ABSTRACT
Molecular evolutionary arms races between viruses and their hosts are important drivers of adaptation. These Red Queen dynamics have been frequently observed in primate retroviruses and their antagonists, host restriction factor genes, such as APOBEC3F/G, TRIM5-α, SAMHD1, and BST-2. Host restriction factors have experienced some of the most intense and pervasive adaptive evolution documented in primates. Recently, two novel host factors, SERINC3 and SERINC5, were identified as the targets of HIV-1 Nef, a protein crucial for the optimal infectivity of virus particles. Here, we compared the evolutionary fingerprints of SERINC3 and SERINC5 to those of other primate restriction factors and to a set of other genes with diverse functions. SERINC genes evolved in a manner distinct from the canonical arms race dynamics seen in the other restriction factors. Despite their antiviral activity against HIV-1 and other retroviruses, SERINC3 and SERINC5 have a relatively uneventful evolutionary history in primates.
IMPORTANCE Restriction factors are host proteins that block viral infection and replication. Many viruses, like HIV-1 and related retroviruses, evolved accessory proteins to counteract these restriction factors. The importance of these interactions is evidenced by the intense adaptive selection pressures that dominate the evolutionary histories of both the host and viral genes involved in this so-called arms race. The dynamics of these arms races can point to mechanisms by which these viral infections can be prevented. Two human genes, SERINC3 and SERINC5, were recently identified as targets of an HIV-1 accessory protein important for viral infectivity. Unexpectedly, we found that these SERINC genes, unlike other host restriction factor genes, show no evidence of a recent evolutionary arms race with viral pathogens.
INTRODUCTION
Evolutionary arms races give rise to intense selective pressures that, while altering both genotype and phenotype, may not result in long-term fitness gains (1). Over the evolutionary history of our primate ancestors, the primacy of viral pathogens is evidenced by evolutionary arms races that have led to rapid evolutionary change and extreme levels of directional and balancing selection on antiviral genes (2, 3).
The compact genomes of primate lentiviruses, a family of retroviruses including HIV, encode overlapping structural and accessory proteins, several of which have evolved to avoid or to counteract specific host proteins that inhibit viral replication (so-called “restriction factors”). In HIV, Vif neutralizes APOBEC3F and APOBEC3G, cytidine deaminases that induce hypermutation in the viral genome (4–7); the viral capsid has evolved to evade recognition by TRIM5-α, which prevents viral-core uncoating (8–10); Vpx (found in simian immunodeficiency viruses and HIV-2, a less prevalent form of HIV) antagonizes SAMHD1 and prevents it from reducing the concentration of cytoplasmic deoxynucleoside triphosphates (dNTPs), crucial for reverse transcription (11, 12); and Vpu or Nef prevents BST-2 (tetherin) from preventing viral-particle release (13–16; for reviews, see references 17 and 18). Many of these host proteins provide barriers to viral cross-species transmission (19–23), supporting the theory that the intense positive selection documented in these host proteins is evidence of an evolutionary arms race (24–28).
In humans, BST-2 has evolved to evade the neutralizing effects of most Nef proteins. BST-2 antagonism in HIV-1 group M, the virus responsible for the global HIV/AIDS pandemic, is instead provided by the Vpu protein; only HIV-1 group O, like most simian lentiviruses, uses Nef (29). Nonetheless, Nef is crucial for efficient viral replication and rapid disease progression in HIV-1 group M infections. One conserved function of Nef is the enhancement of virion infectivity (30). This phenotype had the potential to be explained by antagonism of a cellular protein or proteins that inhibit lentiviral infectivity. Recently, two independent groups identified such proteins: SERINC5 and SERINC3 (31, 32). The SERINC proteins are found in host cell membranes and, in the absence of Nef, incorporate into the virion and seemingly interfere with the transfer of the viral capsid into the newly infected cell. Here, we investigated whether SERINC5 and SERINC3 were involved in arms races over their evolutionary history in primates in a manner similar to those of other restriction factors with antiretroviral functions.
MATERIALS AND METHODS
Evolutionary fingerprinting using FUBAR.
