Abstract
Site-directed spin labeling (SDSL) in combination with Electron Paramagnetic Resonance (EPR) spectroscopy is a well-established method that has recently grown in popularity as an experimental technique, with multiple applications in protein and peptide science. The growth is driven by development of labeling strategies, as well as by considerable technical advances in the field, that are paralleled by an increased availability of EPR instrumentation. While the method requires an introduction of a paramagnetic probe at a well-defined position in a peptide sequence, it has been shown to be minimally destructive to the peptide structure and energetics of the peptide-membrane interactions.
In this chapter, we describe basic approaches for using SDSL EPR spectroscopy to study interactions between small peptides and biological membranes or membrane mimetic systems. We focus on experimental approaches to quantify peptide-membrane binding, topology of bound peptides, and characterize peptide aggregation. Sample preparation protocols including spin-labeling methods and preparation of membrane mimetic systems are also described.
Keywords: Peptide binding, membrane mimetics, accessibility EPR experiments, peptide aggregation, binding isotherms
1. Introduction
Biological activity of many peptides arises from the targeted interactions with cellular membranes. Examples of naturally occurring peptides that bind to biological membranes include bacterial toxins and antimicrobial peptides. These naturally occurring peptides inspired an intensive search for additional natural and synthetic peptides with enhanced cell surface binding properties for more efficient drug targeting, including cancer therapeutics (Aina, Sroka, Chen, & Lam, 2002; Boohaker, Lee, Vishnubhotla, Perez, & Khaled, 2012; Torchilin, 2008). The rational design of such membrane-active peptides, and the needs for understanding the peptide biological functions, stimulated further research on structural changes in the peptide upon membrane binding and elucidating the mode of peptide interactions with the membranes. Currently, a number of experimental biophysical methods are available to investigate the peptide membrane interactions, including not only the binding coefficients, but also orientation and immersion depth of peptides in biological membranes or membrane-mimetic systems. Those methods mainly include solution and solid state nuclear magnetic resonance (NMR), spin labeling electron paramagnetic resonance (EPR), fluorescence spectroscopy, infrared- and oriented circular dichroism spectroscopy, and small angle neutron scattering (SANS). Spin-labeling EPR spectroscopy offers an informative and convenient way to probe interactions of small peptides with biological membranes and to determine the secondary structure and peptide location and orientation in membrane systems. Although spin-labeling EPR approaches discussed here are fully applicable for studies of membrane proteins and complexes, this chapter is focused primarily on experimental methods and data interpretation for binding and association of small peptides with biological membranes and their mimetics.
2. Peptide labeling with EPR active probes and preparation of membrane mimetic systems
Natural and synthetic peptides are EPR-silent (unless chelated to paramagnetic metal ion(s)) and, therefore, labeling of peptides by covalent modification of natural side-chains represents the necessary step in the sample preparation. In this Chapter we restrict our discussion to labeling with stable organic free radicals and, particularly, nitroxides. From a chemical perspective, one can introduce a new unnatural chemical moiety (a nitroxide) through a) peptide synthesis using an unnatural amino acid, b) covalent modification of a natural side chain, and c) covalent modification of an unnatural amino acid. Currently, the first two approaches are more common and will be discussed in further detail.
2.1 Spin-labeling of peptides by covalent modification of natural amino acid side-chains
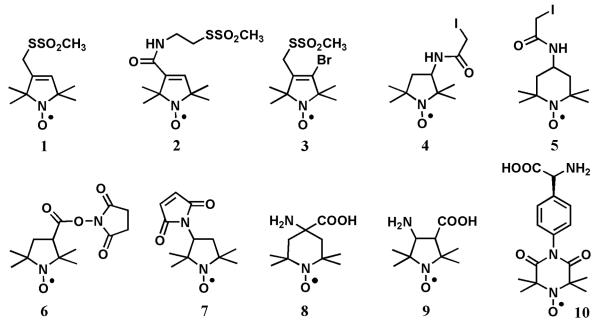
Peptides are generally more stable than proteins and, therefore, are more amenable to chemical modifications including covalent attachment of nitroxides. While site-directed spin-labeling of proteins typically relies on an incorporation of a unique amino acid (cysteine) and the highly specific reactivity of its sulfhydryl group with a methanethiosulfonate-derivatized nitroxide, one can easily employ other chemistries for peptide labeling. For example, the terminal hydroxyl at the peptide C-terminus can be labeled using a modification of the mild one-pot esterification method by Hassner and Alexanian (Hassner & Alexanian, 1978) as was demonstrated by Dzikovski et al. for spin-labeling of a pentadecapeptide antibiotic gramicidin A (Dzikovski, Borbat, & Freed, 2004). Amino groups of a peptide can be readily modified by a reaction with succinimidyl-2,2,5,5-tetramethyl-3-pyrroline-1-oxyl-3-carboxylate (SSL, Figure 1, compound 6). If a peptide has several amino groups, the multiple products of the reaction carried out without excess of the labeling reagent can be separated by HLPC, yielding selectively labeled peptide adducts (e.g., see (Altenbach & Hubbell, 1988)).
Figure 1.
Chemical structures of the representative spin labels: Compound 1, (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate [MTSL]; Compound 2, (1-oxyl-2,2,5,5-tetramethylpyrroline-3-yl)carbamidoethyl methanethiosulfonate; Compound 3, 4-bromo-(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate; Compound 4, 3-(2-iodoacetamido)-2,2,5,5-tetramethyl-1-pyrrolidinyl-1-oxyl [3-(2-iodo acetamido)-PROXYL]; Compound 5, 4-(2-iodoacetamido)-2,2,6,6-tetramethylpiperidine 1-oxyl [4-(2-iodoacetamido)-TEMPO]; Compound 6, 1-oxyl-2,2,5,5-tetramethyl-1-pyrrolidi nyl-3-carboxylate N-hydroxysuccinimide ester; Compound 7, 3-maleimido-2,2,5,5-tetramethyl-1-pyrrolidinyl-1-oxyl [3-maleimido-PROXYL]; Compound 8, 4-amino-4-carboxy-2,2,6,6-tetramethylpiperidine 1-oxyl [TOAC]; Compound 9, 3-amino-1-oxyl-2,2,5,5- tetramethylpyrrolidine-4-carboxylic acid[POAC];and Compound10, 4-(3,3,5,5-tetramethyl-2,6-dioxo-4-oxylpiperazin-1-yl)-l-phenylglycine [TOPP].
The most common method of a site-specific incorporation of a nitroxide into a peptide, however, is based on covalent modification of the Cys sulfhydryl group. Similar to the protein labeling, the essential prerequisite of the sample preparation involves site-directed mutations of the peptide sequence to replace the unwanted native cysteines with another amino acid (typically Ser) and the native amino acid with Cys at desired site(s). Then peptides of the desired sequences are prepared synthetically or expressed in vitro. In order to attain more complete structural information, one may replace the native amino acids in the peptide sequence by cysteine in a systematic way to obtain a library of structurally similar spin-labeled peptides. Such a systematic approach is known as a “nitroxide scan” of a peptide or a protein domain.
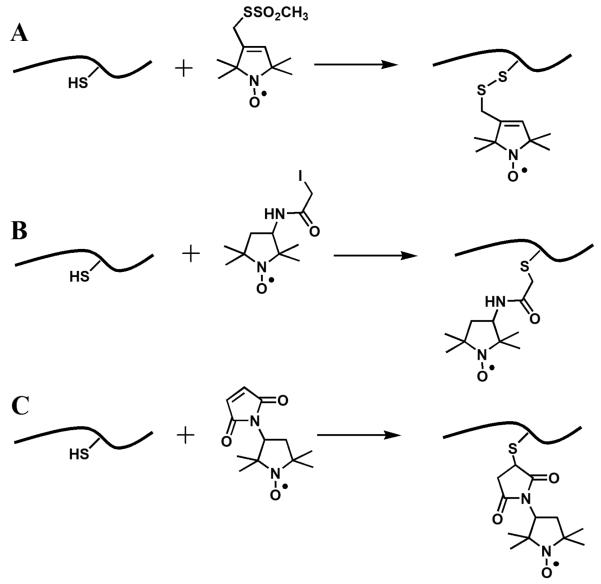
A number of sulfhydryl-reactive nitroxide labels are available commercially. The structures of the most common labels, (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate (MTSL, Figure 1, 1) (Berliner, Grunwald, Hankovszky, & Hideg, 1982), 3-maleimido-2,2,5,5-tetramethyl-1-pyrrolidinyl-1-oxyl (maleimido-proxyl, MSL, Figure 1, 7) (Griffith & McConnel.Hm, 1966), and 3-(2-iodoacetamido)-2,2,5,5-tetramethyl-1-pyrrolidinyl-1-oxyl (iodoacetamido-proxyl, IAP, Figure 1, 4) (Ogawa & McConnel.Hm, 1967; Ogawa, McConnel.Hm, & Horwitz, 1968) and the structures of the resulting attachment linkers are shown in Figures 1 and 2. MTSL (sometimes abbreviated as MTSSL) is the label that is used the most often. The label was first described by Berliner et al. as a reagent to quantify accessible thiols in proteins because of the reversibility of the covalent modification of sulfhydryl groups (Berliner, et al., 1982). Nowadays, the label is primarily used in site-directed spin-labeling EPR because of its high specificity to Cys. The sidechain formed upon the labeling reaction is comparable in molecular volume to phenylalanine or tryptophan, and is commonly abbreviated in literature as R1. A number of methanethiosulfonate spin labels with varied length and structure of the tether were synthesized including commercially available (1-oxyl-2,2,5,5-tetramethylpyrroline-3-yl)carbamidoethyl methane-thiosulfonate (MTS-4-oxyl, Figure 1, 2) and 4-bromo-(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate (4-bromo-MTSL, Figure 1, 3) (Pistolesi, Pogni, & Feix, 2007).
Figure 2.
Structures of the nitroxide side chains produced by reactions of the sulfhydryl group of a cysteine residue with common spin labels. (A) (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate [MTSL], 1; (B) 3-(2-iodoacetamido)-2,2,5,5-tetramethyl-1-pyrrolidinyl-1-oxyl [3-(2-iodoacetamido)-PROXYL], 4; and (C) 3-maleimido-2,2,5,5-tetramethyl-1-pyrrolidinyl-1-oxyl [3-maleimido-PROXYL], 7.
While experimental protocols for covalent modifications of the native amino acids are well-developed and easily reproducible in a lab, the method has some disadvantages. The main disadvantage is in a large conformational space accessible to the reporting NO group. The distance between Cβ and NO group varies between 4 and 8 Å, depending on the label and its conformation, introducing an uncertainty in the position with respect to the peptide backbone and, therefore, affecting data interpretation. A recently-introduced label with two MTS attachment groups reduces the uncertainty in the nitroxide position and is advantageous for distance measurements in proteins. This attachment, however, also can make the peptide structure potentially more rigid and requires two Cys mutations per one spin label attached (Fleissner et al., 2011).
2.1.1 Spin-labeling of peptides by MTSL
For labeling a peptide with MTSL, a stock solution of the label is prepared at 100 to 200 mM concentration in acetonitrile (ACN); however, ethanol (EtOH) or DMSO can also be used as solvents. The stock solution should be kept at or below −20 °C and protected from light. The label from the stock solution is added to a peptide solution at 5- to 10-fold molar excess, gently mixed, and then incubated at room temperature from 1 to 3 hrs, often followed by labeling overnight at 4 °C. With small water-soluble peptides, we were able to achieve >95% cysteine modification by labeling at room temperature for just 1 hr. The unreacted spin label is then separated by HPLC or other methods depending on the nature of the peptide. For some sample preparations, a detachment of MTSL tag is observed upon storage, especially at pH above 8.0. The X-band (9 GHz) EPR spectrum of a free nitroxide label consists of three sharp lines corresponding to each of the three 14N hyperfine transitions. The lines have almost equal peak-to-peak intensities expected for a nitroxide in the fast motional narrowing regime. The EPR signal from free label would overlap with the signal from the labeled peptide in solution and, thus, complicate, for example, the analysis of the membrane binding experiments. The signal from the free label can be difficult to accurately account for, especially in case of small, highly dynamic peptides because of very similar EPR spectra at X-band. This consideration highlights the need for a careful separation of the free label from the labeled peptide.
2.1.2 Spin-labeling of peptides by maleimido- and iodo-derivatives of nitroxides
MSL and IAP nitroxide tags have some advantages over the methanethiosulfonate label for EPR experiments carried out under mild reducing conditions that can lead to the cleavage of the disulfide bond formed by the latter label. Reaction with maleimides is highly specific to Cys at pH 6 to 7.5. At pH above 8, however, the maleimide label can react with the primary amines and modify the lysine side chain or the amino group of the N-terminus (Brewer & Riehm, 1967; Steinhoff, Dombrowsky, Karim, & Schneiderhahn, 1991).
