Abstract
Some organisms can survive exposure to extreme desiccation by entering a state of suspended animation known as anhydrobiosis. The free-living nematode Aphelenchus avenae can be induced to enter the anhydrobiotic state by exposure to a moderate reduction in relative humidity. During this preconditioning period, the nematode accumulates large amounts of the disaccharide trehalose, which is thought to be necessary, but not sufficient, for successful anhydrobiosis. To identify other adaptations that are required for anhydrobiosis, we developed a novel SL1-based mRNA differential display technique to clone genes that are upregulated by dehydration in A. avenae. Three such genes, Aav-lea-1, Aav-ahn-1, and Aav-glx-1, encode, respectively, a late embryogenesis abundant (LEA) group 3 protein, a novel protein that we named anhydrin, and the antioxidant enzyme glutaredoxin. Strikingly, the predicted LEA and anhydrin proteins are highly hydrophilic and lack significant secondary structure in the hydrated state. The dehydration-induced upregulation of Aav-lea-1 and Aav-ahn-1 was confirmed by Northern hybridization and quantitative PCR experiments. Both genes were also upregulated by an osmotic upshift, but not by cold, heat, or oxidative stress. Experiments to investigate the relationship between mRNA levels and protein expression for these genes are in progress. LEA proteins occur commonly in plants, accumulating during seed maturation and desiccation stress; the presence of a gene encoding an LEA protein in an anhydrobiotic nematode suggests that some mechanisms of coping with water loss are conserved between plants and animals.
Although water is essential for life, certain organisms can survive exposure to extreme desiccation by entering into a state of suspended animation known as anhydrobiosis (36). Anhydrobiotic organisms can survive in the dry state for indefinite periods of time, after which, upon rehydration, they are able to resume normal metabolic activity (5, 14, 16). These organisms are found across all biological kingdoms, with baker's yeast, Saccharomyces cerevisiae, being a familiar example. More complex animals, for instance, rotifers, tardigrades, and nematodes such as Aphelenchus avenae, have also been shown to be anhydrobiotic, as have the cysts of the crustacean Artemia salina (14). Certain plant species are also known to be anhydrobiotic, including the angiosperm Craterostigma plantagineum, an example of a “resurrection” plant (35), the bryophyte Tortula ruralis (3), and orthodox plant seeds and pollen (50). One of the best-characterized metabolic changes which occurs during the induction of anhydrobiosis is the accumulation of high concentrations of disaccharides: sucrose is the predominant sugar accumulated in plants, while animals and yeast accumulate trehalose. Trehalose accumulation is believed to protect membranes and proteins from desiccation damage by replacing structural water (12, 19). This sugar may also contribute to the formation of an intracellular glass (17), which might stabilize the cell contents by inhibiting membrane fusion, protein denaturation, and free radical mobility. Recent reports indicate that bdelloid rotifers do not synthesize trehalose or other disaccharides during the induction of anhydrobiosis (42, 56). This finding suggests that the accumulation of trehalose is not necessary for some animals and that there are alternative biochemical and genetic strategies for cellular protection during anhydrobiosis.
The free-living mycophagous nematode A. avenae can be induced to enter the anhydrobiotic state by pre-exposure to moderate reductions in relative humidity prior to extreme desiccation (18). Previous studies have focused on the biochemical changes associated with anhydrobiosis in A. avenae, making it one of the best-characterized anhydrobiotic nematodes. While the accumulation of trehalose in nematodes such as A. avenae is believed to be necessary for anhydrobiotic survival, it is not sufficient. A further period of preconditioning following maximum trehalose accumulation is needed before maximum survival is seen, suggesting that other changes must also occur in A. avenae before it can successfully enter the anhydrobiotic state (33). By analogy with anhydrobiotic plants (35), these adaptations may include changes in primary metabolism, alterations to cell membranes, osmotic adjustments via the accumulation of compatible solutes or hydrophilic proteins, and the synthesis of stress-related proteins such as antioxidants and heat shock proteins, but such adaptations have not been defined for A. avenae or any other anhydrobiotic animal. We have initiated a research program aimed at gaining a deeper understanding of the molecular biology of anhydrobiosis in nematodes. This study presents our investigations into anhydrobiosis in A. avenae and details some of the changes, both biochemical and genetic, that are associated with water loss in this nematode.
MATERIALS AND METHODS
Maintenance and harvesting of A. avenae.
A. avenae was grown at 20°C in the dark on Rhizoctonia solani grown on a substrate of autoclaved wheat. After a growth period of 2 to 3 weeks (before the onset of swarming behavior), nematodes were rinsed off of the substrate with autoclaved tap water. Nematodes were cleaned by flotation on a 30% (wt/vol) sucrose solution followed by three washes in autoclaved tap water and were collected by vacuum filtration onto Supor-450 filters (0.45-μm pore size; Gelman Science) by use of a Sartorius funnel.
Carbohydrate extraction and analysis.
Nematode samples (40,000 nematodes per filter) were exposed to 98% relative humidity (RH) for various times prior to exposure to active dried silica (∼10% RH) for 48 h. The samples were stored frozen at −80°C until they were used for carbohydrate extraction. A saturated solution of potassium dichromate (K2Cr2O7) was used to obtain 98% RH (55, 59), and an active dried silica gel was used to desiccate the samples.
Matched samples exposed to the same dehydration regimen as that used for carbohydrate analysis were prepared and used to determine anhydrobiotic survival. These nematodes were then rehydrated with distilled water (18) and immersed in autoclaved tap water for 20 h. The percentage of survival was assessed by a microscopic observation of motility. Carbohydrate extraction and analysis were performed according to the method of O'Leary et al. (49). In brief, the samples (nematodes and filter) were sonicated in the presence of sand and ethanol, and after filtration (0.22-μm-pore-size Spin-X centrifuge filters; Costar) the filtrate was dried and resuspended in deionized water. The dry weight was determined by weighing duplicate samples of nematodes that had been incubated over a silica gel in a small airtight plastic box and placed at 70°C for 3 days. Carbohydrates were analyzed by high-pressure liquid chromatography on a Spectra Physics HPLC instrument (model SP 8800) attached to a Shodex Rl SE-61 refractive index detector. Samples were run through a carbohydrate H+ column, with 2.5 mM H2SO4 used as the mobile phase. Carbohydrates were identified and quantified by running serial dilutions of known carbohydrate standards through the column.
RNA isolation and first-strand cDNA synthesis.
A. avenae samples (10,000 nematodes per filter) were prepared as described above and exposed to 90% RH for 24 h over a saturated solution of magnesium sulfate (MgSO4 · 7H2O) prior to storage at −80°C. Untreated control nematodes were also prepared. RNAs were isolated by the use of TRIzol reagent (Gibco BRL, Grand Island, N.Y.) according to the manufacturer's instructions. The RNA was resuspended in 20 μl of diethyl pyrocarbonate-H2O. First-strand synthesis was performed by using 1 μl (200 U) of Superscript II reverse transcriptase (Gibco BRL), 10 μl of each RNA sample, and an anchored oligo(dT) primer (dT12MN) according to the manufacturer's instructions. After heat inactivation of the reverse transcriptase, the samples were treated with RNase H (Gibco BRL) and stored frozen at −70°C for subsequent PCRs.
PCR amplification and analysis.
