Abstract
The variegated expression of murine Ly49 loci has been associated with the probabilistic behavior of an upstream promoter active in immature cells, the Pro1 element. However, recent data suggests that Pro1 may be active in mature NK cells and function as an enhancer element. To directly assess if Pro1 transcripts are present in mature Ly49-expressing NK cells, RNA sequencing of the total transcript pool was performed on freshly isolated splenic NK cells sorted for expression of either Ly49G or Ly49I. No Pro1 transcripts were detected from the Ly49a, Ly49c or Ly49i genes in mature Ly49+ve NK cells that contained high levels of Pro2 transcripts. Low levels of Ly49g Pro1 transcripts were found in both Ly49G+ve and Ly49G−ve populations, consistent with the presence of a small population of mature NK cells undergoing Ly49g gene activation, as previously demonstrated by culture of splenic NK cells in IL-2. Ly49 gene reporter constructs containing Pro1 failed to show any enhancer activity of Pro1 on Pro2 in a mature Ly49-expressing cell line. Taken together, the results are consistent with Pro1 transcription playing a role in gene activation in developing NK, and argue against a role for Pro1 in Ly49 gene transcription by mature NK cells.
Keywords: RNA Seq, mouse NK, Ly49, transcription
Introduction
Natural killer (NK) cells are an important element of the immune system allowing for a rapid and innate response against virally infected and tumor cells1. The NK cell cytotoxic response is controlled by a balance of activating and inhibitory signals. In mice, the C-type lectin-related Ly49 receptor family is a major mediator of these signals2. These receptors detect the absence or alteration of specific major histocompatibility complex class I (MHC I) molecules found on most healthy cells. Ly49 receptors are expressed in a variegated and stochastic manner, generating a repertoire of NK cells displaying different combinations of Ly49 receptors, which allows for the detection of deviations in the expression of specific MHC I molecules.
This receptor variegation is achieved at the transcriptional level through the interplay of multiple promoters. The selective activation of Ly49 genes is regulated by Pro13, a promoter upstream of the core Pro24, 5 and Pro36 promoters responsible for the production of Ly49-coding transcripts. The Pro1 promoter is bidirectional, capable of transcribing in either the sense or antisense direction in a probabilistic manner that is controlled by the relative strength of competing transcription-factor binding sites7. Forward transcription from Pro1 produces a spliced sense transcript that traverses the downstream promoter regions, and may play a role in opening up the chromatin of the Pro2 and Pro3 promoters to allow gene transcription. Reverse transcription yields noncoding antisense transcripts that likely play no active role in silencing the gene, as deletion of the Pro1 region in Ly49 transgenes results in no detectable Ly49 expression, indicating that silence is the default state8. The variegated expression of Ly49 genes appears to be regulated primarily at the chromatin level, with expressed genes showing increased acetylation at H3K9 and at multiple residues of H4 in the Pro2 region9, 10. Regulation by DNA methylation is unlikely, due to a low level of CpG residues in the Pro2 region.
Although initial studies of the Ly49a gene identified a single transcriptional start site (TSS) at Pro211, additional studies suggest that there is no single TSS but rather that transcription can begin at various sites within major transcriptional regions throughout the Ly49 genes6, 12. Recent studies of the Ly49 loci have challenged the traditional view of promoters, as transcriptional start sites for the variegated Ly49 genes were not generally found to be associated with regions possessing transcriptional activity in in vitro promoter assays12. Traditional promoters were identified in genes not associated with probabilistic expression, including the activating Ly49d and h genes as well as the non-NK genes Ly49b and q, suggesting that probabilistic expression may be connected with the lack of clearly defined promoter elements. The Pro1 upstream promoter element found in variegated Ly49 genes was shown to be active in mature NK cells and function as an enhancer element, suggesting that it may play a role in Ly49 transcript initiation in mature NK cells13.
In the current study, we assess the total Ly49 transcriptional landscape of mature Ly49-expressing NK cells by RNA sequencing, revealing that Pro1 transcripts are very rare in mature NK cell populations, and further demonstrate that Pro1 lacks enhancer activity. We also investigate the unusual properties of the Ly49i gene, characterizing a novel Ly49i promoter (designated Pro2i) preceding exon -1b and identifying rare antisense transcripts originating from the core promoters Pro2 and Pro3.
Results
RNA sequencing of sorted Ly49G versus Ly49I-expressing splenic NK cells
The majority of previous studies of RNA expression by murine splenic NK cells have made use of gene arrays to assay gene expression profiles, and are therefore lacking information with regard to promoter utilization, alternative splicing, rare transcripts, and do not effectively discriminate between closely related Ly49 gene transcripts. In order to obtain a more precise determination of all Ly49 transcripts present in mature splenic NK cells, RNA was isolated from freshly isolated Ly49G or Ly49I-expressing splenic NK cells, to avoid artifacts associated with culture of NK cells in cytokines. The Ly49G-specific monoclonal antibody 4D1114 and the Ly49C/I –specific monoclonal 5E615, were used to sort 4D11-positive/5E6-negative versus 5E6-positive/4D11-negative NK cell subsets from C57BL/6 mice (Figure 1). These anti-Ly49 antibodies were chosen due to their ability to recognize a substantial fraction of the NK cells present in C57BL/6 spleen (50% of NK for 4D11, 48% for 5E6). The subsets obtained with this combination of antibodies would theoretically represent licensed Ly49C/I+ve NK cells that bind to the MHC H-2b present in C57BL/6 and unlicensed Ly49G+ve cells that do not bind to H-2b16, 17. However, the 5E6 antibody fails to recognize Ly49C in C57BL/6 due to the strong cis interaction of Ly49C with H-2b on the NK cell18, 19. 5E6 was shown to recognize Ly49C in BALB/c mice that express H-2d MHC, which is not a strong Ly49C ligand20. FACS analysis of C57BL/6 NK cells with the Ly49I–specific YLI-90 antibody together with 5E6 revealed a coincident staining pattern, whereas use of the Ly49C-specific 4LO3311 antibody identified the Ly49C-expressing subset18. Therefore, the subsets studied using the 4D11 and 5E6 antibodies in C57BL/6 mice are in fact Ly49G+ve/Ly49I−ve (G+/I−) and Ly49I+ve/Ly49G−ve (I+/G−) and will be referred to as such in this study. The complete lack of Ly49C recognition by the 5E6 antibody in C57BL/6 mice was confirmed by the nearly identical levels of Ly49c transcripts in the I+/G− versus the G+/I− samples (Table 1). The partial masking of Ly49I by the H-2Kb ligand present on the surface of C57BL/6 NK cells is also suggested by the relatively low enrichment of Ly49i transcripts (~14 fold) in the I+/G− population compared to the high enrichment of Ly49g transcripts (~87 fold) in the G+/I− cells. Interestingly, transcripts for the Ly49f gene were enriched approximately 1.8 fold in the G+/I− population, in agreement with previous results from gene array studies that compared unlicensed NK cells to licensed NK cells, and found that Ly49f was the only gene up-regulated in unlicensed NK cells in all mouse models tested21. The G+/I− cells studied here are not a pure population of unlicensed cells, due to the presence of Ly49C-expressing cells that strongly recognize H-2Kb, thus accounting for the modest enrichment of Ly49f transcripts. The high enrichment of Ly49G-expressing cells does however reveal some additional genes associated with this subset. Table 2 lists gene transcripts that were enriched > 2 fold in the G+/I− NK cells. The modest level of enrichment for these genes likely reflects their expression by relatively small subsets of G+/I− cells, such as the presence of thymus-derived NK indicated by the enrichment of IL7r-alpha, CD3-gamma, and CD3-delta transcripts22. It is unlikely that this is due to Ly49G-expressing T cells, since there was no detectable T cell contamination in the sorted populations shown in Figure 1. The relative enrichment of these thymic NK markers in the G+/I− subset is likely due to the significantly higher percentage of thymus-derived NK that express Ly49G (15%) versus Ly49I (2%)22. The high level of enrichment of CD3-epsilon (~14 fold) is consistent with previous reports of intracellular expression of CD3-epsilon by activated NK cells23. The 2.5 fold enrichment of Ly49a transcripts in the G+/I− subset is likely due to the low inhibitory signaling present in these cells, thus allowing a greater window of opportunity for receptors that have a lower probability of activation, such as the increased expression of Ly49f on unlicensed NK cells21.