It should be noted that all common reducing agents, such as tris(2-carboxyethyl)phosphine (TCEP), dithiothreitol (DTT), or mercaptoethanol that are widely used in peptide and protein preparations to prevent Cys oxidation would have to be removed from the peptide solution prior to the reaction with a spin label. Both DTT and TCEP interfere with the labeling by maleimide and methanethiosulfonate reactants, and, to lesser extent, with iodoacetamido derivatives, and the inhibition is more pronounced with DTT (Getz, Xiao, Chakrabarty, Cooke, & Selvin, 1999; D. E. Shafer, Inman, & Lees, 2000). In addition, the presence of reducing agents, even at relatively low concentrations, results in a loss of EPR signal (Getz, et al., 1999). TCEP was shown to react with nitroxides more slowly than DTT, however, these measurements were carried out at 0 °C (Getz, et al., 1999), whereas the majority of EPR studies of biomolecules are conducted at physiological temperatures, which results in faster nitroxide reduction.
2.2 Spin labeling by inserting unnatural amino acid via peptide synthesis
A number of paramagnetic unnatural amino acids were reported in the literature (M. Balog et al., 2003; M. R. Balog et al., 2004; Kalai, Schindler, Balog, Fogassy, & Hideg, 2008; Tominaga et al., 2001; Wright et al., 2007). The most commonly used one is 2,2,6,6-tetramethyl-N-oxyl-4-amino-4-carboxylic acid (TOAC, Figure 1, 8) (Rassat & Rey, 1967; Schreier, Bozelli, Marín, Vieira, & Nakaie, 2012; A. M. Shafer, Nakaie, Deupi, Bennett, & Voss, 2008; Thomas et al., 2005). Typically, TOAC is inserted into a peptide via solid-phase peptide synthesis. Being incorporated into peptides through a peptide bond, TOAC is an achiral and very rigid cyclic molecule with only one degree of freedom - the conformation of the six-member ring. Because of the rigidity and a close position of the nitroxide moiety to the peptide backbone, TOAC has been very useful in studies of backbone dynamics, peptide secondary structure and the orientation of peptides with respect to the membrane normal (Anderson et al., 1999; Ghimire et al., 2012; Marsh, Jost, Peggion, & Toniolo, 2007; McNulty & Millhauser, 2000; Newstadt, Mayo, Inbaraj, Subbaraman, & Lorigan, 2009; Sahu et al., 2014; A. M. Shafer, et al., 2008). However, one cannot exclude a possibility of a distortion of the α-helical structure by the presence of this structurally rigid unnatural amino acid (Elsasser, Monien, Haehnel, & Bittl, 2005). Another disadvantage of TOAC is in a relatively low coupling yield during the peptide synthesis that is caused by a steric hindrance. An alternative beta-amino acid, POAC, (2,2,5,5-tetramethylpyrrolidine-N-oxyl-3-amino-4-carboxylic acid, Figure 1, 9) was shown to provide higher coupling yields compared to TOAC (Tominaga, et al., 2001). Incorporation of another chiral amino acid, TOPP, (4-(3,3,5,5-tetramethyl-2,6-dioxo-4-oxylpiperazin-1-yl)-l-phenylglycine, Figure 1, 10) into peptide had been shown to not perturb the peptide structure in solutions (Stoller, Sicoli, Baranova, Bennati, & Diederichsen, 2011), thus, providing another promising tool for structural studies of peptides and proteins.
2.3 Membrane mimetic: liposomes preparation
While both multilamellar and unilamellar vesicles have been widely used in studies of peptide-bilayer interactions, unilamellar liposomes are generally preferred because of a capability of providing control over the available binding surface and, to some extent, the bilayer curvature. Traditionally, unilamellar vesicles with diameters up to 100 nm are classified as small unilamellar vesicles (SUVs), from 100 nm to few μm as large unilamellar vesicles (LUVs), and larger vesicles, typically with an average diameter of 100 μm are called giant unilamellar vesicles (GUVs). The use of GUVs in EPR is impractical because of a low average lipid concentration in the sample.
A typical liposome preparation protocol involves making stock solutions of lipids in chloroform or other suitable solvents and mixing the solutions to achieve the desired bilayer lipid composition. Organic solvents are then removed at room temperature using a round bottom flask connected to a rotary evaporator to form a thin lipid film on the flask wall. If the solution volume is small, the solvents can be removed under a flow of dry nitrogen gas. The residual organic solvent should be removed from the lipid film by a vacuum pump operated for a few hours or overnight. The lipid film is then hydrated with a buffer by vortexing at a temperature above the gel-liquid crystal phase transition temperature, Tm, of the lipid bilayer. Hydrated lipids are then freeze-thaw cycled by dipping the sample vial into liquid nitrogen and warming it up to a temperature above Tm, up to ten times. This procedure results in a formation of multilamellar vesicles (MLVs).
SUVs with diameters in the range of 15-50 nm can be formed by high power sonication of the MLVs, until the aqueous solution becomes clear (Huang, 1969; Yamaguchi, Nomura, Matsuoka, & Koda, 2009). Depending on the lipid and vesicle diameter, the outer leaflet of the SUV bilayer contains about 60% of lipid molecules (Michaels.Dm, Horwitz, & Klein, 1973). SUVs are not as stable as the large unilamellar vesicles (LUV) and, therefore, should be used in EPR or other experiments as soon as they are prepared, because even charged vesicles may fuse to form larger aggregates.
SUVs with diameters greater than 40-50 nm and LUVs are typically prepared by extrusion of MLVs through two stacked polycarbonate filters of a specific pore diameter using an extruder maintained at a temperature above Tm (Traikia, Warschawski, Recouvreur, Cartaud, & Devaux, 2000). Commercial liposome extruders are available from Avanti Polar Lipids, Inc. (Alabaster, AL) and Northern Lipids, Inc. (Burnaby, BC, Canada). Typically, up to ten passages through the filters are carried out to ensure the best achievable size distribution that is verified by dynamic light scattering (DLS). In our experiments with DOPC, DOPC/DOPG, DOPC/DOPS mixed liposomes, an extrusion through 100 nm pores produces LUVs with mean diameters from 120 to 135 nm, depending on the lipid composition. The final lipid concentration after the extrusion can be determined by microdetermination of phosphorus (P. S. Chen, Toribara, & Warner, 1956; Itaya, 1966 ). LUVs should be stored under argon at temperatures above the Tm of the lipids to avoid rapid vesicle fusion (Larrabee, 1979). Under such conditions, LUVs have an excellent storage stability of up to a few days (Lasic, 1988; Lasic & Martin, 1990), 1:1 leaflet lipid stoichiometry, and the lipid lateral packing density close to that of eukaryotes’ membranes. All these features make LUVs an excellent model system for studies of membrane-peptide interactions.
3. EPR measurements of membrane peptide binding and evaluation of partition coefficients
3.1 Experimental considerations
The general methodology for quantifying binding of spin-labeled (SL) peptide to lipid bilayer membranes is well established. Detection of the peptide binding relies on sensitivity of the nitroxide EPR spectrum to rotational diffusion of the probe. Typically, peptide binding experiments are carried out using conventional X-band (9.5 GHz) spectrometers. Continuous wave (CW) X-band EPR spectra of nitroxides reflect both the rotational anisotropy and correlation time of the label motion at the ps to ns timescale, making this method suitable for characterization of protein secondary structure and dynamics (Hubbell, Gross, Langen, & Lietzow, 1998; Hubbell, McHaourab, Altenbach, & Lietzow, 1996; McHaourab, Lietzow, Hideg, & Hubbell, 1996). X-band CW EPR spectra from SL-peptides in aqueous solutions fall into a fast motional regime with characteristic rotational correlation time of the label in the sub-ns range, regardless of whether the peptide is unstructured or possesses secondary structure. Upon membrane binding, the rotational motion of the peptide and the label become restricted, resulting in an EPR spectrum that is easily distinguishable from the one corresponding to the free peptide in the aqueous phase.
Binding titrations are typically carried out by adding lipids to about 100 to 200 μl sample of a peptide solution. A small volume of peptide/lipid solution, typically about 5 μl, is drawn into a quartz or a glass capillary with inner diameter (i.d.) of 0.5-1.0 mm (or less) that is placed into an EPR resonator. After recording the spectra, the sample is unloaded back into the bulk peptide/lipid mixture, a new portion of lipids is added and a new sample is drawn for EPR spectroscopy. Alternatively, a quartz capillary with plunger, that allows for drawing in and pushing out the sample without removal of the capillary from the resonator can be used (Victor & Cafiso, 2001). If the amount of the available peptide and lipids is not an issue, the samples for each titration point can be prepared separately with a given concentration of the peptide and a varied concentration of the lipids.
Temperature stabilization is essential in the peptide binding experiments, as its variations would not only affect the peptide binding equilibrium to the lipid bilayer, but also the line shape of the EPR spectra. The latter might cause errors in determination of the fractions of free and bound peptide. In our NCSU laboratory for measurements using a Varian Century Series X-band EPR spectrometer (Palo Alto, CA) the temperature is stabilized better than ±0.01 °C using an in-house-built variable temperature accessory comprising from a digitally controlled circulator bath that pumps fluid through high-efficiency aluminum radiators attached to an EPR resonator (Alaouie & Smirnov, 2006). Variable temperature systems capable of temperature stabilization in the physiologically relevant range are commercially available from Bruker.
Peptide concentration in solution should be kept below the critical micelle concentration (CMC). In order to determine the CMC of a peptide, the amplitude of the EPR signal, typically the peak-to-peak amplitude of the central nitrogen hyperfine component, mI=0, is measured as a function of the peptide concentration. Below the CMC, when the peptide is monomeric, the dependence is linear with zero intercept. Above the CMC, the peak-to-peak amplitude of the sharp, three-line spectra becomes essentially concentration-independent. Upon micellization of the peptide, spin-labels get in close contact, providing conditions for strong exchange and dipole-dipole interaction between the electron spins. As a result, the EPR signal from the peptide in a micelle will appear as a very broad single line that would not contribute significantly to the intensity of the central peak of the sharp free component.
Figure 3 illustrates typical changes in X-band CW EPR spectra from a small unstructured peptide observed upon binding to LUVs. NOD3 peptide with amino acid sequence, KKKKKKKFFC(R1)F, was labeled with a threefold excess of MTSL for 4 h at room temperature and separated from the unreacted spin label by a gradient HPLC. EPR spectra from 50 μM NOD3-R1 in a buffer (100 mM MOPS containing 100 mM KCl at pH=7.0), and in the presence of LUVs formed from a mixture of 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphocholine (POPC) and 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-L-serine (POPS) in 24:1 molar ratio (total lipid concentration of 8 mg/mL) are shown in Figure 3.
Figure 3.
Changes in room temperature X-band (9 GHz) CW EPR spectra from 50 μM MTSL-labeled NOD3 peptide upon binding to POPC:POPG (24:1 molar ratio) LUVs. Left: a freely tumbling peptide in an aqueous buffer. Right: EPR spectrum of the peptide in the presence of POPC:POPG LUVs is shown at fivefold amplification. The peak-to-peak intensities, A−1, of the high field (mI=−1) nitrogen hyperfine components are shown by arrows.
3.2 Analysis of peptide binding by a signal amplitude method
For each EPR spectrum obtained in the course of a titration, the fraction of the membrane-bound peptide can be determined from measuring A−1, the peak-to-peak amplitude of the high-field, m1=−1, nitrogen hyperfine component (Cafiso & Hubbell, 1981; Mchaourab, Hyde, & Feix, 1994). The fraction of the bound peptide, fb, can be calculated using the relation:
| Eq.1 |
where and A−1 are the peak-to-peak amplitudes of the high-field line for the peptide free in buffer and the experimental sample containing LUVs, respectively, and is the amplitude of the high-field line when the peptide is fully bound to the membrane (Figure 3). For weakly binding peptides, the condition of the complete binding may be difficult to achieve in the titration experiment as there is a practical limit of how much lipids can be added. For the latter cases, can be treated as an adjustable parameter during the binding isotherm analysis. If the EPR spectrum from the bound peptide is much broader than the one from the free peptide in solution (for example, the spectrum from the bound peptide falls in the slow motional regime), then the bound peptide contributes little to the overall peak-to-peak EPR amplitude and the contribution of the term can be neglected. As an example, in a binding experiment, was estimated to be less than 3% of (Bhargava & Feix, 2004).
