Summary
NHE1 plays an important role in intratumoral microglial activation and microglia–glioma interactions, probably by enhancing microglial secretion of matrix-metalloproteinase 9 and inflammatory cytokines. Inhibition of microglial NHE1 activity significantly attenuated microglial activation and glioma migration/proliferation.
Abstract
Microglia play important roles in extracellular matrix remodeling, tumor invasion, angiogenesis, and suppression of adaptive immunity in glioma. Na+/H+ exchanger isoform 1 (NHE1) regulates microglial activation and migration. However, little is known about the roles of NHE1 in intratumoral microglial activation and microglia–glioma interactions. Our study revealed up-regulation of NHE1 protein expression in both glioma cells and tumor-associated Iba1+ microglia in glioma xenografts and glioblastoma multiforme microarrays. Moreover, we observed positive correlation of NHE1 expression with Iba1 intensity in microglia/macrophages. Glioma cells, via conditioned medium or non-contact glioma-microglia co-cultures, concurrently upregulated microglial expression of NHE1 protein and other microglial activation markers (iNOS, arginase-1, TGF-β, IL-6, IL-10 and the matrix metalloproteinases MT1-MMP and MMP9). Interestingly, glioma-stimulated microglia reciprocally enhanced glioma proliferation and migration. Most importantly, inhibition of microglial NHE1 activity via small interfering RNA (siRNA) knockdown or the potent NHE1-specific inhibitor HOE642 significantly attenuated microglial activation and abolished microglia-stimulated glioma migration and proliferation. Taken together, our findings provide the first evidence that NHE1 function plays an important role in glioma–microglia interactions, enhancing glioma proliferation and invasion by stimulating microglial release of soluble factors. NHE1 upregulation is a novel marker of the glioma-associated microglial activation phenotype. Inhibition of NHE1 represents a novel glioma therapeutic strategy by targeting tumor-induced microglial activation.
Introduction
Malignant gliomas are highly aggressive brain cancers with poor prognosis. Patients with the most aggressive glioblastomas have a median survival of only 14–18 months due to rapid tumor growth and therapeutic resistance to current treatments (1–4). Accumulating evidence indicates that malignant gliomas are heterogeneous masses comprised of tumor cells intermixed with parenchymal cells. This unique tumor microenvironment plays a vital role in glioma progression (5). Most importantly, microglia and infiltrating macrophages account for up to 30% of the glioma mass, and represent important components of the glioma microenvironment (6). Glioma cell survival, growth and metastasis are facilitated by tumor-associated microglia and macrophages (7,8). For example, tumor cells synthesize and release colony stimulating factor 1 (CSF-1), triggering migration of microglia and macrophages to the tumor sites and promoting subsequent microglia/monocyte release of epidermal growth factor and vascular endothelial growth factor (9). Then, the secreted factors activate their respective tumor-expressed receptors and lead to increased tumor cell proliferation, invasion and angiogenesis (9). Bidirectional communication between glioma cells and glioma-associated microglia/macrophages (GAMs) induces GAMs to adopt both the pro-inflammatory and alternative resolving phenotypes, with identification of a specific glioma-associated activation phenotype in GAMs (10–14).
Na+/H+ exchanger isoform 1 (NHE1) protein is over-expressed in gliomas and other cancer cells to maintain alkaline intracellular pH (pHi) by extruding H+ (15,16). We recently reported that NHE1 co-localizes with ezrin at lamellipodia of glioma. Upregulation of NHE1 protein expression in glioma plays a vital role in glioma migration and survival (16). Increased NHE1 also enhances glioma cell resistance to TMZ-mediated apoptosis via activation of extracellular signal-regulated kinase (ERK) signaling pathways (16). Furthermore, microglia and macrophages exhibited increased NHE1 expression during microglia activation and migration in response to lipopolysaccharide (LPS) and the chemotactic hormone, bradykinin (17,18). However, it remains unknown whether NHE1 regulates intratumoral GAM activation and promotes a favorable microenvironment for glioma survival and migration.
In this study, we investigated the role of NHE1 in microglial activation and the interactions between microglia and glioma cells. Glioma induces NHE1 protein upregulation in microglia. NHE1 is required for glioma-induced microglia activation. These functions are probably mediated by enhanced microglial secretion of matrix-metalloproteinase 9 (MMP9) and inflammatory cytokines. Most importantly, inhibition of microglial NHE1 activity with HOE642 or small interfering RNA (siRNA)-mediated protein knockdown significantly attenuated microglial activation as well as glioma migration and proliferation. Taken together, these new findings demonstrate that NHE1 activity plays an important role in intratumoral microglia–glioma communication, and identify NHE1 as a novel therapeutic target to modulate the tumor microenvironment.
Methods
Cell culture
All studies involving human tissues were performed with approval from the University of Wisconsin-Madison and University of Pittsburgh Institutional Review Board with informed consent obtained from patients.
Primary glioma stem cells (GSC22 and GSC99) were acquired from Dr John Kuo in August 2011 and were passaged in serum containing medium no longer than 6 months to yield GC22 and GC99 glioma cells (19,20). Initial validation of cell lines was assessed by formation of spheres, ability to differentiate into both neuron and astrocyte lineages, and by tumor formation after intracranial injection and serial engraftment. GSC22 was last validated for tumor initiation in January 2014 and GSC99 in May 2011. GC22 cells were last validated by intracranial implantation in May 2013 (21). GC99 cells have not been inject intracranially, but grow in serum-containing media to a high passage numbers (>40). The human astrocyte cell line was a kind gift from Dr Clive Svendsen (Cedars-Sinai). These glioma and astrocyte cell lines were grown in adherent cultures and maintained in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals, cat #S11150H). Immortalized human microglia cell line SV40 was purchased from Applied Biological Materials, Inc. and maintained in DMEM/F12 (Gibco) supplemented with 10% FBS in collagen coated vessels. To coat vessels, collagen 1 (Gibco, cat# A10483-01) was diluted in PBS to a final concentration of 0.1mg/ml and incubated on the vessels at 37°C for 1h, followed by two rinses with PBS. Cultures were passaged approximately every 4 days with fresh medium at a density of 106 cells/75cm2 in a culture flask. Passage 20–40 of glioma cells, passage 10–25 of human astrocytes, and passage 2–15 of microglia cells were used in the study.
Cell conditioned medium collection
For the preparation of astrocyte-conditioned medium (ACM) or glioma-conditioned medium (GCM) treated microglia cultures, on day 1, 2×106 astrocytes or glioma cells were seeded in 100mm dishes in 10ml DMEM + 10% FBS media overnight. On day 2, cells were washed twice with serum-free DMEM/F12 medium and then incubated with 10ml fresh DMEM/F12 + 10% FBS at 37°C for 24h. Also, 2×106 microglia cells were seeded in collagen coated 100mm dishes in DMEM/F12 + 10% FBS overnight. On day 3, the medium from astrocyte or glioma cultures (80–90% confluent) was collected and centrifuged at 2000g at 4°C for 10min. The supernatant was collected as ACM or GCM. Fresh conditioned media was prepared for each set of cultures. Microglia cultures were washed twice with serum-free DMEM/F12 medium followed by incubation with 10ml of ACM or GCM plus or minus 1 µM HOE642 (Sanofi-Aventis, Germany), 0.5 µM MK2206 (Selleck Chemical), or 100ng/ml lipopolysaccharide (Sigma) for 24h. On day 4, the medium was then collected from the microglia cultures (70–80% confluent) and centrifuged at 2000 × g at 4°C for 10min. The supernatant was collected and defined as A/MCM or G/MCM. The microglial cells were rinsed twice in PBS, removed with a cell scraper, and collected for further biochemical experiments.
For the noncontact glioma–microglia co-cultures, 2×105 glioma cells were seeded on the surface of 0.4 μm inserts (Millicell) in DMEM/F12 with 10% FBS. 2×105 human microglia/well were seeded on the bottom of a six-well plate in DMEM/F12 with 10% FBS. After 24h, the inserts were placed into the six-well plate containing microglia cultures and cells were simultaneously cocultured for 24h.
Transwell chemotaxis and matrigel matrix invasion assay
Transwell membrane cell culture inserts (8.0 μm pore size, Becton Dickinson) were coated with 0.5 μg/ml poly-d-lysine as described previously (22). Dissociated glioma cells (4×104 cells) in 100 μl MCM were seeded on top of the membrane insert while the lower wells contained 700 µl serum-free DMEM. The plates were incubated for 5h at 37°C and migrated cells were determined according to published methods (22).
The invasion assay was modified from a previously published protocol (23). Inserts (8 µm pore size, Becton Dickinson) were coated with 1mg/ml BD Matrigel Matrix (BD Biosciences, San Diego, CA). Each insert was coated with 100 μl Matrigel Matrix, dried in sterile conditions (37°C) for 3h, and then reconstituted in 200 μl of culture medium for 30min. Glioma cells were subsequently seeded at a density of 4×104 per insert. To establish co-cultures, either scr- or NHE1 siRNA-treated microglia were seeded in the lower compartment 48h prior to the addition of glioma cells. After 16h of co-culturing at 37°C, the Matrigel Matrix was carefully removed using cotton tips. Cells that had migrated through the filters were fixed with 4% formaldehyde and stained with Cresyl Violet (Sigma). Three fields of cells were counted under a 20× phase microscopy as described (22).