Figure 1.
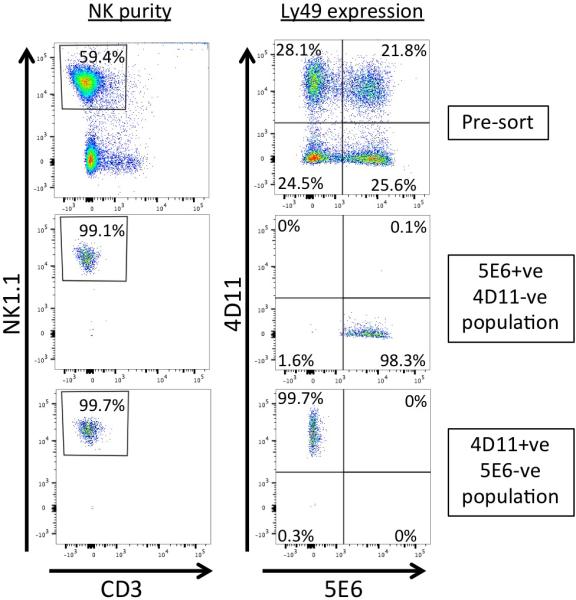
Purity of sorted NK cell populations. Flow cytometry analysis of a representative experiment of 3 independent purifications and sorts performed. Populations were live-gated based on scatter, and analyzed before sorting (Pre-sort; upper panels) or after sorting (middle and lower panels) with antibodies recognizing NK1.1, CD3, Ly49G2 (4D11), and Ly49I (5E6) as noted. Purity of NK1.1+/CD3- cells (NK purity) is shown in the left column. Ly49 expression profiles (Ly49 expression) are shown in the right column. Purity of sorted 5E6+ve/4D11-ve (middle) and 4D11+ve/5E6-ve (bottom) populations are shown in the lower rows. Percentages represent the percent of live cells (left column) or percent of gated NK1.1+/CD3- cells observed with the antibodies (right column).
Table 1.
Relative levels of Ly49 transcripts in G+/I− versus I+/G− NK cells.
| Gene name | Protein | Function | Fold change | p value |
|---|---|---|---|---|
| Klra9 | Ly49I | Inhibitory | −13.64 | 1.89E-12 |
| Klra14-ps | Ly49N | Pseudogene | −1.01 | 0.868 |
| Klra5 | Ly49E | Inhibitory | 1.20 | 0.098 |
| Klra3 | Ly49C | Inhibitory | 1.24 | 0.003 |
| Klra8 | Ly49H | Activating | 1.30 | 0.007 |
| Klra10 | Ly49J | Inhibitory | 1.34 | 0.030 |
| Klra13-ps | Ly49M | Pseudogene | 1.63 | 1.51E-04 |
| Klra4 | Ly49D | Activating | 1.77 | 1.19E-05 |
| Klra6 | Ly49F | Inhibitory | 1.81 | 3.70E-05 |
| Klra1 | Ly49A | Inhibitory | 2.47 | 7.40E-07 |
| Klra7 | Ly49G | Inhibitory | 86.89 | 5.91E-14 |
Table 2.
Genes expressed 2-fold or greater in G+/I− versus I+/G− NK cells.
| Gene | Protein | Function | Fold-change | p value |
|---|---|---|---|---|
| Ikzf2 | IKAROS Family Zinc Finger 2 (Helios) | Transcription factor | 2.10 | 2.17E-06 |
| Csf2 | GM-CSF | granulocyte/macrophage production/differentiation |
2.17 | 1.31E-05 |
| Clip3 | CAP-GLY Domain Containing Linker Protein 3 | links microtubules with cellular organelles. | 2.01 | 1.31E-04 |
| Klra1 | Ly49a | Class I MHC receptor | 2.47 | 7.40E-07 |
| Dll1 | Delta-Like 1 | Notch ligand | 2.31 | 9.82E-05 |
| Sox6 | SRY-box 6 | Transcription factor | 2.21 | 1.83E-05 |
| Chl1 | Cell Adhesion Molecule Homologous to L1CAM | Cell Adhesion | 2.59 | 2.41E-05 |
| Gpr55 | G Protein-Coupled Receptor 55 | Receptor for L-alpha-lysophosphatidylinositol | 2.64 | 1.71E-05 |
| Slamf6 | SLAM Family Member 6 | Coreceptor for NK cell activation | 2.85 | 3.38E-06 |
| Cd3g | CD3-gamma | TCR signaling | 3.24 | 4.83E-06 |
| Il7r | IL-7 Receptor Subunit Alpha | lymphocyte survival | 3.96 | 8.11E-08 |
| Cd3d | CD3-delta | TCR signaling | 2.54 | 3.40E-05 |
| Dpysl5 | Dihydropyrimidinase-Like 5 | Neuron differentiation, interacts with L1CAM | 3.21 | 1.03E-05 |
| Cd3e | CD3-epsilon | TCR signaling | 13.77 | 5.87E-07 |
| Klra7 | Ly49g | Class I MHC receptor | 86.89 | 5.91E-14 |
Ly49 expression is not associated with Pro1 transcription
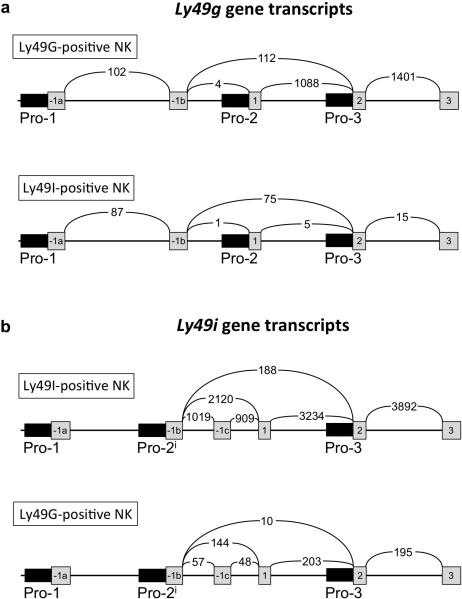
To directly assess if Pro1 transcripts can be found in mature Ly49-expressing NK cells, the RNA sequence data spanning the complete Ly49 gene cluster was analyzed in detail, including a survey of individual sequences to identify novel transcripts that are not annotated in the reference mouse genome. Figure 2 shows a summary of Ly49g and Ly49i sense transcripts detected by RNA sequencing of freshly isolated Ly49G or Ly49I-expressing splenic NK cells. The number of spliced RNA sequences spanning 2 exons as detected by Sashimi analysis are shown. The majority of Ly49i transcripts originated within exon -1b, as previously reported by Gays et al.12, whereas Ly49g transcripts originated primarily in exon 1, indicating that Pro2 is the dominate promoter in splenic NK cells. The partial masking of Ly49I by H-2b is suggested by the presence of a low level of Ly49I-coding transcripts in the G+/I− population (~5% of the transcript level in I+/G−), indicating that a subset of the Ly49I-expressing NK cells are not efficiently recognized by the 5E6 antibody. In contrast, potential Ly49G-coding RNAs were rare in the I+/G− population, and these RNAs did not exceed the level of non-translated Pro1 transcripts, indicating a lack of translatable Ly49g mRNAs in this population, consistent with the efficient detection of Ly49G by the 4D11 antibody.