In experiments when lipids are titrated into a sample with the given amount of the peptide, the total concentration of the peptide may be considered constant during the titration experiment. In such case, the peak-to-peak amplitude of the high-field line can be measured directly from the experimental spectra and need to be corrected only for the signal gain setting of the spectrometer, if the gain was changed from sample to sample. In titrations when the labeled peptide is added to lipids, CW EPR spectra should be intensity-normalized to the same double integral before measuring the signal amplitude. The concentrations of the free and the bound peptides are calculated from fb and the known total concentration of the peptide in the sample.
3.3 Analysis of peptide binding by spectral simulations
Experimental EPR spectra from peptide-liposome mixtures can be decomposed into the individual components, corresponding to the free peptide in solution and bound to the lipids, by using spectral simulation. Typically, the EPR spectrum from the free peptide can be modeled as a superposition of Voigt line shapes (a convolution integral of Lorentzian and Gaussian functions) with different homogeneous line width assigned to each of the three nitrogen hyperfine components. The simulations could be further improved by accounting for additional unresolved hyperfine interactions. For example, the quality of the fit of EPR spectra of NOD3-R1 peptide improved significantly by including a hyperfine interaction with one unique proton and satellite lines due to natural abundance 13C. A fitting program based on a fast convolution algorithm with a Levenberg-Marquardt optimization method (Smirnov, 2008; Smirnov & Belford, 1995; Smirnova et al., 1997) was employed to extract 14N, 1H and 13C isotropic hyperfine coupling parameters, as well as Lorentzian and Gaussian contributions to the linewidth of three nitroxide components. The experimental spectrum from the peptide in the presence of LUVs was modeled as a superposition of two spectra: a free peptide in the aqueous phase and a slower moving bound peptide. The line shape was modeled as described above with an explicit unresolved hyperfine splitting on one 1H and 13C nuclei as determined from simulations of the free peptide spectrum. All the spectral parameters including a small admixture of the dispersion contribution were adjustable for both centers, except for the probability of 13C and the unique hydrogen hyperfine. Figure 4 shows an example of an experimental EPR spectrum and the least-squares simulated components corresponding to the free and the bound peptides. The fraction of the bound peptide can be calculated from the double integrals of the simulated spectra available from the fit as:
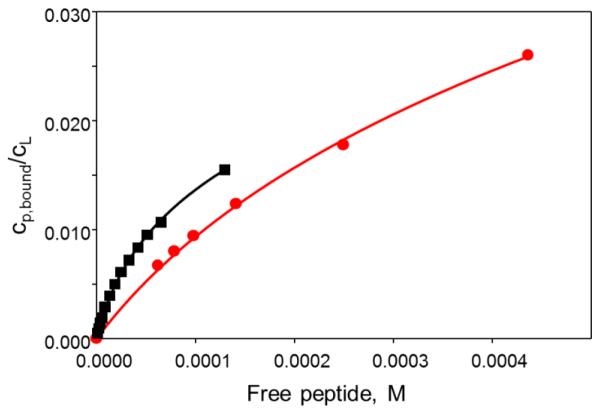
| Eq.2 |
where Ib and If are the double-integrated intensities of the bound and the free component, respectively. Figure 5 shows experimentally measured binding isotherms for an interaction of NOD3 peptide with LUVs prepared from pure POPC lipids and from POPC lipids doped with the anionic lipid POPG at 4 mol%. The fraction of the peptide bound and the concentration of the free peptide were calculated based on the spectral simulations, as described above.
Figure 4.
(A) Experimental room temperature X-band (9 GHz) EPR spectrum from 50 μM NOD3-R1 in the presence of 8 weight% LUVs (POPC:POPG in 24:1 molar ratio) LUVs. Simulated spectra corresponding to the free peptide in solution and bound to LUVs are shown as (B) and (C), respectively; (D) is the fit residual—a difference between the experimental and the sum of the two simulated components.
Figure 5.
Binding of NOD3-R1 peptide to LUVs composed of POPC (red (gray in the print version) circles) and POPC:POPS lipids in 24:1 molar ratio (black squares). A degree of binding, cp, bound/cL (mole of bound peptide per mole of lipid in the outer leaflet), is plotted versus concentration of the free peptide in solution. The solid curves are the best least-squares fits obtained using a model described by Eqs.(7)–(9).
When an EPR spectrum of the bound form of a peptide does not fall into the fast motion regime it cannot be simulated by a Voigt function. This is more typical for binding of basic peptides to liposomes with a high content of anionic lipids, and/or for TOAC-labeled peptides. While one can employ slow-motion models for EPR spectra, such simulations would significantly increase the computation burden, especially when an entire binding series of EPR spectra has to be simulated. An easier and equally effective approach for determination of the peptide binding constant involves modelling of the experimental spectrum in the presence of lipids, F(B), as a superposition of experimentally detected spectra, Ff(B) from the free peptide and Fb(B) from the bound form.
| Eq.3 |
and the fraction of bound form is calculated as:
| Eq.4 |
where a and b are weight coefficients, and Ib and If are double integrated intensities of the experimental spectra Ff(B) and Fb(B), respectively.
If a peptide binds only weakly, obtaining an experimental EPR spectrum from the bound form without any contributions from the free peptide may be problematic. Then the free peptide component could be subtracted manually by adjusting the amplitude of the free peptide signal till the residual spectrum corresponding to the bound form would not contain any sharp components as confirmed by a visual inspection. Spectra manipulation options in Xepr acquisition and data processing suite for the Bruker Biospin ELEXSYS series of EPR spectrometers (Billerica, MA) allows for an easy subtraction of the experimental spectra and evaluation of the double integral intensities. Alternatively, a convolution-based fitting of filtered EPR spectra can be applied for an automatic least-squares separation of fast and broad slow-motion nitroxide components without the explicit simulation of the slow motion spectrum (Smirnov, 2008).
4. Analysis of binding isotherms
When discussing peptide-membrane interactions, the term “binding” is used rather loosely, typically referring to interactions by a physical adsorption and partitioning. Specific peptide-phospholipid interactions, when a peptide forms a well-defined stoichiometric peptide-lipid complex are uncommon (Breukink et al., 1999; Choung et al., 1988; Choung, Kobayashi, Takemoto, Ishitsuka, & Inoue, 1988) and more particular for large membrane proteins. A review of EPR methods for studying lipid-protein interactions is given by (Marsh, 2010).
Once the fraction of the bound peptide, fb, is determined, concentrations of the free and the bound forms of the peptide can be calculated from the known total amount of the labeled peptide added to the sample. In a partitioning model, no assumptions on the peptide-lipid binding stoichiometry are made. The affinity of the peptide to the lipid membrane is described by a partition constant, Kp:
| Eq.5 |
where cp,m is the concentration of the peptide immediately above the membrane surface, cp,bound is the concentration of the peptide bound, a is the fraction of the lipids in the outer leaflet accessible for binding (a=0.5 for LUVs), and cL is the total lipid concentration. In a simple partitioning model, the concentration of the peptide at the membrane surface is the same as the bulk concentration of the free peptide in the aqueous phase, cp,f, and the partition coefficient is obtained from the linear part of the plot of xb vs. concentration of the free peptide. The partitioning model is suitable for the binding driven by hydrophobic interactions at a low peptide-to-lipid ratio. The linear relation is anticipated, for example, for interactions of uncharged peptides with membranes at non-saturating conditions.
The Langmuir adsorption isotherm is often employed to describe peptide-membrane interaction. If an assumption that the binding site is made up of n non-interacting and energetically equivalent lipid molecules is valid, then the Langmuir binding isotherm can be written as
| Eq.6 |
where is the mole fraction of the occupied binding sites. The Eq. 6 is equivalent to the ligand binding model when specific binding between the peptide and n equivalent lipids, forming PLn complex, is assumed.
Electrostatic forces can significantly contribute to the peptide-lipid interactions resulting in deviations from the binding behavior predicted by partitioning or the Langmuir models. For an electrostatically-driven peptide binding, the Eq. 5 is still valid; however, the contribution of the electrostatic interaction would change the effective concentration of the peptide at the surface of the membrane, cp,m, compared to that in the bulk solution (Seelig, 2004; Wieprecht & Seelig, 2002). Effective cp,m depends on the peptide charge and the membrane surface electrostatic potential, ψ0, and can be described using the Boltzmann distribution:
| Eq.7 |
where zp is the electric charge associated with the peptide, e0 is the elementary charge, T is the temperature, and k is the Boltzmann constant. The corresponding surface potential, ψ0, can be determined by solving the Gouy-Chapman equation (Aveyard, 1973; Mathias, McLaughlin, Baldo, & Manivannan, 1988; McLaughlin, 1977, 1989) :
| Eq.8 |
where σ is the two-dimensional density of electric charge on the surface, ε0 is the permittivity of the free space, κ is the dielectric constant of the solution, R is the universal gas constant, F is the Faraday constant, ci is the concentration of the i-th electrolyte in the bulk aqueous phase, and zi is the signed valency of the i-th species. The use of Eq. 8 requires an evaluation of the surface charge density, σ, for the given lipid bilayer system, as well as taking into account changes in σ from binding of cationic peptides. Surface charge density can be calculated from the known fraction of the charged lipids and literature data for the average area per lipid, or measured experimentally as described by (Voinov, Rivera-Rivera, & Smirnov, 2013). The presence of cations, such as Na+ or K+, that may specifically bind to anionic lipids, could reduce the effective membrane surface charge. This effect can be accounted for by modeling the ion-membrane interaction using the Langmuir binding isotherm. To describe binding of a cationic peptide with an electric charge z to a membrane containing xL mole fraction of an anionic lipid with a charge zL, in the presence of salt NaCl, we assume that:
| Eq.9 |
where A is the surface area per lipid, cNa and KNa are concentration of cations and its binding constant to the anionic lipid, respectively, and e is the elementary charge (Seelig, 2004; Wieprecht & Seelig, 2002). Binding of a charged peptide to an electrically neutral lipid membrane generates a positive electric charge at the membrane surface. From the extent of binding, xb, the surface charge density can be calculated assuming the first term in Eq. 9 to be zero. Eq. 9 can be modified to include a correction factor to take into account the possible penetration of peptide into the lipid membrane and the resulting increase in the membrane surface area (Kuchinka & Seelig, 1989); however, at a low peptide to lipid ratio this effect could be neglected.
Combining Eqs. 7 and 8, and calculating σ from the experimental values of xb, a numerical solution for K, ψ0, and cp,m for each of the experimental data pairs of xb and cp,f can be found. The partition coefficient, Kp, describes a hydrophobic binding or an adsorption of the peptide and should be independent of the peptide concentration for the given lipid system, as long as the model is applicable. Figure 5 shows experimental binding isotherms of NOD3 peptide to neutral POPC LUVs and to POPC LUVs containing 4 mol% of negatively charged POPG lipid and the best fit of the experimental data to the electrostatic binding model (Eqs. 5, 7-9). During the fit, Kp and the peptide charge, z, were used as adjustable parameters. As expected, the cationic NOD3 peptide binds stronger to LUVs containing anionic lipids than to zwitterionic DOPC lipids. Least-squares fitting of the binding isotherm for DOPC:DOPG liposomes, however, led to z estimated to be +2.5. This was unexpected as the peptide contains seven lysine residues. It is possible that not all the lysine side chains are protonated because of close proximity to each other.
5. Topology of membrane-associated peptides – paramagnetic relaxers accessibility EPR experiments
Determination of the topology of membrane-associated peptides relies on measurements of molecular accessibility of the spin-labeled sidechains to paramagnetic relaxation agents with differential solubility in water and in lipid environments. The method is well-established and was reviewed on multiple occasions (Fanucci & Cafiso, 2006; Klug & Feix, 2008). In such experiments molecular oxygen is served as a non-polar relaxer that partitions preferably into a lipid phase with a gradient of an increasing concentration towards the center of the membrane. Ni(II) ethylenediaminediacetate (NiEDDA) is a neutral solute that, while partitioning primarily into water, would also penetrate into the lipid head group region and slightly into the hydrophobic section of the bilayer. Another commonly employed relaxer is a chromium oxalate (Crox) negatively-charged complex that cannot penetrate deeply into the bilayer (Altenbach, Froncisz, Hubbell, & Hyde, 1988; Altenbach, Froncisz, Hyde, & Hubbell, 1989). Experimental data indicate that that the Heisenberg exchange is the dominant mechanism for magnetic interactions of nitroxide spin labels with both molecular oxygen and transition metal ions, as the spin-lattice relaxation time of these paramagnetic relaxers is much shorter than the spin-lattice relaxation time of the nitroxide. The contribution from the dipole-dipole interaction can be neglected in most cases. Effect of the Heisenberg exchange on the nitroxide spin-lattice relaxation time T1e is directly proportional to the nitroxide-relaxer collisional rate and, therefore, can be used as a measure of the nitroxide accessibility to the relaxer. Effect of the relaxer on T1e of a nitroxide label can be measured by either a power saturation CW EPR experiment or a direct pulsed saturation recovery method.