RNA interference knockdown of NHE1 in microglia
The scrambled siRNA (Silencer® Negative Control No. 1 siRNA, Cat. No. AM4635) and siRNAs targeting human NHE1 (ID: s13023) were purchased from Invitrogen. A final concentration of 15nM siRNA was used for inhibiting NHE1 protein expression. Dissociated microglia were seeded in six-well plates (105 cells/well/2ml) in DMEM/F12 with 10% FBS at 24h prior to transfection. Lipofectamine RNAiMAX/siRNA complexes were prepared by adding the siRNA and 5 μl of Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) in 500 μl serum-free Opti-MEM (Gibco). Complexes were allowed to form at room temperature (RT) for 10min and added to each well. The cells were incubated at 37°C and subjected to experiments 48h after transfection.
Cell viability assays
For the PrestoBlue viability assay, 2×104 glioma cells in serum-free culture medium (180 µl) were cultured in 96-well plates for 24h. Then, glioma cultures were treated with C/MCM, A/MCM or G/GCM plus or minus HOE642 (1 μM) for 24–48h. PrestoBlue dye (20 µl/well) was added and the cultures were incubated for 30min at 37°C. Subsequently, absorbance at 564nm was recorded according to the manufacturer’s protocol (Invitrogen). Cell viability was expressed as a percentage relative to the cells treated with C/MCM+Veh.
For assessing glioma proliferation, the BrdU assay (Millipore) was performed per the manufacturer’s instructions. 1×104 glioma cells were seeded in 96-well plates for 24h and cells were cultured for 48h in a serum-free DMEM/F12 media in the presence or the absence of 1 µM HOE642 or 0.5 µM MK2206. Proliferation rate was assayed with 4h of BrdU incorporation. Thereafter, a dual absorbance of 450/550nm was read and recorded. Relative changes of cell proliferation were calculated using normalized values in the untreated cells.
Immunofluorescence staining
Immunofluorescence staining on xenografts of NOD-SCID mouse brains implanted with glioma was performed as described (22). Briefly, tissue sections were deparaffinized and rehydrated in water, and microwaved in antigen unmasking solutions (Vector laboratories) for 20min to retrieve epitopes. Sections were then blocked with 5% normal goat serum for 1h at RT, followed by overnight incubation in mouse anti-NHE1 antibody (Santa Cruz #136239, 1:100) and rabbit anti-Iba1 antibody (Wako #019-19741, 1:100) at 4°C. Fluorescence-labeled secondary antibodies (Invitrogen, 1:200) were applied for 1h at RT. For immunofluorescence staining on cultured cells, cells were fixed then blocked with 5% normal goat serum as described (22). Primary antibodies included mouse anti-NHE1 (Santa Cruz, 1:100), rabbit anti-Iba1 (Wako, 1:100), rabbit anti-iNOS (Abcam #15323, 1:100) and mouse anti-Arg1 (Sigma #AV45672, 1:100). Fluorescence images were captured with a Leica DMIRE2 confocal microscope (×40) and Leica confocal software (Leica Microsystems, Mannheim, Germany).
A tissue microarray (TMA) from glioblastoma multiforme (GBM) patients diagnosed between 1999 and 2009 was created from the University of Wisconsin Department of Pathology and Laboratory Medicine archives as described (20). Diagnosis and tissue punch location were defined by neuropathology prior to incorporation into the microarray. Mouse anti-NHE1 antibody and rabbit anti-Iba1 antibody were used to label the TMA. Fluorescence images were captured with a Leica DMIRE2 confocal microscope (×40) and Leica confocal software. Two to three images were obtained from each punch. After background subtraction with the Leica confocal software, ImageJ was utilized to quantify the integrated intensity of NHE1. Iba1+ cells were manually counted in each image.
Gelatinase zymography
Gelatinase zymography was performed on 10% pre-cast zymogram gels (Bio-Rad) under nonreducing conditions. Culture media (20 µl) was mixed with 2× sample buffer and loaded on the gel in tris glycine SDS buffer, as suggested by the manufacturer (Bio-Rad). Following electrophoresis, proteins were renatured by placing the gels in renaturing solution (Bio-Rad) for 30min at RT. Then gels were developed at 37°C overnight, followed by staining with Coomassie Brilliant Blue R-250 staining solution (Bio-Rad) for 1h at RT. Destaining was accomplished by washing gels in 100ml of destain solution (Bio-Rad) for 60min with gentle rocking. Gels were digitally imaged and analyzed densitometrically with ImageJ.
Immunoblotting assay
Samples (25 µg total protein) were denatured and electrophoretically separated on 4–15% SDS gels (Bio-Rad). Blots were blocked (with 5% nonfat milk) and incubated with a primary antibody at 4°C. Primary antibodies included mouse anti-NHE1 (Santa Cruz, 1:500), rabbit-anti-Iba1 (Wako, 1:500), rabbit anti-p-Akt (Cell Signaling #S473, 1:1000), or rabbit anti-t-Akt (Cell Signaling #C67E7, 1:1000). After rinsing, the blots were incubated with horseradish peroxidase-conjugated secondary immunoglobin G (1:2000, Vector) for 1h at RT. Bound antibody was visualized with an enhanced chemiluminescence assay (Amersham, Piscataway, NJ). Signal intensities were analyzed using ImageJ and were normalized to beta-actin or alpha-tubulin loading controls. Full size blot scans are available in the Supplementary Figure 7, available at Carcinogenesis Online.
Real-time PCR
Total RNA was isolated from microglial cultures using RNeasy kit (Qiagen) and served as a template to synthesize cDNA by extension of oligo(dT)15 primers (2.5 mmol/L) with 200U of M-MLV reverse transcriptase (Sigma–Aldrich). Real-time PCR amplifications were performed in duplicates in a 20-μl reaction volume containing 2x Taqman PCR MasterMix (Applied Biosystems) and a set of probes (listed in Supplementary Table 1, available at Carcinogenesis Online). The amount of target mRNA was first normalized to the expression level of the GAPDH rRNA amplified from the same sample and then to untreated controls. Data were analyzed by the Relative Quantification (ΔΔC t) method using 7300 System SDS software (Applied Biosystems).
Cytokine enzyme-linked immuno assay
Cell culture medium from the glioma-stimulated microglia cultures was collected and centrifuged at 2000 × g at 4°C for 10min. Medium concentrations of IL-6, IL-10 and TGF-β were measured with commercial enzyme-linked immuno assay (ELISA) quantification kits (R&D Systems), according to the manufacturer’s instructions.
Statistical analysis
The results are expressed as the mean ± standard error of the mean (SEM). Comparisons between groups were made by Student’s t-test or one-way analysis of variance using the Bonferroni post hoc test in the case of multiple comparisons (SigmaStat, Systat Software, Point Richmond, CA). A significance level of P < 0.05 was considered statistically significant in all tests and the n values represent the number of independent cultures or tissue samples. Pearson correlation coefficient was calculated using online statistics software (24).
Results
NHE1 protein is abundantly expressed in intratumoral microglia/macrophages
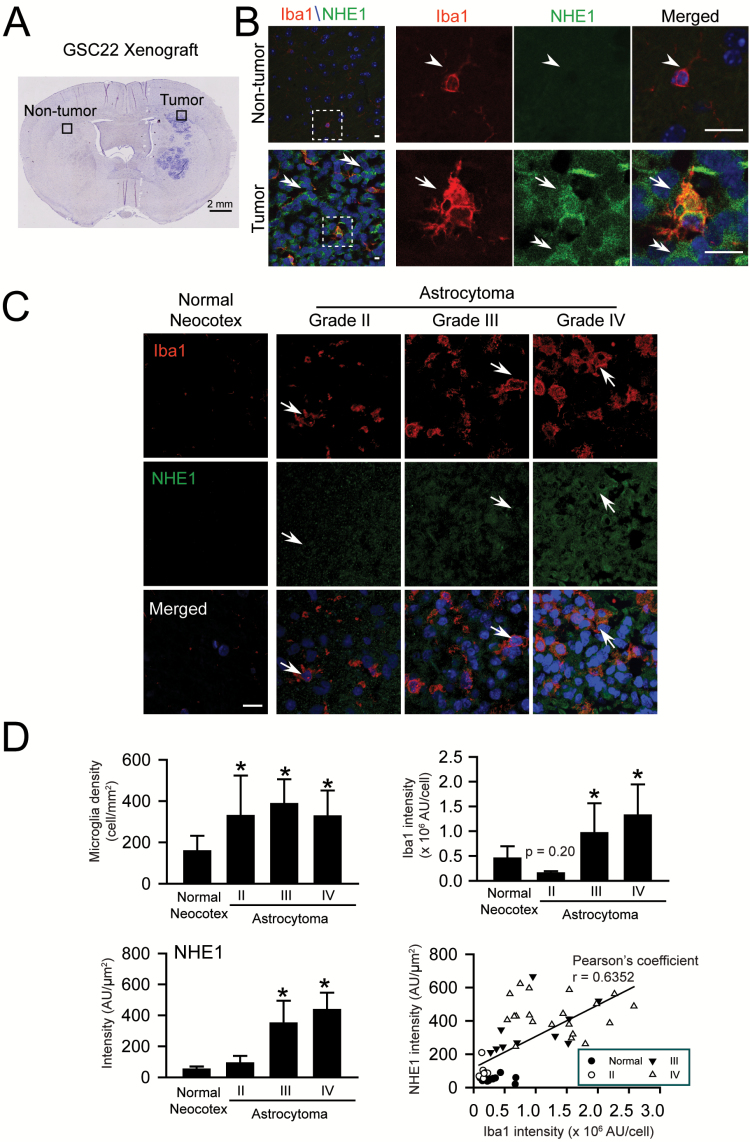
We first characterized GAM activation and NHE1 protein expression in xenograft tissues in SCID mouse brains derived from human GC22 as established before (20). Only a few cells stained positively for either Iba1 or NHE1 proteins in the nontumor region (Figure 1A and B, arrowheads). In contrast, the number of Iba+ cells (microglia/microphages) was markedly increased within the glioma xenograft (arrow). They expressed strong NHE1 signals and displayed the ‘amoeboid’ activated morphology. In accordance with our previous findings (16), glioma cells (Iba1− cells, double-arrow heads in Figure 1B) also exhibited abundant NHE1 protein expression.