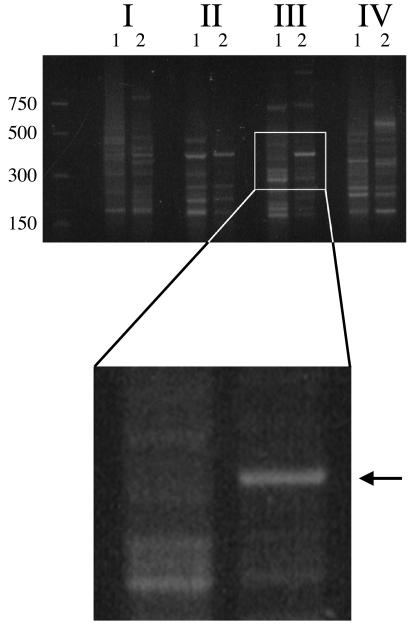
PCR amplification was performed with 0.5 μl of first-strand cDNA template in a 15-μl PCR mixture consisting of 1.5 μl of 10× buffer, 0.3 μl of deoxynucleoside triphosphate mix (10 mM [each]), and 0.125 μl of KlenTaq LA polymerase (1 U) (Sigma). The reaction was primed with the same oligo(dT12MN) primer used during first-strand synthesis and with a nematode 22-nucleotide (nt) SL1 sequence (GGTTTAATTACCCAAGTTGAG) with additional selective nucleotide extensions at its 3′ end (see Fig. 2). An initial denaturation step at 95°C for 5 min was followed by 30 cycles of 50°C for 1 min, 72°C for 3 min, and 94°C for 1 min and then a final extension of 10 min at 72°C and a 4°C hold. After PCR amplification, the products were mixed with 5 μl of 6× loading dye (Sigma) and analyzed in a 3% high-resolution agarose gel (NuSieve GTG agarose; FMC). The gel was run for 6 h with Tris-acetate-EDTA buffer at 4°C in order to maximize resolution. The gel was stained with ethidium bromide and visualized under UV light. Bands that appeared to be upregulated in the treated samples but not in the controls were excised directly from the gel.
FIG. 2.
Agarose gel analysis of PCR products obtained from control and desiccated A. avenae nematodes by SL1-based mRNA differential display technique. The top panel shows the amplification products obtained by using four different primer combinations, as follows: I, SL1+GC and oligo(dT12MN); II, SL1+GCG and oligo(dT12MN); III, SL1+GTA and oligo(dT12MN); and IV, SL1+T and oligo(dT12MN) (where M is A, G, or C and N is A, G, C, or T). Lanes 1, untreated control; lanes 2, A. avenae exposed to 90% RH for 24 h. The bottom panel shows a detail of one of the cDNA fragments that was up-regulated in the desiccated samples, with an arrow indicating the up-regulated band.
Cloning and reverse Northern blot analysis.
PCR-amplified bands of interest were cloned into the pCR2.1-TOPO plasmid by use of a TA cloning kit (Invitrogen) according to a low-melting-temperature-agarose method. Briefly, a slice of low-melting-temperature agarose containing the band of interest was heated at 65°C until the slice melted and then was maintained at 37°C. Four microliters of melted agarose containing the PCR product of interest was cloned directly according to the supplier's instructions. Twelve recombinant clones were selected for each cloned band, and these were PCR amplified by using the M13 forward and reverse primer pair. The PCR products were blotted and fixed in duplicate onto a positively charged nylon membrane (Roche) by using a slot blot apparatus (Schleicher and Schuell) according to the manufacturer's instructions. The membranes were probed with RNA from either untreated or desiccated nematodes that had been labeled with an AlkPhos direct kit (Amersham Pharmacia Biotech). Clones that were confirmed as being upregulated in response to desiccation stress were selected for further analysis.
Northern blot analysis.
Nematode samples were treated with a variety of stresses for 24 h. These stresses were exposure to 90% RH (desiccation), 30°C (heat), 4°C (cold), 100 mM paraquat (oxidative), and 500 mM sucrose (osmotic). All treatments were performed with sterile tap water, except for desiccation stress, for which vacuum-filtered nematodes were placed on 1.5-cm-long Millipore filter papers (40,000 nematodes per filter) and desiccated over a saturated solution of magnesium sulfate (MgSO4 · 7H2O). A small aliquot of each nematode sample was used to determine survival; nematodes were placed in sterile tap water at 20°C in the dark for 24 h prior to the assessment of survival by a microscopic observation of motility. Total RNAs (40 μg) were extracted from treated and untreated A. avenae nematodes as described above. Fresh untreated nematodes were used as controls in all experiments. Ten micrograms of RNA was set aside for quantitative PCR analysis (see below), and the remaining RNA (30 μg) was electrophoresed, blotted, and hybridized according to the instructions of an AlkPhos direct kit (Amersham Pharmacia Biotech). DNA probes were generated with cleaned PCR products (QIAprep Spin Miniprep kit; Qiagen). Labeling and detection of the probes were performed by using an AlkPhos direct kit (Amersham Pharmacia Biotech).
Genomic DNA analysis.
Genomic DNA was isolated from freshly harvested nematodes (0.5 g) by standard techniques, and PCR primers were designed to amplify the genomic copy of each gene; similarly, fresh RNA was isolated as described previously and used to generate a cDNA copy of each gene. PCRs with the genomic copies of the genes were performed by using Red Hot polymerase (ABGene, Epsom, United Kingdom) as specified by the manufacturer (94°C for 30 s, 57°C for 30 s, and 72°C for 45 s for 30 cycles). Reverse transcription-PCRs (RT-PCR) were performed by using the Access RT-PCR system (Promega, Southampton, United Kingdom) according to the manufacturer's protocols, with primer pairs designed to amplify full-length genes (Table 1). Cloning and sequencing were performed as described above by using a TA cloning kit (Invitrogen).
TABLE 1.
Primers used for this study
| Primer no.a | Primer name | DNA sequence (5′-3′) |
|---|---|---|
| 1 | Aav-lea-1 forward | CACTACCGCTTACAACCAATC |
| 2 | Aav-lea-1 reverse | ACAGGAATATCACTGACAGAT |
| 3 | Aav-ahn-1 forward | CAAATCAACAACAATGCCACC |
| 4 | Aav-ahn-1 reverse | AGAGGATCATTGCACGGAATT |
| 5 | Aav-glx-1 forward | GCGAACATGGGAAAAGTCAAC |
| 6 | Aav-glx-1 forward | CAGAGATGCCTTTGGACATTA |
| 7 | Ama-F degenerate | GARTTYTTYTTYCAYGCNATG |
| 8 | Ama-R degenerate | NGTCATYTGNGTNGCNGGYTC |
| 9 | Aavlea1 Q-PCR F | GATGGAGGAGTACAAGCAGCA |
| 10 | Aavlea1 Q-PCR R | TCATGAAGGTGGAACAAGGTC |
| 11 | Aavahn1 Q-PCR F | GGACAGTACGAGCCGAAAGTA |
| 12 | Aavahn1 Q-PCR R | CACTTTCGACGTGATGAA |
| 13 | Aavglx1 Q-PCR F | GATCCAGGACTATCTCGCTCA |
| 14 | Aavglx1 Q-PCR R | AATTCGGTGAGCTTCTTCTCC |
| 15 | Aavama1 Q-PCR F | GTGTAGAGCCGCCTTAGCTG |
| 16 | Aavama1 Q-PCR R | ATGTGGGTGGAGGATCAGAC |
Primers 1 to 6, primer pairs used to amplify the full-length genomic and cDNA copies of genes induced during desiccation; primers 7 and 8, degenerate primer pair used to amplify the Aav-ama-1 gene; primers 9 to 16, primer pairs used to perform real-time quantitative PCR analyses.
Digested DNAs (5 μg) were separated in a 0.75% agarose gel and transferred by capillary action in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) onto a Hybond N+ membrane (Amersham Pharmacia Biotech) overnight (54). Once the transfer was complete, the membrane was dried in an oven for 30 min at 60°C, and the DNAs were fixed to the membrane by UV cross-linking in a Stratalinker (Statagene). DNA probes were generated with cleaned PCR products, and labeling and detection of the probes were performed by using an AlkPhos direct kit (Amersham Pharmacia Biotech).
Bioinformatic analyses.
