Abstract
Allosteric proteins bind an effector molecule at one site resulting in a functional change at a second site. We hypothesize that networks of contacts altered, formed, or broken are a significant contributor to allosteric communication in proteins. In this work, we identify which interactions change significantly between the residue-residue contact networks of two allosteric structures and then organize these changes into graphs. We perform the analysis on 15 pairs of allosteric structures with effector and substrate each present in at least one of the two structures. Most proteins exhibit large, dense regions of contact rearrangement, and the graphs form connected paths between allosteric effector and substrate sites in five of these proteins. In the remaining ten proteins, large-scale conformational changes such as rigid-body motions are likely required in addition to contact rearrangement networks to account for substrate-effector communication. On average, clusters which contain at least one substrate or effector molecule comprise 20% of the protein. These allosteric graphs are small worlds; that is, they typically have mean shortest path lengths comparable to those of corresponding random graphs and average clustering coefficients enhanced relative to those of random graphs. The networks capture 60 to 80% of known allostery-perturbing mutants in three proteins, and the metrics degree and closeness are statistically good discriminators of mutant residues from non-mutant residues within the networks in two of these three proteins. For two proteins, coevolving clusters of residues which have been hypothesized to be allosterically important differ from the regions with the most contact rearrangement. Residues and contacts which modulate normal mode fluctuations also often participate in the contact rearrangement networks. In summary, residue-residue contact rearrangement networks provide useful representations of the portions of allosteric pathways resulting from coupled local motions.
Keywords: allosteric mechanism, small-world network, graph theory, conformational change, signal propagation
Introduction
Allosteric regulation is a major mechanism of control in many biological processes, including cell signaling, gene regulation, and metabolic regulation.1 Allosteric proteins bind an effector molecule at one site resulting in a functional change at a second site.2 Recently there has been much interest in allosteric-like communication. Thermodynamic theories explain allostery via population shifts in conformational ensembles,3–5 and there is experimental evidence that alternate allosteric states are simultaneously populated in solution.6 Nonetheless, mechanical transitions in individual molecules must underlie population shifts of ensembles of conformations.7,8 That is, in individual molecules, energetic pathways of spatially contiguous, physically coupled structural changes and/or dynamic fluctuations must link substrate and effector sites.
Crystal structures have revealed that most allosteric proteins are complex systems with both tertiary and quaternary structure changes.9 Thus, to quantitatively describe mechanisms of allosteric communication, one would need to account for multiple levels of conformational changes in both the positions of and the interactions between the elements of protein structures. Recently, we compiled a database of 51 proteins with both inactive (I) and active (A) crystal structures, and we quantitatively characterized differences in local structure between the two states.10 Here, we extend our previous work by taking a first step toward quantifying allosteric mechanisms from structure. Specifically, we calculate contact rearrangement networks (CRNs) from differences in the contact network between the two structures to describe one way in which communication through tertiary structure might arise from the kinds of local motions that we identified in the allosteric benchmark paper.10 We do not explicitly account for large-scale rigid-body (quaternary structure) motions in constructing CRNs; thus, we do not expect CRNs to completely describe allosteric mechanisms in the proteins we analyze. Thus, we use the terms communicate, pathway, and couple generally to refer to allosteric coupling between any two points (residues) in an allosteric protein rather than specifically to refer to substrate site-effector site communication unless explicitly specified. We expect that these CRN analyses will provide detailed, useful, and quantitative descriptions of a phenomenon which has previously observed in manual analyses and predicted by the Koshland-Nemethy-Filmer (KNF) model,11 which describes allosteric signaling primarily through rearrangement of tertiary structure.
Many previous computational approaches to protein allostery incorporate theoretical models which are likely to influence the results (e.g. techniques like Gaussian Network Model,12 normal mode analysis,13 molecular dynamics,14 and ensemble computation and energetic analysis15). In contrast, we seek to learn as much as possible about allosteric pathways in proteins through direct, model-free analyses of crystal structures. In addition, by utilizing the data available in crystal structures, a structural analysis approach can provide insight into allostery orthogonal to that from experimental mutational studies (e.g. refs. 16,17).
Networks are natural representations for studying complex systems, and several studies on protein residue-residue contact networks have revealed functionally interesting information not immediately accessible from the atomic coordinates themselves. Protein structure contact networks display the small-world phenomenon;18 that is, they are tightly clustered yet have short paths between residues.19 Highly central residues in such contact graphs as identified by closeness, betweenness, or change in the mean shortest path of the graph upon removal often correspond to known functionally important positions such as key residues in folding,20 active sites,21,22 hotspots of protein-protein interfaces,23 and residues important to allosteric communication.24 In addition, one previous work has found biologically significant changes in the intersubunit contact network between two structures of lac repressor that are very close in Cartesian space,25 and distance-difference matrices have been used to postulate coupling mechanisms in hemoglobin.26
We calculate residue-residue contact rearrangement networks for 15 allosteric proteins from our benchmark set,10 and we characterize the graphical and functional properties of these networks. For each protein, we identify which residue-residue interactions rearrange and organize these changes into a graph. Since this graph includes information from both allosteric structures, it necessarily provides more useful information about allosteric coupling in the protein than does a contact network analysis of either end-state structure (e.g. ref. 24). Such a network representation of changes in the contact map is useful because it allows identification of coupling relationships among residues in the tertiary structure and identification of critical residues. We describe the range of structures of CRNs in the 15 proteins, and for each protein, we calculate the extent of the CRN and assess the small-worldness of its connected components. The metrics degree and closeness identify graphically important residues which may also be functionally important. We use known allostery-perturbing mutations in three proteins to assess the ability of CRNs to capture functionally important regions of allosteric structures and the ability of degree and closeness to rank residues within CRNs by functional importance. Finally, for two of the 15 proteins, we compare CRNs to statistical coupling analysis, a sequence-based algorithm for identifying putative allosteric networks in proteins,27 and we compare CRNs from two other proteins to published elastic network analyses, which can give insight into dynamic fluctuations.13,28 CRN analysis may identify principles about allosteric communication which could aid in the rational design of allosteric regulation into non-allosteric proteins.
Results
We select 15 heterotropically allosteric proteins from our benchmark set10 for which the two structures together contain at least one small-molecule substrate and at least one small-molecule effector. The names and Protein Data Bank (PDB)29 codes of these proteins are given in Table I.
Table I. Allosteric test set and substrate-effector coupling parameters in applicable proteins.