Figure 1.
Intra-tumor microglia express abundant NHE1 protein. (A) Representative SCID mouse xenograft brain tumor tissue section derived from glioma stem cell GC22. Black boxes: locations for data collection. (B) Representative immunofluorescence images illustrate increased expression of NHE1 protein in Iba1+ microglia and Iba1− glioma cells in xenograft tumor tissues. White boxes: magnified areas. Arrowhead: low expression of NHE1 or Iba1. Arrow: increased NHE1 or Iba 1 expression. Scale bars: 20 μm. (C) Concurrent increase in expression of NHE1 protein and Iba1+ cell density in a tissue microarray from glioma patients with different tumor grades. Arrows: increased NHE1 protein expression in Iba1+ cells. Scale bars: 25 μm. (D) Upregulation of NHE1 and Iba1 expression in glioma tissues. Histograms illustrate intensity of NHE1 protein expression, numbers of Iba1+ cells, and Iba1 intensity in GAM in normal or glioma tissues, normal neocotex (n = 9), grade II (n = 10), grade III (n = 9) and grade IV (n = 20). *P < 0.05 versus normal neocotex. Pearson’s coeffeciency was calculate between NHE1 protein expression and Iba1 intensity in GAM, r = 0.6352, P < 0.01.
We further investigated NHE1 protein expression using a human GBM TMA. A sparse number of Iba1+ cells with low NHE1 protein expression were detected in normal neocotical region (Figure 1C). In contrast, the number of Iba1+ cells and NHE1 protein expression were concurrently increased with astrocytomas (Figure 1C and D). NHE1 protein was particularly abundant in both Iba1+ GAMs (arrows) and Iba1− glioma cells in grade III and IV astrocytomas. In addition, a moderate correlation was found between NHE1 protein expression and Iba1 intensity in GAMs (Pearson’s coefficiency r = 0.6352, P < 0.01). Therefore, NHE1 protein expression is up-regulated in both glioma cells and Iba1+ GAMs.
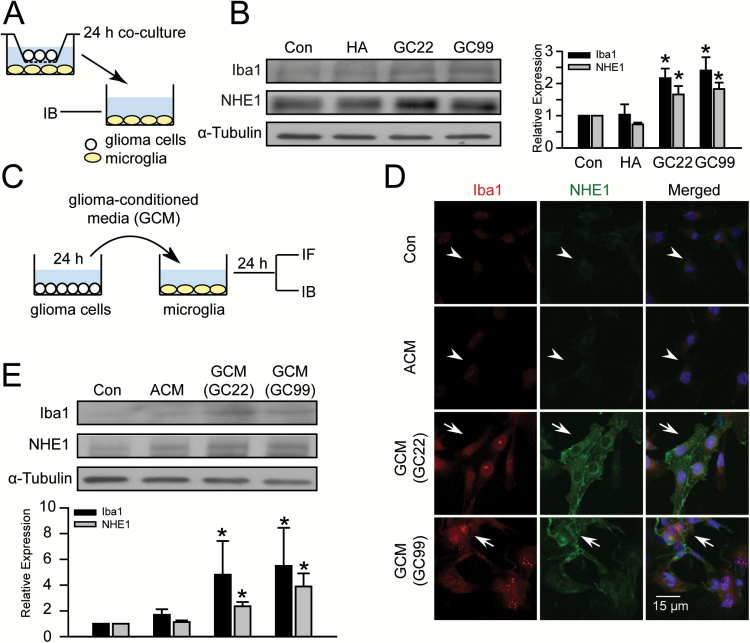
Glioma stimulates concurrent upregulation of Iba1 and NHE1 protein expression in microglia
We hypothesized that the concomitant upregulation of NHE1 in both microglia and glioma cells may play a role in the pro-tumoral interactions between these cell types. An experimental protocol was established to study glioma–microglia interactions by glioma–microglia co-culture system (Figure 2A). Microglia cultured in fresh culture medium (Con) or co-cultured with human astrocytes were used as controls. After 24h incubation, co-culture of microglia with glioma cells (either GC22 or GC99) up-regulated microglial NHE1 and Iba1 protein expression, but co-culturing with human astrocytes failed to trigger this response (Figure 2B). To fully rule out the possibility of any contact-dependent signals, we conducted additional experiments by exposing the microglia to glioma-conditioned media (GCM) (Figure 2C). Microglial exposure to fresh culture medium (Con) or human ACM were used as controls. After 24h treatment, neither control condition had an effect on Iba1 or NHE1 expression in microglial cells (Figure 2D, arrowheads). In contrast, GCM from two primary glioma lines (GC22, GC99) led to concurrent upregulation of Iba1 and NHE1 protein expression in microglia (arrows, Figure 2D). These findings were further validated by immunoblotting assays. Low expression of Iba1 and NHE1 protein was detected in microglial cells treated with either Con or ACM. Upon stimulation, NHE1 protein level increased by 3.3±0.4 fold in the GCM (GC22)-treated microglia and 3.9±0.3 folds in the GCM (GC99)-treated cells. In the same samples, Iba1 protein levels were also elevated by 2.1±0.5 fold in the GCM (GC22)-treated and 1.8±0.4 fold in the GCM (GC99)-treated groups (Figure 2E). These findings indicate that soluble factors secreted by glioma cells encourage microglial activation and increase NHE1 protein levels, further supporting our hypothesis that NHE1 activation in microglia is involved in glioma-mediated microglia activation.
Figure 2.
Glioma stimulates concurrent upregulation of Iba1 and NHE1 protein in microglia. (A) Illustration of the transwell co-culture system allowing noncontact interactions between glioma and microglia cells through diffusible factors. Microglia were placed in the bottom compartment in medium alone (Con), or co-cultured with human astrocyte (HA), GC22 or GC99 in the upper compartment for 24h. (B) Changes in NHE1 or Iba1 proteins in microglia are shown. Data represent mean ± SEM. n = 4, *P < 0.05 versus Con. (C) GCM was harvested from glioma cultures (GC22 or GC99) and applied to microglia cultures for 24h before immunofluorescence or immunoblotting assays. Fresh culture media (Con) and astrocyte-conditioned media (ACM) were used as controls. (D) Representative immunofluorescent images of GCM-treated microglia cultures exhibiting upregulation of Iba1 (red) and NHE1 expression (green). Arrowheads: low expression of Iba1 or NHE1. Arrows: increased NHE1 or Iba 1 expression. Scale bar: 15 μm. (E) Representative immunoblots of Iba1 and NHE1 protein in microglia. Data are mean ± SEM. n = 4, *P < 0.05 versus Con.