5.1 Accessibility measurements by EPR
In the power saturation experiment, the peak-to-peak amplitude of the central line of the first derivative nitroxide EPR signal, App, is measured as a function of the incident microwave power and then fitted to the following function:
| Eq.10 |
where I is a scaling factor, P1/2 is the microwave power required to reduce the resonance amplitude to half of its maximum unsaturated value, and ε is a measure of the homogeneity of the saturation of the resonance line, ε=1.5 for the homogeneous and 0.5 for the inhomogeneous saturation limits. From the least-squares fitting of the experimental CW EPR “roll-over” curves, the P1/2 parameter is obtained. Then, in order to measure the relaxer accessibility, values for P1/2 are determined for the sample equilibrated with N2, air (20.95% O2), and N2 but in presence of a certain concentration of a hydrophobic relaxer such as, for example, NiEDDA. The collision rate with a fast relaxing agent is directly related to the difference in P½ in the presence and absence of the relaxer:
| Eq.11 |
The accessibility parameter, Πrelaxer, for a given relaxer is determined as (Farahbakhsh, Altenbach, & Hubbell, 1992).:
| Eq.12 |
where ΔHp–p is the peak-to-peak linewidth of the central nitrogen hyperfine component of the nitroxide EPR spectrum and ΔHp–p(DPPH) is the peak-to-peak linewidth of the solid DPPH (2,2-diphenyl-1-picrylhydrazyl) reference sample. Division by the peak-to-peak linewidth makes ΔP′½ independent upon , and normalization by the data for the reference compound DPPH compensates for variations in the EPR resonator quality factor Q.
The location of the spin label within a bilayer membrane and at the water-membrane interface can be characterized by the depth parameter, Φ, defined as:
| Eq.13 |
The parameter Φ is directly related to the difference in the standard state chemical potentials of the reagents at any depth, independent of viscosity or steric constraints imposed by the environment and also independent of the EPR line shape.
The calibration curve for the depth parameter as a function of the label position can be easily obtained using lipid bilayers doped with n-doxyl PC (1-palmitoyl-2-stearoyl-(n-doxyl)-sn-glycero-3-phosphocholine) lipids labeled at a position n along the alkyl chain. n-Doxyl PC lipids with n=5, 7, 10, 12, 14, and 16 and Tempo PC, a lipid molecule with a nitroxide tethered to the lipid headgroup, are all commercially available from Avanti Polar Lipids, Inc. The dependence of Φ upon the distance is approximately linear for labels within the bilayer interior, as measured using spin-labeled lipids (Qin & Cafiso, 1996), a spin-labeled membrane protein (Altenbach, Greenhalgh, Khorana, & Hubbell, 1993), and a transmembrane peptide WALP (Nielsen, Che, Gelb, & Robinson, 2005). Although the gradients in concentration of both oxygen and metal ions level off quickly in the aqueous phase, the calibration curve was extended to the aqueous phase using a membrane-bound spin-labeled protein, (Frazier, Wisner, Falke, & Cafiso, 2000) allowing for determination of distances up to about 5 Å in the aqueous side of the membrane-solution interface (Victor & Cafiso, 2001).
Alternatively, molecular accessibility parameters can be measured using pulsed saturation recovery (SR) experiment, which allows for direct measurement of the electronic spin-lattice relaxation time T1e (Eaton & Eaton, 2005; Pyka, Ilnicki, Altenbach, Hubbell, & Froncisz, 2005). Changes in the spin-lattice electronic relaxation rates, ΔR1, for oxygen and NiEDDA can be used as the time domain analogues of the accessibility parameter:
| Eq.14 |
where T1e(N2) is the nitroxide spin-lattice relaxation time measured for the nitrogen-equilibrated sample, and T1e(relaxer) is the effective relaxation time in the presence of a relaxation agent, and [relaxer] is the concentration of the relaxer. The depth parameter is defined as:
| Eq.15 |
Experimental SR traces are typically least-squares fitted to a single exponential decay to extract the relaxation rate . However, for spin-labeled systems with two distinctive states, the saturation recovery data should be analyzed in terms of a double exponential decay (Nielsen, et al., 2005; Pyka, et al., 2005; Yin, Feix, & Hyde, 1990). The main advantage of the SR method is in the ability to resolve multiple nitroxide populations with different molecular accessibilities to the relaxer molecules (Eaton & Eaton, 2005; Kusumi, Subczynski, & Hyde, 1982; Pyka, et al., 2005; Subczynski, Widomska, Wisniewska, & Kusumi, 2007). The use of the SR method in peptide binding and other biophysical EPR experiments, however, is limited by the need for specialized instrumentation. Although commercial spectrometer systems capable of SR EPR are available from Bruker BioSpin (Billerica, MA), the installations are not as common as CW EPR spectrometers.
5.2 Experimental considerations
CW EPR power saturation method has been the method of choice for most spin-labeling accessibility studies as it could be performed using a standard CW spectrometer typically equipped with a loop-gap resonator to achieve the maximum microwave power on the sample required for at least a partial signal saturation. Accessibility measurements require a strict control of molecular oxygen concentration in the sample that is typically achieved by drawing a solution into gas-permeable TPX (polymethylpentene or PMP) capillaries (available from L&M EPR Supplies, Milwaukee, MI, or from Bruker, Karlsruhe, Germany) and continuously flushing nitrogen gas through the resonator space. Alternatively, liquid samples can be drawn into a disposable gas-permeable thin-wall PTFE (polytetrafluoroethylene) capillary, such 0.81 mm i.d., 0.86 mm outer diameter (o.d.) tubing available from Zeus Industrial Products, Inc. (Orangeburg, SC). Such thin and flexible tubing allows for the ends of the capillary to be closed by crimping that will stay in place when the capillary is inserted into an open-ended quartz tube (3 mm i.d.). The quartz tube is then placed inside a variable temperature dewar of the X-band EPR or and EPR resonator that is continuously flashed with a gas mixture with a controlled oxygen concentration (Smirnov, Clarkson, & Belford, 1996).
The optimal concentration of NiEDDA in EPR accessibility experiments varies depending on the location of the nitroxide label within the membrane. Typically, 20 mM concentration is sufficient while twice as high concentrations of NiEDDA may be needed for peptides with the nitroxide chains deeply embedded into a bilayer or located at sterically protected sites. The latter are expected for aggregated membrane peptides. For nitroxide labels exposed to the aqueous phase, concentrations as low as 3 mM NiEDDA may be sufficient to measure changes in the nitroxide relaxation properties. Accessibility data can be easily normalized to 20 mM NiEDDA concentration for the comparison purposes.
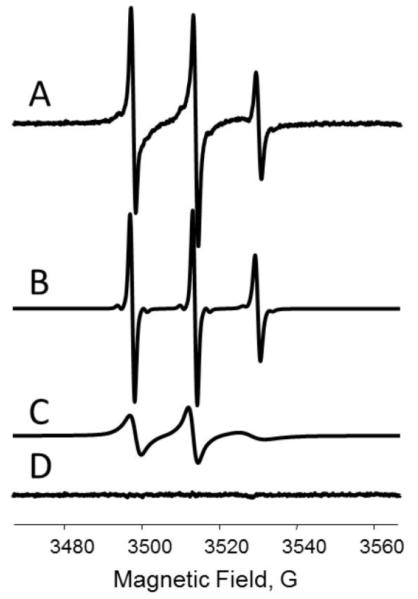
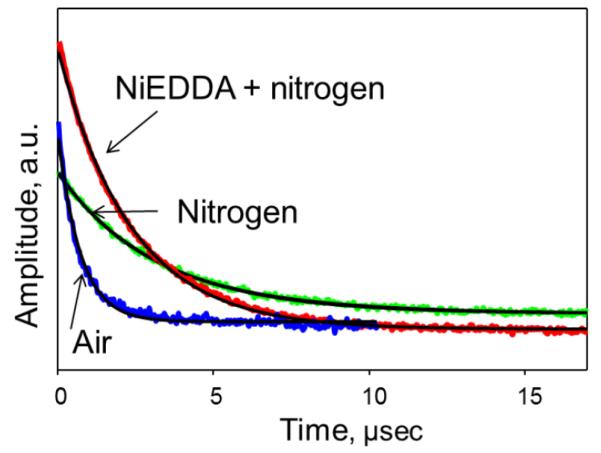
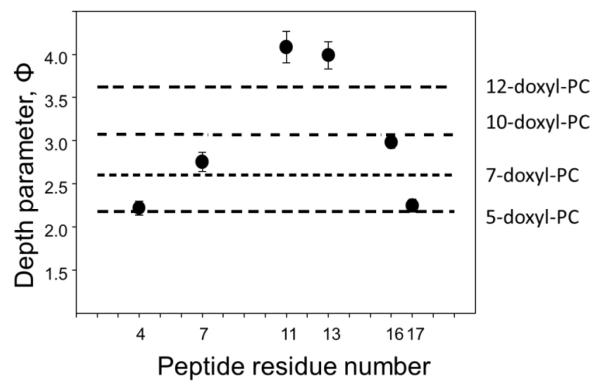
Figures 6 and 7 demonstrate an application of the depth parameter approach to elucidate the transmembrane location of STM23, a 23 amino acid peptide containing a highly hydrophobic 16 amino acid segment. Figure 6 shows experimental saturation recovery traces for the peptide labeled with MTSL at residue 4 (STM23-4R1) and incorporated into LMVs composed of 1,2-dipalmitoleoyl-sn-glycero-3-phosphoethanolamine (DPoPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid in 2:3 molar ratio. Two samples were prepared, one using the buffer of choice and the second one using a buffer containing 20 mM NiEDDA. Saturation recovery traces were obtained for a nitrogen equilibrated sample prepared with 20 mM NiEDDA, and for air- and nitrogen-equilibrated samples without the metal complex. The corresponding fits with a single-exponential decay function are superimposed on experimental curves. The depth parameter as a function of the label position is presented in Figure 7 and was calculated using Eqs. 14 and 15. The membrane depth calibration data were obtained using LMVs of the same composition labeled with n-doxyl PC (n=5, 7, 10, and 12) in the presence of an unlabeled STM23, and are shown in Figure 7 as horizontal lines. The data demonstrates a transmembrane insertion of the STM23 peptide into DPoPE/POPC bilayers.
Figure 6.
Effects of paramagnetic broadening agents on experimental X-band (9 GHz) room temperature saturation recovery traces for a MTSL-labeled peptide (STM23-4R1) bound to DPoPE/POPC LUVs with position of the nitroxide-labeled chain approximately 8 Å below the lipid phosphate groups: green (light gray in the print version), nitrogen equilibrated sample; blue (dark gray in the print version), air-equilibrated sample; red (gray in the print version), nitrogen-equilibrated sample containing 20 mM NiEDDA. Least-squares fits to single-exponential decays are shown as solid black lines.
Figure 7.
The depth parameter Φ as a function of the nitroxide label position along the STM23 peptide incorporated into LUVs consisting of DPoPE and DOPC lipids in 2:3 molar ratio. The horizontal lines represent the experimental depth parameters measured for the LMVs of the same composition doped with n-doxyl PC (n=5, 7, 10, and 12) at 1 mol%. The depth parameter clearly indicates a transmembrane insertion of the peptide.
Dependence of the depth parameter upon the label position in the peptide sequence can provide information on the secondary structure of the peptide bound to phospholipid bilayers. Such an approach requires measurements of the EPR accessibility parameter for an extended set of consecutive spin-labeled sites and it is known to work best for the surface associated peptides. For a peptide forming an α-helix with ≈3.6 residues per turn, a spin label positioned at every third or fourth residue would have about the same accessibility to a specific paramagnetic relaxer. That is, the surface-exposed residues would exhibit a higher accessibility to a water-soluble relaxer, like NiEDDA. The corresponding sites on the other side of the helix would be buried deeper into the bilayer, showing a lower accessibility to NiEDDA, but a higher accessibility to oxygen. The plot of the corresponding depth parameter vs. the residue number would than reveal the characteristic periodicity of 3.6 for an α-helical and 2 for a β-sheet structure. Highly illustrative examples of this approach can be found in the following references (Klug, Su, & Feix, 1997; Sato & Feix, 2006; Turner, Braide, Mills, Fanucci, & Long, 2014).