Figure 2.
Evaluation of Pro1, Pro2 and Pro3 transcription in G+/I− versus I+/G− splenic NK cells. The exon structures of the 5’ region of the Ly49g and Ly49i genes are shown, with exons depicted as gray boxes, and promoter regions shown as thin black rectangles with the name of the exon or promoter element listed underneath. The arched lines containing numbers indicate the number of RNA sequences found that span the two exons linked by the line. Numbers are the average of three separate cell-sorting experiments. (a) Ly49g transcripts detected in either sorted G+/I− NK cells (Ly49G+ve NK, upper panel), or I+/G− NK cells (Ly49I+ve NK, lower panel). (b) Ly49i transcripts detected in either sorted I+/G− NK cells (Ly49I+ve NK, upper panel), or G+/I− NK cells (Ly49G+ve NK, lower panel).
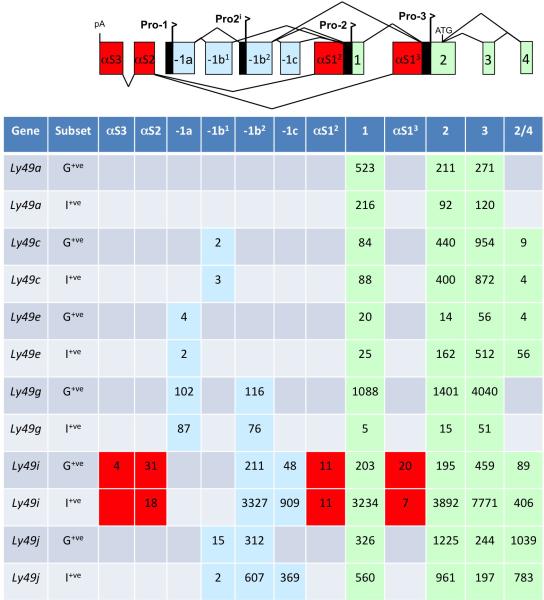
A summary of the spliced 5’ RNA sequences observed in inhibitory Ly49 genes is shown in Figure 3. Ly49 Pro1 transcripts containing exon -1a were only found for 2 genes, Ly49e and Ly49g. The complete lack of Pro1 transcripts in Ly49I-expressing cells indicates that Pro1 transcriptional activity is not required for gene expression. The presence of Pro1 transcripts from the Ly49e and Ly49g genes in Ly49-expressing splenic NK cells may be related to the ability of these genes to be activated in mature NK cells by stimulation with IL-224, 25, suggesting that the Pro1 transcripts could be derived from cells that have received a stimulatory signal, and are in the process of gene activation. Notably, there is no significant difference in the level of Pro1 transcripts between G+/I− and I+/G− subsets, even though there are 87-fold more Ly49g transcript reads in the G+/I− population. The low level of Ly49g Pro1 transcription likely indicates a small population of cells that are in the process of activating the Ly49g gene. In the G+/I− cells, this would represent activation of a second Ly49g allele, as previously observed for purified monoallelic Ly49GB6-expressing NK cells after culture in IL-225.
Figure 3.
Ly49 Pro1 transcripts are not associated with gene expression in mature NK cells. A schematic depicting all of the 5’ Ly49 exons revealed by RNA sequencing analysis is shown. Antisense exons are shown as red rectangles. The first antisense exons originating from either Pro2 or Pro3 are labeled αS12 and αS13 respectively. The second and third antisense exons are labeled αS2 and αS3. The first 4 exons contained in Ly49 Pro2 transcripts are shown as labeled green rectangles. Upstream exons derived from either Pro1 or Pro2i are shown as labeled light blue rectangles. The exon labeled -1a is the first exon of Pro1 transcripts. The exon labeled -1b1 is the upstream exon found in Ly49c transcripts, whereas the exon labeled -1b2 is used by Ly49g Pro1 transcripts and is the first exon of Pro2i transcripts. Exon -1c is an alternative exon used by some Pro2i transcripts. The table below lists spliced exons observed for either the G+/I− (Ly49G+ve) or I+/G− (Ly49I+ve) NK cell populations. Numbers listed are the average number of events from 3 independent cell-sorting experiments.
Detection of novel Ly49i transcripts
RNA sequencing of Ly49I-expressing splenic NK cells revealed the unexpected presence of antisense Ly49i transcripts originating from the Pro2 and Pro3 regions spliced to two antisense exons upstream of Pro1 that will be referred to as antisense exons 2 and 3 (asEx2 and asEx3; Figure 3). Antisense transcripts were not detected for any other Ly49 genes, indicating that the presence of antisense transcripts may be due to the use of a promoter upstream of exon -1b as the primary site of transcription in the Ly49i gene, as recently reported by Gays et al12. We have named this novel promoter Pro2i, and the transcriptional activity of this element is analyzed in the next section. To confirm the continuity of antisense transcripts identified by RNA sequencing, RT-PCR was performed to isolate antisense transcripts originating from promoters Pro2 and Pro3, as well as novel alternatively spliced sense transcripts originating in exon -1b and containing a previously unreported 132 bp non-coding exon (-1c) from both C57BL/6 and BALB/c mice. Novel Ly49i transcripts originating in exon -1b and containing exon -1c have been deposited in GenBank (KU645200-KU645202). The Ly49i Pro2 and Pro3 antisense transcripts utilized alternative exon 2 splice acceptor sites, resulting in either a 198 or 279 bp antisense exon 2 (GenBank#s KU645199, KU662345-KU662346). Pro2i transcripts were isolated from BALB/c spleen and bone marrow cDNA, indicating that Pro2i activity is not specific to the C57BL/6 Ly49i gene. Many of the antisense transcripts found in B6 mice could not be confirmed in BALB/c mice, since the splice acceptor for Ly49i antisense exon 3 is not present in the BALB/c Ly49i gene26. Only one Ly49i antisense transcript, asPro2-Ex2, was confirmed in BALB/c.
