Abstract
In response to various extracellular signals, the morphology of the human fungal pathogen Candida albicans switches from yeast to hypha form. Here, we report that GPR1 encoding a putative G-protein-coupled receptor and GPA2 encoding a Gα subunit are required for hypha formation and morphogenesis in C. albicans. Mutants lacking Gpr1 (gpr1/gpr1) or Gpa2 (gpa2/gpa2) are defective in hypha formation and morphogenesis on solid hypha-inducing media. These phenotypic defects in solid cultures are suppressed by exogenously added dibutyryl-cyclic AMP (dibutyryl-cAMP). Biochemical studies also reveal that GPR1 and GPA2 are required for a glucose-dependent increase in cellular cAMP. An epistasis analysis indicates that Gpr1 functions upstream of Gpa2 in the same signaling pathway, and a two-hybrid assay reveals that the carboxyl-terminal tail of Gpr1 interacts with Gpa2. Moreover, expression levels of HWP1 and ECE1, which are cAMP-dependent hypha-specific genes, are reduced in both mutant strains. These findings support a model that Gpr1, as well as Gpa2, regulates hypha formation and morphogenesis in a cAMP-dependent manner. In contrast, GPR1 and GPA2 are not required for hypha formation in liquid fetal bovine serum (FBS) medium. Furthermore, the gpr1 and the gpa2 mutant strains are fully virulent in a mouse infection. These findings suggest that Gpr1 and Gpa2 are involved in the glucose-sensing machinery that regulates morphogenesis and hypha formation in solid media via a cAMP-dependent mechanism, but they are not required for hypha formation in liquid medium or during invasive candidiasis.
Many signaling processes are mediated by G-protein-coupled receptors (GPCRs) in eukaryotic cells. Typically, a ligand-bound GPCR activates or inhibits heterotrimeric G-protein complexes and transmits signals to downstream effectors, including adenylate cyclase, phospholipases, or protein kinases (35). The activity of a G protein is controlled by the kind of guanine nucleotide bound to the protein. A GDP-GTP exchange on the Gα subunit causes dissociation of the α subunit from the βγ complex, and either the free α subunit or the free βγ complex, or in some cases both, regulates downstream effectors. Then, GTP hydrolysis with its intrinsic GTPase activity promotes the reassociation of the heterotrimer and attenuation of signaling.
In the yeast Saccharomyces cerevisiae, two Gα subunits, Gpa1 and Gpa2, have been characterized. Gpa1 regulates the mating response by coupling with Ste2 (α-factor receptor) and Ste3 (a-factor receptor) (26). In this pathway, the free βγ complex transmits the signal to a conserved mitogen-activated protein (MAP) kinase cascade (14, 42). In contrast, Gpa2 was shown to regulate the intracellular cyclic AMP (cAMP) level in response to glucose stimulation through interaction with a putative GPCR, Gpr1 (22, 32, 49). This signaling pathway is independent of the function of the small G protein, Ras2 (47), which regulates both the MAP kinase cascade and the cAMP signaling pathway. Recent genetic evidence suggests that this Gpr1-Gpa2 interaction controls the morphological transition from yeasts to pseudohyphae through the cAMP pathway in response to nutritional conditions (25, 31, 32, 45), and Gpr1 specifically appears to be a nutritional sensor in the pathway.
Candida albicans is a major fungal pathogen of humans and produces both irritating mucosal infections and life-threatening invasive disease. C. albicans can grow in several morphological forms, ranging from the yeasts to pseudohyphae to true hyphae. Many environmental conditions are known to induce the filamentous growth of C. albicans, and some of these inducers of hypha formation, such as mammalian serum, neutral pH, N-acetylglucosamine, nutrient deprivation, and temperature, have been characterized (36). However, the mechanism used by C. albicans to sense these environmental cues and create these morphogenetic changes is still unknown.
The strong molecular conservation between C. albicans and S. cerevisiae in many cellular processes and a genome sequencing program have enabled the identification of many signaling pathways and regulators involved in the hypha formation of these ascomycetes. Recent studies have revealed that the morphological switching in C. albicans is regulated by both the MAP kinase cascade and the cAMP pathway. Signaling proteins in these pathways, including the small G protein Ras1 (28), MAP kinases, Cek1 (12), Hst7 and Cst20 (21, 27), adenylate cyclase Cdc35 (38), and cAMP-dependent protein kinases Tpk1 and Tpk2 (6, 41), were shown to be well conserved in function with those of S. cerevisiae. These findings strongly suggest that the multiple signaling systems regulating hypha formation in C. albicans correspond to the signaling mechanisms controlling the morphological transition from the yeasts to pseudohyphae in S. cerevisiae. In the present study, we demonstrate that Gpr1 and Gpa2 regulate morphogenesis and hypha formation in C. albicans through the cAMP signaling pathway, but they are not associated with the virulence composite of this pathogen.
MATERIALS AND METHODS
Strains and media.
Yeast strains used in the present study are listed in Table 1. Strains were grown on either YPD medium (1% yeast extract, 2% Bacto Peptone, and 2% glucose) or synthetic dextrose (SD) medium (2% glucose, 0.67% yeast nitrogen base without amino acids [Difco], and appropriate supplements) prepared as previously described (40). For the selection of Ura auxotrophic clones, SD (Ura−) medium was supplemented with 0.01% uridine and 0.1% 5′-fluoroorotic acid (20). For specific experiments involving hyphal growth, Spider medium was prepared as described previously (29). For serum-mediated filamentation, YPD medium containing 10% fetal calf serum was used. An embedded condition was used as previously described (10).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| Candida albicans | ||
| CAI4 | Δura3::imm434/Δura3::imm434 | 15 |
| YTC028 | Δura3::imm434/Δura3::imm434 TRP1/TRP1::URA3 | This study |
| YTC014 | Δura3::imm434/Δura3::imm434 TRP1/TRP1::ADH1prGPR1-URA3 | This study |
| HTC7 | Δura3::imm434/Δura3::imm434 TRP1/TRP1::ADH1prGPA2Q355L-URA3 | This study |
| TCA05 | Δura3::imm434/Δura3::imm434 GPR1/gpr1::hisG | This study |
| YTC068 | Δura3::imm434/Δura3::imm434 GPR1/gpr1::hisG TRP1/TRP1::URA3 | This study |
| TCA07 | Δura3::imm434/Δura3::imm434 gpr1::hisG/gpr1::hisG | This study |
| YTC049 | Δura3::imm434/Δura3::imm434 gpr1::hisG/gpr1::hisG TRP1/TRP1::URA3 | This study |
| YTC108 | Δura3::imm434/Δura3::imm434 gpr1::hisG/gpr1::hisG TRP1/TRP1::GPR1-URA3 | This study |
| YTC104 | Δura3::imm434/Δura3::imm434 gpr1::hisG/gpr1::hisG TRP1/TRP1::ADH1prGPR1-URA3 | This study |
| YTC100 | Δura3::imm434/Δura3::imm434 gpr1::hisG/gpr1::hisG TRP1/TRP1::ADH1prGPA2Q355L-URA3 | This study |
| TCA08 | Δura3::imm434/Δura3::imm434 GPA2/gpa2::hisG | This study |
| YTC072 | Δura3::imm434/Δura3::imm434 GPA2/gpa2::hisG TRP1/TRP1::URA3 | This study |
| TCA12 | Δura3::imm434/Δura3::imm434 gpa2::hisG/gpa2::hisG | This study |
| YTC032 | Δura3::imm434/Δura3::imm434 gpa2::hisG/gpa2::hisG TRP1/TRP1::URA3 | This study |
| YTC109 | Δura3::imm434/Δura3::imm434 gpa2::hisG/gpa2::hisG TRP1/TRP1::GPA2-URA3 | This study |
| YTC020 | Δura3::imm434/Δura3::imm434 gpa2::hisG/gpa2::hisG TRP1/TRP1::ADH1prGPR1-URA3 | This study |
| HTC12 | Δura3::imm434/Δura3::imm434 gpa2::hisG/gpa2::hisG TRP1/TRP1::ADH1prGPA2Q355L-URA3 | This study |
| Saccharomyces cerevisiae PJ69-4Aa/αa | MATa/α trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | 19 |
For two-hybrid analysis.