BLAST searches of the Swiss-Prot TrEMBL database, Wormbase (http://www.wormbase.org, release WS115), and the Drosophila melanogaster database (SIB BLAST Network Service) were performed to identify a set of genes related to Aav-lea-1. Repeat motifs in these genes were identified visually and with the aid of the Dotlet program (http://www.isrec.isb-sib.ch/java/dotlet/Dotlet.html). Repeat motifs were aligned with the T-Coffee program (48). Incomplete motifs or motifs which introduced gaps were removed from the alignments. The physical and chemical parameters of the selected proteins were calculated with the ProtParam tool (2; http://ca.expasy.org/tools/protparam.html). Secondary structure analysis was performed with the PELE program of the SDSC Biology Workbench (http://workbench.sdsc.edu/) and with the PONDR (Predictors of Natural Disordered Regions) algorithm.
Quantitative real-time RT-PCR.
A real-time PCR analysis of the Aav-lea-1, Aav-ahn-1, and Aav-glx-1 genes after the exposure of nematodes to different stresses was performed by using the SYBR Green I detection system (Qiagen) and a LightCycler PCR machine (Roche Molecular Biochemicals, Mannheim, Germany). The copy number of each gene was calculated by using a gene-specific standard curve, and the RNA polymerase II gene (Aav-ama-1) was used as an external control.
The ama-1 gene was cloned by using a redundant primer pair based on the alignment of several nematode ama-1 protein sequences. A first-strand cDNA was prepared and PCR amplified under standard conditions with the degenerate primer pair Ama-F and Ama-R. The PCR product was analyzed in a low-melting-point agarose gel, and a band of the correct estimated size (approximately 1 kb) was excised from the gel, cloned, and sequenced. BLAST searches of the National Center for Biotechnology Information nucleotide database confirmed that the gene, Aav-ama-1 (AY357257), encoded the large subunit of RNA polymerase II.
Gene-specific real-time PCR standards were generated as follows. PCR primers were designed to amplify a small region (approximately 125 bp) of the following control and target genes: Aav-lea-1, Aav-ahn-1, Aav-glx-1, and Aav-ama-1 (Table 1). Each of these primer pairs was used to perform RT-PCR with an RNA template (500 ng) which had been extracted from nematodes that were exposed to 98% RH for 24 h. The resulting PCR product was cloned into the PCR II vector (Invitrogen), introducing a T7 primer site upstream of the desired gene fragment. Each cloned fragment was PCR amplified with the M13 forward and reverse primer pair. The PCR product was cleaned (QIAprep Spin Miniprep kit; Qiagen) and was used for an in vitro transcription reaction (Megascript kit; Ambion). The resulting cRNA was treated with DNase I, quantified, and used to generate cRNA standards (in the range of 10−9 to 10−3 copies per μl). These standards were stored at −80°C and used within 6 to 8 weeks.
For real-time PCRs, total RNAs (10 μg, as for Northern blot analysis) were treated with DNase I (RNase free; Invitrogen) and quantified in a spectrophotometer, and each RNA sample was standardized to 500 ng per μl and stored at −80°C. Real-time PCRs were performed by the use of a Quantitect SYBR Green I RT-PCR kit (Qiagen) according to the manufacturer's protocol, with 1.0 μM (each) primers, 500 ng of sample RNA template, and 1 U of heat-labile uracil-N-glycosylase per reaction. A gene-specific external standard curve was generated by using cRNA standards that were run simultaneously with the experimental samples. Thermal cycling was performed in accordance with the Quantitect kit's instructions for a total of 40 cycles at an annealing temperature of 58°C for each primer pair. Real-time PCR analysis was performed with Lightcycler software, the threshold cycle was automatically calculated by the second-derivative-maximum method, and the copy number of the specific mRNA in the experimental samples was calculated by extrapolation from the gene-specific standard curve.
RESULTS
Effect of preconditioning time on carbohydrate content and desiccation tolerance of A. avenae.
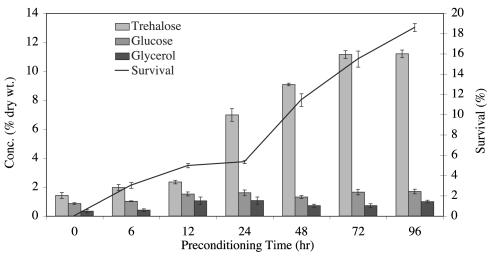
Changes in the concentrations of the carbohydrates trehalose, glucose, and glycerol in response to exposure to 98% RH for various time periods (0 to 96 h) followed by exposure to dried silica gel (∼10% RH) for 24 h were measured in A. avenae (Fig. 1). The influence of the length of preconditioning time at 98% RH on the nematodes' ability to survive exposure to silica gel for 24 h was also investigated. The only carbohydrate whose concentration altered significantly in response to preconditioning was trehalose (P < 0.001 [Kruskal-Wallis]). The initial concentration of trehalose was low (1.5% dry weight), but it rose gradually with preconditioning time to a maximum of 11% dry weight after 72 h, remaining constant thereafter until the experiment was terminated after 96 h. The rise in trehalose concentration correlated with a similar rise in nematode survival levels. A. avenae nematodes were unable to survive a direct exposure to dried silica gel without preconditioning, but survival rose to almost 15.5% after 72 h at 98% RH. While trehalose levels peaked at 72 h, nematode survival values continued to rise, reaching 18.5% after 96 h of preconditioning, at which point the experiment was terminated. The levels of both glucose and glycerol did not change significantly during the preconditioning period. These results are consistent with the findings of Higa and Womersley (33) and suggest that other factors in addition to trehalose accumulation are required for survival.
FIG. 1.
Changes in levels of trehalose, glucose, and glycerol in A. avenae. Nematodes were preconditioned at 98% RH for various time periods (0 to 96 h) followed by dehydration over dried silica for 24 h before carbohydrates were extracted. Survival data were obtained in an identical fashion, except that after dehydration the nematodes were prehydrated for 24 h and then rehydrated for 24 h in water before estimations of survival. Each value is the mean ± standard error of three replicates.
Identification of genes that are upregulated in A. avenae in response to desiccation stress.
We developed a novel molecular genetic approach to identify genes that are upregulated in A. avenae in response to dehydration. The development of mature mRNAs in nematodes involves the trans-splicing of a small highly conserved sequence to the 5′ ends of the developing mRNAs (40). Several splice leader sequences have been identified, but the most common trans-spliced sequence is referred to as SL1: in Caenorhabditis elegans >80% of transcripts are SL1 spliced (6). The 22-nt SL1 sequence is highly conserved across the phylum Nematoda and can be used as a primer in conjunction with an oligo(dT) primer at the 3′ end to amplify full-length cDNAs (46). The addition of extra nucleotides to the 3′ ends of these primers reduces the number of transcripts that are amplified, permitting the visualization of individual bands (58).
Using this approach, we compared cDNAs from untreated A. avenae and from nematodes exposed to 90% RH for 24 h by utilizing four different primer combinations. PCR products were analyzed by agarose gel electrophoresis (Fig. 2). Changes in the expression levels of transcripts could be seen clearly by using an SL1-based differential display technique. The addition of extra selective nucleotides to the SL1 primer generated different PCR amplification profiles. DNA fragments apparently corresponding to differentially regulated mRNAs were excised from the gel and then cloned. Of six such fragments, three were shown to correspond to genes that were upregulated in response to desiccation stress: they were Aav-lea-1 (GenBank accession no. AF423069 and SP:Q95V77), Aav-ahn-1 (AY340998), and Aav-glx-1 (AY340999). Aav-glx-1 (glutaredoxin 1) encodes a small predicted protein of 107 amino acid residues (11.6 kDa) that is a member of the glutaredoxin family and exhibits strongest homology (e−30) to the C. elegans glutaredoxin (WP:CE25238). The two other genes are described below.