Pro2i promoter activity
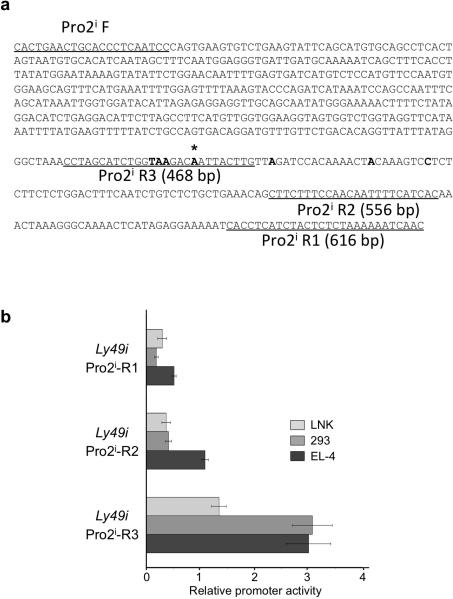
Given the large number of transcripts originating from exon -1b, and the previously characterized dominant start site in this region12, the region preceding exon -1b was investigated for the presence of a novel promoter element (Pro2i). To evaluate the promoter activity the Pro2i region, a series of pGL3 luciferase constructs spanning this sequence was generated (Figure 4a). The shortest fragment tested, R3, which ended at the dominant transcriptional start site12, showed weak promoter activity in the mature mouse NKT cell line EL-4 that expresses Ly49A/G27 and demonstrates Pro2 promoter activity3, but not in the immature NK cell line LNK that lacks Ly49 expression but supports Pro1 transcription3 (Figure 4b). The low activity of the putative Pro2i promoter in EL-4 cells indicated that either this promoter has a cellular specificity that is distinct from the Ly49a and Ly49g Pro2 elements, or that a distal enhancer element might be required to produce substantial activity.
Figure 4.
Promoter activity of Ly49i Pro2i constructs. (a) Sequences of Pro2i fragments. Underlined regions indicate sequence of primers used to generate each fragment. Bold nucleotides indicate TSSs identified by Gays et al. (12), with an asterisk signifying the dominant TSS. (b) Activity of pGL3 reporter constructs transfected into LNK, HEK293 (293), or EL-4 cells. The average fold activity of constructs relative to empty pGL3 vector from at least 3 independent experiments is shown. Error bars represent +/− 1 SEM, n=5.
The Ly49i Pro1 element does not exhibit enhancer activity
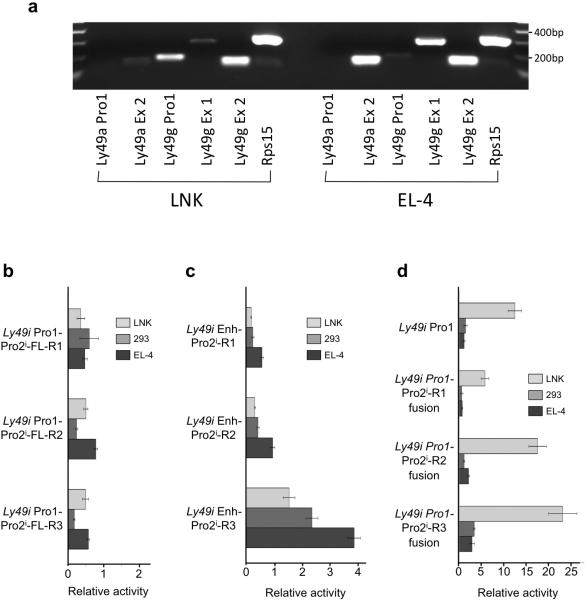
As it has recently been suggested that the distal upstream element Pro1 may act to enhance transcriptional activity of the downstream Ly49 promoters13, a series of Pro1:Pro2i reporter constructs were generated to test for interaction of these two elements. To test the ability of Pro1 to enhance transcription from the novel promoter in its native configuration, we generated pGL3 constructs containing the full Ly49i sequence spanning the ~ 3.5 kb region from Pro1 to Pro2i and measured their activity in either LNK cells that support Pro1 transcription or EL-4 cells that support Pro2 transcription. The transcriptional specificity of these two cells lines was confirmed by RT-PCR of Pro1 transcripts from LNK cells and Pro2 transcripts from EL-4 (Figure 5a). Specific Ly49g Pro1 transcripts were detected in LNK, but not in EL-4. Although only weak signals were present in LNK cells for exon 2 of Ly49a and exon 1 of Ly49g that is preferentially used by Pro2 (Figure 2a), EL-4 clearly showed amplification of these exons, indicating the presence of Pro2 activity. The LNK and EL-4 cell lines are therefore appropriate lines for the in vitro study of Pro1 and Pro2 transcriptional activity. All of the full-length Ly49i Pro1-Pro2i constructs tested showed less activity than the background activity seen in the pGL3-Basic negative control (Figure 5b). This suggests that there is some inhibitory element in the intervening sequence between Pro1 and Pro2i. Alternatively, a high level of antisense Pro1 activity could produce antisense RNA spanning the luciferase gene, thus inhibiting its activity. To rule out this possibility, Pro1:Pro2i reporter constructs were linearized at a restriction enzyme site downstream of the luciferase gene prior to transfection. Linearized reporter constructs did not shown any improvement in promoter activity (data not shown), therefore Pro1 antisense transcripts are not responsible for the decreased activity of Pro1:Pro2i reporter constructs.
Figure 5.
The Pro1 element does not possess enhancer activity. (a) Detection of Pro1 transcripts in LNK and Pro2 transcripts in EL-4. RT-PCR analysis of RNA purified from either LNK (left panel) or EL-4 (right panel) with Ly49a and Ly49g primers specific for Pro1 (Ly49a/g Pro1) or downstream exons is shown. Exon 2 (Ly49a/g Ex 2) of Ly49a or Ly49g is found in both Pro1 and Pro2 transcripts, however Ly49g exon1 (Ly49g Ex1) is preferentially used by Pro2 transcripts. Amplification of the ribosomal S15 protein (Rps15) is shown as a positive control. (b) Activity of constructs containing the full-length genomic sequence of the Ly49i gene from Pro1 to Pro2i, ending at each of the 3’ primers shown in Figure 3 (Ly49i Pro1-Pro2i-FL-[R1-R3]) were transfected into LNK, HEK293 (293), or EL-4 cells. (c) Activity of constructs containing Pro1 in the pGL3 enhancer site and Pro2i in the promoter site. (d) Activity of Pro1-Pro2i fusion constructs with intervening sequence between Pro1 and Pro2i removed. The average fold activity of constructs relative to empty pGL3 vector from at least 3 independent experiments is shown. Error bars represent +/− 1 SEM, n=5.
To test for possible Pro1 enhancer function without the intervening Ly49i sequence between Pro1 and Pro2i, Pro1 was cloned into the distal enhancer site of the pGL3 vector. There was no significant difference in activity between the Pro2i constructs with and without the Pro1 element in the enhancer site of pGL3 in any cell line tested (compare Figure 4b with Figure 5c).
To test for possible interaction between Pro1 and Pro2i if the two elements were in close proximity to each other, PCR-SOEing28 was used to generate Pro1-Pro2i fusion constructs. The Pro1-Pro2i fusion constructs showed no difference in activity in the mature EL-4 cell line, but greatly increased promoter activity, above either Pro2i-R3 or Pro1 alone, in the immature LNK cell line (Figure 5d). As Pro2i-R3 alone showed no activity in LNK, we performed a 5’RACE to confirm that transcripts were originating from the Pro2i element and that Pro2i was not acting as an enhancer for Pro1. The site of transcript initiation identified in the Pro1-Pro2i fusion corresponded to the major site identified by Gays et al.12. It appears that the Pro1 element can contribute to Pro2i activity in the immature LNK cell line, but as this relationship is dependent on the proximity of Pro1 and Pro2i, it does not function as an enhancer, and represents the addition of transcription factor binding sequences that conveys LNK cell-specific activity to the promoter complex.