6. Detecting membrane-induced aggregation and agglomeration of peptides
6.1 CW EPR method – a diamagnetic dilution experiment
Spin-labeling EPR provides an easy and informative way to detect formation of peptide-peptide clusters or aggregation that sometimes is promoted by peptide-membrane interactions. The approach is based on measuring a broadening of a CW EPR signal from the labeled peptide due to magnetic interactions between the labels brought into close proximity by aggregation. At a low peptide-to-lipid ratio, and in the absence of peptide-peptide interactions, peptides are distributed randomly through the membrane, providing a sufficiently long distance between the nitroxide labels to neglect spin-spin magnetic interactions. The EPR spectra for such peptides are determined by the label dynamics. If two or more labels are located at distances < 25 Å, the CW EPR signal is broadened and has lower peak-to-peak amplitude than the spectrum from the same concentration of the non-interacting spins. Typically, peptide aggregates are more tightly packed, causing a slower rotational dynamics of the nitroxide and an additional spectral broadening. Then, the on-set of the aggregation or cluster formation is detected as a decrease in the amplitude of the CW EPR signal, normalized for the given number of spins. Thus, the peptide aggregation or cluster formation can be readily detected by comparing the EPR signal from the cluster sample with the signal from a diamagnetically “diluted” sample taken under the same conditions (Altenbach & Hubbell, 1988; Martin Margittai & Langen, 2006).
Firstly, let us consider the types of magnetic interactions responsible for the spectral broadening. For two nitroxide labels separated by a distance in the range from 8-10 to 25 Å, only the dipole-dipole interaction contributes to the observed broadening (Altenbach, Oh, Trabanino, Hideg, & Hubbell, 2001; Hubbell, Cafiso, & Altenbach, 2000) while at distances below 8 Å, the Heisenberg spin-exchange starts to play a measurable role (Molin, 1980). At a very close contact between the labels, the spin exchange results in a collapse of the nitrogen hyperfine spectral features, resulting in an observation of a single line spectrum. Although such spectra are atypical for a membrane-associated peptide aggregation, exchange-narrowed single line spectra were reported for spin-labeled amyloids (M. Chen, Margittai, Chen, & Langen, 2007; Cobb, Sönnichsen, Mchaourab, & Surewicz, 2007) and tau filaments (M. Margittai & Langen, 2004). The broadening effect due to the dipole-dipole interactions is detectable at both cryogenic and physiological temperatures although in the latter case the dipole-dipole interactions can be partially averaged by translational motion of spin-labels or a rotation of the dipolar vector with respect to the magnetic field.
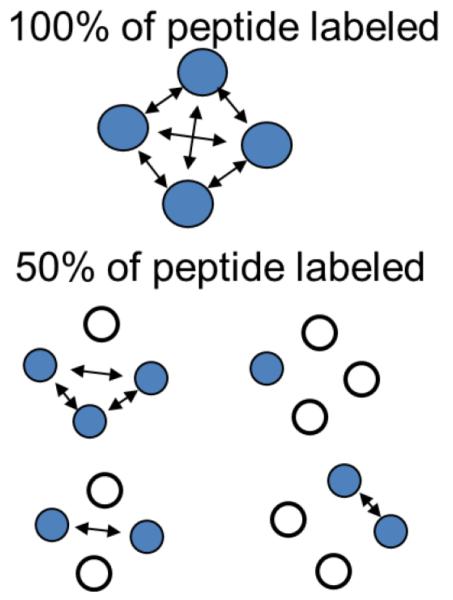
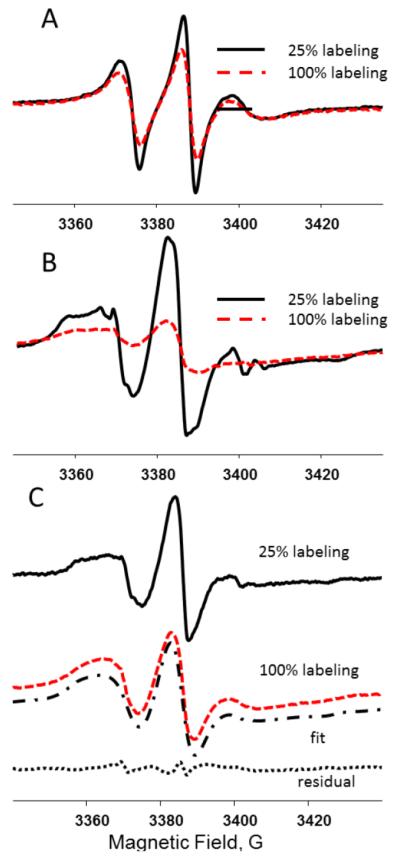
In a typical experiment a diamagnetically diluted sample is prepared by mixing labeled and unlabeled peptides at various ratios. Alternatively, a sample could be labeled with a mixture of MTSL and its diamagnetic analog methyl methanethiosulfonate (MMTS, available from Toronto Research Chemicals Inc. (Toronto, ON, Canada) (Gross, Columbus, Hideg, Altenbach, & Hubbell, 1999) mixed at different ratios and, therefore, producing samples with the required magnetic dilution. The use of the diamagnetic analog ensures that the steric interactions due to the presence of the nitroxide labels are preserved in the diamagnetically diluted sample. Preserving such interactions could be of significance, especially if information on local dynamics of the peptide in an aggregate or a cluster is of interest. Figure 8 schematically shows a reduction in the number of the close-contact spin pairs in an aggregated sample upon the diamagnetic dilution. Figure 9A depicts an example of spectral changes observed upon diamagnetic dilution in the case of an aggregation of STM25, a 25 amino acid peptide containing a highly hydrophobic transmembrane segment of 18 amino acids. STM25 was labeled at various positions, n, with MTSL (STM25-nR1) and incorporated in LMVs composed of POPC and POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine) lipids in the 1:2 mole ratio containing varying amounts of cholesterol. The spectrum shown in dashed line was recorded for a sample containing 100%-labeled STM25-11R1 peptide while the spectrum in solid line is from a magnetically diluted sample containing the peptide at 25 mol% labeling. In both samples the peptide-to-lipid ratio was 1:100. Figure 9B shows analogous spectra from the same peptide incorporated in POPC/POPE LUVs containing 30 mol% of cholesterol. The aggregation-induced broadening is readily observable in both cases as a decrease in the amplitude of these intensity-normalized spectra.
Figure 8.
A cartoon illustrating a reduction in the number of close-contact spin pairs (indicated by arrows between filled circles depicting spin-labeled peptides) in aggregated peptide samples upon a diamagnetic dilution with EPR-silent peptides (open circles).
Figure 9.
Intensity normalized room temperature CW X-band (9 GHz) EPR spectra of STM25-11R1 in POPE:POPC 1:2 LMVs in the presence of 10 mol% (A) and 30 mol% (B) cholesterol and STM25-11R1 in POPE:POPC 1:2 LMVs in presence of 25 mol% (C): 100% labeled peptide (red, light gray in the print version dashed line) and 25% labeled peptide (black solid line). C: The spectrum from 100% labeled peptide was least squares fitted to a model assuming a homogenous (Lorentzian) broadening due to magnetic interactions. The result of the fit is shown as a dash-dot line. The fit residual, i.e., difference between the experimental and the simulated spectra, is shown at the bottom as a dotted line.
In order to estimate the broadening effect we have applied a one-linewidth parameter fitting model that was previously developed for fitting inhomogeneously broadened EPR spectra by (Smirnov & Belford, 1995), and was used by us to extract relaxer-induced broadening and changes in linewidth due to dynamic effects (Smirnova, Smirnov, Clarkson, & Bedford, 1995; Smirnova, Smirnov, Clarkson, & Belford, 1995). A similar approach was described by Mchaourab et al. to extract the spectral broadening from pairwise spin-spin interactions (McHaourab, Oh, Fang, & Hubbell, 1997). This model is based on simulating the CW EPR spectrum from the aggregated sample by using a convolution integral given by Eq. 16, in which F0(B′) is the spectrum in the absence of magnetic interactions (the infinitely diluted sample), F(B) is the predicted spectrum from an aggregated sample, and m(B) is the Lorentzian broadening, measured as the δ(ΔBLp-p) peak-to-peak line width:
| Eq.16 |
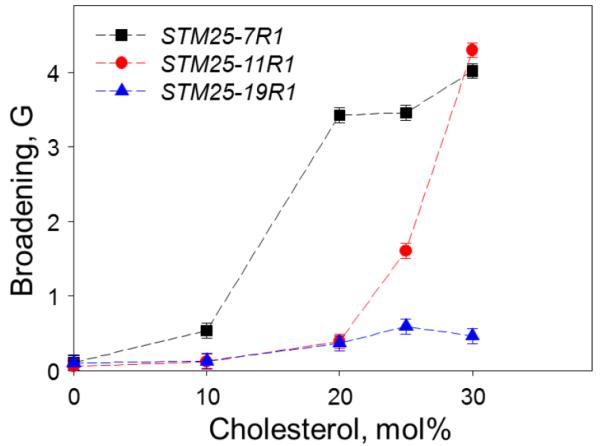
In practice, the spectrum F0(B′) from the “infinitely diamagnetically diluted sample” could be difficult to obtain experimentally because of the vanishingly low signal intensity. Thus, the signal-to-noise consideration would typically dictate a compromise value for the diamagnetic dilution. In our analysis, we took an experimental EPR spectrum from the sample containing a peptide with 25 mol% of labeling as an approximation for the “infinitely diluted” spectrum F0(B′). In this case the broadening is measured relative to the one already present in the diamagnetically diluted sample. The approach is illustrated in Figure 9C showing experimental spectrum for a sample containing 100%-labeled STM25-11R1 peptide incorporated in POPC/POPE LUVs containing 25 mol% of cholesterol shown in dashed line, spectrum from a magnetically diluted sample containing the peptide at 25 mol% labeling is shown in solid line, and the result of the fit using equation 16 in dash-dot line. The fit residual, i.e., difference between the experimental and the simulated spectra, is shown at the bottom in dotted line. The aggregation broadening was determined to be 0.31 G for the fully-labeled STM25-11R1 in a lipid bilayer containing 10 mol% of cholesterol, and 1.6 G and 4.7 G in a membrane containing 25 and 30 mol% of cholesterol correspondently. The broadening can be used as an empirical parameter to characterize the extent of the aggregation as the larger broadening corresponds to shorter distances between the labels. Figure 10 displays an aggregation-induced broadening for STM25-11R1 peptide in POPC/POPE LMVs a function of the cholesterol content. A clear onset of the aggregation for STM25-11R1 peptide is observed at a cholesterol concentration above 20 mol %. An interesting dependence of the broadening is observed for labels located at different positions along the peptide chain.
Figure 10.
Lorentzian broadening for room temperature X-band (9 GHz) EPR spectra of STM25-nR1 peptide fully labeled with the MTSL side chain (R1) at positions n = 7, 11, and 19 as a function of cholesterol content in POPE:POPC (1:2 molar ratio) LMVs. The samples of identical lipid and peptide composition but diamagnetically diluted with unlabeled STM25 to 25% of STM25-nR1 were used to obtain the reference unbroadened EPR spectra. See text for further details.
If the peptide-peptide interactions are highly specific, some information on the mode of the peptide-peptide interaction can be elucidated by comparing the broadening effects for the labels located at different positions of the peptide chain. This approach is demonstrated in Figure 10 by comparing the broadening observed for the labels located at the positions 7, 11, and 19 of the STM25 peptide. Although the aggregation effect is clearly detected by the labels at the positions 7 and 11 at 25 mol% of cholesterol, the relative broadening is much higher for the position 7 than for the position 11. Unexpectedly, the difference disappears in the samples with 30 mol% of cholesterol. The observed broadening is, however, much smaller for the peptide labeled at the position 19, even for the sample with the highest cholesterol content. This broadening trend can be explained by considering that 7R1 and 11R1 are located in a highly hydrophobic region of the peptide, while 19R1 is close to the hydrophilic tail of the peptide containing five consecutive lysine residues that are exposed to the aqueous phase. The presence of positive charges on the Lysine sidechain prevents the aggregation of the hydrophilic tail. Combined with the accessibility measurements that point to STM25 being located at the membrane surface in the bilayer preparations with cholesterol content above 20 mol%, the broadening data suggest a “star-like” arrangement of the peptide at the membrane surface: the hydrophobic tails of the peptide aggregate to minimize the contact with the aqueous phase forming the center of the “star,” while the hydrophilic tails show a large separation forming the “arms”. It is not clear however, how many peptides participate in the formation of an individual aggregate.
