently far apart that cytokinesis will put one set into each daughter.
Anaphase A saw the disappearance of the KMTs as the chromosomes approached the poles. The elongated interpolar spindle of anaphase B, in contrast, can persist, and in some cells it actually grows additional MTs as the chromosomes segregate. Animal (Uehara and Goshima 2010) and plant (Nakaoka et al. 2012) cells develop a concentration of the augmin complex near the minus ends of the interpolar MTs, especially as those ends become separated from the poles by the reforming nuclei. The resulting concentration of γ-tubulin initiates many new MTs, with their plus ends pointing toward the cell equator. These two MT families interdigitate at the spindle midplane, increasing the number of MTs in the zone between the separating chromosomes to the point that the MT plus ends of the peripheral bundles extend to the cell cortex. It is intriguing that similar increases in MT number are found in higher plant and animal cells, although the mechanisms for the subsequent cytokinesis are completely different. In animal cells, these MTs bind a kinesin-6, whose plus end–directed motor activity conveys the “central-spindlin complex” to the MT tips and thus to the cell cortex, where its activation of a Rho GTPase contributes to the regulation of cleavage (Pavicic-Kaltenbrunner et al. 2007). The incoming furrow then bundles many of these MTs to form the “midbody,” which can persist for some time into the subsequent interphase. Ultimately, the midbody is lost through a combination of proteolysis and completed cleavage, which pinches it off from one cell or the other, sometimes even both (Byers and Abramson 1968). As the spindle disappears, the two spindle poles begin to function as centrosomes, initiating new interphase MTs to help establish the normal interphase cytoskeleton.
2.8. Telophase Is the Time for Restoring a Functional Interphase Nucleus
While the chromosomes are segregating, important changes are occurring in the kinase activities that control the cell cycle. CDK1 activity is dropping, which alters the activity of other kinases that had helped the cell get into mitosis—the cell is preparing to go back into interphase. Chromosome arms contract, perhaps as a result of renewed condensin activity (Renshaw et al. 2010), which draws the two chromatin masses into smaller spaces immediately next to the spindle poles. Proteins of the inner nuclear envelope associate with the chromosomes, perhaps lamins as well, and pieces of the nuclear envelope begin to rebind to the still-condensed chromosomes (Guttinger et al. 2009). When these membranes contact one another they fuse, building up a new envelope that can define a nuclear compartment. At some point, nuclear pore complexes reassemble into the envelope, allowing the cell to establish the normal relationship between the nucleoplasm and cytoplasm. At about this time, chromatin decondensation begins. As it proceeds, transcription can restart, and nucleoli can re-form. These processes appear to be independent of cytoskeletal action, but they are all under cell cycle control. To this end, there is a mitosis exit pathway that controls the relationship between the end of mitosis and initiation of cytokinesis (Gupta et al. 2013). This helps to keep the nuclear events in concert with the disassembly of the spindle and the reestablishment of an interphase cytoplasm in each of the daughter cells.
3. CONCLUSION
During mitosis, a cell organizes its many components for equipartition at cytokinesis. The MT cytoskeleton plays an essential role through its assembly into the mitotic spindle, a machine for the organization and segregation of both chromosomes and centrosomes. The study of mitosis has illuminated many features and subtleties of the MT cytoskeleton and its control by site-specific nucleation, protein–protein interactions, protein phosphorylation, and proteolysis. Much has been learned about mitotic mechanisms, providing insight into the workings of MTs and their many associated proteins. Many details remain to be determined, but the broad outlines of how mitosis works are now well understood. They form an elegant example of cytoplasmic engineering and a testimony to the skill and persistence of the scientific community. Nonetheless, there are several important unanswered questions about the mitotic mechanism. Examples include the details of the pathways for chromosome condensation and the modes of interaction between MT ends and both kinetochores and spindle poles. We know that tubulin can add and dissociate from MT plus ends at kinetochores, and it can dissociate from MT minus ends at the poles during anaphase, even when these connections are under the load imposed by mitotic force generators. The pathways for these tubulin–MT exchanges, the roles that MT dynamics play in mitotic force generation, and how all these processes are regulated remain a mystery. Another example of an unsolved problem is the spindle mechanism for error correction. Chromosomes segregate accurately most of the time, but this is due in part to the ability of the spindle to correct mistakes that occur by chance in bipolar chromosome attachment during prometaphase. How errors are recognized and corrected is still poorly understood, although it is an important issue for the health of all higher animals and plants. Another unsolved issue relates to the orientation of the spindle—this feature of mitosis is regulated in many cells, helping to define the positions of daughter cells as they form. The mechanisms that sense body coordinates and orient spindles correctly are not yet well understood. Finally, a more complete description of mitosis than the one presented here would include the variety in mitosis that has been described in cells from a wide range of phylogeny—for example, trypanosomes, dinoflagellates, and primitive algae. A better understanding of this diversity would probably help us to understand fundamental issues about the mitotic mechanism and perhaps even something about the evolution of mitosis.