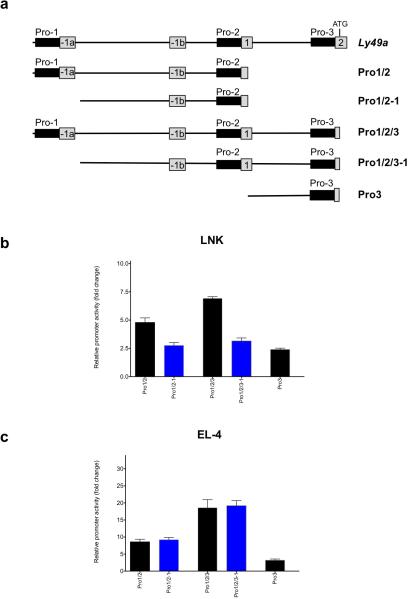
Ly49a Pro2 promoter activity is independent of Pro1 in Ly49-expressing cells
In order to identify additional Ly49 genomic regions that may contribute to promoter activity, promoter activity of the full Ly49a Pro1/Pro2/Pro3 region was analyzed. Figure 6a shows the constructs generated that contained various combinations of the Ly49 upstream elements. Figure 6b shows the activity of these constructs in the immature LNK cell line, and Figure 6c shows the activity in the mature Ly49-expressing EL-4 cells. The Pro2 promoter has been shown to be the major promoter used for the transcription of Ly49a in mature NK cells, as confirmed by the RNA sequence data presented in Figure 3. Constructs containing Pro2 and Pro3 had twice the activity of constructs containing Pro2 alone when studied in the EL-4 cell line. This enhancement was not observed in the LNK cell line. Although the Pro3 region had low activity when analyzed in isolation, it demonstrated substantial activity in EL-4 cells when combined with Pro2, possibly due to enhanced production of spliced transcripts originating from Pro2. Pro1 did not enhance Pro2/Pro3 activity in EL-4 cells, however it did increase the activity of constructs in LNK cells, consistent with Pro1 activity being limited to immature NK cells.
FIGURE 6.
Ly49a Pro2/Pro3 promoter activity is independent of Pro1 in Ly49-expressing cells. (a) Schematic of Ly49a promoter fragments analyzed. The upper line shows the complete Ly49a control region, with exons shown as numbered boxes, and promoter regions indicated by black boxes. The specific regions contained in each Ly49a-pGL3 reporter construct are shown below, with the name of the construct indicated to the right of each line. (b) Activity of constructs in LNK cells. (c) Activity of constructs in EL-4 cells. Blue-colored bars indicate constructs lacking the Pro1 region. The average fold activity of constructs relative to empty pGL3 vector from at least 3 independent experiments is shown. Error bars represent +/− 1 SEM, n=5.
Discussion
The current study represents the first comprehensive analysis of Ly49 transcription in mature Ly49-expressing splenic NK cells. The expression of Ly49 proteins represents the final stage of NK differentiation that determines their ability to detect missing self29 .It is therefore important to study a population of purified Ly49-positive NK cells to identify transcripts associated with gene expression. The results obtained in this study confirm the activity of the previously identified Ly49 promoters, and do not reveal any previously unknown promoter regions, however, novel spliced antisense transcripts originating from the Ly49i Pro2 and Pro3 promoters were detected.
With regard to the recent suggestion that Pro1 may function as an enhancer in mature NK cells, multiple lines of evidence presented here demonstrate that Pro1 transcripts are not associated with gene expression, and the Pro1 element likely plays no active role in protein expression in mature Ly49 expressing cells. In vitro promoter assays suggest that Pro1 does not enhance downstream Ly49 promoter activity. Luciferase reporter assays using large genomic fragments of the Ly49a gene revealed no enhancement of downstream promoter activity by Pro1 in its native genomic context in the Ly49-expressing EL-4 cell line. Some enhancement of transcriptional activity by Pro1 was seen in the LNK cell line, but it is likely due to translation of spliced Pro1 transcripts generated in these cells. RNA-sequencing analysis of freshly isolated ex vivo Ly49-expressing cells shows that transcripts originating from Pro1, which would be expected if it were acting as a promoter/enhancer in these cells, are totally absent in most variegated Ly49 genes and very rare in the genes where they are found.
An important difference between the current study and the recent results from Gays et al.13 is the use of freshly isolated, Ly49-expressing ex vivo NK cells for the analysis of Ly49 gene transcription. The purified NK cells used by Gays et al. were not selected for Ly49 expression, and were cultured for 12 days in IL-2, and thus represent an activated NK cell population. The Ly49g gene has been shown to be activated during culture of NK cells in IL-2, as has the Ly49e gene23, 24. In addition, Ly49g Pro1 transcripts were highly induced by treatment of mice with IL-2, and this was correlated with a rapid expansion of Ly49G+ve NK cells30. Therefore, the detection of Pro1 transcripts from these genes in purified NK cells cultured in IL-2 by Gays et al. is consistent with a role for Pro1 transcripts in the initial opening of the downstream Pro2 and Pro3 promoter region, resulting in Ly49 gene expression by mature NK cells. Furthermore, the use of sorted Ly49-expressing cells in the current study ensured that only mature NK cells that are producing Ly49 protein were assayed, and not immature NK that are in the process of activating the Ly49 genes.
Paradoxically, although the Gays et al. study detects Pro1 forward transcripts in multiple cell lines, they suggest that there is little to no Pro1 forward transcriptional activity, and very strong reverse activity. In addition, RT-PCR experiments revealed that in all cases where Pro1 forward transcripts were detected, reverse transcripts were detected at a lower level13. This may be due to an inherent instability of the non-coding antisense transcript, however it is important to note that very high levels of Pro1 forward transcripts are detected, in contrast to the results obtained with in vitro reporter assays. Several possible factors that may contribute to this discrepancy are as follows: 1) backbone sequences in the pGL series of reporter plasmids can affect activity, and we have chosen to use the pGL3 vector since we have found that the modifications made to reduce background transcriptional activity in the pGL4 vector had the opposite effect in the human and mouse NK cell lines we study; 2) changing the relative amount of flanking sequence in bidirectional promoter systems will change the relative strength of the competing promoters, and we have chosen core promoter fragments that have relatively balanced forward and reverse activity in order to study the switching properties; 3) the promoter activities in the Gays et al. study are reported relative to a very strong promoter, making Pro1 forward activity seem inconsequential; 4) Pro1 forward transcriptional activity is decreased or lost after extended periods of culture of the LNK line, making it important to use early passage cells for promoter analysis.
The model of Pro1 function suggests that forward transcripts originating from this element traverse the downstream promoters, displacing histones and allowing access of transcription factors required to activate the gene. The opening of the downstream promoter region may not require high levels of forward transcripts. The relatively low promoter activity found in Ly49 Pro1 is comparable to the activities detected in distal KIR promoter elements that have been associated with gene activation31, 32.