Isolation of GPR1 gene.
In a search of the C. albicans sequence database (http://alces.med.umn.edu/Candida.html), we found a partial sequence, which showed some homology to the GPR1 gene of S. cerevisiae. The probe for C. albicans GPR1 was prepared by PCR with the primer pair GPR1-1+ and GPR1-1− (see Table 3) based on the partial sequence. Genomic DNA of C. albicans was digested with several restriction enzymes and Southern blot analysis was performed with the probe. We found an ∼5.7-kb fragment by HindIII digestion that hybridized with the probe (data not shown). Genomic DNA was then digested with HindIII, and a genomic library was constructed in the pUC19 cloning vector. The GPR1 gene was isolated by colony hybridization. A plasmid pMS30 carrying the 5.7-kb HindIII fragment containing the GPR1 gene was subjected to DNA sequencing. The nucleotide sequence has been deposited in the GenBank/EMBL/DDBJ database under accession number AB084519.
TABLE 3.
Oligonucleotides used in this study
| Primer | Sequencea |
|---|---|
| GPR1-1+ | TCCTACAAGAATATTGACTCAT |
| GPR1-1− | GGGTAATTGGAGCAATGG |
| GPR1-2+ | AA CCGGACCTAATATCAATAGC CCGGACCTAATATCAATAGC |
| GPR1-2− | CTGGCAACAATCAATGGGACAAA |
| GPR1-3+ | TG GGAAAAATGCTGCTGGTAATGG GGAAAAATGCTGCTGGTAATGG |
| GPR1-3− | G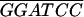 AATAATTCTTTGTCGTCTTCGG AATAATTCTTTGTCGTCTTCGG |
| GPR1-4+ | CTATG GTGCAGGCATCAATTG GTGCAGGCATCAATTG |
| GPR1-4− | T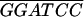 CATTGGGGGTCCTTTTTTGAG CATTGGGGGTCCTTTTTTGAG |
| GPA2+ | GTATCTGTTTCACTTGCAACAGTC |
| GPA2− | ACAACAGCTTGAATACGTATCGAC |
| Q355L+ | ATTTATTTGATGTTGGTGGTttAAGGTCAGAAAGAAAAAAATGG |
| Q355L− | CCATTTTTTTCTTTCTGACCTTaaACCACCAACATCAAATAAAT |
| ADH1-NcoI+ | CAAATCGTCTACCAGAAGTTATGC |
| ADH1-NcoI− |
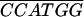 CGAGAACTCGGAAAAAAAAAGTGC CGAGAACTCGGAAAAAAAAAGTGC |
| ADH1-NdeI+ | ACACTTCTAAACAAACCAATCAAT |
| ADH1-NdeI− | AA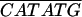 AATTGTTTTTGTATTTGTTGTTGTTGTTG AATTGTTTTTGTATTTGTTGTTGTTGTTG |
| TRP1+ | CGTTGAGGTTTCCGTTGTTTTC |
| TRP1− | CTGCCACCATAGTCGTAAGAGG |
| HWP1+ | GTGAAACAGAGGAAGCTCTTATTC |
| HWP1− | TTCAGAAGTAGTAGTTGTAGCAGG |
| ECE1+ | TATGGGTATTCTTGGCAACATTCC |
| ECE1− | TCAATACCGACAGTTTCAATGCTC |
| ACT1+ | AACATCCAGTTTTGTTGACCGAAG |
| ACT1− | ACTTCATGATGGAGTTGAAAGTGG |
The introduced restriction site is underlined; changed nucleotides are in lowercase.
Construction of plasmids.
For the construction of plasmids, PCRs were performed by using high-fidelity DNA polymerase KOD Plus (Toyobo). Fragments generated by PCR were always sequenced to ensure that no unanticipated mutations were introduced during the amplification. All plasmids and oligonucleotides used in the present study are listed in Tables 2 and 3, respectively. Plasmid pMS58 contains a GPR1 gene disruption cassette with URA3 as a selection marker. To create pMS58, the 5-kbp HindIII fragment of pMS30 was subcloned into the HindIII site of pUC19 (PstI−) in which the PstI site was disrupted. The plasmid was then cleaved with PstI and MscI to remove two-thirds of the GPR1 open reading frame (ORF), and a 3.9-kbp BamHI (blunted)-PstI fragment of a hisG-URA3-hisG cassette derived from a pCaU1 plasmid was ligated. The plasmid pCaU1 was a gift from Chugai Pharmaceutical. In this plasmid, a HindIII fragment of pNKY51 (1) containing S. cerevisiae URA3 was replaced by C. albicans URA3. The resultant plasmid was digested with BamHI-BglII, and the hisG-URA3-hisG fragment was inserted into the BamHI site of pUC19 to generate pCaU1.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Source |
|---|---|---|
| Plasmids for phenotypic characterization | ||
| pCaU1 | hisG-URA3-hisG | Chugai Pharma |
| pHT225 | URA3 in pUC19 | This study |
| pHT258 | TRP1 in pUC19 | This study |
| pHT244 | URA3 TRP1 in pUC19 | This study |
| pMS30 | 5.7-kb GPR1 HindIII in pUC19 | This study |
| pYT098 | 5.7-kb GPR1-TRP1 | This study |
| pYT100 | 5.7-kb GPR1-URA3-TRP1 | This study |
| pMS58 | GPR1 PstI-PstIhisG-URA3-hisGBamHI-MscIGPR1 | This study |
| pTM235 | 4.4-kb GPA2 fragment in pGEM-T easy | This study |
| pTM247 | GPA2NcoI-PstIhisG-URA3-hisGBamHI-XbaIGPA2 | This study |
| pHT200 | 5.6-kb GPA2 HindIII in pUC19 | This study |
| pYT099 | 5.6-kb GPA2-TRP1 | This study |
| pYT101 | 5.6-kb GPA2-URA3-TRP1 | This study |
| pTM249 | ADH1 promoter (NcoI) in pUC19 | This study |
| pHT256 | ADH1pr-GPA2Q355L-URA3 | This study |
| pHT257 | ADH1pr-GPA2Q355L-URA3-TRP1 | This study |
| pHT229 | ADH1 promoter (NdeI) in pUC19 | This study |
| pHT231 | ADH1pr-GPR1-URA3 | This study |
| pHT232 | ADH1pr-GPR1-URA3-TRP1 | This study |
| Plasmids for two-hybrid analysis | ||
| pAS2-1 | BD | Clontech |
| pACT2 | AD | Clontech |
| pYT025 | BD-Gpr1 (301-433) | This study |
| pYT029 | BD-Gpr1 (660-823) | This study |
| pYT030 | AD-Gpa2 (1-503) | This study |
Numbers in parentheses are amino acid numbers.