One way to characterize selection acting on a gene is to examine the distribution of selection coefficients across all codon sites in that gene: an evolutionary fingerprint (33). FUBAR, a popular tool for inferring positive or purifying selection at individual sites, also infers the full joint distribution of synonymous (α) and nonsynonymous (β) rates for the entire gene (34). The possible values for α and β are finely discretized, forming a 20 by 20 grid of α-β pairs; the probability for the ith pair is denoted θi, and α-β at each site is modeled as an independent draw from θ. Using a symmetric Dirichlet prior distribution, P(θ), FUBAR infers the posterior distribution of θ, given the sequence alignment, P(θ|S), using Bayes theorem and Markov chain Monte Carlo (MCMC) sampling. The posterior mean, θ̂, can be plotted as a surface to visualize the inferred distribution of selection coefficients.
FUBAR was originally designed for identifying sites under positive selection, and the accuracy of θ̂ is relatively unimportant for site-specific inference, which governed some of the design choices behind FUBAR. However, here, θ̂ is the quantity of interest, so we modified FUBAR to use a smoother grid and we sampled much more extensively: 10 MCMC chains with 10 million samples each, discarding the first million as burn-in. We obtained a smooth grid with α and β values ranging from 0 to 50 (to ensure extreme selective regimes were covered) but with progressively increasing spacing, using the function (50 × k5)/195 where k is equal to {0, 1, …, 19}. Computing θ̂ under these conditions produces smooth and accurate evolutionary fingerprints.
We can use these evolutionary fingerprints to assess the similarity of selective forces acting upon two genes. Since θ̂ is a vector with 400 weights, there are a number of ways to compute the similarity between θ̂j and θ̂k for two genes, j and k. When an alignment is short (i.e., the number of codon sites is small), the regularization from the Dirichlet prior over θ sustains nonnegligible support for α-β classes that are nevertheless unsupported by any sites, lowering the peaks and raising the troughs of θ̂, causing artifactual divergence in many standard distribution similarity metrics. We thus used the Pearson correlation between θ̂j and θ̂k, cor(θ̂j, θ̂k), which can quantify how similar their shapes are without being affected by different degrees of regularization.
Using 1 − cor(θ̂j, θ̂k) as the distance between two genes, j and k, we compute a distance matrix between all pairs of genes. Very small distances indicated that the distributions over α and β were very similar. We performed average-linkage hierarchical clustering to identify nested clusters of genes with similar evolutionary fingerprints. Clustering and visualization were performed with the HierarchicalClustering package in Mathematica 10 (https://www.wolfram.com/mathematica).
Sequence data.
Alignments were downloaded from the University of California, Santa Cruz, genome browser (35) from the alignment of 19 mammalian (16 primate) genomes with humans (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/multiz20way/). These alignments are available in Data Set S1 in the supplemental material. Only the 14 Simiiformes (New World monkeys, Old World Monkeys, and apes) were included in the analysis to account for the loss of signal for the evolutionary fingerprint deeper in the phylogeny: Papio anubis (olive baboon), Callithrix jacchus (common marmoset), Chlorocebus sabaeus (African green monkey), Gorilla gorilla gorilla (western lowland gorilla), Homo sapiens (humans), Macaca mulatta (rhesus macaque), Macaca fascicularis (crab-eating macaque), Nasalis larvatus (proboscis monkey), Nomascus leucogenys (northern white-cheeked gibbon), Pan paniscus (bonobo), Pan troglodytes (chimpanzee), Pongo pygmaeus abelii (orangutan), Rhinopithecus roxellana (golden snub-nosed monkey), and Saimiri boliviensis (squirrel monkey).
We analyzed five restriction factor genes (APOBEC3F [human NCBI reference NM_145298], APOBEC3G [NM_021822], BST-2 [NM_004335], TRIM5-α [NM_033034], SAMHD1 [NM_015474]); two SERINC genes [SERINC3 (NM_006811] and SERINC5 isoform 1 [NM_001174072], SERINC5 isoform 2 [NM_001174071]); a canonically positively selected gene (lysozyme [NM_000239]); and nine well-characterized genes (AVPR1a [NM_000706], FITM2 [NM_001080472], FOXP2 isoform 1 [NM_014491], OPN1LW [NM_020061], oxytocin [NM_000916], Pokemon [NM_001256455], RB1 [NM_000321], rhodopsin [NM_000539], and SonicHedgehog isoform 2 [NM_000193]) (ideonexus; http://ideonexus.com/2008/05/13/the-top-10-human-genes/).