All structures are determined by x-ray crystallography with resolutions ranging from 1.8 to 2.9 Å. Details for the inactive and active structures of the 15 proteins, including resolution and ligands bound to each state, can be found in supplementary table II of our allosteric benchmark paper.10 For connectivity, P (partial) means that at least one substrate node connects to at least one effector node through the graph, whereas ‘G’ (global) means that a single cluster connects all substrate and effector nodes. LSE is the length of the shortest substrate-effector path, including protein-ligand contacts, where applicable.
| Protein |
T = 0.20
|
T = 0.30
|
T = 0.40
|
|||
|---|---|---|---|---|---|---|
| Connect? | L | Connect? | L | Connect? | L | |
| anthranilate synthase | N | - | N | - | N | - |
| ATCase | Y | 10 | N | - | N | - |
| ATP sulfurylase | Y | 7 | N | - | N | - |
| ATP-PRT | N | - | N | - | N | - |
| DAHP synthase | N | - | N | - | N | - |
| FBPase-1 | Y | 7 | Y | 8 | Y | 9 |
| glcN-6-P deaminase | Y | 6 | N | - | N | - |
| glycogen phosphorylase | Y | 9 | N | - | N | - |
| GTP cyclo-hydrolase I | Y | 5 | Y | 6 | Y | 7 |
| lactate DH | Y | 5 | Y | 5 | N | - |
| NAD-malic enzyme | Y | 7 | N | - | N | - |
| phosphofructokinase | Y | 3 | Y | 3 | Y | 3 |
| phosphoglycerate DH | N | - | N | - | N | - |
| PTB1B | Y | 6 | N | - | N | - |
| uracil PRT | Y | 4 | Y | 4 | N | - |
We calculate an undirected, weighted contact rearrangement graph for each protein where the nodes are all the residues present in both structures, and the weight of an edge between two residues i and j that form a contact in one or both structures is the rearrangement factor R(i j). R(i,j) captures the change in the composition of the set of atoms which form the interaction between residues i and j (Figure 1) by the fraction of atoms which are lost or gained from the interface between the two residues. Finally, we determine the connected components or clusters of a graph from all edges in the graph with weights above a threshold T. We set T = 0.3 in this work to exclude 99% of all possible edges in a control set of 14 proteins not exhibiting allosteric motions (details in Methods).
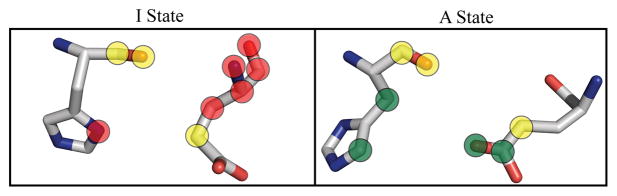
Figure 1. Rearrangement of a residue-residue interaction in phosphofructokinase.
Left panel: interaction between E241 and H160 of chain A in the inactive state; right: this interaction in the active state. Red circles mark six atoms unique to the residue-residue interface in the I state, green circles mark four atoms unique to the A state, and yellow circles mark three atoms present in both states. In these two residues, there are a total of 19 atoms, so the rearrangement factor R(i,j) = max(6, 4)/19 = 0.32 (see Methods for the details of calculating R(i, j)).
Overview of graphs
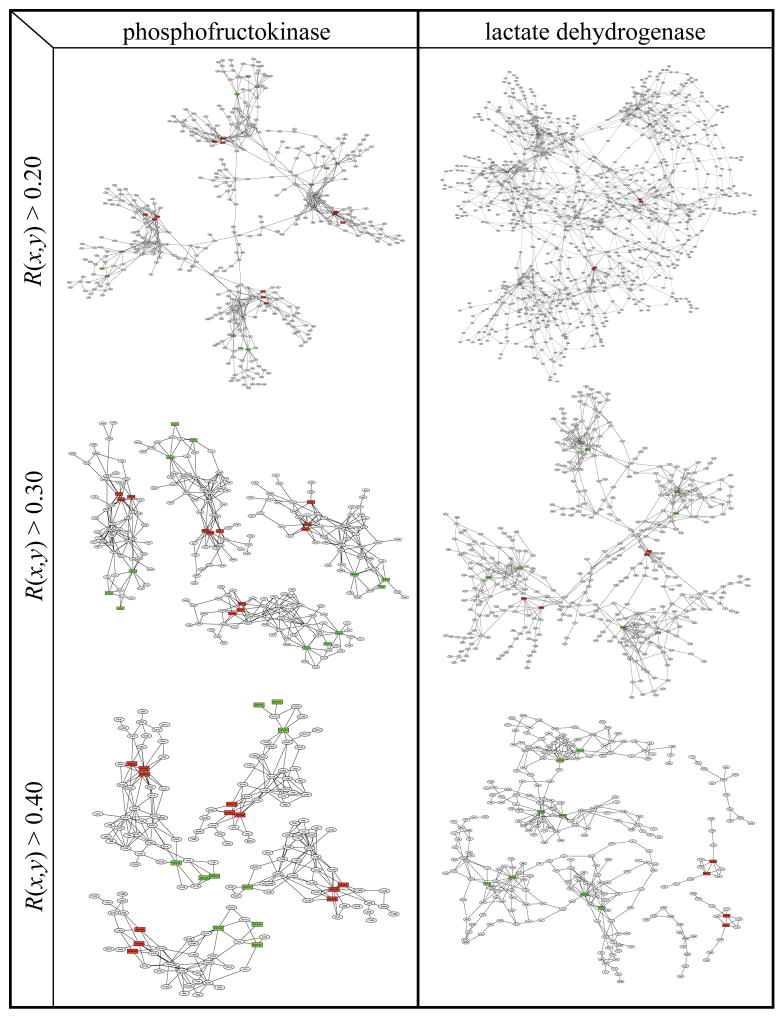
Figure 2 shows the range of CRN structures in six representative proteins (the CRNs of the remainder of the 15 proteins are in Supplementary Figure 1). In NAD-malic enzyme, most of the contact rearrangement occurs in the immediate vicinity of substrate and effector sites, with two clusters each connecting two nearby effector sites to one another. Phosphofructokinase (PFK) shows slightly larger clusters which each connect one effector site to the nearest substrate site with a dense web of paths. The graph of glycogen phosphorylase (GYP) links two distant substrate sites together through a large and dense cluster, with two smaller clusters surrounding each of the effector sites. The lactate dehydrogenase (LDH) graph comprises four dense regions surrounding substrate sites which are loosely linked to form one large ‘globally connected’ cluster linking all substrate and effector sites together. The globally connected graph of fructose bisphosphatase (FBPase) also contains regions of high and low density, though the high-density regions are more strongly linked to one another than in LDH. The graph of GTP cyclohydrolase I (GCH) links all substrate and effector sites among one catalytic decamer and two regulatory pentamers. Table I shows that the graphs of five of 15 proteins form one or more connected paths between substrate and effector sites with a distance of 3 to 8 links. Furthermore, in all graphs, there is significant contact rearrangement density in the vicinity of substrate and/or effector sites.