LEA proteins in A. avenae and other nematodes.
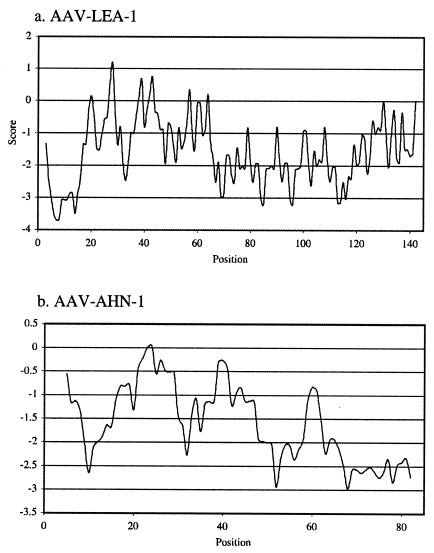
One cDNA isolated in this study encoded a protein which exhibited strong homology to a group of proteins associated with water deficits in plants, the late embryonic abundant (LEA) proteins (7, 35). The cDNA corresponding to the gene Aav-lea-1 was shown to be full length, encoding a predicted protein of 143 amino acid residues with five 11-mer repeats that are typical of the group 3 LEA proteins (9). A hydropathy plot of the Aav-LEA-1 protein, calculated according to the method of Kyte and Doolittle (41), is presented in Fig. 3a. The virtually unbroken hydrophilicity of this protein, as indicated by the negative score values, makes it unlikely that it spans membranes or forms a globular structure with a buried hydrophobic core. Indeed, members of our laboratories have recently shown that the Aav-LEA-1 protein seems to be natively unfolded in solution, with a high degree of hydration and a low level of compactness (29).
FIG. 3.
(a) Hydropathy analysis of predicted protein Aav-LEA-1 based on Kyte-Doolittle values, using a three-residue window. Those values below the zero line are negative and are therefore hydrophilic. The region from positions 69 to 102 contains three of the five 11-mer motifs arranged consecutively. (b) Hydropathy analysis of predicted protein Aav-AHN-1 based on Kyte-Doolittle values, using a three-residue window. The predicted protein is highly hydrophilic, as indicated by the negative score values.
A BLASTP analysis of the Aav-lea-1 sequence against the Swiss-Prot and TrEMBL databases revealed that this sequence has highest identity to a group 3 LEA protein (SP:Q06431) from the white birch, Betula verrucosa (pendula), and to several other group 3 LEA sequences from plants, but two C. elegans sequences, TR:Q9XTH4 and TR:O16527, were detected with less significant E values (Table 1). These C. elegans sequences were used in turn to search the Swiss-Prot and TrEMBL databases, yielding a large number of hits for additional group 3 LEA proteins from plants. The highest similarities were observed for an LEA protein from Glycine max (2e−31) and for a hypothetical LEA-like protein (TR:Q9FKV7) from Arabidopsis thaliana (2e−26). Sequences with similarity to the C. elegans LEA proteins were also detected in a wide range of bacteria and protozoa, and related sequences were also obtained from insects and vertebrates, but with less significant E values. BLASTP searches in Wormbase identified homologous sequences in Caenorhabditis briggsae.
Table 2 presents an analysis of the 11-mer repeat motifs and some physical and chemical parameters for the deduced plant and nematode proteins that are most closely related to Aav-LEA-1 as well as for some more distant sequences from other organisms. The consensus sequence for the 11-mer repeat motif in the eight nematode and two plant proteins is AWDA(TA)KDKAGD, which is in good agreement with the general form of the motif found in plant LEA-3 proteins (22). Positions 1, 2, 5, and 9 are hydrophobic; positions 3, 7, and 11 are negative; and positions 6 and 8 are positive. Positions 4 and 10 are not specified in plant LEA-3 motifs (21, 22), but in the data set presented here these positions contain hydrophilic amino acids. The low estimated pIs obtained for all of the nematode sequences indicate that these proteins are negatively charged at a physiological pH, but each 11-mer repeat has predominately lysine residues at positions 6 and 8 which are positively charged at a physiological pH. All of these proteins are highly hydrophilic, as indicated by the large numbers of charged amino acids in their sequences and by their negative hydropathicity scores (Table 2).
TABLE 2.
Characteristics of LEA proteins from nematodes and related sequences from other organisms
| Species | Identity | E value | No. of amino acids | No. of 11-mer repeats | Consensus at amino acida
|
pI | % negative amino acid residues | % positive amino acid residues | Hydropathicityb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |||||||||
| A. avenae | SP:Q95V77 | 143 | 5 | K | A | A | E | Ft | K | Q | R | A | G | E | 5.34 | 21 | 18.2 | −1.585 | |
| B. verrucosa (pendula) | SP:Q06431 | 8e−18c | 474 | 15 | YS | At | A | E | K | A | Kr | E | TY | K | D | 5.56 | 24.5 | 22.6 | −1.399 |
| C. elegans | TR:Q9XTH4 | 1e−08c | 497 | 14 | A | W | D | SA | A | K | De | K | A | S | De | 4.59 | 23.7 | 12.9 | −1.054 |
| TR:O16527 (lea-1) | 3e−08c | 733 | 23 | A | Wy | D | ST | T | K | D | K | A | G | D | 4.8 | 21.6 | 16 | −1.126 | |
| WP:CE35290 (dur-1) | 9e−06c | 790 | 18 | A | V | AV | D | G | A | K | A | A | G | DG | 5.91 | 11.9 | 10.5 | −0.654 | |
| C. briggsae | WP:CBP15141 | 1.5e−104d | 775 | 12 | A | W | Ed | As | A | K | E | K | A | K | Ed | 4.45 | 25.7 | 11.7 | −0.899 |
| WP:CBP01222 | 2.3e−60d | 732 | 19 | A | Wy | Edn | A | Ta | K | D | K | A | Qg | De | 4.9 | 21.5 | 15.9 | −1.104 | |
| WP:CBP06823 | 4.2e−38d | 324 | 9 | A | Aw | D | A | A | K | D | K | A | As | G | 5.6 | 18.5 | 17.3 | −0.833 | |
| WP:CBP15139 | 1.5e−35d | 431 | 14 | A | W | De | Sa | Ta | K | A | K | A | X | D | 6.44 | 16.2 | 15.3 | −0.815 | |
| G. max | TR:Q39873 | 4e−26e | 458 | 24 | A | A | Q | K | T | K | D | Y | A | G | D | 7.08 | 17.3 | 17.3 | −1.223 |
| Deinococcus radiodurans | SP:Q9RV58 | 5e−18e | 298 | 10 | A | A | D | Q | A | K | D | K | A | Q | D | 6.22 | 15.4 | 15.1 | −0.817 |
| D. melanogaster | TR:9VA86 | 2e−13f | 463 | 19 | A | Gn | Ad | Av | Ga | N | Va | A | G | Dn | VA | 4.34 | 9.9 | 2.8 | 0.086 |
| A. avenae | AAQ20894 | 86 | 0 | 10.06 | 14 | 22.1 | −1.564 | ||||||||||||
The consensus sequence for each motif represents the most frequent amino acid at each position. When more than one amino acid occurs frequently, additional amino acids are listed if their frequency is >25%. Amino acids shown in the same font size at a single position indicate that those amino acids are equally frequent, while a smaller font indicates less frequent amino acids.
Grand average of hydropathicity (41).
BLASTP analysis of Q95V77 against the Swiss-Prot TrEMBL database.
BLASTP report for WPCE31359 (Q9XTH4) (www.wormbase.org, release WS115).
BLASTP analysis of Q9XTH4 against the Swiss-Prot TrEMBL database.
BLASTP analysis of O16527 against the D. melanogaster database (SIB BLAST Network Service).
A novel hydrophilic gene, Aav-ahn-1, is upregulated in A. avenae in response to desiccation.