ACKNOWLEDGMENTS
I thank G. Goshima for his critical reading of this paper, Mary Morphew and Eileen O’Toole, who contributed electron micrographs, Jennifer DeLuca of Colorado State University, who generously provided all of the light micrographs, and the National Institutes of Health (NIH), which supported this work through GM033787.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is in this collection.
- Aist JR, Bayles CJ, Tao W, Berns MW. 1991. Direct experimental evidence for the existence, structural basis and function of astral forces during anaphase B in vivo. J Cell Sci 100: 279–288. [DOI] [PubMed] [Google Scholar]
- Akiyoshi B, Gull K. 2014. Discovery of unconventional kinetochores in kinetoplastids. Cell 156: 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Silkworth WT, Nardi IK, Nicholson JM, Compton DA, Cimini D. 2014. The mitotic origin of chromosomal instability. Curr Biol 24: R148–R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA. 2002. Inheritance of the endoplasmic reticulum and Golgi apparatus. Curr Opin Cell Biol 14: 496–499. [DOI] [PubMed] [Google Scholar]
- Barr FA. 2004. Golgi inheritance: Shaken but not stirred. J Cell Biol 164: 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125: 1375–1386. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. 1990. Real-time visualization of cell cycle–dependent changes in microtubule dynamics in cytoplasmic extracts. Cell 62: 579–589. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Hunt AJ. 2005. Microtubule movements on the arms of mitotic chromosomes: Polar ejection forces quantified in vitro. Proc Natl Acad Sci 102: 13903–13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Scholey JM. 2011. Mitotic motors and chromosome segregation: The mechanism of anaphase B. Biochem Soc Trans 39: 1149–1153. [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. 2009. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell 20: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Abramson DH. 1968. Cytokinesis in HeLa: Post-telophase delay and microtubule- associated motility. Protoplasma 66: 413–435. [DOI] [PubMed] [Google Scholar]
- Chakravarty A, Howard L, Compton DA. 2004. A mechanistic model for the organization of microtubule asters by motor and non-motor proteins in a mammalian mitotic extract. Mol Biol Cell 15: 2116–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. 2014. The kinetochore. Cold Spring Harb Perspect Biol 6: a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YH, Bischoff JR, Beach D, Goldman RD. 1990. Intermediate filament reorganization during mitosis is mediated by p34cdc2 phosphorylation of vimentin. Cell 62: 1063–1071. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Scholey JM. 2007. Mitotic motors: Kinesin-5 takes a brake. Curr Biol 17: R544–R547. [DOI] [PubMed] [Google Scholar]
- Coue M, Lombillo VA, McIntosh JR. 1991. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol 112: 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca KF, Lens SM, DeLuca JG. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci 124: 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mey J, Lambert AM, Bajer AS, Moeremans M, De Brabander M. 1982. Visualization of microtubules in interphase and mitotic plant cells of Haemanthus endosperm with the immuno-gold staining method. Proc Natl Acad Sci 79: 1898–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU. 2004. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428: 93–97. [DOI] [PubMed] [Google Scholar]
- Dumont S, Mitchison T. 2012. Mechanical forces in mitosis. In Comprehensive biophysics (ed. Egelman E), Vol. 4, pp. 298–320. Elsevier, Amsterdam. [Google Scholar]
- Elting MW, Hueschen CL, Udy DB, Dumont S. 2014. Force on spindle microtubule minus ends moves chromosomes. J Cell Biol 206: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74: 504–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G. 2006. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J 25: 4897–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J, Carpi N, Betz T, Betard A, Chebah M, Azioune A, Bornens M, Sykes C, Fetler L, Cuvelier D, et al. 2011. External forces control mitotic spindle positioning. Nat Cell Biol 13: 771–778. [DOI] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. 2012. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 14: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea BB, Ruderman JV. 2006. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proc Natl Acad Sci 103: 4493–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. 2002. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol 156: 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Glotzer M. 2016. Cytokinesis in fungi and metazoa. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Goodson HV, Jonasson EM. 2016. Microtubules and MAPs. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR. 2006. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J 25: 4888–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Mana-Capelli S, McLean JR, Chen CT, Ray S, Gould KL, McCollum D. 2013. Identification of SIN pathway targets reveals mechanisms of crosstalk between NDR kinase pathways. Curr Biol 23: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Laurell E, Kutay U. 2009. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol 10: 178–191. [DOI] [PubMed] [Google Scholar]
- Hannak E, Kirkham M, Hyman AA, Oegema K. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol 155: 1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Murray AW. 1995. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol 131: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Smith D. 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays TS, Salmon ED. 1990. Poleward force at the kinetochore in metaphase depends on the number of kinetochore microtubules. J Cell Biol 110: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays TS, Wise D, Salmon ED. 1982. Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. J Cell Biol 93: 374–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425. [DOI] [PubMed] [Google Scholar]