NHE1 activation is required for the glioma-induced microglial polarization
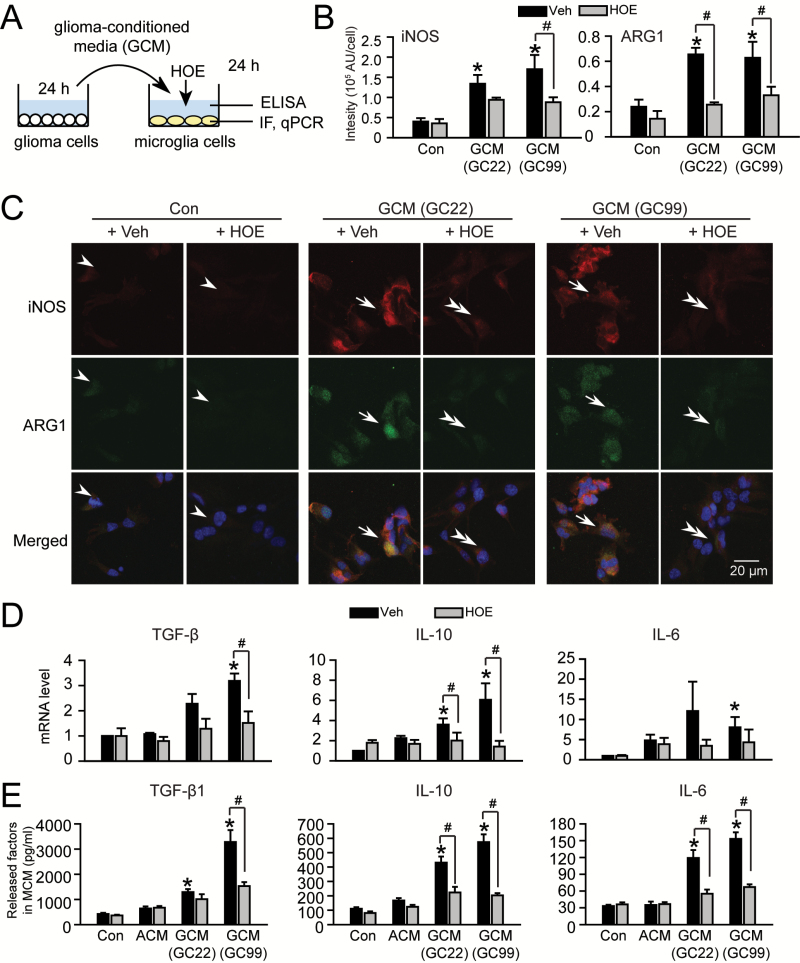
To determine if NHE1 plays a direct role in the induction of the activated microglial phenotype we characterized the function of microglial NHE1 in microglia activation in response to glioma stimulation (Figure 3A). GCM was harvested from glioma cultures (GC22 or GC99) and then added to microglia cultures for 24h either in the presence or absence of NHE1 inhibitor HOE642 (1 μM). Low expression levels of iNOS and Arg1 proteins were detected in unstimulated microglia (Con, arrowheads, Figure 3B and C). GCM stimulation triggered transformation of microglia from ‘ramified’ into ‘amoeboid’ cellular morphology (arrows, Figure 3C). Interestingly, GCM exposure up-regulated the expressions of both iNOS and Arg1 in microglia, which are generally considered as markers of anti-tumorigenic and pro-tumorigenic, respectively (arrows, Figure 3B and C). However, blocking microglial NHE1 with its inhibitor HOE642 during the GCM exposure period nearly abolished the GCM-induced upregulation of iNOS and Arg1 in microglia (double-arrowheads, Figure 3C), as well as amoeboid-like changes in microglia morphology (double-arrowheads, Figure 3B). Of note, ACM exposure appeared to stimulate expressions of iNOS and Arg1 in microglia as well, but these changes did not reach statistical significance within four sets of experiments (P = 0.66 for iNOS and P = 0.60 for Arg1, Supplementary Figure 1 is available at Carcinogenesis Online). These findings indicate that microglia stimulation in response to glioma-secreted factors depends on NHE1 activity in microglia.
Figure 3.
Glioma concurrently stimulates NHE1 activation and the glioma-associated activation phenotype in microglia. (A) GCM was harvested from glioma cultures (GC22 or GC99) and then added to microglia cultures for 24h either in the presence or absence of NHE1 inhibitor HOE642 (1 μM). Microglia or MCM were collected for immunofluorescence staining (IF), qPCR assays and ELISA. (B) Summary in changes of iNOS and Arg1 IF intensity under different conditions as listed in C. Data are mean ± SEM. n = 4, *P < 0.05 versus Con. (C) Representative immunofluorescence images showing expression of inducible nitric oxide synthase (iNOS, red) and arginase1 (ARG1, green) in microglia cultures under different conditions (Con ± HOE or GCM ± HOE). Scale bar: 10 μm. (D). Changes in mRNA expression of microglial activation markers under different conditions (Con ± HOE, ACM ± HOE or GCM ± HOE). Data are mean ± SEM. n = 4, *P < 0.05 versus Con. (E) Changes of released factors in MCM under different conditions evaluated by ELISA as listed in D. Data are mean ± SEM. n = 4, *P < 0.05 versus Con.
We further evaluated microglial cytokine expression in this glioma-mediated activation. Quantitative PCR assay revealed that the mRNA levels of interleukin-6 (IL-6), IL-10 and TGF-β in microglia were increased after either GC22-GCM or GC99-GCM stimulation (Figure 3D). These changes are glioma-specific. Most importantly, inhibition of microglial NHE1 activity by HOE642 significantly suppressed all three mRNA upregulations (Figure 3D) and nearly abolished the release of these factors in the microglia-conditioned media (MCM) (Figure 3E). Taken together, these findings strongly suggest that microglial NHE1 function is involved in glioma-mediated microglial activation.
To determine whether the cytokines measured in G/MCM were truly secreted by microglia and not residual factors present in GCM, we compared TGF-β and IL-6 contents in GCM (GC99 GCM), MCM or glioma-stimulated microglial conditioned medium (G/MCM). As shown in Supplementary Figure 2A and B, available at Carcinogenesis Online, TGF-β levels were 2-fold higher in G/MCM than GCM (3294.9±184.2 versus 1655.5±272.1 pg/ml, P < 0.05), indicating the majority of TGF-β measured in MCM came from microglia. IL-6 was not detected in GCM, demonstrating that the IL-6 measured in G/MCM was released by microglia (Supplementary Figure 2C is available at Carcinogenesis Online).
Furthermore, we tested the role of glioma NHE1 in the production of cytokines that stimulate microglial cytokine release. GC99 released similar amounts of TGF-β in the absence or presence of the NHE1 inhibitor HOE642 (1 µM, 24h, Supplementary Figure 2A and B, available at Carcinogenesis Online). Blocking of glioma NHE1 activity in GC99 cells with HOE642 prior to the GCM-mediated microglial stimulation decreased the microglial TGF-β release in G/MCM by ~30% (although it did not reach statistical significance). In contrast, inhibiting glioma NHE1 had no effects on IL-6 release in G/MCM (Supplementary Figure 2C is available at Carcinogenesis Online). These findings suggest that glioma cells release soluble factors that stimulate microglial TGF-β production in a manner partially depending on glioma NHE1 activity, whereas microglial IL-6 expression is independent of glioma NHE1 function. Of note, blocking NHE1 in glioma cells decreases baseline proliferation as determined by BrdU assay, which may also influence the composition of secreted cytokines in this model (Supplementary Figure 2D is available at Carcinogenesis Online). Together, our data indicate that NHE1 activity in glioma is involved in distinct signaling pathways in regulating microglial activation.
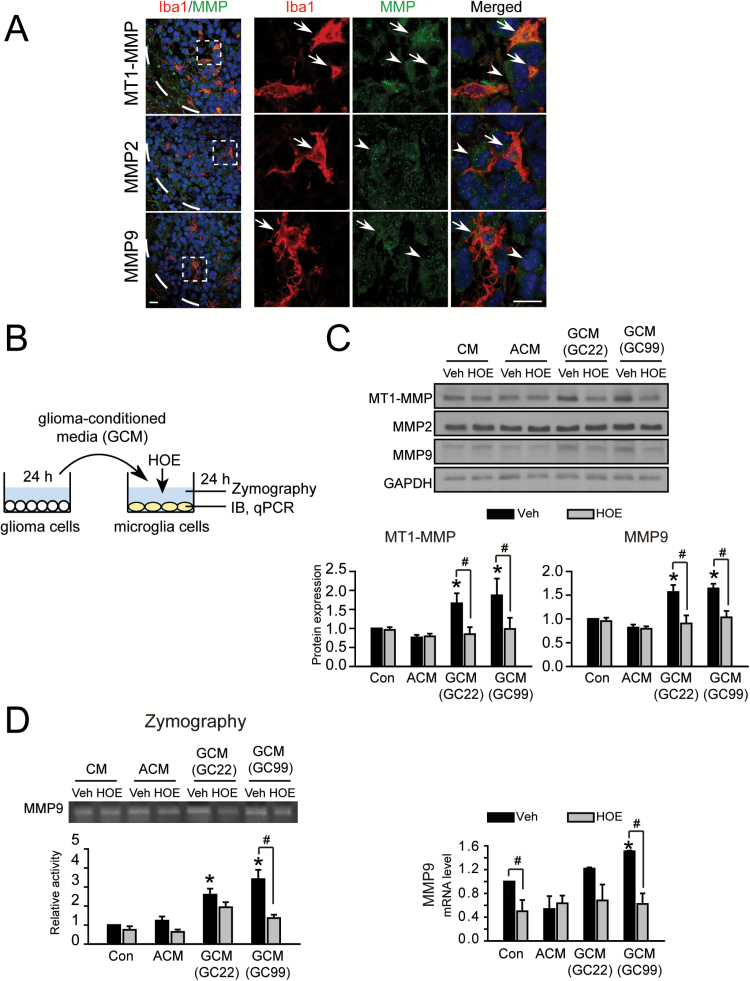
Microglial NHE1 activity promotes expression and activation of matrix metalloproteinases
Matrix metalloproteinases (MMPs) are involved in the expansion of malignant gliomas by facilitating their penetration of anatomical barriers and migration within the neuropil. We further investigated roles of microglial NHE1 activity in modulation of expression and activation of different MMPs. First, we observed that GC22 xenograft tissues exhibited abundant expression of membrane type 1 metalloprotease (MT1-MMP), MMP2 and MMP9 (Figure 4A). Type 1 metalloprotease (MT1-MMP), MMP2 and MMP9 are abundantly expressed in GC22 xenograft tissues. MT1-MMP appears more abundant in Iba1+ cells, which is consistent with previous reports, while MMP2 and MMP9 are present in both Iba1− (arrowheads) and Iba1+ cells (arrows) (13). Next, we applied GCM to microglia which resulted in increased expression of MT1-MMP and MMP9 proteins, but not MMP2 (Figure 4B–E and Supplementary Figure 1C–F, available at Carcinogenesis Online). Furthermore, the enzymatic activity of MMP9 was increased in the stimulated microglial cells by zymography. Interestingly, this increased MT1-MMP and MMP9 expression and MMP9 enzymatic activity were abolished after microglial incubation with HOE642, indicating NHE1 activity is required for these changes.