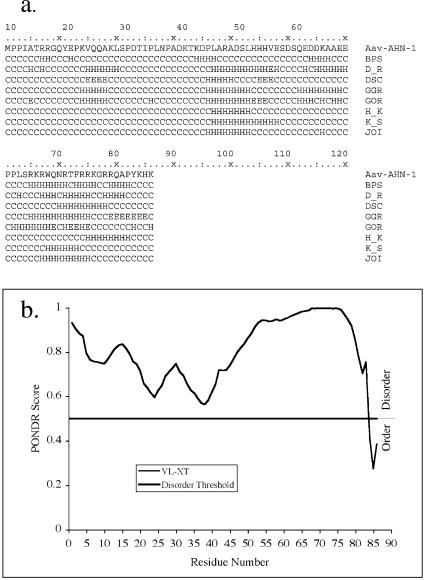
The cDNA sequence corresponding to Aav-ahn-1 (GenBank no. AY340998) failed to reveal homologies to any known DNA or protein sequence in the GenBank or Wormbase database. The transcript encodes a protein of 86 amino acid residues with an estimated molecular mass of 10 kDa; we have named this novel protein anhydrin. A hydropathy plot revealed that anhydrin is highly hydrophilic, with no significant hydrophobic regions (Fig. 3b). Three independent computational methods suggested that anhydrin is largely or wholly unstructured in solution. First, the consensus of seven secondary structure prediction programs was that the majority of the protein will assume a random coil structure with two small regions of alpha helix (Fig. 4a). Secondly, the PONDR (Predictors of Natural Disordered Regions) algorithm (44, 52) predicted that anhydrin is disordered throughout its length (Fig. 4b). The final method used was that of Uversky et al. (57), who have developed an algorithm that compares the mean net hydrophobicity (<H>) of a protein with its mean net charge (<R>). A low mean hydrophobicity and relatively high overall charge are associated with a lack of compactness in proteins under physiological conditions, resulting in a natively unfolded structure. If, for a given value of <R>, <H> is calculated to be lower than a specific boundary value, <H>b, then the protein is probably natively unfolded. For anhydrin, the values of <H> and <H>b are 0.33 and 0.45, respectively, which is consistent with the protein being natively unfolded.
FIG. 4.
(a) Secondary structure analysis of predicted protein Aav-AHN-1 performed with the PELE program available on the SDSC Biology Workbench (http://workbench.sdsc.edu/). Seven different structure predictions are shown, with the most likely structural feature at each residue indicated by H (α-helix), E (β-sheet), or C (random coil). The programs used are denoted BPS (10), D_R (20), DSC (38), GGR (27), GOR (26), H_K (34), and K_S (37). The “winner-takes-all” joint prediction was given by the JOI program. (b) PONDR analysis of Aav-AHN-1 protein predicts that it is disordered throughout its length.
Organization and expression of desiccation-responsive genes in A. avenae.
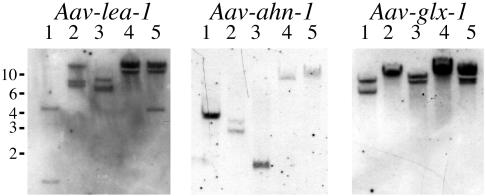
Southern hybridization of genomic A. avenae DNA digested with several restriction enzymes was performed with probes for each of the three genes (Fig. 5). For Aav-lea-1, the hybridization pattern suggested that the genome of A. avenae contains at least two, and possibly three, closely related copies of the gene. PCR isolation (data not shown) of the gene and further cDNA clones corresponding to the original cDNA clone (pJB2.1) confirmed its sequence and established the presence of two small introns in Aav-lea-1; neither of the related genes detected by Southern hybridization was isolated in these PCR experiments, perhaps indicating significant sequence divergence in the region to which the primers anneal. The hybridization pattern for Aav-ahn-1 was less complex and was consistent with a single gene being present in the genome. The sequence of the original cDNA (pJB3.3) clone was confirmed by amplification and sequencing of the gene and RT-PCR products; a single, short intron was detected in Aav-ahn-1. Hybridization data for Aav-glx-1 suggested that there are two closely related copies of the glutaredoxin gene and were consistent with PCR experiments in which two similar but different sequences were identified from both genomic DNA and cDNA templates. Of four RT-PCR clones sequenced, two corresponded exactly to the original cDNA and two contained the same 10 sequence differences in the coding region. Interestingly, all of these differences occurred at the third codon position and had no effect on the predicted amino acid sequence. Two different genes, each with a single, small intron, were isolated, and they corresponded to the two types of cDNA sequence; the second gene was named Aav-glx-2.
FIG. 5.
Southern blot analysis of the genes Aav-glx-1, Aav-lea-1, and Aav-ahn-1. Each lane contains the products of individual restriction digests with the endonucleases HindIII, BamHI, XbaI, EcoRI, and EcoRV, respectively.
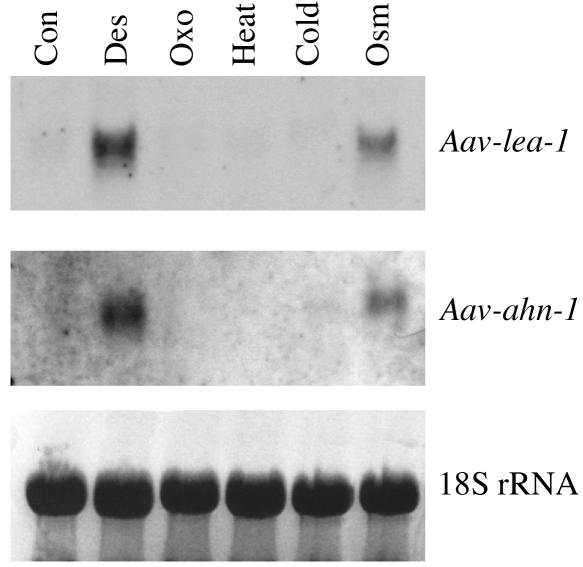
To gain further information on the regulation of these genes, we analyzed RNAs extracted from nematodes exposed to various environmental stresses (desiccation, oxidative, heat, cold, and osmotic stress) for 24 h by performing Northern blotting (Fig. 6). When the RNAs were hybridized with probes derived from the genes Aav-lea-1 and Aav-ahn-1, a marked increase in signal intensity was observed in the lanes containing RNAs extracted from nematodes exposed to both desiccation stress and osmotic stress, but not in those for the other tested stresses. The Aav-glx-1 gene appeared to be equally expressed in response to all stresses, except for heat, in which case it appeared to be down-regulated (data not shown).
FIG. 6.
Northern blot analysis of Aav-lea-1 and Aav-ahn-1 gene expression using RNAs extracted from nematodes exposed to various stresses for 24 h. Lanes: 1, control; 2, desiccation; 3, oxidative; 4, heat; 5, cold; and 6, osmotic.