Figure 2. Contact rearrangement networks for six selected proteins.
The CRNs of the remaining nine proteins are shown in Supplementary Figure 1. Circles in each graph represent protein residues, and red and green squares represent substrate and effector molecules, respectively. Lines connect pairs of residues with R(i,j) ≥ 0.3 and residues in the graph with any ligands which are adjacent (within 5.0Å) in either structure. All connected components which include at least one substrate or effector molecule are shown. Graphs are plotted with yEd graph editor.
While all of the proteins we examined show extensive contact rearrangement, these networks are likely to be more important for biological function in some proteins than in others. The CRN probably plays an important role in substrate-effector communication in each protein with large, dense regions of contact rearrangement, whether or not the graph links substrate and effector sites. In each graph with connected substrate-effector paths except that of GCH, the CRN indicates significant physical substrate-effector coupling through the tertiary structure because most or all effector sites are linked to their nearest respective substrate sites by at least two non-overlapping paths. However, in both proteins for which the graphs exhibit connected substrate-effector paths and proteins without such paths, allosteric coupling between sites might depend not only on CRNs, but also upon other mechanisms like rearrangements of interactions between rigid bodies.
Scope and network characteristics of CRNs
Table II shows that on average among the 15 proteins, 35% of residues occur in any cluster, while 20% of residues occur in clusters containing at least one substrate or effector, which we hereafter refer to as allosteric clusters. The extent of all clusters varies from 22% of the residues for DAHP synthase and phosphoglycerate dehydrogenase (PGDH) to 55% for ATP-PRT, while the extent of allosteric clusters varies from 5% for PGDH to 34% for LDH. Average degree, where degree is the number of other nodes connected to a node, measures the density or redundancy of a network. The value of this metric ranges from 2.3 for PGDH (nearly nonredundant) to 4.0 and 4.1, respectively, for FBPase and PFK (moderately redundant). However, in the graphs in Figure 2, nodes of degree 1 and/or long chains of nodes which project away from the main bodies of the clusters depress the observed average degree relative to that of the core network.
Table II. Graphical characteristics of contact rearrangement networks.
For each protein, average degree is calculated over all nodes in all clusters connected to one or more ligands.
| Protein | fraction of protein
|
clusters with ≥ 1 ligands
|
|||
|---|---|---|---|---|---|
| any cluster | ≥1 ligands | Avg Degree | Avg Cluster Coefficient | Mean Shortest Path | |
| phosphoglycerate DH | 0.23 | 0.06 | 2.3 | 0.11 | 3.4 |
| ATP sulfurylase | 0.30 | 0.21 | 2.6 | 0.11 | 7.5 |
| glcN-6-P deaminase | 0.49 | 0.21 | 2.6 | 0.14 | 8.1 |
| DAHP synthase | 0.23 | 0.08 | 2.7 | 0.23 | 3.2 |
| ATP-PRT | 0.55 | 0.24 | 2.8 | 0.14 | 8.2 |
| GTP cyclo-hydrolase I | 0.43 | 0.33 | 2.9 | 0.14 | 14.4 |
| glycogen phosphorylase | 0.39 | 0.16 | 2.9 | 0.12 | 6.3 |
| PTB1B | 0.24 | 0.17 | 2.9 | 0.26 | 3.7 |
| NAD-malic enzyme | 0.27 | 0.11 | 3.1 | 0.24 | 3.5 |
| uracil PRT | 0.27 | 0.20 | 3.1 | 0.19 | 7.6 |
| ATCase | 0.44 | 0.31 | 3.2 | 0.13 | 12.4 |
| lactate DH | 0.55 | 0.35 | 3.3 | 0.16 | 10.9 |
| anthranilate synthase | 0.36 | 0.18 | 3.3 | 0.19 | 7.6 |
| FBPase-1 | 0.36 | 0.29 | 4.0 | 0.18 | 9.8 |
| phosphofructokinase | 0.28 | 0.18 | 4.1 | 0.23 | 3.5 |
|
| |||||
| average | 0.36 | 0.21 | |||
| standard deviation | 0.11 | 0.09 | |||
|
|
|||||
| w/o cc and path length data
| |||
|---|---|---|---|
| Protein | fraction of protein
|
avg degree (≥1 ligands) | |
| any cluster | ≥1 ligands | ||
| phosphoglycerate DH | 0.23 | 0.06 | 2.3 |
| ATP sulfurylase | 0.30 | 0.21 | 2.6 |
| glcN-6-P deaminase | 0.49 | 0.21 | 2.6 |
| DAHP synthase | 0.23 | 0.08 | 2.7 |
| ATP-PRT | 0.55 | 0.24 | 2.8 |
| GTP cyclo-hydrolase I | 0.43 | 0.33 | 2.9 |
| glycogen phosphorylase | 0.39 | 0.16 | 2.9 |
| PTB1B | 0.24 | 0.17 | 2.9 |
| NAD-malic enzyme | 0.27 | 0.11 | 3.1 |
| uracil PRT | 0.27 | 0.20 | 3.1 |
| ATCase | 0.44 | 0.31 | 3.2 |
| lactate DH | 0.55 | 0.35 | 3.3 |
| anthranilate synthase | 0.36 | 0.18 | 3.3 |
| FBPase-1 | 0.36 | 0.29 | 4.0 |
| phosphofructokinase | 0.28 | 0.18 | 4.1 |
|
| |||
| average | 0.36 | 0.21 | |
| standard deviation | 0.11 | 0.09 | |
|
|
|||
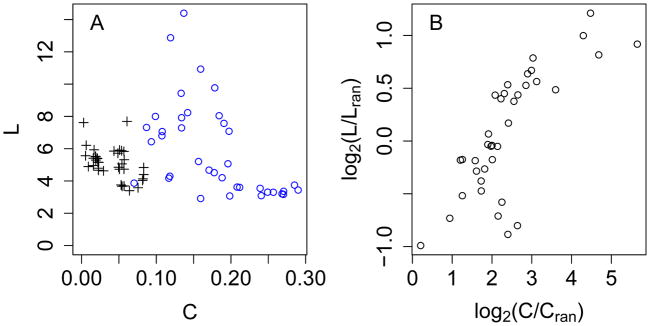
Relative to a corresponding random graph, a small-world network (SWN) exhibits an enhanced average clustering coefficient (C) but a similar mean shortest path (L).18 For a random network with N nodes and average degree k, Lran ≈ ln N/ln k and Cran ≈ k/N 18 while for a corresponding 1-lattice regular network, and .20,30 Figure 3 shows that among 34 allosteric clusters in 15 proteins, C ranges from 0.07 to 0.30, intermediate between the ranges of Cran and Creg, and L ranges from 3 to 14, which is considerably closer to the range of Lran than to that of Lreg. For a few regular network points, Creg ranges from 0.05 to 0.2, which overlaps with the allosteric clusters; however, these low C result from the artificially low k of some networks (see above paragraph). Thus, CRNs are small worlds, which exhibit both high density and efficient communication between points and should be robust to random mutations.31 Furthermore, the distributions of degree k in these 34 clusters (data not shown) are not Poisson as expected for random networks.32 Rather, the number of nodes N(k) at degree k decreases monotonically as k increases from 1 to a maximum of 5 to 15 depending on the cluster, which means that a few nodes act as key hubs. These distributions are based on limited data, so it is not possible to determine if they are more consistent with scale-free33 or single-scale34 behavior.