To construct a GPA2 disruption cassette, we amplified the GPA2 gene by PCR by using the oligonucleotide primers GPA2+ and GPA2− based on the GPA2 sequence from the Proteome BioKnowledge Library Database (Incyte) and then cloned it into pGEM-T Easy (Promega) to construct the plasmid pTM235. To yield the GPA2 disruption plasmid pTM247, pTM235 was cleaved with NcoI and XbaI to remove the GPA2 ORF, and then a hisG-URA3-hisG cassette was inserted. We also isolated a GPA2 genomic clone (pHT200) from the HindIII-digested C. albicans genomic library constructed in pUC19, which contained the 5.7-kb HindIII fragment. A dominant-active mutation of GPA2 was made by changing Gln-355 to Leu by using a Quick Change site-directed mutagenesis kit (Stratagene). Site-directed mutagenesis was performed by using pHT200 as a template with the primers Q355L+ and Q355L− in which the changed nucleotide is underlined (see Table 3). The promoter region of the alcohol dehydrogenase gene ADH1 of C. albicans (4) was amplified by PCR by using the primers ADH1-NcoI+ and ADH1-NcoI− to introduce an NcoI site and then inserted into the SmaI site of pUC19 to obtain the plasmid pTM249. A 3.9-kb NcoI fragment containing the GPA2 gene with the dominant-active mutation was introduced into pTM249, and a 1.1-kb HindIII (blunted) fragment containing URA3 from pCaU1 was cloned into the PstI (blunted) site to yield the plasmid pTM256 in which GPA2Q355L was designed to be expressed under the control of the ADH1 promoter.
The promoter region of ADH1 of C. albicans was also amplified by PCR with primers ADH1-NdeI+ and ADH1-NdeI− to introduce a NdeI site and then inserted into the SmaI site of pUC19 to obtain the plasmid pHT229.
A 0.7-kb sequence of the 5′ region of GPR1 ORF was amplified by PCR with primers GPR1-2+ and GPR1-2− to introduce a NdeI site at the start codon, digested with NdeI and PstI, and then ligated into pUC19. The resultant plasmid was digested with NdeI, and the NdeI fragment from pHT229 containing the ADH1 promoter was inserted to form ADH1prGPR1-5′/19. A 3.2-kb PstI-SphI fragment containing the 3′ region of GPR1 from pMS30 was inserted into the PstI-SphI site of pHT229, the resultant plasmid was digested with PstI, and then a 0.3-kb PstI fragment from ADH1prGPR1-5′/19 was inserted to form ADH1prGPR1/19. An HindIII fragment containing the URA3 gene from pCaU1 was ligated into pUC19 to form pHT225, and a KpnI-SphI fragment containing ADH1prGPR1 was ligated into pHT225 to form pHT231. A 1.4-kb fragment containing the TRP1 gene was amplified by PCR with the primers TRP1+ and TRP1− and inserted into the KpnI (blunted) site of pHT225, pHT256, pHT231, and EcoRI (blunted) site of pUC19 (NdeI−) to generate pHT244, pHT257, pHT232, and pHT258, respectively.
The 5.7-kb HindIII fragment containing the GPR1 gene and the 5.6-kb HindIII fragment containing the GPA2 gene were inserted into the HindIII site of pHT258 to obtain pYT098 and pYT099, respectively. A 1.1-kb HindIII (blunted) fragment containing URA3 from pCaU1 was also inserted into the BamHI-SacI site (blunted) of pYT098 and pYT099 to form pYT100 and pYT101, respectively.
The vectors pAS2-1 and pACT2 for the two-hybrid assay were purchased from Clontech. Plasmids for examining the interaction between Gpr1 and Gpa2 were constructed as follows. The fragment encoding the third cytoplasmic loop of Gpr1 (residues 301 to 433) was amplified by PCR with the primers GPR1-3+ and GPR1-3− to introduce NcoI and BamHI sites, and cloned into the SmaI site of pUC19. This NcoI-BamHI fragment was cloned into the NcoI-BamHI site of pAS2-1 to yield the plasmid pYT025. The fragment encoding the carboxyl-terminal tail of Gpr1 (residues 660 to 823) was amplified by PCR with primers GPR1-4+ and GPR1-4− to introduce NcoI and BamHI sites and cloned into the SmaI site of pUC19. The NcoI-BamHI fragment was cloned into the NcoI-BamHI sites of pAS2-1 to yield the plasmid pYT029. To obtain pYT030 for the expression of the GAL4 binding protein fused with the full Gpa2, a 2-kb NcoI-AflII (blunted) fragment from pHT200 was inserted into the NcoI-SmaI site of pACT2.
Strain construction.
The deletion of GPR1 alleles in strain CAI4 (15) was carried out by homologous recombination by a multiple-step procedure. To disrupt one of the GPR1 genes, pMS58 was linearized by SpeI digestion and transformed into C. albicans strain CAI4. Ura prototroph strains were selected on SD (Ura−) medium, and integration of the hisG-URA3-hisG cassette into the GPR1 allele was confirmed by PCR. To remove the URA3 gene by intrachromosomal hisG recombination, a small number of the transformed cells were spread onto SD medium containing 0.01% uridine and 0.1% 5′-fluoroorotic acid, and viable colonies were further analyzed by Southern blot analysis to obtain the strain TCA05. The other functional GPR1 gene was disrupted by repeating the same procedure to create a gpr1/gpr1 strain (TCA07). To delete the GPA2 genes, we carried out homologous recombination with a multiple-step procedure as described above with SphI-digested pTM247. As a result of the first recombination, we produced a GPA2/gpa2 strain (TCA08), and a gpa2/gpa2 strain (TCA12) was obtained after the second recombination. To create GPR1-overexpressing strains YTC014, YTC104, and YTC020, the plasmid pHT232 was digested with KpnI to linearize it and then introduced into strains CAI4, TCA07, and TCA12, respectively. Also, CAI4, TCA07, and TCA12 were transformed with pHT257 digested with KpnI to produce strains HTC7, YTC100, and HTC12, respectively, which overexpress the constitutively activated allele of the GPA2 gene. Strains CAI4, TCA05, TCA07, TCA08, and TCA12 were transformed with KpnI-digested pHT244, which integrates the URA3 gene into the TRP1 locus, to obtain YTC028, YTC068, YTC049, YTC072, and YTC032, respectively.
For the complementation test, the gpr1/gpr1 mutant (TCA07) and the gpa2/gpa2 mutant (TCA12) strains were transformed with KpnI-digested pYT100 and pYT101, which contained GPR1 and GPA2 genes with their native promoter, to obtain YTC108 and YTC109, respectively.
Microscopy.
Hypha formation in embedded conditions and cell morphology in liquid culture were examined by using ×4 and ×40 objective lenses, respectively, with a BX51 phase-contrast microscope (Olympus), and images were obtained with an Evolution LC digital camera system (Media Cybernetics). Colonial morphology was examined by using a VH-8000 Digital HF microscope with a VH-Z05 ×5-40 zoom lens (Keyence). Photographed images were processed by using Adobe Photoshop 7.0.
cAMP assay.