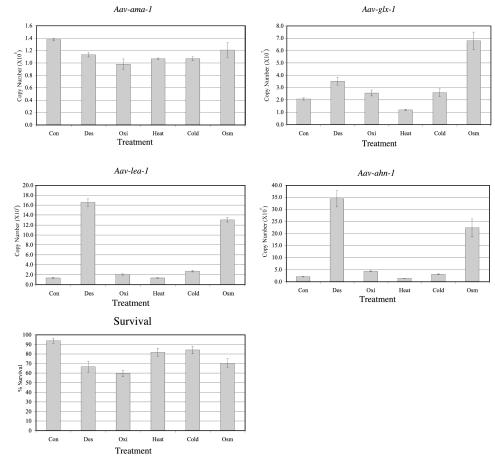
Quantitative PCR experiments were performed to estimate mRNA copy numbers for the genes Aav-lea-1, Aav-ahn-1, and Aav-glx-1 after exposure to several stresses (desiccation, oxidative, heat, cold, and osmotic stress); Aav-ama-1, encoding a subunit of RNA polymerase II, was included as an internal reference (Fig. 7). An absolute quantification method using a gene-specific external standard curve was used to determine the copy number of each gene per microgram of total RNA. The level of gene expression for both Aav-lea-1 and Aav-ahn-1 did not alter significantly for any of the treatments relative to the control, except for desiccation and osmotic stress. Aav-lea-1 gene expression was shown to increase >12-fold in response to desiccation stress and by a factor of almost 10 in response to osmotic stress. Similarly, the expression of Aav-ahn-1 increased 17- and 11-fold in response to desiccation stress and osmotic stress, respectively. The expression of the Aav-glx-1 gene was seen to increase slightly in response to desiccation stress and more significantly in response to osmotic stress. Importantly, the expression of the internal control gene, Aav-ama-1, did not appear to vary significantly in response to any of the treatments relative to the untreated control. The survival of A. avenae exposed to each of the stresses was also determined, and the small changes in survival cannot account for the large changes in the levels of gene expression observed. In summary, genes for LEA protein and anhydrin are specifically upregulated by dehydration induced by both drying and an increased osmotic potential. However, the glutaredoxin gene seems to be only weakly responsive to several stimuli, with the possible exception of osmotic stress, and therefore is not considered a significant contributor to the desiccation stress response in A. avenae.
FIG. 7.
Quantitative PCR analysis of expression of the genes Aav-ama-1, Aav-lea-1, Aav-ahn-1, and Aav-glx-1 performed by an absolute quantitative method. Each value represents the mean ± standard deviation of four replicates. Nematode samples were exposed to the treatment regimens for 24 h prior to RNA isolation, and matched survival data were determined for nematode samples treated in an identical manner, but survival was assessed after a 24-h recovery period in sterile tap water at 20°C.
DISCUSSION
In the anhydrobiotic nematode A. avenae, both trehalose accumulation and survival of complete desiccation increase with preconditioning time at a reduced relative humidity. However, as first shown by Higa and Womersley (33), an additional period of preconditioning following maximum trehalose accumulation is needed before maximal survival is seen. This suggests that other changes must also occur in A. avenae before it can successfully enter the anhydrobiotic state. Extensive research in drought-tolerant and anhydrobiotic plants also implicates factors besides the accumulation of disaccharides for dehydration tolerance (15, 35, 51). In addition, Lapinski and Tunnacliffe (42) found that two species of bdelloid rotifers do not synthesize trehalose or other disaccharides during the induction of anhydrobiosis and yet exhibit excellent desiccation tolerance. The present study was developed with the aim of identifying some of the additional molecular mechanisms involved in the successful induction of anhydrobiosis in A. avenae. Our data indicate that these additional factors include two highly hydrophilic proteins, specifically the LEA group 3 protein Aav-LEA-1 and a novel protein we have called anhydrin, and the antioxidant enzyme glutaredoxin.
The identification of a LEA protein gene, Aav-lea-1, in A. avenae suggests that at least some of the mechanisms used by anhydrobiotic nematodes are shared with plants undergoing desiccation stress. LEA proteins were first identified during seed maturation (23) and subsequently were found to be expressed in other plant tissues undergoing various forms of water stress, including drought, freezing, and osmotic stress (7). LEA proteins have a biased composition, being composed predominantly of hydrophilic amino acids, and their sequences display significant regularities, resulting in proteins of low complexity (60). Six groups of LEA proteins are recognized (8), and a revised class numbering system has recently been proposed (60). lea genes are consistently represented in differential screens for transcripts with increased expression during desiccation stress for a variety of plants. Dure (22) highlighted the occurrence of lea genes in nonplant genomes, including the nematode C. elegans and the prokaryotes Deinococcus radiodurans, Bacillus subtilis, and Haemophilus influenzae. Our analysis (Table 2) indicates that several lea genes occur in the nematodes C. elegans and C. briggsae. One of these genes, dur-1 (dauer upregulated), is expressed in C. elegans dauer larvae (13), an environmentally resistant stage of the nematode. An lea gene which is upregulated upon desiccation has also been identified in the nematode Steinernema feltiae (24). Garay-Arroyo et al. (25) designed an algorithm to search bacterial and fungal databases for proteins with a physicochemical resemblance to LEA group 1 and 2 proteins with regard to extreme hydrophilicity and a high percentage of glycine residues. These authors identified five candidate genes in Escherichia coli and twelve in S. cerevisiae as well as other sequences from Bacillus subtilis and Neurospora crassa. The accumulation of several of these “hydrophilin” transcripts was demonstrated in E. coli and S. cerevisiae in response to osmotic stress. The LEA protein which we have identified in A. avenae belongs to the group 3 family of LEAs, which are characterized in plants by the presence of a conserved 11-mer repeat of the general form TA(EQ)AAK(EQ)KAXE (21, 22). Representatives of this group of genes also occur in prokaryotes (22), protozoans (our unpublished database analyses), nematodes, and rotifers (56). It therefore seems likely that LEA proteins and hydrophilins will prove to be widespread among anhydrobiotic organisms.
Although the exact functional role of LEA proteins has yet to be determined, several functions have been proposed for them, including acting as molecular chaperones, hydration buffers, membrane stabilizers, and an ion sink (1, 16, 21). It has also been shown that a group 3 LEA protein from Typha latifolia pollen stabilizes a sucrose glass in vitro (62), although the significance of this is unclear since it can also be achieved by poly-l-lysine (63). Wise (60) used an algorithm that allows proteins to be compared based on similarities in their peptide compositions and that uses peptide profiles to interrogate a database of proteins with known function. This search identifies key words associated with each peptide group, thereby suggesting possible functions for LEA proteins which can be tested experimentally (61). Recent work by our groups has begun to investigate the structural changes induced by desiccation of the Aav-LEA-1 protein. Hydrodynamic and spectroscopic analyses indicate that Aav-LEA-1 is normally unstructured but that the protein undergoes a dramatic but reversible increase in folding in response to desiccation, with the development of a significant α-helical component and possibly coiled-coil structures. The dehydration of proteins is usually associated with their denaturation, and the observation that Aav-LEA-1 becomes more structured in response to desiccation may have significant relevance to its functional role (29).
The second gene discovered, Aav-ahn-1, encodes a small protein of 86 amino acid residues. Extensive searching of the various sequence databases failed to identify any significant homologies; similarly, a search for protein domains and motifs failed to provide any insights into its functional role. Looking at the hydropathy profile for the Aav-AHN-1 protein, we noted the extensive hydrophilic nature of this protein, with a grand average hydropathicity value of −1.564, which suggests that this protein is highly hydrated in aqueous solutions. Secondary structural analysis indicated that the protein will exist predominately as a random coil with a small proportion of α-helices, while the PONDR algorithm also suggested that the protein lacks any significant secondary structure. In this regard, anhydrin resembles LEA proteins and therefore could share similar hypothetical functions. For example, it might act as a hydration buffer during the dehydration process, slowing the rate of water loss and maintaining a minimal level of bound water within the cell even at low relative humidities. However, further analysis will be needed in order to elucidate the exact biochemical role of this novel protein.
The third gene isolated during this study, Aav-glx-1, encodes a member of the glutaredoxin family of proteins, also known as the thioltransferases. Aav-glx-1 expression levels increased 1.7-fold in response to desiccation stress and tripled in response to osmotic stress; negligible increases in Aav-glx-1 gene expression were seen in response to cold and oxidative stress. A significant decrease in the expression of Aav-glx-1 was seen in response to heat treatment; this decrease correlated with a drop in the level of nematode survival upon exposure to heat. However, this decrease in Aav-glx-1 gene expression in response to heat cannot be explained solely by the reduced survival, as the survival rate was lower in the sample exposed to oxidative stress.