No Pro1 transcripts were found in mature Ly49-expressing NK cells for the remaining variegated inhibitory genes, Ly49a, Ly49c, Ly49i, and Ly49j. This supports the hypothesis that Pro1 activity is important for the process of gene activation but does not play a role in the expression of the Ly49 protein by mature NK cells. However, it is possible that the Pro1 element plays a role in maintaining an open chromatin configuration in a transcription-independent manner, and this would not be detected by in vitro reporter assays.
Initial studies of the specificity of the Pro1 promoter were performed before the antisense activity of the region was known. The extensive work of Gays et al. shows that reverse Pro1 activity is substantial and present in many cell types13. Conversely, the forward activity detected is weaker and more restricted. These findings are consistent with a model in which forward transcription from the Pro1 element is required for opening of the downstream regions required for gene expression. The production of reverse transcripts represents the default “off“ state, and therefore may not be as tightly regulated as forward transcription.
It remains unclear whether the rare antisense transcripts detected originating from the Ly49i Pro2 and Pro3 regions play any functional role in gene regulation at any point in NK development or whether they are merely a consequence of an inability to transcribe efficiently in the forward direction due to the competition from the upstream Pro2i element33. Studies of the human transcriptome have revealed that approximately 8% of promoters are bidirectional with start sites separated by less than 300 bp34. It may be that many more promoters have the capacity to transcribe in the antisense direction, but the dominant function of factors generating sense transcripts prevents antisense transcription, and inhibition of sense transcription is required to reveal antisense activity. The ability of upstream promoters to inhibit transcription from downstream promoters provides an explanation for the silencing of Pro1 in mature NK. Although Pro1 transcripts are required to open up the Pro2/Pro3 region of the gene, their continued presence would inhibit transcription factor binding to these promoters and reduce expression.
In summary, the data presented here confirm the immature NK cell-specificity of the Pro1 element and show that for each variegated Ly49 gene, expression can occur in the absence of Pro1 transcripts. It will be of interest to determine the exact molecular events that determine the “window of opportunity” for Ly49 gene activation and how they affect Pro1 activity.
Materials and Methods
Animals
C57BL/6 and BALB/c mice were maintained and bred in the NCI Frederick animal Breeding Facility. Animal care was provided in accordance with the procedures in, “A Guide for the Care and Use of Laboratory Animals”. Ethical approval for the animal experiments detailed in this manuscript was received from the Institutional Animal Care and Use Committee (Permit Number: 000386) at NCI-Frederick.
RNA-Seq library preparation and sequencing
For each of 3 independent experiments, single cell suspensions were prepared from the spleens of 20 C57BL/6Ncr mice. NK cells were initially enriched using the MACS NK Cell purification system (Miltenyi Biotec, San Diego, CA) as described by the manufacturer. The Fc receptors of the resulting populations of enriched NK cells were blocked with 2.4G2 and the cells were stained with antibodies to CD3, NK1.1 (eBioscience Inc, San Diego CA), Ly49G2 (4D11), and Ly49C/I (5E6). CD3−, NK1.1+,Ly49G+, Ly49C/I− and CD3−, NK1.1+,Ly49G−, Ly49C/I+ cells were sorted on a FACS Aria (BD Biosciences, San Jose, CA). Resulting populations were greater that 96% pure by post-sort analysis. Sorted NK cells were immediately processed for extraction of total RNA.
Stranded RNA-seq libraries were constructed from 0.4 μg total RNA using the TruSeq Stranded Total RNA Sample Prep Kits (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. The library insert sizes were approximately 175bp. Unique barcode adapters were applied to each library. Equal volumes of individual libraries were pooled and run on a MiSeq benchtop DNA sequencer (Illumina). The libraries were then repooled to equimolar concentrations based on the MiSeq demultiplexing results. The final pooled library was sequenced on a HiSeq2000 sequencer (Illumina) using TruSeq Version 3 chemistry and a bioinformatics pipeline built on RTA version 1.13.48 and CASAVA 1.8.2 software (Illumina). A minimum of 40 million 100 base read pairs were generated for each library. The RNA-seq dataset has been submitted to the GEO database, and are available under accession number GSE83153.
Raw Fastq files were aligned to mouse genome (mm10) using STAR (v. 2.3.0)35. Genes were subsequently counted using Rsubread36, and further analyzed for differential expression using limma-voom37 to obtain the fold-change values shown in Tables 1 and 2. Splice junctions were visualized with the Sashimi function of the Integrated Genome Viewer (IGV-v2.3.67). Only uniquely mapped reads were included in the analyses.
Cell collection, RNA Isolation, and cDNA Generation
Bone marrow and spleen cells were collected from two C57BL/6 (B6) and two BALB/c mice. Spleens were nicked with a pair of scissors to puncture the outer capsule and were then dissociated into a single cell suspension using a Dounce homogenizer. Bone Marrow was collected by cutting the ends of the femur and tibia bones and flushing out the marrow with phosphate buffered saline using a 23 gauge needle. Red blood cells were removed using ACK lysing buffer (Lonza, Walkersville, MD, USA) and NK cells were enriched by removal of non-NK cells using the EasySep Mouse NK Cell Enrichment Kit (STEMCELL Technologies, Vancouver, BC, Canada). RNA was extracted from the NK enriched cells using the Qiagen RNeasy Plus Mini Kit with an additional on-column DNase I digest using an RNase-Free DNase Set (Qiagen, Valencia, CA, USA) to reduce DNA contamination. cDNA copies of RNA transcripts were synthesized from isolated RNA with the TaqMan Reverse Transcription Reagents Kit (Applied Biosystems, Carlsbad, CA, USA) using the included random hexamer primers.
Ly49i Transcript identification by RT-PCR
PCR of cDNA was used to isolate copies of transcripts from Pro2i using a Pro2i exon forward primer (5’-GGACTTTCAATCTGTCTTTGCTG) and Exon 3 reverse primer (5’-GAGTTGCCAGGATACTGAACAC). Rare antisense (as) transcripts were isolated using a nested PCR scheme first using a set of outer primers: asPro2 forward 1 (5’-GAAGATAGAGAAGACACACTCCTG), asPro3 forward 1 (5’-CAAAAACCCTGACATGGACTGATCC), asExon3 reverse 1 (5’-GTATAGTCATGTGTAGTTTCCTGTG) and as Exon2 reverse (5’-CCTCATGACTAAGGATGCTGAAC). The ChargeSwitch PCR Clean-Up Kit (Invitrogen, Carlsbad, CA, USA) was used to remove these primers from the products of first round of the nested PCR and these products were then used as the template for a second round of PCR using an inner primer set: asPro2 forward 2 (5’-GAAGACACACTCCTGCAGAGAGG), asPro3 forward 2 (5’-GGACTGATCCCAATTTCCATCAC), asExon3 reverse 2 (5’-CTGTGATCTTCAGTTTTGAAGTGG), and the same asExon2 reverse primer as the first round. Extension steps of PCRs for antisense transcripts were performed at 65°C to prevent the polymerase from prematurely detaching from a long poly-A region in antisense exon 2. The identities of transcripts were confirmed by cloning PCR products into the TOPO TA Cloning PCR 2.1 vector (Invitrogen) and sequencing.