Figure 3. Small-world characteristics of allosteric clusters.
Data are derived from 34 allosteric clusters from 15 proteins which contain 20 or more nodes and for which average degree is greater than 2. Blue circles: observed allosteric clusters. Black crosses: random counterparts with the same number of nodes and average degree. Red triangles: regular counterparts (four points with mean shortest path > 50 have been excluded for clarity).
We identify ‘key residues’ within the graph of each protein as those nodes which rank among the top five by degree or closeness among all allosteric clusters (Supplementary Table I). Closeness measures centrality by the inverse of the average distance of a node to all other nodes in a cluster. While degree identifies locally critical nodes, closeness and other centrality measures identify globally critical nodes which mediate efficient communication between points in the network and thus are most important to its small-world behavior.18
Two proteins in detail
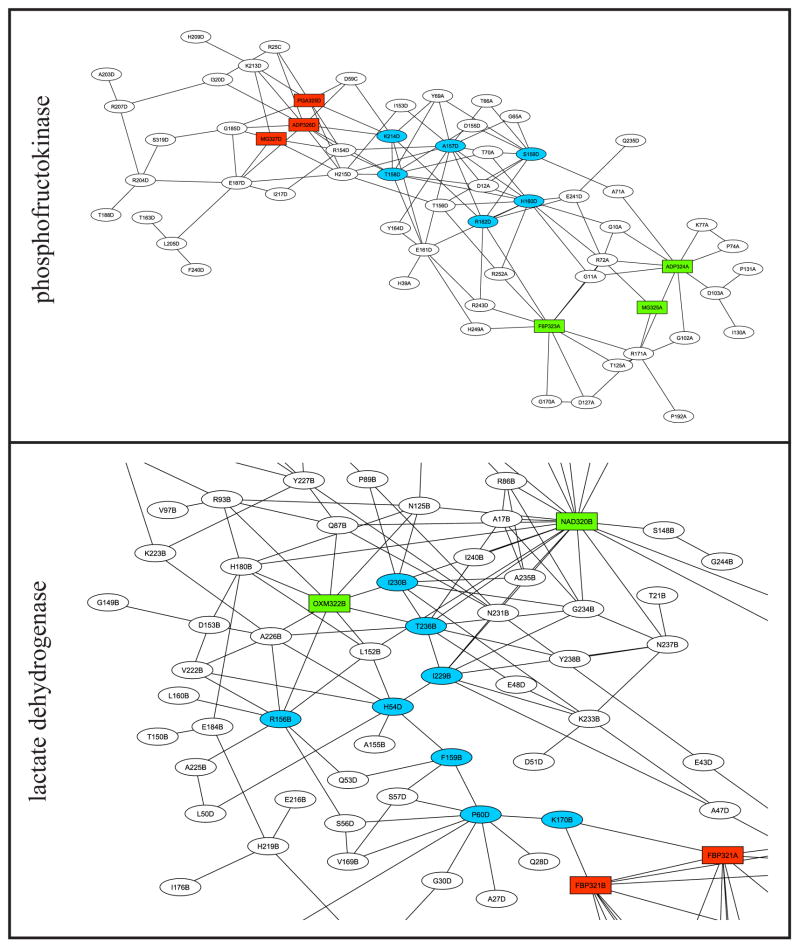
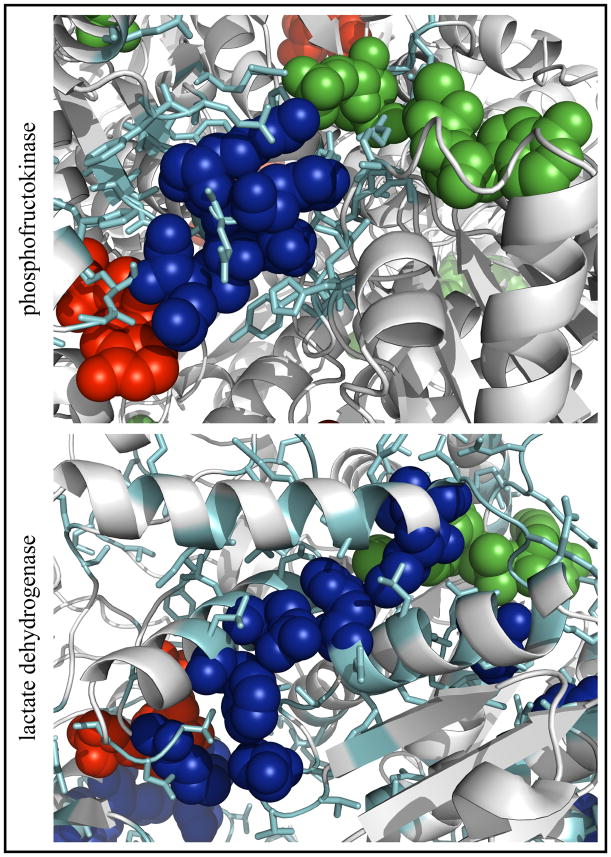
Figure 4 shows subsets of the graphs of PFK and LDH and these graphs mapped onto the respective three-dimensional structures, highlighting key residues in each protein. The PFK network (Figure 4A) is tightly clustered and contains multiple paths between substrate and effector sites, and the key residues cluster graphically between these sites. Figure 4B shows that in the three-dimensional structure, the key residues lie along the line between the two sites in the center of the cluster. The LDH network (Figure 4C) is tightly clustered around the substrate site but compared to PFK, there are fewer short paths between substrate and effector, though all of the key residues lie along paths between the two sites. However, Figure 4D shows that in the three-dimensional structure, in the middle of a large cluster, seven of these eight residues lie along a nearly straight line between the two sites. Thus, Figure 4B and D suggest that in applicable CRNs, degree and closeness identify residues which lie along short paths between substrate and effector sites in the three-dimensional structure.