The procedure to determine the intracellular cAMP levels was as described previously (49). Briefly, C. albicans cells were inoculated in YPD at an optical density at 600 nm (OD600) of 0.1, grown at 30°C for 24 h, and then washed once with water and once with MES buffer (10 mM MES [morpholineethanesulfonic acid] containing 0.1 mM EDTA; pH 6). Cells were suspended with MES buffer to an OD600 of 8 and then incubated for 2 h at 30°C. Glucose was added to a final concentration of 2%, and 500-μl aliquots were taken from the culture before and after glucose addition at the indicated time points. Samples were transferred to 2-ml microcentrifuge tubes containing 0.5 g of glass beads and 500 μl of 10% trichloroacetic acid, briefly vortexed, and frozen immediately in liquid nitrogen. The samples were thawed on ice and sonicated under chilled conditions (twice at 130 W for 2.5 min) by using an INSONATOR 201 M (Kubota). After centrifugation, trichloroacetic acid was extracted four times with water-saturated ether. The cAMP content was measured with the BIOTRAK cAMP enzyme immunoassay system (Amersham Biosciences) according to the manufacturer's instructions.
Two-hybrid assay.
To examine the interactions between Gpr1 and Gpa2, a two-hybrid assay was performed by using MATCHMAKER Two-Hybrid System2 (Clontech). S. cerevisiae strain PJ69-4Aa/α (19) was cotransformed with plasmids in which the GAL4 activation domain or GAL4 binding domain was fused with Gpa2 or Gpr1 as shown in Table 2. Transformants were selected on SD (Leu− Trp−) medium. The interaction of the proteins was examined by both Ade prototrophy and a β-galactosidase assay.
Northern blot analysis.
C. albicans strains were grown under the indicated conditions, and total RNA was isolated by using a FastRNA Kit-Red (Bio 101). Each RNA sample was separated by electrophoresis on a 1% agarose gel containing formaldehyde and blotted onto a Hybond-N+ membrane (Amersham Biosciences) by capillary action. Hybridization and washing were performed as described previously (45). DNA probes for HWP1, ECE1, and ACT1 were amplified by PCR with the primers HWP1±, ECE1±, and ACT1±, respectively (Table 3). Each probe was purified with a Wizard SV column (Promega) and labeled with [32P]dATP by using a Strip-EZ DNA random priming kit (Ambion).
Virulence studies.
The virulence of the gpr1/gpr1 or the gpa2/gpa2 mutant strains was tested in a murine tail vein injection model. Strains were grown overnight in Sabouraud agar, and a light suspension of each was made in Sabouraud broth. One drop of this suspension was added to 50 ml of Sabouraud broth, which was then shaken at 250 rpm overnight at 35°C. Cells were washed three times, resuspended in phosphate-buffered saline, and counted with a hemocytometer. ICR outbred mice were inoculated by tail vein injection with ∼3 × 106 blastospores. There were 10 mice in each group; the groups included animals injected with YTC028, YTC049, YTC068, YTC032, YTC072, and HTC7 strains and were observed for survival. All mice were observed twice daily, and any animal that displayed lethargy or an inability to maintain grooming was sacrificed.
RESULTS
Isolation and characterization of C. albicans GPR1.
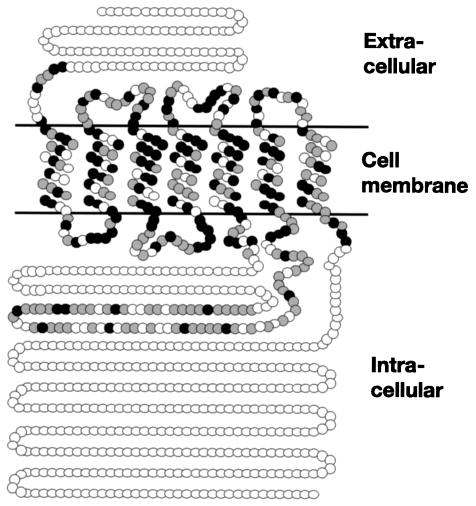
The C. albicans GPR1 gene was isolated from a genomic library from strain CAI4 as described in Materials and Methods. The GPR1 ORF was predicted to encode 823 amino acids with a calculated molecular mass of 92,418 Da. The deduced amino acid sequence shows seven putative transmembrane regions, indicating that it does belong to the GPCR family. Like Gpr1 in S. cerevisiae (47, 48), the C. albicans Gpr1 has a long third intracellular loop from amino acid positions 287 to 438 and also a long carboxyl-terminal tail in the cytoplasm from positions 496 to 823 (Fig. 1). The amino acid sequence of C. albicans Gpr1 showed 43% sequence identity with S. cerevisiae Gpr1 and 23% sequence identity with Git3 of Schizosaccharomyces pombe (Fig. 1). The transmembrane regions and latter half of the third cytoplasmic loop are well conserved in C. albicans Gpr1 and S. cerevisiae Gpr1, and these observations suggest that the two proteins probably have a closely related role in intracellular signaling.
FIG. 1.
Predicted two-dimensional model of C. albicans Gpr1. Closed circles indicate identical amino acids, and shaded circles indicate conserved amino acids compared to S. cerevisiae Gpr1. The amino acid positions of the transmembrane regions are as follows: 100 to 122, 135 to 157, 177 to 199, 220 to 242, 264 to 286, 439 to 458, and 473 to 495. The secondary structure of Gpr1 was analyzed by using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/).
GPR1 and GPA2 are required for morphogenesis in C. albicans in a cAMP-dependent manner.
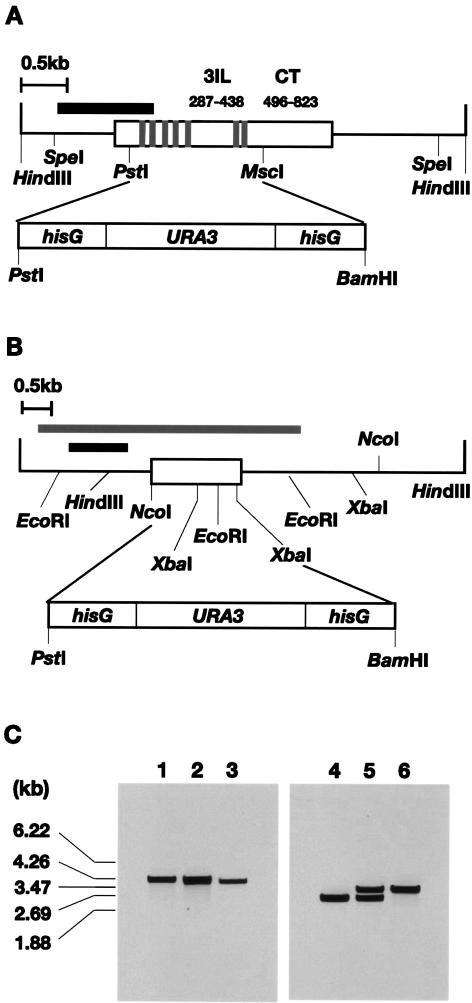
At first, we examined whether Gpr1 plays a role in the hyphal development in C. albicans. GPR1 and GPA2 genes were deleted in strain CAI4 by homologous recombination by using a multiple step procedure (Fig. 2A and B). The deletion of each gene was verified by Southern blot analysis (Fig. 2C). The wild type and the two mutant strains were grown on either solid FBS or Spider medium and colony morphology was observed (Fig. 3). On the solid FBS medium, the wild-type strain showed a heaved central region with filaments surrounded by a wide fringe of agar-invading filaments. In the gpr1 mutant (gpr1/gpr1) and gpa2 mutant (gpa2/gpa2) strains, the fringe was distinctly smaller than that of the wild-type strain, and no filaments were observed in the central region. This difference in phenotype between wild-type and mutant strains was especially apparent on the solid Spider medium. On this medium, the wild-type strain had a large wrinkled center with a fringe of filaments, whereas both mutant strains showed colonies with a flat and smooth surface. We observed that the gpa2 mutant strain had an even smoother surface than the gpr1 mutant. The reduction in the number of filaments in the mutant strains on FBS medium, as well as the wrinkling on Spider medium, was suppressed by 10 mM N6,2′-O-dibutyryl-cAMP (dbcAMP) (Fig. 3), which is a nonmetabolic derivative of cAMP. Also, the degree of hypha formation in embedded condition was examined. When cells were embedded in YPS agar matrix, the wild-type strain showed extensive filament formation but the mutant strains did not. Similarly, dbcAMP suppressed these defects, and the defects found in the mutant strains were also complemented by introducing corresponding genes with their native promoters (Fig. 3).