Figure 4.
Microglial NHE1 activity links to upregulation and activation of microglial MMPs. (A) Expression of different MMPs (MT1-MMP, MMP2 and MMP9) as characterized by immunofluorescences stainings in GC22 xenograft tissues. Representative immunofluorescence images show expression of microglia marker Ibal (red) and different MMPs (green). White boxes: areas were magnified in the right panels. White dash lines: tumor boarders. Arrow: expression of MMPs in Iba1+ cells. Arrowheads: expression of MMPs in Iba1− cells. Scale bar: 10 μm. (B) Protocol. GCM was harvested from glioma cultures (GC22 or GC99) and added to microglia cultures for 24h either in the presence or absence of NHE1 inhibitor HOE642 (1 μM). Microglia were subjected to qPCR assays and immunoblotting. MCM was harvested for gelatin zymography assays. (C) Representative immunoblots showing MMP expression in microglia under different conditions. Data represent mean ± SEM. n = 6, *P < 0.05 versus C/MCM + Veh; # P < 0.05 versus G/MCM + HOE. (D) Representative zymography showing activity of MMP9 in MCM in the presence or absence of HOE642. Data are mean ± SEM. n = 4, *P < 0.05 versus C/MCM + Veh; # P < 0.05 versus G/MCM + HOE. (E) Changes in mRNA expression of MMP9. Data are mean ± SEM. n = 4, *P < 0.05 versus Con.
Microglial NHE1 activity plays a role in microglia-induced glioma proliferation
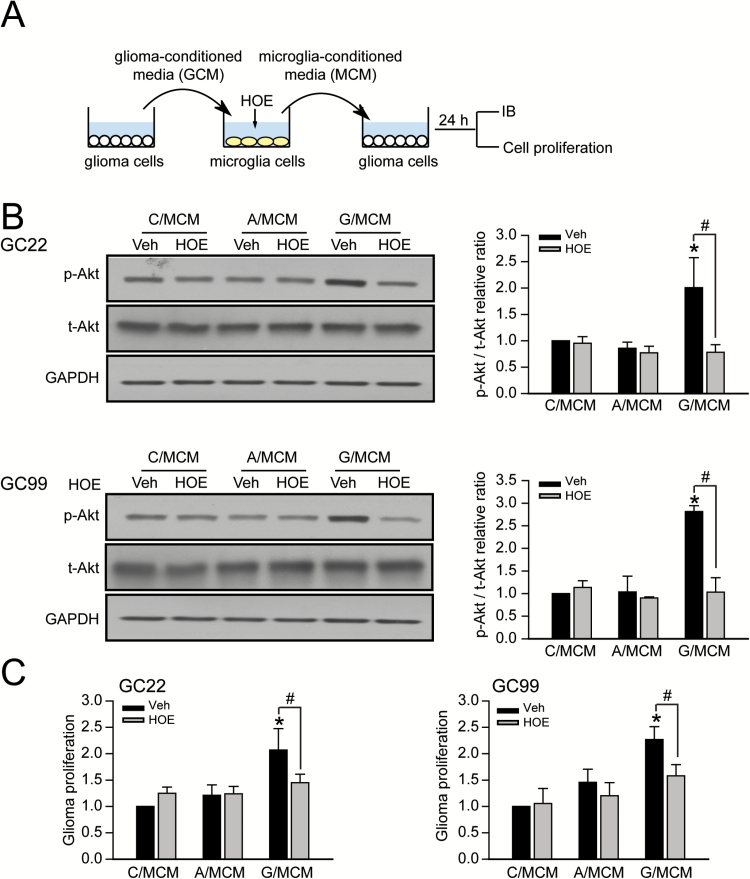
We then investigated whether blocking NHE1 activity in microglia would affect glioma cell function. Microglia were incubated in either GCM, ACM or unconditioned medium for 24h. Conditioned medium was collected from glioma-stimulated microglia (G/MCM), astrocyte-stimulated microglia (A/MCM), or control medium-stimulated microglia (C/MCM) and subsequently applied to glioma cells. NHE1 inhibitor HOE642 was added into microglia cultures of each condition to specifically inhibit microglial NHE1 activity during inoculation of MCM, prior to exposure to glioma cells (Figure 5A). After exposing glioma cells to MCM for 24h, expression of total Akt (t-Akt) and phosphorylated Akt (p-Akt), a marker of proliferation, were evaluated. The levels of total Akt protein remained unchanged in all experimental groups whereas G/MCM significantly increased p-Akt protein expression in both glioma cell lines (2.08±0.27 fold in GC22 and 2.21±0.34 fold in GC99) (Figure 5B). Neither C/MCM nor A/MCM affected p-Akt levels. Notably, inhibition of microglial NHE1 activity with HOE642 significantly reduced glioma cell’s p-Akt protein expression (2.00±0.56 fold change in GC22 and 2.81±0.12 fold change in GC99), showing that microglial-derived factors stimulate Akt-signaling cascades in glioma cells (Figure 5B).
Figure 5.
Microglial NHE1 activation stimulates glioma proliferation. (A) Protocol. GCM were harvested from glioma cultured in a FBS-free medium for 24h (GC#22 or GC#99). Microglia cultures were then exposed to GCM for 24h either in the presence or absence of NHE1 inhibitor HOE642 (1 μM). G/MCM were subsequently harvested and added to glioma cultures for 24h. Regular microglia culture media (C/MCM) and astrocyte-conditioned media (A/MCM) were used as controls. (B) Representative immunoblots showing increased expression of p-Akt in glioma cells in response to G/MCM. Data are mean ± SEM. n = 4, *P < 0.05 versus C/MCM+Veh; # P < 0.05 versus G/MCM + HOE. (C) Histograms summarizing glioma cell viability after 48h of MCM stimulations. Data are mean ± SEM. n = 4, *P < 0.05 versus C/MCM + Veh; # P < 0.05 versus G/MCM + HOE.
Akt may be involved in the regulation of glioma and microglial cytokine release in light of its binding and phosphorylation of NHE1’s c-terminus (25). To explore this, we used the specific Akt inhibitor MK2206 at 0.5 µM, a concentration with no toxicity or inhibition of cell survival (26). Supplemental Figure 3A–C, available at Carcinogenesis Online, shows that treatment of GC99 with MK2206 elevated TGF-β release in GCM by 71% but did not affect IL-6 release. Moreover, MK2206 failed to block GCM-induced TGF-β or IL-6 release from microglia. The opposite effects of blockade of glioma NHE1 and Akt on release of TGF-β from glioma suggest that function of glioma NHE1 in stimulating TGF-β release is not regulated by Akt. However, we did observe that blocking glioma Akt with MK2206 abolished glioma proliferation and also decreased glioma-stimulated microglia-induced glioma proliferation (Supplementary Figure 3D, available at Carcinogenesis Online). This suggests that glioma Akt is involved in glioma–microglia communication, although it is not dependent on TGF-β.
We next conducted a PrestoBlue cell viability assay. There was a modest increase in proliferation of GC22 and GC99 cells (~20%, P > 0.05) after exposure to the G/MCM for 24h as compared with cells exposed to C/MCM (data not shown). Increasing the exposure to 48h stimulated cell viability by 1.8-fold in GC22 and 2.1-fold in GC99 (Figure 5C). In contrast, inhibition of NHE1 activity with HOE642 significantly reduced the G/MCM-mediated glioma viability (Figure 5C). These findings indicate that microglial NHE1 activity in microglia stimulated by GCM is required for the production of soluble factors that in turn enhance glioma viability.