Figure 4. Detail of phosphofructokinase and lactate dehydrogenase contact rearrangement networks.
A, C: subsets of the graphs of PFK and LDH, respectively, containing one effector site and one substrate site, plotted as in Figure 2. For each node, the first group of characters is the one-letter amino acid code or the ligand name as appropriate, and the last letter is the chain identifier. Light blue: nodes in the top five by degree or closeness (key residues). B, D: the same networks mapped onto the three-dimensional structures (A state structure 4PFK for PFK and I state structure 1LTH(T) for LDH). Cyan: residues in the cluster. Blue: key residues in largest cluster. Green: substrate. Red: allosteric effector. The PFK cluster contains effector molecules from both the I state (PGA) and the A state (MgADP) structures and substrates (F6P and MgADP) from the A state structure. The subset of the globally connected cluster of LDH contains the effector (FBP) and substrates (cofactor NAD and substrate OXM) from the A state structure. The two molecules of FBP shown in the LDH graph represent pseudosymmetrically related orientations of FBP present in the crystal structure of LDH.
Networks capture experimentally known allosteric residues
Published studies (see Supplementary Table II) have identified six allostery-perturbing mutants of PFK, 13 of FBPase, and 30 of aspartate transcarbamoylase (ATCase). These mutants perturb allosteric coupling by such metrics as Ki of an allosteric inhibitor,35 relative activity with versus without effector bound,36 and coupling free energy between effector and substrate.37 All of these studies were targeted rather than exhaustive, so calculation of the sensitivity and specificity of our algorithm from these data is not possible. For each protein, Supplementary Table III shows the presence or absence of each mutant in the allosteric clusters of the CRN. Table IIIA summarizes that allosteric clusters capture good (60–70% for FBPase and ATCase) to excellent (83% for PFK) fractions of allostery-perturbing mutations. Fisher’s exact test shows that these results are highly statistically significant for PFK and ATCase (p ≤ 10−3) and significant for FBPase (p = 0.011); that is, mutants are distinctly more likely to be found in the allosteric clusters than in the protein structure as a whole. These results show that CRNs identify functionally important regions whether or not they form connected substrate-effector paths.
Table III. Statistics of mutants occurring in contact rearrangement networks of three proteins.
A: Hit rates are averaged over asymmetric units, and p-values are calculated by Fisher’s exact test55 from the number of residues in the protein, number of network residues, total number of mutants and total number of mutants captured, all divided by the number of asymmetric units. B: Degree and closeness values for network residues are averaged over multiple occurrences in different asymmetric units as appropriate. p-values are determined from the differences in the distributions of degree and closeness values between non-mutant and mutant residues by the one-sided two-sample Wilcoxon rank-sum test.55
| A. Mutants occuring in network
| ||||
|---|---|---|---|---|
| Protein | number of mutants | number of monomers | hit rate | p-value |
| PFK | 6 | 4 | 20/24 (83%) | 4.10E-13 |
| FBPase-1 | 13 | 4 | 32/52 (62%) | 2.40E-07 |
| ATCase | 30 | 6 | 121/180 (67%) | 2.20E-16 |
| B. Wilcoxon rank-sum tests
| ||
|---|---|---|
| Protein (cluster #) | p-value degree | closeness |
| PFK (1–4) | 7.90E-04 | 2.10E-04 |
| FBPase-1 (1) | 7.70E-05 | 3.30E-05 |
| ATCase (1) | 0.81 | 1 |
| ATCase (2–4) | 0.095 | 4.20E-03 |
Furthermore, we test the ability of degree and closeness to rank residues within these large networks by functional importance. The two-sample Wilcoxon rank-sum test shows that both degree and closeness give a significantly higher average rank to known mutant residues than to non-mutants for PFK and FBPase (p ≤ 0.03). Previous works have shown that closeness identifies active site residues from contact maps of static structures more effectively than degree;21,22 for these allosteric graphs, closeness is slightly more effective at ranking residues by functional importance than is degree for both PFK and FBPase. Both degree and closeness fail to discriminate allostery-perturbing mutant residues from non-mutant residues within ATCase clusters. However, the central core of one ATCase cluster, which contains the residues with highest closeness, may be functionally important even though it has not been previously tested for allostery-perturbing mutations (Supplementary Figure 2A).
Comparison with statistical coupling analysis
The statistical coupling analysis (SCA) algorithm27,38 identifies putative allosterically coupled networks of residues from correlated sequence perturbations in large protein families. The CRN algorithm provides a useful way of assessing how SCA networks relate to the changes in three-dimensional structure which are likely the primary mechanism of allosteric coupling in proteins. We compare results between SCA and CRNs for two proteins in this work, PFK and FBPase, for which the sequence families are sufficiently large for SCA and for which we collected allostery-perturbing mutants (see methods for SCA details).
Supplementary Figure 3 shows that for PFK and FBPase, CRN residues and SCA residues occupy mostly non-overlapping regions of the structures. Fisher’s exact test shows that this overlap is moderately significant for PFK (p=0.0031) but insignificant for FBPase (p=0.78). However, the correlation between R(i,j) and the SCA parameter ΔΔEstat for those residue pairs with R(i,j) ≥ 0.05 and ΔΔEstat ≥ 0.01 is 0.12 for PFK and −0.06 for FBPase, indicating a lack of a significant quantitative relationship between the two putative allosteric coupling parameters. In addition, of the mutants listed in supplementary table II, SCA only captures 2 of 6 for PFK (p=0.18) and 5 of 13 for FBPase (p=0.33). These results challenge the hypothesis that evolutionary coupling of positions in a protein structure reflects a particularly allosteric role for those residues. A recent work suggests that an evolutionarily coupled group of residues in a protein might specify a stably folding sequence.39
Comparison with elastic network models
Two recent studies have employed normal mode analysis (NMA) on elastic network models of proteins toward identifying residues important to allostery. These approaches elicit large-scale collective motions from a network of springs connecting Cα positions of locally contacting residues, and thus they are different from the small-scale local structural rearrangements analyzed in this manuscript.