RESULTS AND DISCUSSION
We adapted FUBAR (34), a rapid Bayesian selection analysis tool, to characterize the evolutionary fingerprints (33) of the SERINC5 and SERINC3 genes across 14 primate species and compared their evolutionary profiles with (i) the five above-mentioned restriction factor genes; (ii) the lysozyme gene, a canonical, positively selected gene in primates; and (iii) nine other genes with diverse, well-characterized functions.
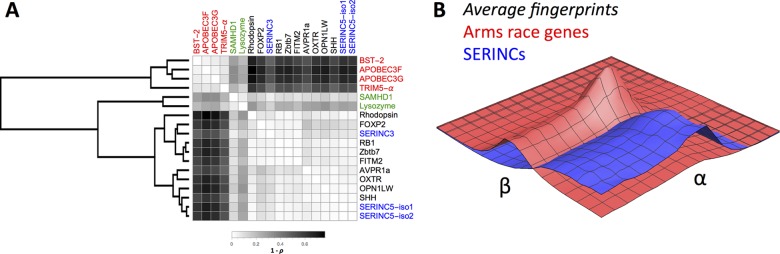
Average-linkage hierarchical clustering of these evolutionary fingerprints revealed three distinct clusters (Fig. 1A). One cluster contained the canonical APOBEC3G, APOBEC3F, BST-2, and TRIM5-α restriction factor genes, each of which has been previously documented to be engaged in an evolutionary arms race (17). These results suggest the existence of an evolutionary fingerprint common to restriction factor genes involved in evolutionary arms races. A second cluster contained the SAMHD1 and lysozyme genes, two genes documented to be under strong positive selection in some primate lineages (24, 36). The third cluster contained the rest of the included genes, with no especially notable signatures of positive selection. Importantly, the third cluster contained both the SERINC5 and SERINC3 genes (31, 32).
FIG 1.
Evolutionary fingerprints of restriction factors and other well-characterized genes. (A) Evolutionary fingerprinting analysis detected three clusters: canonical arms race genes (red), positively selected genes (green), and SERINC/other genes (blue/black). Notably, the SERINC genes do not cluster with the canonical arms race genes. (B) Average evolutionary fingerprints for the SERINC genes (blue) and the canonical arms race genes (red), confirming that the canonical arms race genes have strong support for sites with high nonsynonymous (β)-to-synonymous (α) rate ratios, whereas the SERINC genes do not.
We found a high proportion of positively selected sites (i.e., the nonsynonymous-substitution rate [β] was greater than the synonymous-substitution rate [α]) in the restriction factor genes engaged in evolutionary arms races: 12.2% to 23.9% of codon sites (Table 1). SAMHD1 and lysozyme also had signals for positive selection: 5.6% and 7.4% of codon sites, respectively. The SERINC proteins had lower proportions of codon sites under positive selection: 0.7% to 2.5% (Table 2 lists specific sites). By this crude metric, the evolutionary history of SERINC proteins in primates looks more like that of the other well-characterized genes than that of the other restriction factor genes or a canonical positively selected gene.
TABLE 1.
Proportions of positively and negatively selected sites using FUBAR
| Functional category | Gene name | No. of sites under positive selection/totala | % sites under positive selection | No. of sites under negative selection/totalb | % sites under negative selection |
|---|---|---|---|---|---|
| Restriction factor | APOBEC3F | 55/373 | 14.7 | 47/373 | 12.6 |
| APOBEC3G | 80/383 | 20.9 | 40/383 | 10.4 | |
| BST-2 | 22/180 | 12.2 | 22/180 | 12.2 | |
| SAMHD1 | 35/626 | 5.6 | 85/626 | 13.6 | |
| TRIM5-α | 118/493 | 23.9 | 62/493 | 12.6 | |
| SERINC | SERINC3 | 12/473 | 2.5 | 66/473 | 14.0 |
| SERINC5-iso1c | 3/461 | 0.7 | 102/461 | 22.1 | |
| SERINC5-iso2c | 5/420 | 1.2 | 92/420 | 21.9 | |
| Other | AVPR1a | 2/418 | 0.5 | 114/418 | 27.3 |
| FITM2 | 2/262 | 0.8 | 52/262 | 19.8 | |
| FOXP2 | 0/715 | 0.0 | 70/715 | 9.8 | |
| Lysozyme | 11/148 | 7.4 | 17/148 | 11.5 | |
| OPN1LW | 3/364 | 0.8 | 91/364 | 25.0 | |
| OXTR | 0/389 | 0.0 | 97/389 | 24.9 | |
| RB1 | 8/928 | 0.9 | 134/928 | 14.4 | |
| Rhodopsin | 0/348 | 0.0 | 78/348 | 22.4 | |
| SHH | 3/462 | 0.6 | 96/462 | 20.8 | |
| Zbtb7 | 5/539 | 0.9 | 98/539 | 18.2 |
The number of codon sites in which the probability that β was greater than α was greater than 0.80.