Ly49 Pro1 and Pro2 transcript identification in LNK and EL-4 cell lines
Total cellular RNA was isolated from LNK or EL-4 cells with the RNeasy Plus Mini Kit (Qiagen), and further purified using DNase I according to the manufacturer's instructions. cDNA was synthesized using Random Hexamer primer with the TaqMan® Reverse Transcription Reagents Kit (Applied Biosystems). PCR was performed with primers in exons –1a and –1b that are specific for the Pro1 transcripts of Ly49a or Ly49g, and primers in exons 1, 2, and 3 to detect Pro2 transcripts. Ly49g exon 1 is primarily found in Pro2 transcripts, but a low level (<4%) of Pro1 transcripts contain this exon (Figure 2a). Primers used were: Ly49a Pro1-Forward, 5’- GTCCAAGGGTGTGACTGGAAGG; Ly49a Pro1-Reverse, 5’- GAGAAGTGAGGACTTTGTAGTG; Ly49a Exon2-Forward, 5’- GCAGAAACAAGTGAGACCTGAGGAG; Ly49a Exon3-Reverse, 5’-CCAACACTGAAACAGCTACCAGAAG; Ly49g Pro1-Forward, 5’-CAAGTGATCAGCCTATTCTTGTG; Ly49g Pro1-Reverse, 5’-CTTGTGTGAGTTTTGTACTTCAG; Ly49g Exon1-Forward, 5’-TTCTCCACAGGAATCACTTCTCAG; Ly49g Exon2-Forward, 5’-GAGTCTTCAAGGTTGCAGAAACTAG; Ly49g Exon3-Reverse, 5’-GAGCTTCCAGGGGACTGAATACTC; Rps15-Forward, 5′-CCGAAGTGGAGCAGAAGAAG; Rps15-Reverse, Rev 5′-CTCCACCTGGTTGAAGGTC. Amplification was carried out using the ZymoTaq™ DNA Polymerase (Zymo Research, Irvine, CA) at 95°C for 10 minutes, and 38 cycles at 95°C for 15 seconds, 58°C for 45 seconds and 72°C for 20 seconds in a total volume of 50 μl in a SimpliAmp™ Thermal Cycler (Applied Biosystems). Products were separated on a 1% agarose gel stained with ethidium bromide and run for 90 minutes at 100 volts in 0.5x TBE buffer, then visualized under UV light.
Generation of promoter constructs
Fragments of progressively shorter length spanning the Pro2i region were generated by PCR using a forward primer (5’-CACTGAACTGTACCCTCAATCC) and a set of reverse primers: Pro2i reverse 1 (5’-GTTGATTTTTTAGAGAGTAGATGAGGTG), Pro2i reverse 2 (5’-GTGATGAAAATTGTTGGAAAGAAG), Pro2i reverse 3 (5’-CAAGTAATTGTCTTACCAGATGCTAGG) from a template plasmid generated from a previous study of the Ly49i region (GenBank AF173846)5. The Ly49i Pro1 construct was generated using primers Pro1 forward (5’-CATCTTCTCATTTCCTTGCTCTC) and Pro1 reverse (5’- GCAGCAAGCTGCTCTTCCTGTCC). Fragments spanning the full Pro1-Pro2i region were generated using the Pro1 forward primer and the Pro2i reverse primer set. Pro1-Pro2i fusion constructs were generated using PCR splicing by overlap extension24 using the Pro1 forward primer and Pro2i reverse primer set along with fusion primers Pro2i Fusion F (5’-GGACAGGAAGAGCAGCTTGCTGCCATCTGAGGACATTCTTAGCCTTC) and Pro1 Fusion R (5’-GAAGGCTAAGAATGTCCTCAGATGGCAGCAAGCTGCTCTTCCTGTCC).
Ly49a promoter constructs were generated by PCR of BAC DNA with the following primers: Pro1-F (5’-GAAGGACTATGTGTTTAGGC); Pro1-del-F (5’-CTGGAAGGTTTGAATGTGGG); Pro3-F (5’-CACCAGAACCACTTCTTGCT); Pro2-R (5’-AGCAAGAAGTGGTTCTGGTG); Pro3-R (5’- CTCAGTTCAAATGGTGCCTC).
Identities of fragments were confirmed by TA TOPO cloning PCR products into PCR 2.1 vector and sequencing. The genomic Ly49 gene fragments were too large for TA TOPO cloning; the StrataClone PCR Cloning Kit (Agilent Technologies, Santa Clara, CA, USA) was used instead. Fragments confirmed by sequencing were transferred from PCR 2.1 or StrataClone vector to pGL3 Basic (Promega, Madison, WI, USA) luciferase reporter vector using either SacI/XhoI or XhoI/HindIII restriction enzymes depending on orientation. Pro1 enhancer constructs were generated by cloning Pro1 into the StrataClone vector and then transferring to pGL3 Basic using BamHI/SalI restriction enzymes.
Cell culture
The mature mouse natural killer T (NKT) cell line EL-4 was cultured in RPMI-1640 medium supplemented with 10% Fetal bovine serum (FBS) and Penicillin-Streptomycin-Glutamine (PSG). Human Embryonic Kidney (HEK) 293T cells were cultured in DMEM with 10% FBS and PSG. The immature mouse NK cell line LNK was cultured in RPMI-1640 medium supplemented with 10% FBS, PSG, sodium pyruvate, non-essential amino acids, 2-mercaptoethanol, and 500 IU/mL IL-2.
Transfections and luciferase assays
Cells were plated at a density of 50,000 cells per well in 24 well plates. Cells were co-transfected with a control pRL-SV40 Renilla Reporter Vector (Promega) to normalize luciferase readings. HEK293 cells and LNK cells were transfected using Lipofectamine 2000 (Invitrogen). LNK cells were transfected with 1000 ng of pGL3 construct and 50 ng of Renilla control using a ratio of 5 μl Lipofectamine to 1 μg DNA. 293 cells were transfected with 200 ng of pGL3 construct and 10 ng of Renilla control using a 0.6 μl Lipofectamine to 1 μg DNA for 293. EL-4 cells were transfected with 1500 ng pGL3 construct and 100 ng Renilla control using TrueFect-Max (United BioSystems, Herndon, VA, USA) at a ratio 2.5 μl TrueFect to 1 μg DNA. After 48 hours of incubation, cells were lysed and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Measurement of the firefly luciferase activity of the Ly49 promoter constructs was normalized relative to the activity of the Renilla luciferase produced by the pRL-SV40 control vector in order to control for differences in transfection efficiency, and each construct was tested in triplicate in at least three independent experiments
5’ RACE of Pro1-Pro2i R3 fusion
LNK cells were transfected with the Pro1-Pro2i R3 fusion pGL3 construct as described above. RNA was isolated using the RNeasy Plus Mini Kit (Qiagen). 10 μg of isolated RNA was used to carry out 5’ rapid amplification of cDNA ends (5’RACE) using the FirstChoice RLM-RACE kit (Ambion, Austin, TX) following the included “5’ RLM-RACE Protocol” using a 5’ outer primer (5’-GCTGATGGCGATGAATGAACACTG) and a 5’ inner primer (5’-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG).
Statistical analysis
T-tests were carried out using Microsoft Excel 2010 for Windows with the Analysis Toolpak add-in. A two-tailed paired T-test was used with a p value less than 0.05 considered significant.