- Hoyt MA. 2001. A new view of the spindle checkpoint. J Cell Biol 154: 909–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Aravamudhan P. 2016. How the kinetochore switches off the spindle assembly checkpoint. Cell Cycle 15: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. 2008. The RanGTP gradient—A GPS for the mitotic spindle. J Cell Sci 121: 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina I, Nandi A, Delivani P, Chacón MR, Klemm AH, Ramunno-Johnson D, Krull A, Lindner B, Pavin N, Tolić-Nørrelykke IM. 2013. Pivoting of microtubules around the spindle pole accelerates kinetochore capture. Nat Cell Biol 15: 82–87. [DOI] [PubMed] [Google Scholar]
- Kamasaki T, O’Toole E, Kita S, Osumi M, Usukura J, McIntosh JR, Goshima G. 2013. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J Cell Biol 202: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon D, McEwen BF, Khodjakov A. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science 311: 388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. 2009. Mitosis: Too much of a good thing (can be bad). Curr Biol 19: R1032–R1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol 10: 59–67. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. 1986. Beyond self-assembly: From microtubules to morphogenesis. Cell 45: 329–342. [DOI] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Komoto S, Kitamura Y, Antony C, Tanaka TU. 2010. Kinetochores generate microtubules with distal plus ends: Their roles and limited lifetime in mitosis. Dev Cell 18: 248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig P, Braunfeld MB, Sedat JW, Agard DA. 2007. The three-dimensional structure of in vitro reconstituted Xenopus laevis chromosomes by EM tomography. Chromosoma 116: 349–372. [DOI] [PubMed] [Google Scholar]
- Kubai DF. 1975. The evolution of the mitotic spindle. Int Rev Cytol 43: 167–227. [DOI] [PubMed] [Google Scholar]
- LaFountain JR, Cohan CS, LaFountain DJ. 2004. Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol Biol Cell 15: 5724–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. 2010. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol 21: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Nigg EA. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol 135: 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka E, Gadeyne A, Stockle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Muller S. 2014. The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26: 2617–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. 1995. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol 128: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Biggins S. 2014. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev 28: 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HT, Poon RY. 2011. How protein kinases co-ordinate mitosis in animal cells. Biochem J 435: 17–31. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol 2: 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P, Desai A, Oegema K, Mitchison TJ, Salmon ED. 2002. Poleward microtubule flux is a major component of spindle dynamics and anaphase A in mitotic Drosophila embryos. Curr Biol 12: 1670–1674. [DOI] [PubMed] [Google Scholar]
- Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, McEwen BF, Mogilner A, Khodjakov A. 2015. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol 17: 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller JL. 1986. Mitogenic signalling and protein phosphorylation in Xenopus oocytes. J Cyclic Nucleotide Protein Phosphor Res 11: 543–555. [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83: 29–38. [DOI] [PubMed] [Google Scholar]
- Matos I, Pereira AJ, Lince-Faria M, Cameron LA, Salmon ED, Maiato H. 2009. Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J Cell Biol 186: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR. 1991. Structural and mechanical control of mitotic progression. Cold Spring Harb Symp Quant Biol 56: 613–619. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, McDonald KL, Edwards MK, Ross BM. 1979. Three-dimensional structure of the central mitotic spindle of Diatoma vulgare. J Cell Biol 83: 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Volkov V, Ataullakhanov FI, Grishchuk EL. 2010. Tubulin depolymerization may be an ancient biological motor. J Cell Sci 123: 3425–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Molodtsov M, Ataullakhanov FI. 2012. Biophysics of mitosis. Quart Rev Biophys 45: 147–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. 1989. Polewards microtubule flux in the mitotic spindle: Evidence from photoactivation of fluorescence. J Cell Biol 109: 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Evans L, Schulze E, Kirschner M. 1986. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45: 515–527. [DOI] [PubMed] [Google Scholar]
- Mitsushima M, Aoki K, Ebisuya M, Matsumura S, Yamamoto T, Matsuda M, Toyoshima F, Nishida E. 2010. Revolving movement of a dynamic cluster of actin filaments during mitosis. J Cell Biol 191: 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. 2003. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell 12: 1477–1487. [DOI] [PubMed] [Google Scholar]
- Nakaoka Y, Miki T, Fujioka R, Uehara R, Tomioka A, Obuse C, Kubo M, Hiwatashi Y, Goshima G. 2012. An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell 24: 1478–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. 1997. How cells get the right chromosomes. Science 275: 632–637. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Campbell MS, Ward SC, Gorbsky GJ. 1998. Tension-sensitive kinetochore phosphorylation in vitro. J Cell Sci 111: 3189–3196. [DOI] [PubMed] [Google Scholar]