Zheng and Brooks measured dynamic correlation between residues under NMA fluctuations to identify ‘hinge’ residues correlated to the most other residues in the protein.13 The hinge residues of myosin (the 10% most correlated residues) can be compared to the CRN obtained from 1Q5G and 1VOM (Supplementary Figure 4A). The CRN captures 53 of 73 dynamically correlated residues (73%), a very high overlap (Fisher’s exact test p < 10−16). This strong result implies that not only do CRNs communicate allosteric signals through tertiary structure, but also that they might modulate large-scale fluctuations captured by low-frequency normal modes and couple them to small-scale changes in the vicinities of ligand sites.
In another NMA study, Gu and Bourne used a method called PIVET to directly assess the effect of contact changes on dynamic fluctuations by removing single contacts in elastic network models to identify interactions which perturb the protein’s fluctuation.28 Comparison of CRNs with this analysis for four functional transitions of the protein CDK2 revealed mixed results. For the ATP binding transition (1HCL→1HCK), there was little conformational change detectable by the CRN. For cyclin binding (1HCK→1FIN, Supplementary Figure 4B), the 8 CRN key residues (top five by degree and/or closeness) capture 4 of the 15 residues in the 10 pairs with the greatest influence on global fluctuation (50%, p = 2.8·10−4 by Fisher’s exact test). In addition, several of the contacts ranked highly by PIVET overlap three high-CRN-degree residues and form a cluster in a portion of the CRN near the cyclin-binding site. Phosphorylation (1FIN→1JST) and peptide binding (1JST→1QMZ) cause smaller conformational changes than cyclin binding, and the CRN key residues do not overlap with the residues in the top 10 pairs according to PIVET. These results suggest that in a large allosteric transition, the densest regions of contact rearrangement are likely to be important for modulating dynamic fluctuations.
Discussion
Theoretical implications
In our previous survey of motions in allosteric protein structures,10 the clustering of moving residues in the three-dimensional space and correlation functions of residue motions strongly suggested that some or all of these proteins communicate allosteric signals through mechanically contiguous clusters of conformational changes. In this work, CRNs describe mechanical allosteric coupling through tertiary structure in detail. Such coupling can extend over long distances through the tertiary structure, and in some cases substrate and effector sites are coupled through tertiary structure. Clusters of changes in dynamics such as those observed upon ligand binding to a PDZ domain40 or mutation of eglin C41 are beyond the scope of our observations but could add a significant additional dimension to allosteric network descriptions for these proteins. Furthermore, the concept of motions giving rise to long-range coupled networks of changes in the interactions among protein structure elements could in principle be generalized beyond the CRN, which quantifies allosteric coupling arising from tertiary structure changes, to describe how higher levels of motion, such as rigid-body motions of domains and subunits, give rise to allostery.
This analysis of contact rearrangement networks suggests that communication between points in allosteric proteins operates via a complex, redundant web of interdependent conformational changes. Such complex signal propagation has been observed in a molecular dynamics simulation of CheY.8 Furthermore, the observation that CRNs are small-world networks with skewed connectivity distributions suggests that communication depends on preferred paths with certain residues playing critical roles in the transmission of signals between points.
We previously observed correlation of motions at up to 20 Å distance between residues, which is equivalent to a series of several atom-atom contacts.10 The CRN model suggests that signals can propagate considerably farther than this, given that the mean shortest path in allosteric clusters varies from about 3 to 14 contacts. The anisotropy of some clusters in the simulation of heat propagation through protein structure42 may account for this observation.
Possibilities beyond the CRN
Proteins which do not exhibit connected substrate-effector paths via CRNs likely rely on other kinds of motion in addition to CRNs to form mechanical linkages between these two sites. For example, in aspartate transcarbamoylase43 and glycogen phosphorylase,44 which show extensive CRNs but not connected substrate-effector paths, the original manual analyses of the crystal structures revealed domain and subunit motions critical to creating a global cooperative transition. In addition, in phosphoglycerate dehydrogenase, which has only a small amount of contact rearrangement around the substrate site and none at the effector site, manual comparison of inactive and active structures suggests that domain and subunit motions are the primary substrate-effector coupling mechanism.45 Thus, while CRNs provide a useful representation of allosteric signal propagation and connectivity through tertiary structure, more complex models of allosteric systems integrating large-scale rigid-body motions would likely be necessary to increase the 1/3 substrate-effector connectivity ratio observed using CRNs alone. These rigid-body motions are consistent with allosteric communication through quaternary structure changes as predicted by the Monod-Wyman-Changeux (MWC)46 model of allostery.
Network topology and key residues
Like contact networks of static protein structures,19,20 networks of contact rearrangements in allosteric proteins exhibit small-world character, which provides efficient communication between points18 and robustness of communication against random structural or mutational perturbation.31 Small-world character of CRNs might not arise directly from small-world character of the contact networks of the underlying structures. Small-worldness is driven by nodes which are well-connected and more importantly, centrally located in the graph.18,33 In a static graph, highly connected nodes are well-constrained by many connections and thus not likely to move, while in a CRN the most highly connected nodes interact with many other nodes in at least one state but also have significantly different optimal sets of interactions in the two respective states. In a static graph, central nodes are typically positioned near the center of mass of the protein,21 while in a CRN they are usually near the geometric center of an allosteric cluster (Figure 4), which may not be near the center of the protein. In fact, our previous analysis revealed that exposed residues in proteins are more likely than buried residues to undergo contact rearrangement.10 Thus, in many allosteric proteins, the CRNs may evolve separately from or in tension with the contact network of the protein as a whole.
Comparison with allosteric mutations
Mutational analysis of CRNs show that CRNs capture functionally important regions of allosteric structures and that degree and closeness effectively rank residues within CRNs by functional importance. In addition, the CRN algorithm might be useful for predicting functionally critical residues in allosteric proteins for further testing by mutation or targeting for therapeutic or engineering purposes. It is possible that previous structure-based algorithms, such as our calculations of local motions,10 the most central residues in a static contact graph of either state,24 or hub and messenger nodes in clusters formed by a hierarchical static contact network decomposition algorithm47 might predict allosteric mutations as well or better than CRNs. Assessment of the general utility of CRNs and other approaches for predicting allosteric mutations by comparison may be a practical subject for future research.