We also examined LPS-mediated stimulation of microglia, a classically M1 type microglial activation, and its subsequent impact on glioma proliferation. Supplementary Figure 4A and B, available at Carcinogenesis Online, shows that exposure of GC99 cultures to basal MCM significantly stimulated glioma proliferation. However, LPS-activated microglia (evidenced by ~4000 pg/ml increased release of IL-6, data not shown) did not trigger additional stimulation of glioma proliferation. Blocking LPS-stimulated microglial NHE1 with HOE642 had no effects on GC99 proliferation. These data suggest that LPS-stimulated microglia do not affect glioma proliferation and the glioma-stimulated microglial phenotype is distinct from classical polarization types. This view is supported by a similar report that LPS-stimulated microglia failed to increase C6 glioma proliferation (27).
Activation of microglial NHE1 enhances glioma migration and invasion
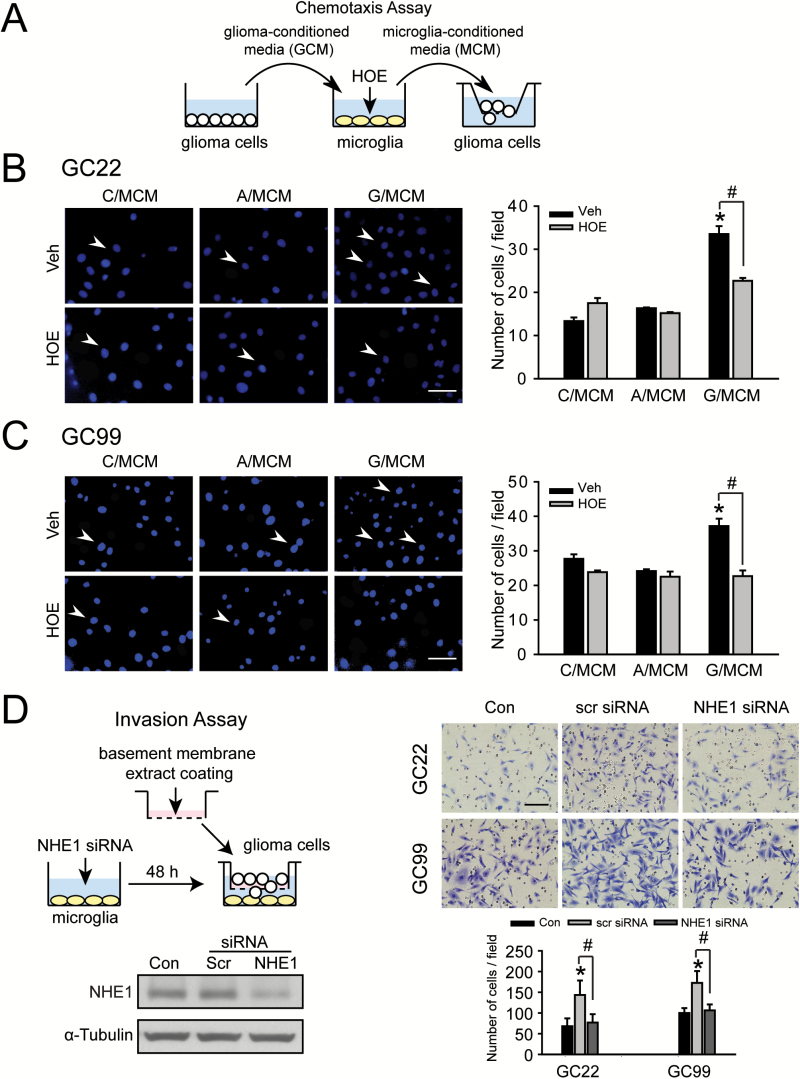
Lastly, we examined the role of microglial NHE1 activity on glioma cell migration and invasion (Figure 6A). Consistent with our previous finding (22), in the transwell chemotaxis assay, GC22 exhibited a lower basal cell migration level (13.3±0.8 cells/field) compared with GC99 (29.1±2.1 cells/field). In response to G/MCM stimulation, migratory ability of GC22 cells increased ~120% from a control of 13.3±0.8 cells/field to 29.3±4.3 cells/field (Figure 6B). Despite the higher basal migratory ability of GC99 cells, G/MCM exposure further stimulates their migration by ~40% (Figure 6C). Blocking NHE1 activity with HOE642 in the microglia cultures abolished their ability to stimulate either GC22 or GC99 migration (Figure 6B and C). To rule out the possibility of any off-target HOE642 effects, endogenous microglial NHE1 was silenced by siRNA, which reduced NHE1 expression by ~70% prior to the microglia–glioma co-culture. Glioma cell invasion through Matrigel (a matrix extract of non-crosslinked ECM macromolecules) was evaluated (Figure 6D). Under control conditions (no cells in the lower compartment), both GC22 and GC99 cells displayed the ability to invade through the Matrigel in 16h, but in the presence of microglia stimulation, the numbers of invading glioma cells (GC22 and GC99) were significantly increased (~110% increase in GC22 and ~70% in GC99) (Figure 6D). Knockdown of microglial NHE1 expression significantly reduced glioma cell invasion by ~40% in GC22 and ~55% in GC99 (P < 0.05, Figure 6D). Taken together, these findings clearly demonstrate that NHE1 activity is required for microglia-stimulated glioma migration and invasion.
Figure 6.
Glioma-mediated activation of microglia subsequently promotes glioma cell migration and invasion. (A) Protocol. GCM was harvested from glioma cultures (GC22 or GC99). Microglia cultures were exposed to GCM for 24h either in the presence or absence of NHE1 inhibitor HOE642 (1 μM). G/MCM was subsequently harvested from the microglia cultures to stimulate glioma migration for 5h. (B, C) Chemotaxis of GC22 or GC99 determined under six different treatment conditions: C/MCM + Veh, C/MCM + HOE642, A/MCM + Veh, A/MCM + HOE642, G/MCM + Veh and G/MCM + HOE642. Left panel: representative images of glioma cells that have migrated through an 8 µm transwell barrier after 5h. Scale bar: 40 μm. Right panel: summary data of migrated cells in the different treatment groups. Data are mean ± SEM. n = 5, *P < 0.05 versus C/MCM + Veh; # P < 0.05 versus G/MCM + Veh. (D) Glioma cells plated on a matrigel-coated Boyden chamber were cultured for 16h in the presence of microglia. Microglia were previously treated with either scr or NHE1 siRNA for 48h prior to the co-culture. Left panel: immunoblots show NHE1 protein was effectively reduced after NHE1 siRNA treatment. Right panel: knockdown expression of microglial NHE1 with siRNA significantly reduced the glioma cell invasion through matrigel-coated Boyden chamber in the co-culture system. Scale bar: 80 μm. Histograms summarize number of invading glioma cells under the different treatment groups. Data are mean ± SEM. n = 5, * P < 0.05.
We also investigated whether glioma NHE1 is required in response to microglia-mediated stimulation of glioma invasion. Invasion of glioma cells plated on Matrigel coated inserts was measured after exposure to either fresh media or G/MCM with or without HOE642 (Supplementary Figure 5A, available at Carcinogenesis Online). G/MCM treatment significantly increased glioma cell invasion by ~160% (P < 0.05, Supplementary Figure 5B, available at Carcinogenesis Online). Treatment with HOE642 reduced glioma cell invasion by ~35%, although this effect did not reach statistical significance. These data suggest that NHE1 in both microglia and glioma cells is involved in the tumor supportive bi-directional communication within the tumor microenvironment.
Discussion
Glioma-associated microglial activation requires NHE1
In malignant glioma, both the resident microglia and infiltrating macrophages derived from circulating monocytes are major participants in glioma progression (5,8). Classically activated microglia/macrophages assume an M1 phenotype capable of antigen presentation and phagocytosis and thus impair tumor progression (28,29). Microglia of the alternatively activated pathway, M2, prevent production of cytokines required to support various T-cells and have been described as tumor supportive (28–30). Although M1 cells are characterized by the expression of the signal transducer and activator of transcription 1 (STAT-1) and the production of iNOS, M2 cells express a different set of marker proteins including CD206, signal transducer and activator of transcription 3 and Arg1 (8,31,32). In our study, we found that microglia upregulate expression of the M1 marker iNOS and the M2 marker Arg1. This is congruent with emerging data from other groups that GAMs cannot clearly be categorized as polarized towards either classical activation profile, but rather they adopt a specific glioma-associated activation phenotype consisting of protein expression characteristic of various microglial activation subclassifications (10,11,14).
Robust NHE1 protein expression was observed in GAMs of the glioma xenografts and human GBM TMAs, and positively correlated with Iba1 levels in GAMs. Our observed upregulations of iNOS and Arg1 in microglia were abolished when NHE1 activity was attenuated either by inhibition with HOE642 or by siRNA knockdown, together indicating an important role of NHE1 in glioma-mediated microglia activation. This is consistent with our previous findings of NHE1 activation in stimulated microglia. We recently reported that NHE1 activity is a vital H+ extrusion mechanism during NADPH oxidase-mediated ‘residuary burst’ when microglia are challenged with oxygen and glucose deprivation as well as LPS stimulation (17,33). The activity and expression of NHE1 is also extensively involved in microglia activation and subsequent neuroinflammation after cerebral ischemia (33). This denotes an important role for NHE1-mediated microglial activation in a wide variety of neurological insults.