The number of codon sites in which the probability that α was greater than β was greater than 0.80.
Different transcript variants (isoforms) of SERINC5 have been reported.
TABLE 2.
Codons in SERINC genes with evidence of positive selection based on FUBAR analysis
| Gene | Codona | α | β | Prob[α < β]b | Bayes factor |
|---|---|---|---|---|---|
| SERINC3 | 50 | 0.771 | 3.888 | 0.879 | 12.200 |
| 102 | 1.026 | 4.417 | 0.870 | 11.240 | |
| 166 | 1.886 | 4.770 | 0.808 | 7.083 | |
| 170 | 0.823 | 7.717 | 0.949 | 30.994 | |
| 253 | 0.919 | 10.022 | 0.953 | 34.127 | |
| 274 | 0.808 | 4.977 | 0.897 | 14.687 | |
| 346 | 0.841 | 4.566 | 0.889 | 13.413 | |
| 380 | 0.985 | 8.810 | 0.946 | 29.361 | |
| 383 | 0.930 | 4.216 | 0.873 | 11.495 | |
| 406 | 0.824 | 4.347 | 0.885 | 12.941 | |
| 449 | 0.772 | 4.840 | 0.901 | 15.217 | |
| 469 | 0.925 | 4.212 | 0.873 | 11.536 | |
| SERINC5-iso1c | 225 | 0.780 | 3.120 | 0.852 | 12.992 |
| 241 | 0.378 | 2.479 | 0.838 | 11.645 | |
| 251 | 1.192 | 4.669 | 0.861 | 13.906 | |
| SERINC5-iso2c | 215 | 0.523 | 2.301 | 0.804 | 8.732 |
| 225 | 0.710 | 2.970 | 0.857 | 12.751 | |
| 241 | 0.359 | 2.539 | 0.848 | 11.873 | |
| 251 | 1.430 | 4.672 | 0.848 | 11.866 | |
| 417 | 1.129 | 3.310 | 0.831 | 10.500 |
Codon sites correspond to alignments available in Data Set S1 in the supplemental material.
Prob[α < β], probability that α was less than β.
Different transcript variants (isoforms) of SERINC5 have been reported.
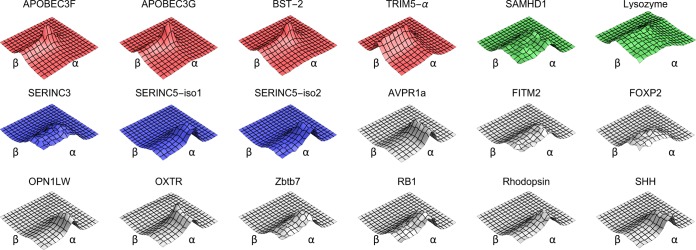
To highlight the nature of the difference between the canonical arms race genes and the SERINC genes, we plotted the average evolutionary fingerprint surface for the restriction factor arms race genes and the SERINC genes (Fig. 1B). The arms race genes have a large proportion of sites with higher nonsynonymous-substitution rates (β), whereas the SERINC genes show a predominance of purifying selection. The same pattern can be seen in the fingerprint surfaces for individual genes within each cluster, which exhibit substantial within-cluster uniformity (Fig. 2).
FIG 2.
Evolutionary fingerprint surfaces of all analyzed genes. The genes are colored according to their evolutionary fingerprint clusters: canonical arms race (red), positive selection (green), and other (gray) genes. The SERINC genes are highlighted in blue.