CRNs, SCA, and NMA
CRNs, SCA, and NMA provide three different perspectives on allosteric communication. CRNs directly measure contact changes from crystal structures and group adjacent rearrangements together, but they are limited to tertiary structural changes and do not directly probe dynamics. SCA exploits the sequence database to identify coupled residues. NMA captures the fluctuations inherent in the elasticity of the three-dimensional structure of the protein. Although our comparisons are limited to a few systems, it is clear that these methods can be complementary. SCA and CRN seem to identify different networks: SCA finds pathways through the core of the protein perhaps relating to the folding nucleus, while CRN networks contain more surface-exposed residues and are directly tied to the average allosteric conformational change. Interestingly, residues which are highly correlated to the rest of the protein in NMA and contacts which most perturb the fluctuations of the elastic network are often part of the CRN, which suggests that key structural changes in allosteric proteins also play an important role in modulating dynamics.
Conclusions
The mechanochemical basis of allosteric coupling in proteins has remained elusive even though the subject has been studied for decades. We have introduced a simple calculation to elicit a network of residues coupled via tertiary structure changes from the difference in the residue-residue contact network between inactive and active state structures of an allosteric protein. The contact rearrangement networks typically show significant localized response in the substrate and/or effector binding site regions. In most proteins, they extend through significant regions of the protein structures, and they form connected substrate site-effector site paths in 5 of 15 proteins; in the remaining 10 proteins (and possibly also in the connected 5), additional coupling mechanisms (e.g. rigid-body motions) will be necessary to fully describe the mechanism of communication between substrate and effector. These results offer strong evidence for propagation of information through protein structure, although propagation is a complex web-like phenomenon not immediately obvious from a cursory examination of the allosteric structures. The observed properties of contact rearrangements may be useful to understand allostery-related diseases, guide allosteric drug design, or design novel allosteric communication in proteins.
Methods
Contact rearrangement
We define a contact between two residues i and j as at least one atom-atom distance between them under 5.0 Å, not counting hydrogens or non-protein atoms. For each contact ij which exists in the I or the A state, we calculate a rearrangement factor which quantifies the difference in that contact between the two states. This rearrangement factor is the maximum of the number of atoms which are unique to the ij interface (the set of atoms defining the ij interaction) in the I and A state structures, respectively, normalized by the total number of atoms in residues i and j (Figure 1). That is, if is the ij interface in the I state, is the ij interface in the A state, and Ni and Nj are the total numbers of atoms in residues i and j, respectively, then the rearrangement factor R(i,j) is
where |𝒞| denotes the number of atoms in set 𝒞. The normalization by Ni + Nj accounts for the size of the component residues i and j, although we have found that CRNs constructed without this normalization are qualitatively similar.
Connected components
For determining connected components, a threshold for R(i,j) separates the most biologically significant contact rearrangements from those which represent crystallographic uncertainty of independently solved structures (< 1 Å RMSD for independently solved crystal structures).48 As a reference for such uncertainty, we use a previously compiled control set of 14 pairs of biologically equivalent crystal structures which includes five non-allosteric proteins and nine allosteric proteins with two structures in the same state.10 The resolutions of the structures in this set range from 1.1 to 2.8 Å. Figure 5 shows the distribution of R(i,j) for all edges in the graphs of all proteins in the control and allosteric sets, respectively. At R(i,j) > 0.1, the density of edges is higher in allosteric graphs than in non-allosteric graphs; however, the density of edges in the control graphs is significant up to at least T=0.2. To exclude approximately 99% of the control distribution and highlight the most significant contact rearrangements, we set T=0.3, above which lies 0.78% of the control distribution, compared with 2.5% at T=0.2 and 0.29% at T=0.4. By contrast, 5.6% of the allosteric R(i,j) distribution lies above T=0.3, which is seven times the corresponding fraction of the distribution in the control set.
Figure 5. Distributions of R(i,j) in graphs for control and allosteric set proteins.
The control and allosteric set distributions are the sum of the normalized distributions of all proteins in these respective sets. Normalization of each protein’s distribution by the number of asymmetric units accounts for symmetry-related edges. Black crosses: control set distribution; blue circles: allosteric set distribution; red: T = 0.3, the threshold used for all calculations in this work.
A breadth-first search49 on all nodes determines the connected components of a graph at a given T. In addition, connected components at a given T include any ligands present in either state which in that state are within 5.0 Å of any residue in a connected component. If a ligand is present in both I and A state structures, then one structure serves as a reference for identifying the neighbors of that ligand. The I state structure is the reference for allosteric inhibitors, which preferentially bind this state, and the A state structure is the reference for allosteric activators and substrates for corresponding reasons. The raw data for the 15 allosteric proteins, including the graphs in GML format and PyMOL scripts for mapping the calculated clusters onto the three-dimensional structure of a protein, are available at http://graylab.jhu.edu/allostery/networks.
Network statistics
Path calculation
The Floyd-Warshall algorithm calculates the shortest path between two nodes in a graph49 for the calculation of mean shortest path of a graph and closeness of a node.
Small world network parameters
The clustering coefficient Ci for node i in an undirected graph is the ratio of the number of connections among neighbors of i (ci) to the maximum possible number of such connections; that is, where ki is the degree of i. The mean shortest path length L for a network is the average among all unique pairs of nodes of the length of the shortest path between the nodes.18
Closeness
The closeness Oi for node i in a graph is the inverse of the average shortest path length between i and all other nodes j in the graph, or
where N is the total number of nodes in the graph and lij is the shortest path between two nodes i and j.50,51
Statistical coupling analysis
For each protein, PFAM full sequence sets for the corresponding domain family52 provided an initial alignment which clustalw re-aligned.53 We then narrowed this alignment to a set of sequences in which all are 80% or more of the query length and no two are 90% or more identical to one another. This resulted in 392 sequences for PFK and 163 for FBPase. Finally, PCMA refined these raw clustalw alignments.54 Software provided by Rama Ranganathan analyzed these two families and identified the most strongly coupled clusters in each protein.27
Supplementary Material
Acknowledgments
We would like to thank Joel Bader, Bob Schleif, and Sidhartha Chaudhury for manuscript comments and helpful discussions, Rama Ranganathan for providing the software for performing statistical coupling analysis, and Bill Russ for instruction regarding the use of this software. TJU was supported by NSF EEC 0139643 REU-Site Program in Chemical, Cell and Tissue Engineering. JJG was supported by NIH awards K01-HG02316 and RO1-GM078221 and a Beckman Young Investigator Award.