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute (NCI), National Institutes of Health (NIH), under contract HHSN261200800001E. This research was supported in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Footnotes
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Saleh A, Makrigiannis AP, Hodge DL, Anderson SK. Identification of a novel Ly49 promoter that is active in bone marrow and fetal thymus. J Immunol. 2002;168:5163–5169. doi: 10.4049/jimmunol.168.10.5163. [DOI] [PubMed] [Google Scholar]
- 4.Kubo S, Nagasawa R, Nishimura H, Shigemoto K, Maruyama N. ATF-2-binding regulatory element is responsible for the Ly49A expression in murine T lymphoid line, EL-4. Biochim Biophys Acta. 1999;1444:191–200. doi: 10.1016/s0167-4781(98)00284-x. [DOI] [PubMed] [Google Scholar]
- 5.Gosselin P, Makrigiannis AP, Nalewaik R, Anderson SK. Characterization of the Ly49I promoter. Immunogenetics. 2000;51:326–331. doi: 10.1007/s002510050626. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelm BT, McQueen KL, Freeman JD, Takei F, Mager DL. Comparative analysis of the promoter regions and transcriptional start sites of mouse Ly49 genes. Immunogenetics. 2001;53:215–224. doi: 10.1007/s002510100313. [DOI] [PubMed] [Google Scholar]
- 7.Saleh A, Davies GE, Pascal V, Wright PW, Hodge DL, Cho EH, et al. Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity. 2004;21:55–66. doi: 10.1016/j.immuni.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Tanamachi DM, Moniot DC, Cado D, Liu SD, Hsia JK, Raulet Genomic Ly49A transgenes: basis of variegated Ly49A gene expression and identification of a critical regulatory element. J Immunol. 2004;172:1074–1082. doi: 10.4049/jimmunol.172.2.1074. [DOI] [PubMed] [Google Scholar]
- 9.Rouhi A, Gagnier L, Takei F, Mager DL. Evidence for epigenetic maintenance of Ly49a monoallelic gene expression. J Immunol. 2006;176:2991–2999. doi: 10.4049/jimmunol.176.5.2991. [DOI] [PubMed] [Google Scholar]
- 10.Rouhi A, Brooks CG, Takei F, Mager DL. Plasticity of Ly49g expression is due to epigenetics. Mol Immunol. 2007;44:821–826. doi: 10.1016/j.molimm.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Kubo S, Itoh Y, Ishikawa N, Nagasawa R, Mitarai T, Maruyama N. The gene encoding mouse lymphocyte antigen Ly-49: structural analysis and the 5’-flanking sequence. Gene. 1993;136:329–331. doi: 10.1016/0378-1119(93)90489-p. [DOI] [PubMed] [Google Scholar]
- 12.Gays F, Koh AS, Mickiewicz KM, Aust JG, Brooks CG. Comprehensive analysis of transcript start sites in Ly49 genes reveals an unex- pected relationship with gene function and a lack of upstream promoters. PLoS ONE. 2011;6:e18475. doi: 10.1371/journal.pone.0018475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gays F, Taha S, Brooks CG. The Distal Upstream Promoter in Ly49 Genes, Pro1, Is Active in Mature NK Cells and T Cells, Does Not Require TATA Boxes, and Displays Enhancer Activity. J Immunol. 2015;194:6068–6081. doi: 10.4049/jimmunol.1401450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason L, Giardina SL, Hecht T, Ortaldo J, Mathieson BJ. LGL-1: a non-polymorphic antigen expressed on a major population of mouse natural killer cells. J Immunol. 1988;140:4403–4412. [PubMed] [Google Scholar]
- 15.Sentman CL, Hackett J, Kumar V, Bennett M. Identification of a subset of murine natural killer cells that mediates rejection of Hh-ld but not Hh-lb bone marrow grafts. J Exp Med. 1989;170:191–202. doi: 10.1084/jem.170.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanke T, Takizawa H, McMahon CW, Busch DH, Pamer EG, Miller JD, et al. Direct assessment of MHC class I binding by seven Ly49 inhibitory NK cell receptors. Immunity. 1999;11:67–77. doi: 10.1016/s1074-7613(00)80082-5. [DOI] [PubMed] [Google Scholar]
- 17.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11:321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacFarlane AW, 4th, Yamazaki T, Fang M, Sigal LJ, Kurosaki T, Campbell KS. Enhanced NK-cell development and function in BCAP-deficient mice. Blood. 2008;112:131–140. doi: 10.1182/blood-2007-08-107847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes CA, Scalzo AA, Degli-Esposti MA, Coudert JD. Ly49C-dependent control of MCMV Infection by NK cells is cis-regulated by MHC Class I molecules. PLoS Pathog. 2014;10:e1004161. doi: 10.1371/journal.ppat.1004161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan J, Lemieux S, Freeman JD, Mager DL, Takei F. Heterogeneity among Ly-49C natural killer (NK) cells: Characterization of highly related receptors with differing functions and expression patterns. J Exp Med. 1996;184:2085–2090. doi: 10.1084/jem.184.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guia S, Jaeger BN, Piatek S, Mailfert S, Trombik T, Fenis A, et al. Confinement of activating receptors at the plasma membrane controls natural killer cell tolerance. Sci Signal. 2011;4:ra21. doi: 10.1126/scisignal.2001608. [DOI] [PubMed] [Google Scholar]
- 22.Vosshenrich CA, García-Ojeda ME, Samson-Villéger SI, Pasqualetto V, Enault L, Richard-Le Goff O, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 23.Renard V, Ardouin L, Malissen M, Milon G, Lebastard M, Gillet A, et al. Normal development and function of natural killer cells in CD3 epsilon delta 5/delta 5 mutant mice. Proc Natl Acad Sci USA. 1995;92:7545–7549. doi: 10.1073/pnas.92.16.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gays F, Martin K, Kenefeck J, Aust JG, Brooks CG. Multiple cytokines regulate the NK gene complex-encoded receptor repertoire of mature NK cells and T cells. J Immunol. 2005;175:2938–2947. doi: 10.4049/jimmunol.175.5.2938. [DOI] [PubMed] [Google Scholar]
- 25.Makrigiannis AP, Rousselle E, Anderson SK. Independent control of Ly49g alleles: implications for NK cell repertoire selection and tumor cell killing. J. Immunol. 2004;172:1414–1425. doi: 10.4049/jimmunol.172.3.1414. [DOI] [PubMed] [Google Scholar]
- 26.Anderson SK, Dewar K, Goulet ML, Leveque G, Makrigiannis AP. Complete elucidation of a minimal class I MHC natural killer cell receptor haplotype. Genes Immun. 2005;6:481–492. doi: 10.1038/sj.gene.6364232. [DOI] [PubMed] [Google Scholar]
- 27.Gays F, Unnikrishnan M, Shrestha S, Fraser KP, Brown AR, Tristram CM, et al. The mouse tumor cell lines EL4 and RMA display mosaic expression of NK-related and certain other surface molecules and appear to have a common origin. J Immunol. 2000;164:5094–5102. doi: 10.4049/jimmunol.164.10.5094. [DOI] [PubMed] [Google Scholar]
- 28.Horton RM, Cai Z, Ho SM, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 29.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 30.Barao I, Alvarez M, Ames E, Orr MT, Stefanski HE, Blazar BR, et al. Mouse Ly49G2+ NK cells dominate early responses during both immune reconstitution and activation independently of MHC. Blood. 2011;117:7032–7041. doi: 10.1182/blood-2010-11-316653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 32.Cichocki F, Hanson RJ, Lenvik T, Pitt M, McCullar V, Li H, et al. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 34.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed and vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]