Despite the biological interaction between the SERINCs and Nef, we did not detect a signal of arms race dynamics typical of other restriction factors. This finding seems to reflect a rather uneventful evolutionary history of the SERINC5 and SERINC3 genes. Why would these genes fail to show evidence of strong positive selection despite their apparently broad antiviral spectrum, which encompasses genetically distant retroviruses, including HIV-1, murine leukemia virus, and equine infectious anemia virus (A. Chande, presented at the 2016 meeting on retroviruses, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 23 to 28 May 2016)? One possibility is that evolutionary constraints on the SERINCs imposed by their other cellular functions make escape, and a subsequent arms race, impossible. Alternatively, the escape dynamics may be limited to a small fraction of sites in the SERINCs, masking the signal for detecting arms race dynamics; in other words, the relatively few sites in the SERINCs under positive selection might indeed be important for interaction with viral antagonists. Another possibility is that an arms race may have occurred, but not one that involved changes at the codon level. For example, BST-2 experienced a 5-amino-acid deletion in humans that counteracts its restriction by most lentiviral Nef proteins (25), and the TRIM genes have undergone gene fusion to acquire a novel protein domain with novel capsid specificity (Trim-Cyp) (37). Finally, the importance of the antiretroviral function of the SERINC proteins may be a relatively novel evolutionary advance, and the arms race between these cellular proteins and viral countermeasures is just about to begin.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases: (AI115701 [J.O.W.], K01AI110181 [J.O.W.], K99AI120851 [B.M.], and R37AI081668 [J.G.]).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00972-16.
REFERENCES
- 1.Van Valen L. 1973. A new evolutionary law. Evol Theory 1:1–30. [Google Scholar]
- 2.Enard D, Cai L, Gwennap C, Petrov DA. 2016. Viruses are a dominant driver of protein adaptation in mammals. eLife 5:e12469. doi: 10.7554/eLife.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worobey M, Bjork A, Wertheim JO. 2007. Point, counterpoint: the evolution of pathogenic viruses and their human hosts. Annu Rev Ecol Evol Systematics 38:515–540. doi: 10.1146/annurev.ecolsys.38.091206.095722. [DOI] [Google Scholar]
- 4.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 5.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 6.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 8.Song B. 2009. TRIM5alpha. Curr Top Microbiol Immunol 339:47–66. [DOI] [PubMed] [Google Scholar]
- 9.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 10.Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med 9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 11.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DT, Serra-Moreno R, Singh RK, Guatelli JC. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol 18:388–396. doi: 10.1016/j.tim.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 15.Tokarev A, Skasko M, Fitzpatrick K, Guatelli J. 2009. Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res Hum Retroviruses 25:1197–1210. doi: 10.1089/aid.2009.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 18.Duggal NK, Emerman M. 2012. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc Natl Acad Sci U S A 101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O'Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, Kaur A, Hirsch VM, Johnson WE. 2010. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol 8:e1000462. doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangeat B, Turelli P, Liao S, Trono D. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J Biol Chem 279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- 22.Schrofelbauer B, Chen D, Landau NR. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc Natl Acad Sci U S A 101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap MW, Nisole S, Stoye JP. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol 15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 24.Laguette N, Rahm N, Sobhian B, Chable-Bessia C, Munch J, Snoeck J, Sauter D, Switzer WM, Heneine W, Kirchhoff F, Delsuc F, Telenti A, Benkirane M. 2012. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11:205–217. doi: 10.1016/j.chom.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim ES, Malik HS, Emerman M. 2010. Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J Virol 84:7124–7134. doi: 10.1128/JVI.00468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog 5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawyer SL, Emerman M, Malik HS. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol 2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol 68:2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pond SL, Scheffler K, Gravenor MB, Poon AF, Frost SD. 2010. Evolutionary fingerprinting of genes. Mol Biol Evol 27:520–536. doi: 10.1093/molbev/msp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, Heitner S, Hickey G, Hinrichs AS, Hubley R, Karolchik D, Learned K, Lee BT, Li CH, Miga KH, Nguyen N, Paten B, Raney BJ, Smit AF, Speir ML, Zweig AS, Haussler D, Kuhn RM, Kent WJ. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 37.Price AJ, Marzetta F, Lammers M, Ylinen LM, Schaller T, Wilson SJ, Towers GJ, James LC. 2009. Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat Struct Mol Biol 16:1036–1042. doi: 10.1038/nsmb.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.