FIG. 2.
Deletions of GPR1 and GPA2 in C. albicans. (A) Restriction endonuclease map of GPR1. The white rectangle indicates the coding region of the gene. The predicted transmembrane regions (gray boxes), the third intracellular loop (3IL), and the cytoplasmic tail (CT) are indicated. The black box shows the amplified region of the probe for Southern blot analysis. (B) Restriction endonuclease map of GPA2. The white rectangle indicates the coding region of the gene. The amplified fragments of the probe for Southern blot analysis and the construction of the gene disruption cassette are indicated by the black box and the gray rectangle, respectively. (C) Southern blot analysis of wild-type and mutant strains. The genomic DNA samples were prepared from strains CAI4 (wild type; lanes 1 and 4), TCA05 (GPR1/gpr1; lane 2), TCA07 (gpr1/gpr1; lane 3), TCA08 (GPA2/gpa2; lane 5), and TCA12 (gpr1/gpr1; lane 6); digested with SpeI (left) or EcoRI (right); and probed with the GPR1 (left) or GPA2 (right) DNA fragment described above.
FIG. 3.
Defects in morphogenesis and hypha formation under solid or embedded conditions caused by deletion of the GPR1 and GPA2 genes. The wild-type strain YTC028 (WT), an isogenic strain with both alleles of GPR1 (YTC049; gpr1/gpr1) or of GPA2 (YTC032; gpa2/gpa2) deleted, and mutant strains with restored corresponding genes (YTC108 [gpr1/gpr1+GPR1] and YTC109 [gpa2/gpa2+GPA2]) were grown on solid YPD medium containing 10% FBS or solid Spider medium at 37°C for 7 days or grown in YPS agar matrix (Embedded) at 25°C for 54 h. The mutant strains (YTC049 [gpr1/gpr1] and YTC032 [gpa2/gpa2]) were also grown with the same medium containing 10 mM dbcAMP.
In contrast, when cells were grown in liquid FBS medium, hypha formation was observed in both the wild-type and the mutant strains at the same level (Fig. 4). These results indicate that neither GPR1 nor GPA2 is required for FBS-induced hypha formation in the liquid culture. However, when cells were grown in liquid Spider medium, reduced hypha formation was observed in the mutant strains (Fig. 4). The gpa2 mutant strain showed primarily a yeast form when incubated in liquid Spider medium for 6 h at 30°C or for 2 h at 37°C. On the other hand, the gpr1 mutant strain showed hypha formation similar to that of the wild-type strain after 2 h of incubation at 37°C and slightly fewer hyphae than the wild-type strain when incubated at 30°C for 6 h. Taken together, these results indicate that Gpr1 and Gpa2 are of importance to the morphogenesis of C. albicans mainly on solid medium. Although they are not required for FBS-induced hypha formation in liquid medium, a minor contribution to hypha formation was also observed in liquid Spider medium.
FIG. 4.
Defects in hypha formation in the liquid medium caused by deletion of the GPR1 and GPA2 alleles. The wild-type strain YTC028 and the isogenic strain with both alleles of GPR1 (YTC049; gpr1/gpr1) or of GPA2 (YTC032; gpa2/gpa2) deleted were grown in liquid YPD medium containing 10% FBS at 37°C for 2 h or grown in liquid Spider medium either at 37°C for 2 h or at 30°C for 6 h.
Genetic interaction between GPR1 and GPA2.
Since both GPR1 and GPA2 were shown to be involved in the hypha formation of C. albicans, we examined the genetic interaction between them. A dominant-active mutation that produces attenuated GTPase activity was introduced into the GPA2 gene by a mutation substituting Gln-355 with Leu (GPA2Q355L) (39). The GPA2Q355L and GPR1 genes were ligated under the ADH1 promoter to form overexpression vectors pHT257 and pHT232, respectively. These constructs or the vector only was introduced into either wild-type or mutant strains, and the hypha formation of each strain was observed under embedded conditions (Fig. 5). Filamentous growth in the wild-type strain was enhanced when either GPA2Q355L or GPR1 was overexpressed and GPA2Q355L had a stronger effect. The defect in hypha formation found in the gpr1 mutant was reversed by overexpression of GPR1 as well as GPA2Q355L. The defect in the gpa2 mutant was reversed by overexpression of GPA2Q355L but not GPR1. These results indicate that Gpa2 acts downstream of Gpr1 in the same signaling pathway, which is consistent with the previous finding that Gpa2 plays roles downstream of Gpr1 in S. cerevisiae (32, 47).
FIG. 5.
Epistasis analysis of GPR1 and GPA2. The wild-type (CAI4), gpr1/gpr1 (TCA07), and gpa2/gpa2 (TCA12) strains were transformed with the integration vector pHT244 containing the URA3 gene (control), pHT257 expressing the hyperactive GPA2Q355L allele under the control of the ADH1 promoter, or pHT232 expressing GPR1 under the control of the ADH1 promoter. The resulting strains were grown in embedded conditions at 25°C for 2 days.
Carboxyl-terminal tail of Gpr1 interacts with Gpa2.
Since our findings indicate that Gpr1 and Gpa2 are both involved in morphogenesis in C. albicans and genetic analysis revealed that Gpr1 plays roles upstream of Gpa2, we examined the physical interaction between Gpr1 and Gpa2 by using the yeast two-hybrid assay. A carboxyl-terminal region of Gpr1 (amino acids 660 to 823) fused with GAL4-BD interacted with Gpa2 fused with GAL4-AD (Fig. 6), as was reported in the case of S. cerevisiae (48). In this experiment, interaction was not observed between Gpa2 and the third intracellular loop of Gpr1 (amino acids 301 to 433) although it has been shown in S. cerevisiae (3, 48). The physical interaction between Gpr1 and Gpa2 strengthens their critical role and direct interaction in regulating filamentous growth in C. albicans.
FIG. 6.