Microglial NHE1 is involved in microglia-stimulated glioma growth
Release of the soluble factors from microglia promotes glioma progression (34,35). Among them, microglia-derived TGF-β promotes upregulation of its cognate receptors TβRI and TβRII on glioma cells and activates signaling cascades that lead to tumor growth and invasion (8,36,37). Our study also detected increased transcriptional expression and release of TGF-β1 by glioma-stimulated microglia, which was inhibited by HOE642. Furthermore, misfolding or mislocalization of NHE1 protein can lead to reduction of active TGF-β release from fibroblasts (38). This suggests that microglial NHE1 may promote glioma cell proliferation through the production and release of TGF-β. IL-6 is another microglia-derived factor correlated with glioma growth and invasiveness (8). GBMs display a significantly higher level of IL-6 expression than normal brain tissue (39). Targeting the IL-6 pathway in glioma stem cells reduces their growth and survival (40). It has been hypothesized that activated microglia synthesize IL-6 following NF-κB activation, which in turn may stimulate transcription factors signal transducer and activator of transcription 3 and NF-κB in glioma cells that initiate pathways of glioma progression such as angiogenesis, migration and apoptosis inhibition (39,41).
In the current study, we detected the release of IL-6 and TGF-β from microglia in response to glioma-mediated stimulation. However, only TGF-β release depends on glioma NHE1 activity. Considering the respective roles of IL-6 and TGF-β in microglial classically pro-inflammatory and alternatively activated polarization, our data suggest that glioma NHE1 activity may promote M2 type protumorigenic polarization in microglia via increasing release of TGF-β. The current study is limited and more extensive cytokine profile analysis of glioma and microglia secretomes in relation to their NHE1 function should be conducted in the future studies. In addition, the possible soluble factors secreted by glioma that regulate microglial NHE1 expression and/or its activation remain unknown. There are two possible candidates, glioblastoma-derived macrophage colony stimulating factor and monocyte chemoattractant protein-1 (MCP1). Macrophage colony stimulating factor plays an important role in regulating the formation and release of microglial secretome substrates (42). Activation of CSF1-R by Macrophage colony stimulating factor promotes microglia survival and proliferation by stimulating ERK1/2 phosphorylation (43), whereas MCP1-CCR2 activation leads to phosphorylation of Ezrin/Radixin/Moesin (ERM) proteins during microglia migration (44). The speculation of these two factors are supported by well-defined binding sites for ERK and ERM on the c-terminus of NHE1 (25), as well our previous findings on their associations to NHE1 activation in neurons and microglia (18,45).
Microglial NHE1 function promotes glioma cell migration and invasion
Degradation of extracellular matrix by membrane-bound and secreted metalloproteases facilitates glioma invasion (46). In particular, efficient digestion of extracellular matrix proteins by membrane-bound metalloproteases like MT1-MMP are pivotal for tumor invasion. They also activate secreted metalloproteases like MMP-2 and MMP-9, which are major proteases involved in glioma invasion in mouse models and probably also in humans (47). MT1-MMP in mouse and human gliomas is expressed predominantly in microglia closely associated with the tumors (13).
We hypothesized that NHE1 is involved in GAM MT1-MMP expression. We detected an upregulation of microglial MT1-MMP after GCM treatment or in microglia-glioma co-culture (Figure 4A and Supplementary Figure 1F, available at Carcinogenesis Online), which was abolished when NHE1 activity was inhibited by HOE642. In breast cancer cells, NHE1 activity increases expression of MT1-MMP via ERK1/2 and p38 mitogen-activated protein kinase (MAPK) signaling pathways (48). We previously reported that inhibition of NHE1 activity with HOE642 reduces expression of p-ERK in glioma cells (16). We therefore speculate that NHE1 inhibition in microglia promotes glioma migration and invasion by downregulation of MT1-MMP expression which may also be regulated by ERK1/2 related signaling. NHE1/p38 MAPK signaling to control MT1-MMP expression is also a possibility. p38 MAPK pathway activation has been shown to be NHE1-independent under certain conditions and will thus require further investigation (13,49,50).
We and others have observed increased expression of MT1-MMP and MMP9 in microglia in response to glioma-derived factors (13,51). However, it has also been reported that exposure of microglia to glioma-derived factors increased microglia MMP2 activity, which we did not observe (30). Moreover, we found that NHE1 inhibition decreased MT1-MMP and MMP9 activity, whereas MMP2 remained unchanged in GC22 and GC99 cells. This contrasts with results obtained in breast tumor cells where NHE1 inhibition decreased expression of MT1-MMP, MMP9 and MMP2 (52). This discrepancy probably results from the use of different tumor types and/or different glioma cell lines (GL261 used by (13) versus GC22 and GC99 used here). These findings suggest that although the variation in molecular mechanisms underlying tumor-associated matrix degradation in different cell types may pose a clinical treatment challenges, it also offers the opportunity for more targeted and personalized therapies for specific molecular tumor subtypes.
Regulation of signaling cascades by the NHE1 protein scaffolding function
In addition to being an ion transporter, NHE1 also functions as a cytoskeletal scaffold. NHE1 binds to the cytoskeletal protein ezrin and is localized to lamellopodia, and regulates cell motility (16). Furthermore, its intracellular C-terminal domain also serves as a scaffold to recruit and position components of various signaling cascades (53). Notably, binding of heat-shock protein 70 to NHE1 in microglia mediates the production of iNOS in response to LPS treatment via the NFΚB signaling pathway (54). In addition, a recent study revealed that the NHE1 C-terminal contains two D-domains, which directly interact with ERK. Mutating these D-domains abolished epidermal growth factor-dependent ERK activation, which suggests NHE1–ERK interaction may be required for full ERK activity (55).
The protein scaffolding functions of NHE1 are at least partially independent of its ion transport function as mutated forms without functional ionic translocation still properly localize to lamellopodia membrane (56). However, regulation of local intracellular pH by NHE1 may still be important for downstream signaling as intracellular pH changes can alter binding affinities of signaling molecules (53). Therefore, the effects of pharmacological NHE1 inhibition observed in our study could also be mediated through the NFKB pathway or other signaling mechanisms. Additional studies are needed to investigate proteins that interact with NHE1 protein, and how they regulate microglial activation and microglia–glioma interactions in the tumor environment.
Conclusion
In summary (Supplementary Figure 6, available at Carcinogenesis Online), we report that NHE1 protein is highly expressed in glioma-associated microglia and macrophages. NHE1 activity is involved in inducing microglia to adopt a glioma-associated activation phenotype. Moreover, microglial NHE1 activity modulates glioma cell expansion and viability via regulation of microglia-derived factors such as MT1-MMP, MMP-9, TGF-β and IL-6. NHE1 inhibition suppresses glioma cell migration and proliferation. Our data suggest NHE1 promotes tumorigenesis, and therefore supports NHE1 inhibition as a novel strategy to attenuate microglia-mediated glioma progression.