Quantitative interpretation of the observed broadening effects in terms of spin-spin distances, and the aggregate structure, can be difficult to obtain. however, because this would require testing specific models of the peptide arrangements, in order to agree with the observed spectral broadening. The approach is based on considering a specific peptide arrangement, for example, dimer, trimer, hexamer, etc., and calculating possible spin-spin pairwise distances, discarding once above 2.5 nm, as the contribution of these pairs in the CW EPR spectrum broadening can be neglected. Next, the changes in EPR spectrum from non-interacting spins (taken at the high diamagnetic dilution limit) due to the predicted distance distributions can be calculated and compared with the experimentally observed spectrum from a non-diluted sample. If the geometry of peptide arrangement is such that only one distance contributes significantly to the broadening, then the methods for determination of distances between the spin pairs are readily available (Altenbach, et al., 2001; Banham et al., 2008; Hubbell, et al., 2000; Rabenstein & Shin, 1995; Steinhoff et al., 1997) to test the validity of the proposed peptide-peptide arrangement. For example, the convolution approach was employed to extract effective distance and distance distribution to characterize the membrane-induced aggregation of alamethicin in bicelles (Bortolus, De Zotti, Formaggio, & Maniero, 2013). Rigid limit CW EPR linewidth broadening as measured for frozen samples (T=78.5 K) was used to estimate spin-spin distances for membrane imbedded TOAC-labeled trichogin GA IV, a lipopeptide with antibiotic activity, assuming formation of a dimer in a model membrane (Syryamina et al., 2012).
Another way to quantify magnetic interactions is by the second moment analysis of the EPR spectra that is based on the famous work of van Vleck (Van Vleck, 1948). The second moment of the broadening function, μ2, is obtained from the intensity-normalized and baseline-corrected broadened, F0(B), and non-broadened, F0(B′), spectra (Abragam, 1961):
| Eq.17 |
where B is the applied magnetic field, and is the magnetic field corresponding to the mean of the spectrum. The second moment of μ2 can be used as an empirical parameter to characterize the extent of the spin-spin interactions, similar to the Lorentzian broadening in the example above, or be related to the pair-wise distance between nitroxide labels. The μ2 of the powder sample of like spins (case of a strong dipolar coupling) can be calculated using the Van Vleck formula (Van Vleck, 1948), assuming a well-defined distance r, resulting in:
| Eq.18 |
where a=1.56·10−60 T2 m6. Eq. 18 can be solved for the inter-spin distance, r:
| Eq.19 |
This approach has been employed to investigate an aggregation of a model transmembrane WALP23 peptide (Scarpelli et al., 2009) as a function of the lipid composition. The main error source in this analysis comes from the polynomial baseline correction errors in the integration of the first derivative EPR spectra.
6.2 Distance measurements and spin counting by Pulsed Electron Double Resonance
Essential information on number of spins in a peptide aggregate can be obtained using advanced EPR pulse methods. Pulsed electron-electron double resonance (PELDOR, also known as DEER, or double electron-electron resonance) is widely used to measure the dipolar electron-electron coupling, in order to determine distances and distance distributions in the nanometer range, from 1.5 to about 8 nm, as well as to provide information on the mutual orientation of the interacting spins. In addition to providing distance information, PELDOR can be used to count the number of spin-labeled monomers in a complex. In this article, we will not discuss the theoretical basis or experimental details of this approach. Readers are referred to excellent reviews on this topic (Freed, 2014; Jeschke, 2012, 2014). In brief, the approach is based on a relationship between the depth of the modulation, Δ, of the PELDOR signal and the number of spins in the cluster, N:
| Eq.20 |
where λ is the B spin inversion efficiency, a parameter defined by the experimental conditions, and f is the efficiency of the labeling (Jeschke, 2012; A. D. Milov, Maryasov, & Tsvetkov, 1998; A. D. Milov, Ponomarev, & Tsvetkov, 1984). To obtain the number of spins in the cluster, an experimental evaluation of λ is required. The latter can be readily achieved by measurements on a model biradical system. Several studies using model bi-, tri-, and tetra-radicals with known labeling efficiency reported the overall error of PELDOR spin-counting of less than 5% (Bode et al., 2007). The PELDOR spin-counting method was successfully applied to study protein oligomerization (Hilger et al., 2005) and self-assembly and aggregation of peptides (Alexander D. Milov et al., 2009; A. D. Milov et al., 2000; Salnikov et al., 2006).
We note that PELDOR measurement require highly specialized pulse EPR spectrometers, operated by a well-trained staff and this currently limits a wide spread application of this very useful and informative technique. Commercial spectrometers operating at X-, Q- and W- band frequencies offered by Bruker BioSpin and a number of home-built spectrometers show excellent technical specifications. PELDOR experiments typically require 50 μM protein concentration and about 30 μL of sample volume for measurements using commercial X-band EPR spectrometers. Q-band spectrometers offer higher sensitivity and require much smaller samples (down to 5 μL) (Ghimire, McCarrick, Budil, & Lorigan, 2009); however, these spectrometers are even less common that the one operating at X-band EPR frequency.
7. Conclusions and outlook
EPR, in combination with spin labeling methods, offers researchers access to a wide range of structural and functional information on peptide-membrane interactions beyond the data on the binding constant. Although the methods do require highly specialized instrumentation, the experimental protocols are well-developed and the instrumentation is generally accessible to broad groups of researchers from academia as well as industry. Continuous progress in EPR instrumentation, especially the recent introduction of portable, low-cost CW EPR spectrometers from Bruker BioSpin and Active Spectrum (Foster City, CA) to the market, opens up further opportunities for even wider utilization of this method both in academic research, student training and industrial applications. Further development of spin-labeling methods and synthesis of new, specific spin-labels hold a promise to extend the number of biological systems amenable to such studies, and to expand the information content of the method.
Acknowledgments
A part of this work was supported by National Science Foundation grants MCB-0451510 and MCB-0843632 (to T.I.S). A part of this work including development of simulation software and analysis of membrane binding data for charged peptides was supported by grant No. DE-FG-02-02ER153 (to A.I.S.) from the U.S. Department of Energy. EPR instrumentation was supported by grants from the National Institutes of Health (No. RR023614), the National Science Foundation (No. CHE-0840501), and NCBC (No. 2009-IDG-1015). The authors are thankful to Dr. Maxim A. Voinov (NCSU) for his expert help in preparing some of the Figures and numerous discussions and suggestions.
9. References
- Abragam A. The Principles of Nuclear Magnetism. OUP; 1961. [Google Scholar]
- Aina OH, Sroka TC, Chen ML, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66(3):184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- Alaouie AM, Smirnov AI. Ultra-stable temperature control in EPR experiments: Thermodynamics of gel-to-liquid phase transition in spin-labeled phospholipid bilayers and bilayer perturbations by spin labels. Journal of Magnetic Resonance. 2006;182(2):229–238. doi: 10.1016/j.jmr.2006.07.002. doi: http://dx.doi.org/10.1016/j.jmr.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Altenbach C, Froncisz W, Hubbell W, Hyde J. The Orientation of Membrane-Bound, Spin-Labeled Melittin as Determined By Electron-Paramagnetic-Res Saturation Recovery Measurements. Biophysical Journal. 1988;53(2):A94–A94. [Google Scholar]
- Altenbach C, Froncisz W, Hyde JS, Hubbell WL. Conformation of Spin-Labeled Melittin At Membrane Surfaces Investigated by Pulse Saturation Recovery and Continuous Wave Power Saturation Electron-Paramagnetic Resonance. Biophysical Journal. 1989;56(6):1183–1191. doi: 10.1016/S0006-3495(89)82765-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. EPR Depth Measurements of Nitroxides in Membrane Bilayers Using Spin Labeled Mutants of Bacteriorhodopsin. Biophysical Journal. 1993;64(2):A51–A51. [Google Scholar]
- Altenbach C, Hubbell WL. The Aggregation State of Spin-Labeled Melittin in Solution and Bound to Phospholipid-Membranes - Evidence That Membrane-Bound Melittin is Monomeric. Proteins-Structure Function and Genetics. 1988;3(4):230–242. doi: 10.1002/prot.340030404. [DOI] [PubMed] [Google Scholar]
- Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. [Article] Biochemistry. 2001;40(51):15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hanson P, McNulty J, Millhauser G, Monaco V, Formaggio F, Toniolo C. Solution structures of TOAC-labeled trichogin GA IV peptides from allowed (g approximate to 2) and half-field electron spin resonance. Journal of the American Chemical Society. 1999;121(29):6919–6927. doi: 10.1021/ja984255c. [DOI] [Google Scholar]
- Aveyard R, Haydon DA. An introduction to the principles of surface chemistry. Cambridge University Press; London: 1973. [Google Scholar]
- Balog M, Kalai T, Jeko J, Berente Z, Steinhoff HJ, Engelhard M, Hideg K. Synthesis of new conformationally rigid paramagnetic alpha-amino acids. Tetrahedron Letters. 2003;44(51):9213–9217. doi: 10.1016/j.tetlet.2003.10.020. [DOI] [Google Scholar]
- Balog MR, Kalai TK, Jeko J, Steinhoff HJ, Engelhard M, Hideg K. Synthesis of new 2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxyl radicals and 2-substituted-2,5,5-trimethylpyrrolidin-1-yloxyl radicals based alpha-amino acids. Synlett. 2004;(14):2591–2593. doi: 10.1055/s-2004-834806. [DOI] [Google Scholar]
- Banham JE, Baker CM, Ceola S, Day IJ, Grant GH, Groenen EJJ, Timmel CR. Distance measurements in the borderline region of applicability of CW EPR and DEER: A model study on a homologous series of spin-labelled peptides. Journal of Magnetic Resonance. 2008;191(2):202–218. doi: 10.1016/j.jmr.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Berliner LJ, Grunwald J, Hankovszky HO, Hideg K. A novel reversible thiol-specific spin label: Papain active site labeling and inhibition. Analytical Biochemistry. 1982;119(2):450–455. doi: 10.1016/0003-2697(82)90612-1. doi: http://dx.doi.org/10.1016/0003-2697(82)90612-1. [DOI] [PubMed] [Google Scholar]