A carboxyl-terminal region of Gpr1 interacts with Gpa2. S. cerevisiae two-hybrid strain PJ69-4Aa/α expressing BD-Gpr1 (301-433) or BD-Gpr1 (660-823) and AD-Gpa2 was grown on SD+ Ura+ Met+ His+ medium (Ade−) for 5 days at 30°C and then tested for β-galactosidase activity (β-gal). Control growth was on SD+ Ura+ Met+ His+ medium containing Ade. BD, DNA-binding domain of Gal4; AD, activating domain of Gal4.
GPR1 as well as GPA2 is required for a glucose-dependent cAMP spike.
Our results indicated that Gpr1 and Gpa2 regulate morphological switching and hypha formation in C. albicans in a cAMP-dependent manner. So what stimulates this pathway? Since it has been shown that both Gpr1 and Gpa2 are required for the glucose-stimulated increase in cAMP and play critical roles in pseudohyphal development through the cAMP pathway in S. cerevisiae, we examined whether Gpr1 and Gpa2 are also involved in glucose-dependent cAMP regulation in C. albicans. As shown in Fig. 7, when glucose was added to glucose-starved cells, an increase in the cellular cAMP level peaked at 1 min after glucose addition in the wild-type strain, whereas the peak was reduced to less than one-third in both the gpr1 and gpa2 mutant strains. This result indicates the involvement of Gpr1 and Gpa2 in the cAMP regulation in C. albicans and supports our observations that the defect in morphological change and hypha formation in the gpr1 and gpa2 mutant strains is suppressed by exogenous added dbcAMP (Fig. 3). It is hypothesized that Gpr1 plays a role as a glucose sensor coupled to Gpa2, which regulates both cellular cAMP levels and the morphological switching in C. albicans.
FIG. 7.
Gpr1 and Gpa2 are required for the glucose-dependent cAMP spike. The wild type (YTC028 [•]), gpr1/gpr1 mutant (YTC049 [▴]) and gpa2/gpa2 mutant (YTC032 [▪]) were grown in YPD medium for 24 h. Cells were washed and incubated in MES buffer at 30°C for 2 h and starved for glucose. Subsequently, glucose was added to a final concentration of 2%, and samples were taken at the indicated time points prior to (0 min) or after the addition of glucose and subjected to cAMP assay as described in Materials and Methods. Samples were assayed in duplicate and averaged. Error bars indicate the standard deviations for three experiments.
Gpr1 as well as Gpa2 regulates hypha-specific gene expression.
Several genes have been identified as hypha-specific genes in C. albicans. These genes include HWP1 (43) and ECE1 (5), which encode cell surface flocculin and a hypothetical protein, respectively, and their expression is controlled by cAMP-dependent mechanisms (9). Since Gpr1 and Gpa2 were shown to regulate hypha formation through the cAMP pathway, we examined whether they specifically regulated these hypha-specific genes. The wild type and the mutant strains were grown on either liquid or solid medium at 37°C for 2 h or 3 days, respectively, and the total RNA was obtained for Northern blot analysis.
When cells were grown in liquid FBS medium, mostly the same expression level of hypha-specific genes, HWP1 and ECE1, was observed in both wild-type and mutant strains (Fig. 8). This result is in good agreement with our observation that strong hypha formation was observed in both wild-type and mutant strains in the liquid FBS medium (Fig. 4). In the liquid Spider medium, although a high expression level of HWP1 and ECE1 was observed in wild-type and gpr1 mutant strains, it was slightly reduced in the gpa2 mutant strain (Fig. 8). Again, this result correlates with our observation that the gpa2 mutant strain shows an apparent defect in hypha formation in liquid Spider medium at 37°C, whereas the gpr1 mutant does not (Fig. 4).
FIG. 8.
Hypha-specific gene expression. Total RNA from the wild type (YTC028), the gpr1/gpr1 mutant (YTC049), and the gpa2/gpa2 mutant (YTC032) under various growth conditions was probed with HWP1, ECE1, and ACT1. For liquid cultures, cells were grown in either YPD, YPD plus 10% FBS, or Spider medium at 37°C for 2 h. For solid cultures, cells were grown on agar plates of YPD, YPD+FBS, or Spider medium at 37°C for 3 days. HWP1 and ECE1 were used as hypha-specific genes. ACT1 was used as an internal control. (A) Northern blot for wild-type and gpr1/gpr1 mutant strains. (B) The level of HWP1 and ECE1 transcripts under the indicated growth conditions in wild-type (□) and gpr1/gpr1 mutant (▨) cells was evaluated relative to that of the ACT1 transcript. (C) Northern blot for wild-type and gpa2/gpa2 mutant strains. (D) The level of HWP1 and ECE1 transcripts under the indicated growth conditions in wild-type (□) and gpa2/gpa2 mutant (▨) cells was evaluated relative to that of the ACT1 transcript.
When cells were grown on solid FBS and solid Spider medium, we could see a clear difference in hypha-specific gene expression between wild-type and mutant strains. Both mutant strains showed reduced levels of HWP1 and ECE1 gene expression compared to the wild-type strain (Fig. 8). This result also coincides with our findings that mutant strains were defective in both morphological change on solid media and hypha formation within the embedded conditions (Fig. 3). These results indicate that the Gpr1-Gpa2 signaling pathway regulates hyphal specific gene expression primarily on solid medium. Since these genes were strongly induced by FBS in the absence of Gpr1 and Gpa2, we hypothesize that an as-yet-unidentified sensing pathway is activated by FBS.
Gpr1 and Gpa2 are not required for virulence in C. albicans.
Our observation that Gpr1, as well as Gpa2, is required for morphological switching on solid and liquid Spider media but not in the liquid FBS medium led us to examine whether the Gpr1-Gpa2 signaling pathway is required for the virulence composite of C. albicans. We used an isogenic series of prototrophic mutants lacking one or both alleles of either the GPR1 or GPA2 gene or the mutant that overexpressed the constitutively activated allele of GPA2 under the control of the ADH1 promoter (ADH1prGPA2Q355L). Virulence studies were performed in mice with the wild-type strain (YTC028), the gpr1/gpr1 mutant strain (YTC049), the GPR1/gpr1 mutant strain (YTC068), the gpa2/gpa2 mutant strain (YTC032), the GPA2/gpa2 mutant strain (YTC072), or the ADH1prGPA2Q355L mutant strain (HTC7).
About 70% of the mice infected with the wild-type C. albicans strain succumbed to lethal infection by day 11 postinfection. The mice infected with mutant strains lacking either one or both alleles of GPR1 or GPA2 showed mostly the same survival rate as the wild-type strain (Fig. 9). These results indicate that GPR1 and GPA2 were not required for virulence in C. albicans in this invasive infection model. This result is well correlated with our findings that Gpr1 and Gpa2 were not required for FBS-induced hypha formation, as well as hypha-specific gene expression in the liquid FBS medium (Fig. 4 and 8). On the other hand, the ADH1prGPA2Q355L strain showed significantly reduced virulence. Of 10 animals infected with the ADH1prGPA2Q355L mutant strain, 7 survived until the experiment was terminated on day 20 postinfection. Our result that the ADH1prGPA2Q355L mutant had increased ability in hypha formation under embedded conditions but showed reduced virulence led us to examine the growth rate and hypha formation of the mutant strain in the presence of FBS.
FIG. 9.