- O’Toole ET, Giddings TH, McIntosh JR, Dutcher SK. 2003. Three-dimensional organization of basal bodies from wild-type and δ-tubulin deletion strains of Chlamydomonas reinhardtii. Mol Biol Cell 14: 2999–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavicic-Kaltenbrunner V, Mishima M, Glotzer M. 2007. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol Biol Cell 18: 4992–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114. [DOI] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152: 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw MJ, Ward JJ, Kanemaki M, Natsume K, Nedelec FJ, Tanaka TU. 2010. Condensins promote chromosome recoiling during early anaphase to complete sister chromatid separation. Dev Cell 19: 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. 1994. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol 124: 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol 130: 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter H Jr, Inoue S, Kubai D. 1978. Mitosis in Barbulanympha. I. Spindle structure, formation, and kinetochore engagement. J Cell Biol 77: 638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BT, Farr KA, Hoyt MA. 1994. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol 14: 8282–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, McIntosh JR. 1987. Interzone microtubule behavior in late anaphase and telophase spindles. J Cell Biol 105: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR. 1984. Tubulin dynamics in cultured mammalian cells. J Cell Biol 99: 2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S, Lynch M, Needleman D. 2014. Deciphering the evolutionary history of open and closed mitosis. Curr Biol 24: R1099–R1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. 2012. The kinetochore-bound ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell 23: 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. 2000. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol 2: 922–930. [DOI] [PubMed] [Google Scholar]
- Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. 1997. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol 137: 1211–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikirzhytski V, Magidson V, Steinman JB, He J, Le Berre M, Tikhonenko I, Ault JG, McEwen BF, Chen JK, Sui H, et al. 2014. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J Cell Biol 206: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. 2011. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell 23: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan G, Kedersha N, Elroy-Stein O. 2007. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol 27: 6639–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt RA, Alderton J. 1988. Intracellular free calcium rise triggers nuclear envelope breakdown in the sea urchin embryo. Nature 332: 364–366. [DOI] [PubMed] [Google Scholar]
- Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier S, Ohi MD, Hwang W, Lang MJ, Ohi R. 2014. Kinesin-12 Kif15 targets kinetochore-fibers through an intrinsic two step mechanism. Curr Biol 24: 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol 178: 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Uhlmann F, Heeger S. 2012. Condensin, chromatin crossbarring and chromosome condensation. Curr Biol 22: R1012–R1021. [DOI] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol 189: 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Goshima G. 2010. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J Cell Biol 191: 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F. 2001. Chromosome cohesion and segregation in mitosis and meiosis. Curr Opin Cell Biol 13: 754–761. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. 2014. A silent revolution in chromosome biology. Nat Rev Mol Cell Biol 15: 431. [DOI] [PubMed] [Google Scholar]
- Vandre DD, Davis FM, Rao PN, Borisy GG. 1986. Distribution of cytoskeletal proteins sharing a conserved phosphorylated epitope. Eur J Cell Biol 41: 72–81. [PubMed] [Google Scholar]
- Volkov VA, Zaytsev AV, Gudimchuk N, Grissom PM, Gintsburg AL, Ataullakhanov FI, McIntosh JR, Grishchuk EL. 2013. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubule. Proc Natl Acad Sci 110: 7708–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VA, Grissom PM, Arzhanik VK, Zaytsev AV, Renganathan K, McClure-Begley T, Old WM, Ahn N, McIntosh JR. 2015. Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J. Cell Biol. 298: 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell 137: 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandke C, Barisic M, Sigl R, Rauch V, Wolf F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, et al. 2012. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J Cell Biol 198: 847–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. 1997. Microtubule dynamics: Treadmilling comes around again. Curr Biol 7: R369–R372. [DOI] [PubMed] [Google Scholar]
- West M, Zurek N, Hoenger A, Voeltz GK. 2011. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol 193: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440: 565–569. [DOI] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L. 1997. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol 137: 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL. 2007. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol 17: 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wadsworth P, Hepler PK. 1990. Microtubule dynamics in living dividing plant cells: Confocal imaging of microinjected fluorescent brain tubulin. Proc Natl Acad Sci 87: 8820–8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Rogers GC, Buster DW, Sharp DJ. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J Cell Biol 177: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieve GW, Heidemann SR, McIntosh JR. 1980. Isolation and partial characterization of a cage of filaments that surrounds the mammalian mitotic spindle. J Cell Biol 87: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]