Supplementary material
Supplementary Figures 1–7 and Table 1 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (partial support, R01NS075995 and R01NS048216 to D.S. and J.S.K.); HEADRUSH Brain Tumor Research Professorship, Roger Loff Memorial GBM Research Fund, and Department of Neurological Surgery (partial support, R01CA158800 to J.S.K.).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- ACM
astrocyte-conditioned medium
- CSF
colony stimulating factor
- DMEM
Dulbecco’s Modified Eagle Medium
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
- GAM
glioma-associated microglia/macrophage
- GBM
glioblastoma multiforme
- GC
glioma cell
- GCM
glioma-conditioned medium
- GSC
glioma stem cell
- IL
interleukin
- LPS
lipopolysaccharide
- MCM
microglia-conditioned media
- MMP
matrix metalloproteinase
- NHE1
Na+/H+ exchanger isoform 1
- RT
room temperature
- siRNA
small interfering RNA
- TMA
tissue microarray
References
- 1. Stupp R., et al. (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med., 352, 987–996. [DOI] [PubMed] [Google Scholar]
- 2. Zheng H., et al. (2008) p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature, 455, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Purow B., et al. (2009) Advances in the genetics of glioblastoma: are we reaching critical mass? Nat. Rev. Neurol., 5, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang J., et al. (2012) Temozolomide: mechanisms of action, repair and resistance. Curr. Mol. Pharmacol., 5, 102–114. [DOI] [PubMed] [Google Scholar]
- 5. Charles N.A., et al. (2011) The brain tumor microenvironment. Glia, 59, 1169–1180. [DOI] [PubMed] [Google Scholar]
- 6. Kostianovsky A.M., et al. (2008) Astrocytic regulation of human monocytic/microglial activation. J. Immunol., 181, 5425–5432. [DOI] [PubMed] [Google Scholar]
- 7. Glass R., et al. (2014) CNS macrophages and peripheral myeloid cells in brain tumours. Acta Neuropathol, 128, 347–362. [DOI] [PubMed] [Google Scholar]
- 8. Li W., et al. (2012) The molecular profile of microglia under the influence of glioma. Neuro. Oncol., 14, 958–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wyckoff J., et al. (2004) A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res., 64, 7022–7029. [DOI] [PubMed] [Google Scholar]
- 10. Szulzewsky F., et al. (2015) Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One, 10, e0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergamin L.S., et al. (2015) Involvement of purinergic system in the release of cytokines by macrophages exposed to glioma-conditioned medium. J. Cell. Biochem., 116, 721–729. [DOI] [PubMed] [Google Scholar]
- 12. Wu A., et al. (2010) Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro. Oncol., 12, 1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Markovic D.S., et al. (2009) Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion. Proc. Natl. Acad. Sci. U. S. A., 106, 12530–12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vinnakota K., et al. (2013) Toll-like receptor 2 mediates microglia/brain macrophage MT1-MMP expression and glioma expansion. Neuro. Oncol., 15, 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boedtkjer E., et al. (2012) Physiology, pharmacology and pathophysiology of the pH regulatory transport proteins NHE1 and NBCn1: similarities, differences, and implications for cancer therapy. Curr. Pharm. Des., 18, 1345–1371. [DOI] [PubMed] [Google Scholar]
- 16. Cong D., et al. (2014) Upregulation of NHE1 protein expression enables glioblastoma cells to escape TMZ-mediated toxicity via increased H⁺ extrusion, cell migration and survival. Carcinogenesis, 35, 2014–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y., et al. (2010) Activation of microglia depends on Na+/H+ exchange-mediated H+ homeostasis. J. Neurosci., 30, 15210–15220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi Y., et al. (2013) Stimulation of Na(+)/H(+) exchanger isoform 1 promotes microglial migration. PLoS One, 8, e74201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clark P.A., et al. (2012) Activation of multiple ERBB family receptors mediates glioblastoma cancer stem-like cell resistance to EGFR-targeted inhibition. Neoplasia, 14, 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zorniak M., et al. (2012) Differential expression of 2′,3′-cyclic-nucleotide 3′-phosphodiesterase and neural lineage markers correlate with glioblastoma xenograft infiltration and patient survival. Clin. Cancer Res., 18, 3628–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alrfaei B.M., et al. (2013) microRNA-100 targets SMRT/NCOR2, reduces proliferation, and improves survival in glioblastoma animal models. PLoS One, 8, e80865.24244722 [Google Scholar]
- 22. Zhu W., et al. (2014) WNK1-OSR1 kinase-mediated phospho-activation of Na+-K+-2Cl- cotransporter facilitates glioma migration. Mol. Cancer, 13, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Albini A., et al. (1987) A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res., 47, 3239–3245. [PubMed] [Google Scholar]
- 24. Wessa P. (2014) Pearson Correlation (v1.0.9) in Free Statistics Software (v1.1.23-r7). Office of Research Development and Education; http://www.wessa.net/rwasp_correlation.wasp/ (November 2014, date last accessed). [Google Scholar]
- 25.Amith, S.R. et al. (2013) Regulation of the Na+/H+ exchanger (NHE1) in breast cancer metastasis. Cancer Res., 73, 1259–1264. [DOI] [PubMed] [Google Scholar]
- 26.Cheng, Y. et al. (2012) MK-2206, a novel allosteric inhibitor of Akt, synergizes with gefitinib against malignant glioma via modulating both autophagy and apoptosis. Mol. Cancer Ther., 11, 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sliwa M., et al. (2007) The invasion promoting effect of microglia on glioblastoma cells is inhibited by cyclosporin A. Brain, 130, 476–489. [DOI] [PubMed] [Google Scholar]
- 28. Martinez F.O., et al. (2008) Macrophage activation and polarization. Front. Biosci., 13, 453–461. [DOI] [PubMed] [Google Scholar]
- 29. Wei J., et al. (2013) The controversial role of microglia in malignant gliomas. Clin. Dev. Immunol., 2013, 285246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Markovic D.S., et al. (2005) Microglia stimulate the invasiveness of glioma cells by increasing the activity of metalloprotease-2. J. Neuropathol. Exp. Neurol., 64, 754–762. [DOI] [PubMed] [Google Scholar]
- 31. Wang N., et al. (2014) Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol., 5, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sica A., et al. (2012) Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest., 122, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi Y., et al. (2011) Role of sodium/hydrogen exchanger isoform 1 in microglial activation and proinflammatory responses in ischemic brains. J. Neurochem., 119, 124–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wurdinger T., et al. (2014) Mechanisms of intimate and long-distance cross-talk between glioma and myeloid cells: how to break a vicious cycle. Biochim. Biophys. Acta, 1846, 560–575. [DOI] [PubMed] [Google Scholar]
- 35. Coniglio S.J., et al. (2013) Review: molecular mechanism of microglia stimulated glioblastoma invasion. Matrix Biol., 32, 372–380. [DOI] [PubMed] [Google Scholar]
- 36. Kjellman C., et al. (2000) Expression of TGF-beta isoforms, TGF-beta receptors, and SMAD molecules at different stages of human glioma. Int. J. Cancer, 89, 251–258. [DOI] [PubMed] [Google Scholar]
- 37. Hulper P., et al. (2011) Tumor localization of an anti-TGF-beta antibody and its effects on gliomas. Int. J. Oncol., 38, 51–59. [PubMed] [Google Scholar]
- 38. Karydis A., et al. (2009) Mislocalized scaffolding by the Na-H exchanger NHE1 dominantly inhibits fibronectin production and TGF-beta activation. Mol. Biol. Cell, 20, 2327–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi C., et al. (2002) Fas engagement increases expression of interleukin-6 in human glioma cells. J. Neurooncol., 56, 13–19. [DOI] [PubMed] [Google Scholar]
- 40. Wang H., et al. (2009) Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cells, 27, 2393–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karin M. (2009) NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol., 1, a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nijaguna M.B., et al. (2015) Glioblastoma-derived macrophage colony-stimulating factor (MCSF) induces microglial release of insulin-like growth factor-binding protein 1 (IGFBP1) to promote angiogenesis. J. Biol. Chem., 290, 23401–23415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stanley E.R., et al. (2014) CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol., 6, a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yao Y., et al. (2014) Monocyte chemoattractant protein-1 and the blood-brain barrier. Cell. Mol. Life Sci., 71, 683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manhas N., et al. (2010) p90 activation contributes to cerebral ischemic damage via phosphorylation of Na+/H+ exchanger isoform 1. J. Neurochem., 114, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valastyan S., et al. (2011) Tumor metastasis: molecular insights and evolving paradigms. Cell, 147, 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cuddapah V.A., et al. (2014) A neurocentric perspective on glioma invasion. Nat. Rev. Neurosci., 15, 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin Y., et al. (2011) NHE1 mediates MDA-MB-231 cells invasion through the regulation of MT1-MMP. Exp. Cell Res., 317, 2031–2040. [DOI] [PubMed] [Google Scholar]
- 49. Pedersen S.F., et al. (2007) The Na+/H+ exchanger, NHE1, differentially regulates mitogen-activated protein kinase subfamilies after osmotic shrinkage in Ehrlich Lettre Ascites cells. Cell. Physiol. Biochem., 20, 735–750. [DOI] [PubMed] [Google Scholar]
- 50. Gillis D., et al. (2001) Osmotic stimulation of the Na+/H+ exchanger NHE1: relationship to the activation of three MAPK pathways. J. Membr. Biol., 181, 205–214. [DOI] [PubMed] [Google Scholar]
- 51. Hu F., et al. (2014) Glioma-associated microglial MMP9 expression is upregulated by TLR2 signaling and sensitive to minocycline. Int. J. Cancer, 135, 2569–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J., et al. (2015) CIAPIN1 targets Na⁺/H⁺ exchanger 1 to mediate MDA-MB-231 cells’ metastasis through regulation of MMPs via ERK1/2 signaling pathway. Exp. Cell Res., 333, 60–72. [DOI] [PubMed] [Google Scholar]
- 53. Baumgartner M., et al. (2004) Na(+)/H(+) exchanger NHE1 as plasma membrane scaffold in the assembly of signaling complexes. Am. J. Physiol. Cell Physiol., 287, C844–C850. [DOI] [PubMed] [Google Scholar]
- 54. Huang C., et al. (2015) Inhibition of endogenous heat shock protein 70 attenuates inducible nitric oxide synthase induction via disruption of heat shock protein 70/Na(+) /H(+) exchanger 1-Ca(2+) -calcium-calmodulin-dependent protein kinase II/transforming growth factor β-activated kinase 1-nuclear factor-κB signals in BV-2 microglia. J. Neurosci. Res., 93, 1192–1202. [DOI] [PubMed] [Google Scholar]
- 55. Johansen J., et al. (2013) Direct interaction with the Na+/H+ exchanger NHE1 regulates ERK1/2 activity. FASEB J., 27, 730.1. [Google Scholar]
- 56. Denker S.P., et al. (2000) Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H(+) translocation. Mol. Cell, 6, 1425–1436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.