Gpr1 and Gpa2 were not required for the virulence of C. albicans. The wild-type strain YTC028 (WT [♦]), the gpr1/gpr1 mutant YTC049 (▴), the GPR1/gpr1 mutant YTC068 (▪), the gpa2/gpa2 mutant YTC032 (✠), the GPA2/gpa2 mutant YTC072 (×), and the strain overexpressing the constitutively activated allele of GPA2 HTC7 (ADH1prGPA2Q355L [•]) were used to infect groups of ∼10 mice each by lateral tail vein injection, and survival was monitored over time.
The ADH1prGPA2Q355L mutant strain showed apparent delay in hypha formation in FBS medium.
Although wild-type and the ADH1prGPA2Q355L mutant strains did not show an obvious difference in growth rate either in the presence or in the absence of FBS (Fig. 10A), the ADH1prGPA2Q355L mutant strain showed apparent delay in hypha formation in the presence of FBS (Fig. 10B). The wild-type cells form hyphae after 2 h of incubation, whereas the mutant cells do not show long hyphae until 6 h of incubation. Although the mechanism of the delay in hypha formation in response to FBS found in the mutant strain is unknown, its delay might contribute to our finding that the virulence of the ADH1prGPA2Q355L mutant is attenuated (Fig. 9).
FIG. 10.
The ADH1prGPA2Q355L mutant strain showed an apparent delay in hypha formation in FBS medium. The wild-type and ADH1prGPA2Q355L mutant cells were pregrown in YPD medium at30°C overnight, transferred to the liquid FBS or YPD medium, and allowed to grow at 37°C. At the appropriate time point after transfer to FBS or YPD medium, the growth rate was monitored by measuring the absorbance at 600 nm (A), and the cell morphology in FBS medium was observed under a microscope (B).
DISCUSSION
A recent study suggested that Gpa2 is involved in the regulation of C. albicans hypha formation (39). Deletion of both alleles of GPA2 caused defects in morphogenesis in Spider medium, in SLAD medium, and in embedded conditions but not in a medium containing serum. In that study, based on the observation that the defect in hypha formation found in the gpa2 mutant was not reversed by exogenously added dbcAMP, it was concluded that Gpa2 is not involved in the cAMP signaling pathway in C. albicans (39). Although some of those results indicating that Gpa2 is required for hypha formation (39) are similar to the results presented here, the observation that the hyphal defects in gpa2 mutant were not suppressed by dbcAMP conflicts with the results of the present study. We present evidence that the Gpr1 receptor, as well as the Gpa2 Gα subunit, regulates morphogenesis in C. albicans through a cAMP-dependent mechanism. We showed that the mutations in both GPR1 and GPA2 produced defects in hypha formation and morphogenesis in C. albicans and that these defects were reversed by exogenous addition of dbcAMP (Fig. 3). We also demonstrated that GPR1 and GPA2 are required for a glucose-dependent increase in cAMP. Epistasis analysis revealed that Gpa2 acts downstream of Gpr1, and two-hybrid analysis showed binding between the C-terminal tail of Gpr1 and Gpa2. Moreover, Northern analysis revealed that the expression level of cAMP-regulated genes was reduced in the mutant strains; taken together, these data present a compelling case that Gpr1 and Gpa2 interact as a signaling complex through a cAMP pathway and regulate morphogenesis and hypha formation in C. albicans.
The impact of growth medium on this pathway for morphogenesis is clear. When grown on solid media that induce hypha formation, the gpr1 and the gpa2 mutant strains had dramatically reduced changes in morphogenesis, and in liquid Spider medium a reduced level of hypha formation was observed. In contrast, the difference in hypha formation between the wild type and mutant strain was not very clear in liquid FBS medium (Fig. 4), suggesting that the Gpr1-Gpa2 signaling pathway is specifically not required for FBS-induced hypha formation in a liquid medium. Recently, the roles in morphogenesis of two cAMP-dependent protein kinase isoforms, Tpk1 and Tpk2, have been examined (6). The tpk2 mutant did not show a major defect in hyphal morphogenesis on solid medium at 37°C, although a slight defect was observed at lower temperatures. In contrast, the tpk1 mutant was morphologically defective on both solid Spider and FBS media. In the liquid FBS and Spider media, tpk1 mutants showed mostly the same degree of hypha formation as the wild-type strain, whereas tpk2 mutants showed markedly reduced hyphal growth. Based on these findings, it was suggested that Tpk1 is required for morphogenesis mainly on solid hypha-inducing media, whereas Tpk2 plays important roles in hypha formation in liquid media (6). Since both the gpr1 and the gpa2 mutant strains showed defects in morphogenesis on solid media but normal hypha formation in liquid FBS medium, we hypothesize that the Gpr1-Gpa2 signaling pathway regulates hypha formation and morphogenesis through Tpk1 and not Tpk2.
Environmental stimuli, such as pH, temperature, serum, and nutrition, have been shown to prominently affect the hypha formation in C. albicans (36). A serum factor has been shown to be a strong hyphal inducer for C. albicans. Although previously the cAMP signaling pathway was reported to be required for FBS-activated hypha formation, our results showed that Gpr1 and Gpa2 were not required for FBS-induced hypha formation in the liquid medium and excluded FBS as an environmental stimulus that activates the Gpr1-Gpa2 signaling pathway. However, a difference in colony morphology and hypha formation was observed in the mutant strains grown on Spider medium, and this finding suggests that other nutrients might be candidates for the external signal. In fact, we found that both Gpr1 and Gpa2 are required for a glucose-dependent cAMP spike in C. albicans (Fig. 7), and thus glucose is the signal that activates morphological switching through the Gpr1-Gpa2 pathway. However, we could also see clear morphological differences when cells were embedded in the YPS agar with sucrose also as a carbon source (Fig. 3 and 5). Since previous studies indicated that the Gpr1-Gpa2 signaling pathway is also activated by other fermentable sugars such as fructose (32, 49), maltose (49), and sucrose (32) in S. cerevisiae, it is quite possible that sucrose activates this pathway in C. albicans.
The Gpr1-Gpa2 signaling pathway is conserved in both the budding yeast S. cerevisiae and the fission yeast S. pombe. In S. cerevisiae, this pathway was shown to regulate pseudohyphal development, as well as invasive growth (25, 31, 32, 45), suggesting that a similar regulating mechanism may exist in C. albicans. We have also shown that this pathway regulates glucose-dependent cell size in S. cerevisiae (unpublished data). In the fission yeast, many genes, including homologues of Gpr1 (git3) and Gpa2 (gpa2/git8), have been identified by analyzing mutations in the genes that confer the constitutive expression of the fructose-1,6-bisphosphatase gene fbp1 (18, 46). The mutation of these git (for glucose-insensitive transcription) genes was shown to derepress fbp1 transcription, starvation-independent conjugation and sporulation, and shortening of the major axis, all of which can be suppressed by exogenous cAMP (11, 17).
The cAMP pathway is also conserved in pathogenic fungi. Cryptococcus neoformans is one of the most common opportunistic pathogens infecting AIDS patients (33) in the human central nervous system. In C. neoformans, Gα Gpa1 was shown to regulate mating and the production of a virulence factor via a cAMP-dependent mechanism (2), and the involvement of PKA was also shown (13). In the plant pathogen Ustilago maydis, the homologous Gα protein Gpa3 controls the cAMP signaling that is necessary for growth as a budding yeast, and dominant activation of Gpa3 impairs virulence (23, 24, 37). Despite the conserved cAMP signaling pathway related to pathogenicity, the receptor coupled with these Gα subunits has not yet been identified in these pathogenic fungi. In the present study, we present evidence that the Gpr1-Gpa2 signaling pathway regulates morphogenesis and hypha formation through the cAMP pathway in C. albicans, but virulence is not affected by this pathway.