The glutaredoxins are usually small (12 to 14 kDa) with a highly conserved structure, particularly in the region of the active site, and act as glutathione-dependent disulfide oxidoreductases (53). Glutaredoxins can reactivate many oxidized proteins by reducing the mixed disulfides formed during oxidative stress (30). The contribution of glutaredoxins to the stress response has been explored with S. cerevisiae, for which research has shown that the products of the two yeast glutaredoxin genes, GLX1 and GLX2, are required for protection during conditions of oxidative stress (45). Subsequent studies have shown that glutaredoxin expression is also induced in yeast by osmotic and heat stress (30). In A. avenae, glutaredoxin is induced by both desiccation and osmotic stress. It is also expected that oxidative stress would lead to an increase in glutaredoxin expression, but we might have failed to detect this because of the high mortality observed when nematodes were treated with paraquat. The general level of increase in glutaredoxin gene expression was significant but small when compared to the increase in expression of the other two genes, Aav-lea-1 and Aav-ahn-1, suggesting that redundant complementary systems such as the thioredoxins may provide additional protection against oxidative damage. In yeast, GLX1 knockouts are unaffected by osmotic and heat stress, indicating that redundant systems must be able to compensate for the absence of GLX1 during exposure to these stress conditions (30).
The relationship between mRNA and protein expression has been investigated in several recent large-scale studies (28, 32, 43). These studies show that mRNA expression levels cannot be consistently relied on as a predictor of protein abundance (since cells can control the abundance of individual proteins at the transcriptional or translational level). In a study of mRNA and protein correlations in yeast, Greenbaum et al. (31) found that open reading frames that show a large degree of variation in their expression levels are controlled transcriptionally, while open reading frames that show minimal variation in their mRNA expression levels throughout the cell cycle appear to control protein expression by posttranscriptional means. If these conclusions also apply to nematodes, then they would suggest that large increases in Aav-LEA-1 and Aav-AHN-1 protein levels should be observed upon dehydration, corresponding to the upregulation of Aav-lea-1 and Aav-ahn-1 transcript levels. This would be consistent with what is observed in plants, for which several authors have shown that lea gene expression and LEA protein accumulation occur in a coordinated manner (4, 11, 21). Experiments to investigate the relationship between mRNA and protein expression for the dehydration-induced genes described here are in progress.
The reaction to environmental stress in plants is often characterized by a generalized response, with many genes that are inducible by more than one particular stimulus. In the case of Arabidopsis thaliana, several genes, including rd29A and cor15a, are upregulated in response to both cold and dehydration stress (39). Similarly, in spinach a group 2 LEA protein, CAP85, is induced in response to both cold and drought (47). The rationale for this cross talk in plants can be explained by the fact that freezing, drought, and high salt all lead to a reduction in the levels of free water within the cell, so genes involved in protection against water stress will be induced by several different environmental conditions (15). Using the genes Aav-lea-1 and Aav-ahn-1, we have investigated this stimulus cross-talk phenomenon in A. avenae. From both the Northern blot analysis and real-time PCR data, it can be seen that the expression levels of both genes increased significantly upon exposure to desiccation and osmotic stress, although the absolute levels of Aav-lea-1 mRNA exceeded those of Aav-ahn-1 upon induction. This suggests that the nematode A. avenae has a specific sensory and signaling pathway that responds to water loss which is triggered by both desiccation at reduced relative humidities and osmotic water loss. We are currently working to increase the panel of desiccation response genes from A. avenae. As more desiccation response genes from A. avenae are identified, it will be possible to gain a better understanding of the molecular adaptations involved in nematode anhydrobiosis and of the signaling pathways leading to these responses.
Acknowledgments
This work was funded by grants from Enterprise Ireland, The Leverhulme Trust, The Isaac Newton Trust, and a Royal Irish Academy/Royal Society Exchange Scheme. A.M.B. is a Science Foundation Ireland Investigator, and A.T. is the Anglian Water Fellow in Biotechnology of Pembroke College, Cambridge, United Kingdom.
REFERENCES
- 1.Alsheikh, M. K., B. J. Heyen, and S. K. Randall. 2003. Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 278:40882-40889. [DOI] [PubMed] [Google Scholar]
- 2.Appel, R. D., A. Bairoch, and D. F. Hochstrasser. 1994. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem. Sci. 19:258-260. [DOI] [PubMed] [Google Scholar]
- 3.Bewley, J. D. 1973. Desiccation and protein synthesis in the moss Tortula ruralis. Can. J. Bot. 51:203-206. [Google Scholar]
- 4.Bianchi, M. W., C. Damerval, and N. Vartanian. 2002. Identification of proteins regulated by cross-talk between drought and hormone pathways in Arabidopsis wild-type and auxin-insensitive mutants, axr1 and axf2. Func. Plant Biol. 29:55-61. [DOI] [PubMed] [Google Scholar]
- 5.Billi, D., and M. Potts. 2002. Life and death of dried prokaryotes. Res. Microbiol. 153:7-12. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal, T. 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11:132-136. [DOI] [PubMed] [Google Scholar]
- 7.Bray, E. A. 1997. Plant responses to water deficit. Trends Plant Sci. 2:48-54. [Google Scholar]
- 8.Bray, E. A., J. Bailey-Serres, and E. Wewetilnyk. 2000. Responses to abiotic stress, p. 1158-1203. In B. B. Buchannan, W. Gruissem, and R. L. Jones (ed.), Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, Md.
- 9.Browne, J., A. Tunnacliffe, and A. Burnell. 2002. Anhydrobiosis—plant desiccation gene found in a nematode. Nature 416:38. [DOI] [PubMed] [Google Scholar]
- 10.Burgess, A. W., P. K. Ponnuswamy, and H. A. Sheraga. 1974. Analysis of conformations of amino acid residues and prediction of backbone topography in proteins. Isr. J. Chem. 12:239-286. [Google Scholar]
- 11.Campbell, S. A., and T. J. Close. 1997. Dehydrins: genes, proteins, and associations with phenotypic traits. New Phytol. 137:61-74. [Google Scholar]
- 12.Carpenter, J. F., L. M. Crowe, and J. H. Crowe. 1987. Stabilization of phosphofructokinase with sugars during freeze-drying: characterization of enhanced protection in the presence of divalent cations. Biochim. Biophys. Acta 923:109-115. [DOI] [PubMed] [Google Scholar]
- 13.Cherkasova, V., S. Ayyadevara, N. Egilmez, and R. S. Reis. 2000. Diverse Caenorhabditis elegans genes that are unregulated in dauer larvae also show elevated transcript levels in long-lived, aged, or starved adults. J. Mol. Biol. 300:433-448. [DOI] [PubMed] [Google Scholar]
- 14.Clegg, J. S. 2001. Cryptobiosis—a peculiar state of biological organization. Comp. Biochem. Phys. B 128:613-624. [DOI] [PubMed] [Google Scholar]
- 15.Close, T. J. 1996. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plantarum 97:795-803. [Google Scholar]
- 16.Crowe, J. H., F. A. Hoekstra, and L. M. Crowe. 1992. Anhydrobiosis. Annu. Rev. Physiol. 54:579-599. [DOI] [PubMed] [Google Scholar]
- 17.Crowe, J. H., J. F. Carpenter, and L. M. Crowe. 1998. The role of vitrification in anhydrobiosis. Annu. Rev. Physiol. 60:73-103. [DOI] [PubMed] [Google Scholar]
- 18.Crowe, J. H., and K. A. C. Madin. 1975. Anhydrobiosis in nematodes: evaporative water loss and survival. J. Exp. Zool. 193:323-334. [Google Scholar]