- Bhargava K, Feix JB. Membrane binding, structure, and localization of cecropin-mellitin hybrid peptides: A site-directed spin-labeling study. Biophysical Journal. 2004;86(1):329–336. doi: 10.1016/s0006-3495(04)74108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode BE, Margraf D, Plackmeyer J, Duerner G, Prisner TF, Schiemann O. Counting the monomers in nanometer-sized oligomers by pulsed electron - Electron double resonance. Journal of the American Chemical Society. 2007;129(21):6736–6745. doi: 10.1021/ja065787t. [DOI] [PubMed] [Google Scholar]
- Boohaker RJ, Lee MW, Vishnubhotla P, Perez JM, Khaled AR. The Use of Therapeutic Peptides to Target and to Kill Cancer Cells. [Review] Current Medicinal Chemistry. 2012;19(22):3794–3804. doi: 10.2174/092986712801661004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolus M, De Zotti M, Formaggio F, Maniero AL. Alamethicin in bicelles: Orientation, aggregation, and bilayer modification as a function of peptide concentration. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2013;1828(11):2620–2627. doi: 10.1016/j.bbamem.2013.07.007. doi: http://dx.doi.org/10.1016/j.bbamem.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286(5448):2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- Brewer CF, Riehm JP. Evidence for possible nonspecific reactions between N-ethylmaleimide and proteins. Analytical Biochemistry. 1967;18(2):248–255. doi: http://dx.doi.org/10.1016/0003-2697(67)90007-3. [Google Scholar]
- Cafiso DS, Hubbell WL. ELECTRON-PARAMAGNETIC-RES DETERMINATION OF MEMBRANE-POTENTIALS. Annual Review of Biophysics and Bioengineering. 1981;10:217–244. doi: 10.1146/annurev.bb.10.060181.001245. [DOI] [PubMed] [Google Scholar]
- Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. Journal of Biological Chemistry. 2007;282(34):24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- Chen PS, Toribara TY, Warner H. MICRODETERMINATION OF PHOSPHORUS. Analytical Chemistry. 1956;28(11):1756–1758. doi: 10.1021/ac60119a033. [DOI] [Google Scholar]
- Choung SY, Kobayashi T, Inoue J, Takemoto K, Ishitsuka H, Inoue K. HEMOLYTIC-ACTIVITY OF A CYCLIC PEPTIDE RO09-0198 ISOLATED FROM STREPTOVERTICILLIUM. Biochimica Et Biophysica Acta. 1988;940(2):171–179. doi: 10.1016/0005-2736(88)90192-7. [DOI] [PubMed] [Google Scholar]
- Choung SY, Kobayashi T, Takemoto K, Ishitsuka H, Inoue K. INTERACTION OF A CYCLIC PEPTIDE, RO09-0198, WITH PHOSPHATIDYLETHANOLAMINE IN LIPOSOMAL MEMBRANES. Biochimica Et Biophysica Acta. 1988;940(2):180–187. doi: 10.1016/0005-2736(88)90193-9. [DOI] [PubMed] [Google Scholar]
- Cobb NJ, Sönnichsen FD, Mchaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proceedings of the National Academy of Sciences. 2007;104(48):18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikovski BG, Borbat PP, Freed JH. Spin-labeled gramicidin A: Channel formation and dissociation. Biophysical Journal. 2004;87(5):3504–3517. doi: 10.1529/biophysj.104.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Eaton G. Saturation Recovery EPR. In: Eaton S, Eaton G, Berliner L, editors. Biomedical EPR, Part B: Methodology, Instrumentation, and Dynamics. Vol. 24/B. Springer US; 2005. pp. 3–18. [Google Scholar]
- Elsasser C, Monien B, Haehnel W, Bittl R. Orientation of spin labels in de novo peptides. Magnetic Resonance in Chemistry. 2005;43:S26–S33. doi: 10.1002/mrc.1692. [DOI] [PubMed] [Google Scholar]
- Fanucci GE, Cafiso DS. Recent advances and applications of site-directed spin labeling. Current Opinion in Structural Biology. 2006;16(5):644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Farahbakhsh ZT, Altenbach C, Hubbell WL. SPIN LABELED CYSTEINES AS SENSORS FOR PROTEIN LIPID INTERACTION AND CONFORMATION IN RHODOPSIN. Photochemistry and Photobiology. 1992;56(6):1019. doi: 10.1111/j.1751-1097.1992.tb09725.x. &. [DOI] [PubMed] [Google Scholar]
- Fleissner MR, Bridges MD, Brooks EK, Cascio D, Kalai T, Hideg K, Hubbell WL. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(39):16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier A, Wisner M, Falke J, Cafiso D. Determination of calcium-induced structural changes and membrane binding orientation of cPLA2-C2 domain. Biophysical Journal. 2000;78(1):415A–415A. [Google Scholar]
- Freed P. P. B. a. J. H. Pulse Dipolar Electron Spin Resonance: Distance Measurements. In: Harmer C. R. T. a. J. R., editor. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Vol. 152. Springer; Heidelberg New York Dordrecht London: 2014. pp. 1–82. [Google Scholar]
- Getz EB, Xiao M, Chakrabarty T, Cooke R, Selvin PR. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. [Article] Analytical Biochemistry. 1999;273(1):73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- Ghimire H, Hustedt EJ, Sahu ID, Inbaraj JJ, McCarrick R, Mayo DJ, Lorigan GA. Distance Measurements on a Dual-Labeled TOAC AChR M2δ Peptide in Mechanically Aligned DMPC Bilayers via Dipolar Broadening CW-EPR Spectroscopy. The Journal of Physical Chemistry B. 2012;116(12):3866–3873. doi: 10.1021/jp212272d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghimire H, McCarrick RM, Budil DE, Lorigan GA. Significantly Improved Sensitivity of Q-Band PELDOR/DEER Experiments Relative to X-Band Is Observed in Measuring the Intercoil Distance of a Leucine Zipper Motif Peptide (GCN4-LZ) Biochemistry. 2009;48(25):5782–5784. doi: 10.1021/bi900781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OH, McConnel Hm. A NITROXIDE-MALEIMIDE SPIN LABEL. Proceedings of the National Academy of Sciences of the United States of America. 1966;55(1):8. doi: 10.1073/pnas.55.1.8. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, Columbus L, Hideg K, Altenbach C, Hubbell WL. Structure of the KcsA potassium channel from Streptomyces lividans: A site-directed spin labeling study of the second transmembrane segment. Biochemistry. 1999;38(32):10324–10335. doi: 10.1021/bi990856k. [DOI] [PubMed] [Google Scholar]
- Hassner A, Alexanian V. SYNTHETIC METHODS .12. DIRECT ROOM-TEMPERATURE ESTERIFICATION OF CARBOXYLIC-ACIDS. Tetrahedron Letters. 1978;(46):4475–4478. [Google Scholar]
- Hilger D, Jung H, Padan E, Wegener C, Vogel KP, Steinhoff HJ, Jeschke G. Assessing oligomerization of membrane proteins by four-pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E-coli. Biophysical Journal. 2005;89(2):1328–1338. doi: 10.1529/biophysj.105.062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH. STUDIES ON PHOSPHATIDYLCHOLINE VESICLES. FORMATION AND PHYSICAL CHARACTERISTICS. Biochemistry. 1969;8(1):344. doi: 10.1021/bi00829a048. &. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nature Structural Biology. 2000;7(9):735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Current Opinion in Structural Biology. 1998;8(5):649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, McHaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4(7):779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- Itaya K. a. U., M. A new micromethod for the colorimetric determination of inorganic phosphate. Clin. Chim. Acta. 1966;14:361–366. doi: 10.1016/0009-8981(66)90114-8. [DOI] [PubMed] [Google Scholar]
- Jeschke G. DEER Distance Measurements on Proteins. In: Johnson MA, Martinez TJ, editors. Annual Review of Physical Chemistry. Vol 63. 2012. pp. 419–446. Vol. 63. [DOI] [PubMed] [Google Scholar]
- Jeschke G. Interpretation of Dipolar EPR Data in Terms of Protein Structure. In: Harmer C. R. T. a. J. R., editor. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Vol. 152. Springer; Heidelberg New York Dordrecht London: 2014. pp. 83–120. [Google Scholar]
- Kalai T, Schindler J, Balog M, Fogassy E, Hideg K. Synthesis and resolution of new paramagnetic alpha-amino acids. Tetrahedron. 2008;64(6):1094–1100. doi: 10.1016/j.tet.2007.11.020. [DOI] [Google Scholar]
- Klug CS, Feix JB. Methods and applications of site-directed spin Labeling EPR Spectroscopy. In: Correia JJ, Detrich HW, editors. Biophysical Tools for Biologists: Vol 1 in Vitro Techniques. Vol. 84. 2008. pp. 617–658. [DOI] [PubMed] [Google Scholar]
- Klug CS, Su WY, Feix JB. Mapping of the residues involved in a proposed beta-strand located in the ferric enterobactin receptor FepA using site-directed spin-labeling. Biochemistry. 1997;36(42):13027–13033. doi: 10.1021/bi971232m. [DOI] [PubMed] [Google Scholar]
- Kuchinka E, Seelig J. INTERACTION OF MELITTIN WITH PHOSPHATIDYLCHOLINE MEMBRANES - BINDING ISOTHERM AND LIPID HEADGROUP CONFORMATION. Biochemistry. 1989;28(10):4216–4221. doi: 10.1021/bi00436a014. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Subczynski WK, Hyde JS. OXYGEN-TRANSPORT PARAMETER IN MEMBRANES AS DEDUCED BY SATURATION RECOVERY MEASUREMENTS OF SPIN-LATTICE RELAXATION-TIMES OF SPIN LABELS. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1982;79(6):1854–1858. doi: 10.1073/pnas.79.6.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrabee AL. TIME-DEPENDENT CHANGES IN THE SIZE DISTRIBUTION OF DISTEAROYLPHOSPHATIDYLCHOLINE VESICLES. Biochemistry. 1979;18(15):3321–3326. doi: 10.1021/bi00582a019. [DOI] [PubMed] [Google Scholar]
- Lasic DD. THE MECHANISM OF VESICLE FORMATION. Biochemical Journal. 1988;256(1):1–11. doi: 10.1042/bj2560001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasic DD, Martin FJ. ON THE MECHANISM OF VESICLE FORMATION. Journal of Membrane Science. 1990;50(2):215–222. doi: 10.1016/s0376-7388(00)80317-8. [DOI] [Google Scholar]
- Margittai M, Langen R. Template-assisted filament growth by parallel stacking of tau. [Article] Proceedings of the National Academy of Sciences of the United States of America. 2004;101(28):10278–10283. doi: 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Langen R. Spin labeling analysis of amyloids and other protein aggregates. In: Kheterpal I, Wetzel R, editors. Amyloid, Prions, and Other Protein Aggregates, Pt C. Vol. 413. 2006. pp. 122–139. [DOI] [PubMed] [Google Scholar]
- Marsh D. Electron spin resonance in membrane research: protein-lipid interactions from challenging beginnings to state of the art. European Biophysics Journal with Biophysics Letters. 2010;39(4):513–525. doi: 10.1007/s00249-009-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D, Jost M, Peggion C, Toniolo C. TOAC spin labels in the backbone of alamethicin: EPR studies in lipid membranes. Biophysical Journal. 2007;92(2):473–481. doi: 10.1529/biophysj.106.092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RT, McLaughlin S, Baldo G, Manivannan K. THE ELECTROSTATIC POTENTIAL DUE TO A SINGLE FIXED CHARGE AT A MEMBRANE-SOLUTION INTERFACE. Biophysical Journal. 1988;53(2):A128–A128. [Google Scholar]
- Mchaourab HS, Hyde JS, Feix JB. BINDING AND STATE OF AGGREGATION OF SPIN-LABELED CECROPIN AD IN PHOSPHOLIPID-BILAYERS - EFFECTS OF SURFACE-CHARGE AND FATTY ACYL-CHAIN LENGTH. Biochemistry. 1994;33(21):6691–6699. doi: 10.1021/bi00187a040. [DOI] [PubMed] [Google Scholar]
- McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme, correlation with protein structure and dynamics. Biochemistry. 1996;35(24):7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- McHaourab HS, Oh KJ, Fang CJ, Hubbell WL. Conformation of T4 lysozyme in solution. Hinge-bending motion and the substrate-induced conformational transition studied by site-directed spin labeling. Biochemistry. 1997;36(2):307–316. doi: 10.1021/bi962114m. [DOI] [PubMed] [Google Scholar]
- McLaughlin S. ELECTROSTATIC POTENTIALS AT MEMBRANE SOLUTION INTERFACES. 1977.