The morphological change has been associated with virulence in C. albicans. Early studies on a morphogenesis-deficient mutant in C. albicans suggested the importance of the hyphal form, since mutants unable to form filaments are avirulent (30, 44). However, characterization of the tup1 mutant, which is constitutively filamentous but also avirulent (7, 8), has indicated that the ability to switch from the yeast to hyphal form is probably critical for virulence (16). A recent study using C. albicans cells in which the NRG1 gene (a negative regulator of filamentation) was expressed under the control of a tetracycline-regulatable promoter revealed distinct roles for yeast and filamentous forms during infection (34). Our result that Gpr1 and Gpa2 are not required for virulence in C. albicans is consistent with our findings that they are not required for FBS-induced hypha formation in liquid medium (Fig. 4). The ADH1prGPA2Q355L mutant strain showed an apparent delay in hypha formation in the liquid FBS medium despite enhanced hypha formation when embedded in the agar matrix. These observations suggest that there may be at least two independent signaling pathways: one that regulates filamentation in liquid FBS medium and one that regulates filamentation in the embedded condition. The mechanism of the delay in FBS-induced hypha formation in the mutant strain is unclear, but this delay may explain why this mutant strain showed reduced virulence.
Acknowledgments
We thank Toshiyuki Mio for Candida strains, plasmids, and fruitful discussions and Toshiaki Harashima and Joseph Heitman for two-hybrid strains and valuable suggestions.
This study was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan and the New Energy and Industrial Technology Department Organization. Y.T. is supported by the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari, K., S. Martin, M. Farkasovsky, I. M. Ehbrecht, and H. Kuntzel. 1999. Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J. Biol. Chem. 274:30052-30058. [DOI] [PubMed] [Google Scholar]
- 4.Bertram, G., R. K. Swoboda, G. W. Gooday, N. A. Gow, and A. J. Brown. 1996. Structure and regulation of the Candida albicans ADH1 gene encoding an immunogenic alcohol dehydrogenase. Yeast 12:115-127. [DOI] [PubMed] [Google Scholar]
- 5.Birse, C. E., M. Y. Irwin, W. A. Fonzi, and P. S. Sypherd. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bockmuhl, D. P., S. Krishnamurthy, M. Gerads, A. Sonneborn, and J. F. Ernst. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243-1257. [DOI] [PubMed] [Google Scholar]
- 7.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 9.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, D. H., Jr., A. D. Giusani, X. Chen, and C. A. Kumamoto. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651-662. [DOI] [PubMed] [Google Scholar]
- 11.Byrne, S. M., and C. S. Hoffman. 1993. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 105(Pt. 4):1095-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csank, C., K. Schroppel, E. Leberer, D. Harcus, O. Mohamed, S. Meloche, D. Y. Thomas, and M. Whiteway. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect. Immun. 66:2713-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 14.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gow, N. A., A. J. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366-371. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman, C. S., and F. Winston. 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 5:561-571. [DOI] [PubMed] [Google Scholar]
- 18.Isshiki, T., N. Mochizuki, T. Maeda, and M. Yamamoto. 1992. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 6:2455-2462. [DOI] [PubMed] [Google Scholar]
- 19.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly, R., S. M. Miller, M. B. Kurtz, and D. R. Kirsch. 1987. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol. Cell. Biol. 7:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler, J. R., and G. R. Fink. 1996. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl. Acad. Sci. USA 93:13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraakman, L., K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde, and J. M. Thevelein. 1999. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol. Microbiol. 32:1002-1012. [DOI] [PubMed] [Google Scholar]
- 23.Kruger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 260:193-198. [DOI] [PubMed] [Google Scholar]
- 24.Kruger, J., G. Loubradou, G. Wanner, E. Regenfelder, M. Feldbrugge, and R. Kahmann. 2000. Activation of the cAMP pathway in Ustilago maydis reduces fungal proliferation and teliospore formation in plant tumors. Mol. Plant-Microbe Interact. 13:1034-1040. [DOI] [PubMed] [Google Scholar]
- 25.Kubler, E., H. U. Mosch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272:20321-20323. [DOI] [PubMed] [Google Scholar]
- 26.Kurjan, J. 1993. The pheromone response pathway in Saccharomyces cerevisiae. Annu. Rev. Genet. 27:147-179. [DOI] [PubMed] [Google Scholar]
- 27.Leberer, E., D. Harcus, I. D. Broadbent, K. L. Clark, D. Dignard, K. Ziegelbauer, A. Schmidt, N. A. Gow, A. J. Brown, and D. Y. Thomas. 1996. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hypha formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 93:13217-13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schroppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 29.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hypha formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 30.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS: 100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama, H., T. Mio, S. Nagahashi, M. Kokado, M. Arisawa, and Y. Aoki. 2000. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Canidida albicans. Infect. Immun. 68:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neer, E. J. 1995. Heterotrimeric G proteins: organizers of transmembrane signals. Cell 80:249-257. [DOI] [PubMed] [Google Scholar]
- 36.Odds, F. C. 1985. Morphogenesis in Candida albicans. Crit. Rev. Microbiol. 12:45-93. [DOI] [PubMed] [Google Scholar]
- 37.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Bolker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 16:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocha, C. R., K. Schroppel, D. Harcus, A. Marcil, D. Dignard, B. N. Taylor, D. Y. Thomas, M. Whiteway, and E. Leberer. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Martinez, C., and J. Perez-Martin. 2002. Gpa2, a G-protein alpha subunit required for hyphal development in Candida albicans. Eukaryot. Cell 1:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman, F. 1991. Getting started with yeast, vol. 194. Academic Press, Inc., San Diego, Calif.
- 41.Sonneborn, A., D. P. Bockmuhl, M. Gerads, K. Kurpanek, D. Sanglard, and J. F. Ernst. 2000. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 35:386-396. [DOI] [PubMed] [Google Scholar]
- 42.Sprague, G. F., Jr., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, vol. 2. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 43.Staab, J. F., C. A. Ferrer, and P. Sundstrom. 1996. Developmental expression of a tandemly repeated, proline- and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans. J. Biol. Chem. 271:6298-6305. [DOI] [PubMed] [Google Scholar]
- 44.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamaki, H., T. Miwa, M. Shinozaki, M. Saito, C. W. Yun, K. Yamamoto, and H. Kumagai. 2000. GPR1 regulates filamentous growth through FLO11 in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 267:164-168. [DOI] [PubMed] [Google Scholar]
- 46.Welton, R. M., and C. S. Hoffman. 2000. Glucose monitoring in fission yeast via the Gpa2 gα, the git5 Gβ and the git3 putative glucose receptor. Genetics 156:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17:1996-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yun, C. W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1997. G-protein coupled receptor from yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 240:287-292. [DOI] [PubMed] [Google Scholar]
- 49.Yun, C. W., H. Tamaki, R. Nakayama, K. Yamamoto, and H. Kumagai. 1998. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 252:29-33. [DOI] [PubMed] [Google Scholar]