- 19.Crowe, J. H., L. M. Crowe, and D. Chapman. 1984. Preservation of membranes in anhydrobiotic organisms: the role of trehalose. Science 223:701-703. [DOI] [PubMed] [Google Scholar]
- 20.Deléage, G., and B. Roux. 1987. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1:289-294. [DOI] [PubMed] [Google Scholar]
- 21.Dure, L., III. 1993. A repeating 11-mer amino acid motif and plant desiccation. Plant J. 3:363-369. [DOI] [PubMed] [Google Scholar]
- 22.Dure, L., III. 2001. Occurrence of a repeating 11-mer amino acid sequence motif in diverse organisms. Protein Pept. Lett. 8:115-122. [Google Scholar]
- 23.Dure, L., III, S. C. Greenway, and G. A. Galau. 1981. Developmental biochemistry of cottonseed embryogenesis and germination—changing messenger ribonucleic-acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 20:4162-4168. [DOI] [PubMed] [Google Scholar]
- 24.Gal, T. Z., I. Glazer, and H. Koltai. 2003. Differential gene expression during desiccation stress in the insect-killing nematode Steinernema feltiae IS-6. J. Parasitol. 89:761-766. [DOI] [PubMed] [Google Scholar]
- 25.Garay-Arroyo, A., J. M. Colmenero-Flores, A. Garciarrubio, and A. A. Covarrubias. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275:5668-5674. [DOI] [PubMed] [Google Scholar]
- 26.Garnier, J., D. J. Osguthorpe, and B. Robson. 1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 120:97-120. [DOI] [PubMed] [Google Scholar]
- 27.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:97-120. [DOI] [PubMed] [Google Scholar]
- 28.Ghaemmaghami, S., W. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 29.Goyal, K., L. Tisi, A. Basran, J. Browne, A. Burnell, J. Zurdo, and A. Tunnacliffe. 2003. Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J. Biol. Chem. 278:12977-12984. [DOI] [PubMed] [Google Scholar]
- 30.Grant, C. M., S. Luikenhuis, A. Beckhouse, M. Soderbergh, and I. W. Dawes. 2000. Differential regulation of glutaredoxin gene expression in response to stress conditions in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1490:33-42. [DOI] [PubMed] [Google Scholar]
- 31.Greenbaum, D., C. Colangelo, K. Williams, and M. Gerstein. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hegde, P. S., I. R. White, and C. Debouck. 2003. Interplay of transcriptomics and proteomics. Curr. Opin. Biotechnol. 14:647-651. [DOI] [PubMed] [Google Scholar]
- 33.Higa, L. M., and C. Z. Womersley. 1993. New insights into the anhydrobiotic phenomenon: the effects of trehalose content and differential rates of evaporative water-loss on the survival of Aphelenchus avenae. J. Exp. Zool. 267:120-129. [Google Scholar]
- 34.Holley, L. H., and M. Karplus. 1989. Protein secondary structure prediction with a neural network. Proc. Natl. Acad. Sci. USA 86:152-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingram, J., and D. Bartels. 1996. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. 47:377-403. [DOI] [PubMed] [Google Scholar]
- 36.Keilin, D. 1959. The problem of anabiosis or latent life: history and current concepts. Proc. R. Soc. Lond. B 150:149-191. [DOI] [PubMed] [Google Scholar]
- 37.King, R. D., and M. J. E. Sternberg. 1990. Machine learning approach for the prediction of protein secondary structure. J. Mol. Biol. 216:441-457. [DOI] [PubMed] [Google Scholar]
- 38.King, R. D., and M. J. E. Sternberg. 1996. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 5:2298-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight, H., and M. R. Knight. 2001. Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 6:262-267. [DOI] [PubMed] [Google Scholar]
- 40.Krause, M., and D. Hirsh. 1987. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell 49:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyte, J., and R. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 42.Lapinski, J., and A. Tunnacliffe. 2003. Anhydrobiosis without trehalose in bdelloid rotifers. FEBS Lett. 553:387-390. [DOI] [PubMed] [Google Scholar]
- 43.Lee, P. S., L. B. Shaw, L. H. Choe, A. Mehra, V. Hatzimanikatis, and K. H. Lee. 2003. Insights into the relation between mRNA and protein expression patterns. II. Experimental observations in Escherichia coli. Biotechnol. Bioeng. 84:834-841. [DOI] [PubMed] [Google Scholar]
- 44.Li, X., P. Romero, M. Rani, A. K. Dunker, and Z. Obradovic. 1999. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. 10:30-40. [PubMed] [Google Scholar]
- 45.Luikenhuis, S., G. Peronne, I. W. Dawes, and C. W. Grant. 1998. The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell 9:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin, S. A. M., F. J. Thompson, and E. Devaney. 1995. The construction of spliced leader cDNA libraries from the filarial nematode Brugia pahangi. Mol. Biochem. Parasitol. 70:241-245. [DOI] [PubMed] [Google Scholar]
- 47.Neven, L. G., D. W. Haskell, A. Hofig, Q. B. Li, and C. L. Guy. 1993. Characterization of a spinach gene responsive to low-temperature and water-stress. Plant Mol. Biol. 21:291-305. [DOI] [PubMed] [Google Scholar]
- 48.Notredame, C., D. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for multiple sequence alignments. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 49.O'Leary, S. A., A. P. Power, C. M. Stack, and A. M. Burnell. 2001. Behavioral and physiological responses of infective juveniles of the entomopathogenic nematode Heterorhabditis to desiccation. Biocontrol 46:345-362. [Google Scholar]
- 50.Oliver, M. J., and J. D. Bewley. 1997. Desiccation-tolerance of plant tissues: a mechanistic overview. Horticult. Rev. 18:171-214. [Google Scholar]
- 51.Piatkowski, D., K. Schneider, F. Salamini, and D. Bartels. 1990. Characterization of five abscisic acid-responsive cDNA clones isolated from the desiccation-tolerant plant Craterostigma plantagineum and their relationship to other water-stress genes. Plant Physiol. 94:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romero, P., Z. Obradovic, X. Li, E. Garner, C. Brown, and A. K. Dunker. 2001. Sequence complexity of disordered protein. Proteins 42:38-48. [DOI] [PubMed] [Google Scholar]
- 53.Rouhier, N., E. Gelhaye, and J. P. Jacquot. 2002. Exploring the active site of plant glutaredoxin by site-directed mutagenesis. FEBS Lett. 511:145-149. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Solomon, M. E. 1951. Control of relative humidity with potassium hydroxide, sulphuric acid, or other solutions. Bull. Entomol. Res. 42:543-554. [Google Scholar]
- 56.Tunnacliffe, A., and J. Lapinski. 2003. Resurrecting Van Leeuwenhoek's rotifers: a reappraisal of the role of disaccharides in anhydrobiosis. Phil. Trans. R. Soc. B 358:1755-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uversky, V. N., J. R. Gillespie, and A. L. Fink. 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41:415-427. [DOI] [PubMed] [Google Scholar]
- 58.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Vandelee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP—a new technique for DNA-fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winston, P. W., and D. H. Bates. 1960. Saturated solutions for the control of humidity in biological research. Ecology 41:232-237. [Google Scholar]
- 60.Wise, M. J. 2003. LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wise, M. J., and A. Tunnacliffe. 2004. POPP the question: what do LEA proteins do? Trends Plant Sci. 9:13-17. [DOI] [PubMed] [Google Scholar]
- 62.Wolkers, W. F., S. McCready, W. F. Brandt, G. G. Lindsey, and F. A. Hoekstra. 2001. Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim. Biophys. Acta 1544:196-206. [DOI] [PubMed] [Google Scholar]
- 63.Wolkers, W. F., M. G. van Kilsdonk, and F. A. Hoekstra. 1998. Dehydration-induced conformational changes of poly-l-lysine as influenced by drying rate and carbohydrates. Biochim. Biophys. Acta 1425:127-136. [DOI] [PubMed] [Google Scholar]