- McLaughlin S. THE ELECTROSTATIC PROPERTIES OF MEMBRANES. Annual Review of Biophysics and Biophysical Chemistry. 1989;18:113–136. doi: 10.1146/annurev.biophys.18.1.113. [DOI] [PubMed] [Google Scholar]
- McNulty JC, Millhauser GL. TOAC - The rigid nitroxide side chain. Distance Measurements in Biological Systems by Epr. 2000;19:277–307. [Google Scholar]
- Michaels Dm, Horwitz AF, Klein MP. TRANSBILAYER ASYMMETRY AND SURFACE HOMOGENEITY OF MIXED PHOSPHOLIPIDS IN COSONICATED VESICLES. Biochemistry. 1973;12(14):2637–2645. doi: 10.1021/bi00738a014. [DOI] [PubMed] [Google Scholar]
- Milov AD, Maryasov AG, Tsvetkov YD. Pulsed electron double resonance (PELDOR) and its applications in free-radicals research. Applied Magnetic Resonance. 1998;15(1):107–143. [Google Scholar]
- Milov AD, Ponomarev AB, Tsvetkov YD. ELECTRON ELECTRON DOUBLE-RESONANCE IN ELECTRON-SPIN ECHO - MODEL BIRADICAL SYSTEMS AND THE SENSITIZED PHOTOLYSIS OF DECALIN. Chemical Physics Letters. 1984;110(1):67–72. doi: 10.1016/0009-2614(84)80148-7. [DOI] [Google Scholar]
- Milov AD, Samoilova RI, Tsvetkov YD, De Zotti M, Formaggio F, Toniolo C, Raap J. Structure of Self-Aggregated Alamethicin in ePC Membranes Detected by Pulsed Electron-Electron Double Resonance and Electron Spin Echo Envelope Modulation Spectroscopies. Biophysical Journal. 2009;96(8):3197–3209. doi: 10.1016/j.bpj.2009.01.026. doi: http://dx.doi.org/10.1016/j.bpj.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milov AD, Tsvetkov YD, Formaggio F, Crisma M, Toniolo C, Raap J. Self-assembling properties of membrane-modifying peptides studied by PELDOR and CW-ESR spectroscopies. Journal of the American Chemical Society. 2000;122(16):3843–3848. doi: 10.1021/ja993870t. [DOI] [PubMed] [Google Scholar]
- Molin YN, Salikhov KM, Zamaraev KI. Spin Exchange. Springer; Berlin: 1980. [Google Scholar]
- Newstadt JP, Mayo DJ, Inbaraj JJ, Subbaraman N, Lorigan GA. Determining the helical tilt of membrane peptides using electron paramagnetic resonance spectroscopy. [Article] Journal of Magnetic Resonance. 2009;198(1):1–7. doi: 10.1016/j.jmr.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen RD, Che KP, Gelb MH, Robinson BH. A ruler for determining the position of proteins in membranes. Journal of the American Chemical Society. 2005;127(17):6430–6442. doi: 10.1021/ja042782s. [DOI] [PubMed] [Google Scholar]
- Ogawa S, McConnel Hm. SPIN-LABEL STUDY OF HEMOGLOBIN CONFORMATIONS IN SOLUTION. Proceedings of the National Academy of Sciences of the United States of America. 1967;58(1):19. doi: 10.1073/pnas.58.1.19. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, McConnel Hm, Horwitz A. OVERLAPPING CONFORMATION CHANGES IN SPIN-LABELED HEMOGLOBIN. Proceedings of the National Academy of Sciences of the United States of America. 1968;61(2):401. doi: 10.1073/pnas.61.2.401. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistolesi S, Pogni R, Feix JB. Membrane insertion and bilayer perturbation by antimicrobial peptide CM15. Biophysical Journal. 2007;93(5):1651–1660. doi: 10.1529/biophysj.107.104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyka J, Ilnicki J, Altenbach C, Hubbell WL, Froncisz W. Accessibility and dynamics of nitroxide side chains in T4 lysozyme measured by saturation recovery EPR. Biophysical Journal. 2005;89(3):2059–2068. doi: 10.1529/biophysj.105.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin ZH, Cafiso DS. Membrane structure of protein kinase C and calmodulin binding domain of myristoylated alanine rich C kinase substrate determined by site-directed spin labeling. Biochemistry. 1996;35(9):2917–2925. doi: 10.1021/bi9521452. [DOI] [PubMed] [Google Scholar]
- Rabenstein MD, Shin YK. DETERMINATION OF THE DISTANCE BETWEEN 2 SPIN LABELS ATTACHED TO A MACROMOLECULE. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(18):8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassat A, Rey P. NITROXIDES .23. PREPARATION OF AMINO-ACID FREE RADICALS AND THEIR COMPLEX SALTS. Bulletin De La Societe Chimique De France. 1967;(3):815–818. [PubMed] [Google Scholar]
- Sahu ID, Hustedt EJ, Ghimire H, Inbaraj JJ, McCarrick RM, Lorigan GA. CW dipolar broadening EPR spectroscopy and mechanically aligned bilayers used to measure distance and relative orientation between two TOAC spin labels on an antimicrobial peptide. Journal of Magnetic Resonance. 2014;249(0):72–79. doi: 10.1016/j.jmr.2014.09.020. doi: http://dx.doi.org/10.1016/j.jmr.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov ES, Erilov DA, Milov AD, Tsvetkov YD, Peggion C, Formaggio F, Dzuba SA. Location and aggregation of the spin-labeled peptide trichogin GA IV in a phospholipid membrane as revealed by pulsed EPR. Biophysical Journal. 2006;91(4):1532–1540. doi: 10.1529/biophysj.105.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Feix JB. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2006;1758(9):1245–1256. doi: 10.1016/j.bbamem.2006.02.021. doi: http://dx.doi.org/10.1016/j.bbamem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Scarpelli F, Drescher M, Rutters-Meijneke T, Holt A, Rijkers DTS, Killian JA, Huber M. Aggregation of Transmembrane Peptides Studied by Spin-Label EPR. Journal of Physical Chemistry B. 2009;113(36):12257–12264. doi: 10.1021/jp901371h. [DOI] [PubMed] [Google Scholar]
- Schreier S, Bozelli JC, Marín N, Vieira RFF, Nakaie CR. The spin label amino acid TOAC and its uses in studies of peptides: chemical, physicochemical, spectroscopic, and conformational aspects. Biophysical Reviews. 2012;4(1):45–66. doi: 10.1007/s12551-011-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig J. Thermodynamics of lipid-peptide interactions. Biochimica Et Biophysica Acta-Biomembranes. 2004;1666(1-2):40–50. doi: 10.1016/j.bbamem.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Shafer AM, Nakaie CR, Deupi X, Bennett VJ, Voss JC. Characterization of a conformationally sensitive TOAC spin-labeled substance P. Peptides. 2008;29(11):1919–1929. doi: 10.1016/j.peptides.2008.08.002. doi: http://dx.doi.org/10.1016/j.peptides.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Shafer DE, Inman JK, Lees A. Reaction of tris(2-carboxyethyl)phosphine (TCEP) with maleimide and alpha-haloacyl groups: Anomalous elution of TCEP by gel filtration. Analytical Biochemistry. 2000;282(1):161–164. doi: 10.1006/abio.2000.4609. [DOI] [PubMed] [Google Scholar]
- Smirnov AI. Post-processing of IEPR spectra by convolution filtering: Calculation of a harmonics' series and automatic separation of fast-motion components from spin-label EPR spectra. Journal of Magnetic Resonance. 2008;190(1):154–159. doi: 10.1016/j.jmr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Smirnov AI, Belford RL. Rapid Quantitation from Inhomogeneously Broadened EPR-Spectra by a Fast Convolution Algorithm. Journal of Magnetic Resonance Series A. 1995;113(1):65–73. doi: 10.1006/jmra.1995.1057. [DOI] [Google Scholar]
- Smirnov AI, Clarkson RB, Belford RL. EPR linewidth (T-2) method to measure oxygen permeability of phospholipid bilayers and its use to study the effect of low ethanol concentrations. Journal of Magnetic Resonance Series B. 1996;111(2):149–157. doi: 10.1006/jmrb.1996.0073. [DOI] [PubMed] [Google Scholar]
- Smirnova TI, Smirnov AI, Clarkson RB, Bedford RL. W-Band (95 Ghz) EPR Spectroscopy of Nitroxide Radicals with Complex Proton Hyperfine-Structure - Fast Motion. Journal of Physical Chemistry. 1995;99(22):9008–9016. doi: 10.1021/J100022a011. [DOI] [Google Scholar]
- Smirnova TI, Smirnov AI, Clarkson RB, Belford RL. Accuracy of Oxygen Measurements in T-2 (Line-Width) Epr Oximetry. Magnetic Resonance in Medicine. 1995;33(6):801–810. doi: 10.1002/mrm.1910330610. [DOI] [PubMed] [Google Scholar]
- Smirnova TI, Smirnov AI, Clarkson RB, Belford RL, Kotake Y, Janzen EG. High-frequency (95 GHz) EPR spectroscopy to characterize spin adducts. Journal of Physical Chemistry B. 1997;101(19):3877–3885. doi: 10.1021/Jp963066i. [DOI] [Google Scholar]
- Steinhoff HJ, Dombrowsky O, Karim C, Schneiderhahn C. 2-DIMENSIONAL DIFFUSION OF SMALL MOLECULES ON PROTEIN SURFACES - AN EPR STUDY OF THE RESTRICTED TRANSLATIONAL DIFFUSION OF PROTEIN-BOUND SPIN LABELS. European Biophysics Journal with Biophysics Letters. 1991;20(5):293–303. doi: 10.1007/bf00450565. [DOI] [PubMed] [Google Scholar]
- Steinhoff HJ, Radzwill N, Thevis W, Lenz V, Brandenburg D, Antson A, Wollmer A. Determination of interspin distances between spin labels attached to insulin: Comparison of electron paramagnetic resonance data with the x-ray structure. Biophysical Journal. 1997;73(6):3287–3298. doi: 10.1016/S0006-3495(97)78353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoller S, Sicoli G, Baranova TY, Bennati M, Diederichsen U. TOPP: A Novel Nitroxide-Labeled Amino Acid for EPR Distance Measurements. Angewandte Chemie-International Edition. 2011;50(41):9743–9746. doi: 10.1002/anie.201103315. [DOI] [PubMed] [Google Scholar]
- Subczynski W, Widomska J, Wisniewska A, Kusumi A. Saturation-Recovery Electron Paramagnetic Resonance Discrimination by Oxygen Transport (DOT) Method for Characterizing Membrane Domains. In: McIntosh T, editor. Lipid Rafts. Vol. 398. Humana Press; 2007. pp. 143–157. [DOI] [PubMed] [Google Scholar]
- Syryamina VN, De Zotti M, Peggion C, Formaggio F, Toniolo C, Raap J, Dzuba SA. A Molecular View on the Role of Cholesterol upon Membrane Insertion, Aggregation, and Water Accessibility of the Antibiotic Lipopeptide Trichogin GA IV As Revealed by EPR. [Article] Journal of Physical Chemistry B. 2012;116(19):5653–5660. doi: 10.1021/jp301660a. [DOI] [PubMed] [Google Scholar]
- Thomas L, Scheidt HA, Bettio A, Huster D, Beck-Sickinger AG, Arnold K, Zschornig O. Membrane interaction of neuropeptide Y detected by EPR and NMR spectroscopy. Article]. Biochimica Et Biophysica Acta-Biomembranes. 2005;1714(2):103–113. doi: 10.1016/j.bbamem.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Tominaga M, Barbosa SR, Poletti EF, Zukerman-Schpector J, Marchetto R, Schreier S, Nakaie CR. Fmoc-POAC: (9-fluorenylmethyloxycarbonyl)-2,2,5,5-tetramethylpyrrolidine-N-oxyl-3- amino-4-carboxylic acid : A novel protected spin labeled beta-amino acid for peptide and protein chemistry. Chemical & Pharmaceutical Bulletin. 2001;49(8):1027–1029. doi: 10.1248/cpb.49.1027. [DOI] [PubMed] [Google Scholar]
- Torchilin VP. Tat peptide-mediated intracellular delivery of pharmaceutical nanocarriers. Advanced Drug Delivery Reviews. 2008;60(4-5):548–558. doi: 10.1016/j.addr.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Traikia M, Warschawski DE, Recouvreur M, Cartaud J, Devaux PF. Formation of unilamellar vesicles by repetitive freeze-thaw cycles: characterization by electron microscopy and P-31-nuclear magnetic resonance. European Biophysics Journal with Biophysics Letters. 2000;29(3):184–195. doi: 10.1007/s002490000077. [DOI] [PubMed] [Google Scholar]
- Turner AL, Braide O, Mills FD, Fanucci GE, Long JR. Residue specific partitioning of KL4 into phospholipid bilayers. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2014;1838(12):3212–3219. doi: 10.1016/j.bbamem.2014.09.006. doi: http://dx.doi.org/10.1016/j.bbamem.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Van Vleck JH. THE DIPOLAR BROADENING OF MAGNETIC RESONANCE LINES IN CRYSTALS. Physical Review. 1948;74(9):1168–1183. doi: 10.1103/PhysRev.74.1168. [DOI] [Google Scholar]
- Victor KG, Cafiso DS. Location and dynamics of basic peptides at the membrane interface: electron paramagnetic resonance spectroscopy of tetramethyl-piperidine-N-oxyl-4-amino-4-carboxylic acid-labeled peptides. Biophysical Journal. 2001;81(4):2241–2250. doi: 10.1016/S0006-3495(01)75871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinov MA, Rivera-Rivera I, Smirnov AI. Surface Electrostatics of Lipid Bilayers by EPR of a pH-Sensitive Spin-Labeled Lipid. Biophysical Journal. 2013;104(1):106–116. doi: 10.1016/j.bpj.2012.11.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieprecht T, Seelig J. Isothermal titration calorimetry for studying interactions between peptides and lipid membranes. In: Simon T. J. M. Sidney A., editor. Current Topics in Membranes. Volume 52. Academic Press; 2002. pp. 31–56. [Google Scholar]
- Wright K, Sarciaux M, de Castries A, Wakselman M, Mazaleyrat JP, Toffoletti A, Toniolo C. Synthesis of enantiomerically pure cis- and trans-4-amino-l-oxyl-2,2,6,6-tetramethylpiperidine-3-carboxylic acid: A spin-labelled, cyclic, chiral beta-amino acid, and 3D-Structural analysis of a doubly spin-labelled beta-hexapeptide. European Journal of Organic Chemistry. 2007;(19):3133–3144. doi: 10.1002/ejoc.200700153. [DOI] [Google Scholar]
- Yamaguchi T, Nomura M, Matsuoka T, Koda S. Effects of frequency and power of ultrasound on the size reduction of liposome. Chemistry and Physics of Lipids. 2009;160(1):58–62. doi: 10.1016/j.chemphyslip.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Yin JJ, Feix JB, Hyde JS. MAPPING OF COLLISION FREQUENCIES FOR STEARIC-ACID SPIN LABELS BY SATURATION-RECOVERY ELECTRON-PARAMAGNETIC RESONANCE. Biophysical Journal. 1990;58(3):713–720. doi: 10.1016/S0006-3495(90)82414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]