References
- 1.Berg JM, Tymoczko JL, Stryer L. Biochemistry. New York: W.H. Freeman; 2002. [Google Scholar]
- 2.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000;9(1):10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luque I, Leavitt SA, Freire E. The linkage between protein folding and functional cooperativity: two sides of the same coin? Annu Rev Biophys Biomol Struct. 2002;31:235–256. doi: 10.1146/annurev.biophys.31.082901.134215. [DOI] [PubMed] [Google Scholar]
- 5.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57(3):433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 6.Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13(6):748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Yu EW, Koshland DE., Jr Propagating conformational changes over long (and short) distances in proteins. Proc Natl Acad Sci U S A. 2001;98(17):9517–9520. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Formaneck MS, Ma L, Cui Q. Reconciling the “old” and “new” views of protein allostery: a molecular simulation study of chemotaxis Y protein (CheY) Proteins. 2006;63(4):846–867. doi: 10.1002/prot.20893. [DOI] [PubMed] [Google Scholar]
- 9.Jardetzky O. Protein dynamics and conformational transitions in allosteric proteins. Prog Biophys Mol Biol. 1996;65(3):171–219. doi: 10.1016/s0079-6107(96)00010-7. [DOI] [PubMed] [Google Scholar]
- 10.Daily MD, Gray JJ. Local motions in a benchmark of allosteric proteins. Proteins. 2007;67(2):385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- 11.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 12.Tobi D, Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proc Natl Acad Sci U S A. 2005;102(52):18908–18913. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng W, Brooks B. Identification of dynamical correlations within the myosin motor domain by the normal mode analysis of an elastic network model. J Mol Biol. 2005;346(3):745–759. doi: 10.1016/j.jmb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Banavali NK, Roux B. Anatomy of a structural pathway for activation of the catalytic domain of Src kinase Hck. Proteins. 2007;67(4):1096–1112. doi: 10.1002/prot.21334. [DOI] [PubMed] [Google Scholar]
- 15.Liu T, Whitten ST, Hilser VJ. Ensemble-based signatures of energy propagation in proteins: a new view of an old phenomenon. Proteins. 2006;62(3):728–738. doi: 10.1002/prot.20749. [DOI] [PubMed] [Google Scholar]
- 16.Lu G, Giroux EL, Kantrowitz ER. Importance of the dimer-dimer interface for allosteric signal transduction and AMP cooperativity of pig kidney fructose-1,6-bisphosphatase. Site-specific mutagenesis studies of Glu-192 and Asp-187 residues on the 190’s loop. J Biol Chem. 1997;272(8):5076–5081. doi: 10.1074/jbc.272.8.5076. [DOI] [PubMed] [Google Scholar]
- 17.Barrick D, Ho NT, Simplaceanu V, Dahlquist FW, Ho C. A test of the role of the proximal histidines in the Perutz model for cooperativity in haemoglobin. Nat Struct Biol. 1997;4(1):78–83. doi: 10.1038/nsb0197-78. [DOI] [PubMed] [Google Scholar]
- 18.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 19.Greene LH, Higman VA. Uncovering network systems within protein structures. J Mol Biol. 2003;334(4):781–791. doi: 10.1016/j.jmb.2003.08.061. [DOI] [PubMed] [Google Scholar]
- 20.Vendruscolo M, Dokholyan NV, Paci E, Karplus M. Small-world view of the amino acids that play a key role in protein folding. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65(6 Pt 1):061910. doi: 10.1103/PhysRevE.65.061910. [DOI] [PubMed] [Google Scholar]
- 21.Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S. Network analysis of protein structures identifies functional residues. J Mol Biol. 2004;344(4):1135–1146. doi: 10.1016/j.jmb.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 22.Thibert B, Bredesen DE, del Rio G. Improved prediction of critical residues for protein function based on network and phylogenetic analyses. BMC Bioinformatics. 2005;6:213. doi: 10.1186/1471-2105-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.del Sol A, O’Meara P. Small-world network approach to identify key residues in protein-protein interaction. Proteins. 2005;58(3):672–682. doi: 10.1002/prot.20348. [DOI] [PubMed] [Google Scholar]
- 24.del Sol A, Fujihashi H, Amoros D, Nussinov R. Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100063. 2006 0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swint-Kruse L. Using networks to identify fine structural differences between functionally distinct protein states. Biochemistry. 2004;43(34):10886–10895. doi: 10.1021/bi049450k. [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan R, Rose GD. The T-to-R transformation in hemoglobin: a reevaluation. Proc Natl Acad Sci U S A. 1994;91(23):11113–11117. doi: 10.1073/pnas.91.23.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10(1):59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 28.Gu J, Bourne PE. Identifying allosteric fluctuation transitions between different protein conformational states as applied to Cyclin Dependent Kinase 2. BMC Bioinformatics. 2007;8:45. doi: 10.1186/1471-2105-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 61):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 30.Watts DJ. Small worlds : the dynamics of networks between order and randomness. Princeton, N.J: Princeton University Press; 1999. p. xv.p. 262. [Google Scholar]
- 31.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406(6794):378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 32.Erdos P, Renyi A. On the evolution of random graphs. Publ Math Inst Hung Acad Sci. 1960;5:17–61. [Google Scholar]
- 33.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 34.Amaral LA, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci U S A. 2000;97(21):11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gidh-Jain M, Zhang Y, van Poelje PD, Liang JY, Huang S, Kim J, Elliott JT, Erion MD, Pilkis SJ, Raafat el-Maghrabi M, et al. The allosteric site of human liver fructose-1,6-bisphosphatase. Analysis of six AMP site mutants based on the crystal structure. J Biol Chem. 1994;269(44):27732–27738. [PubMed] [Google Scholar]
- 36.De Staercke C, Van Vliet F, Xi XG, Rani CS, Ladjimi M, Jacobs A, Triniolles F, Herve G, Cunin R. Intramolecular transmission of the ATP regulatory signal in Escherichia coli aspartate transcarbamylase: specific involvement of a clustered set of amino acid interactions at an interface between regulatory and catalytic subunits. J Mol Biol. 1995;246(1):132–143. doi: 10.1006/jmbi.1994.0072. [DOI] [PubMed] [Google Scholar]
- 37.Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 2000;97(8):3844–3849. doi: 10.1073/pnas.050588097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286(5438):295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 39.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437(7058):512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 40.Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J Mol Biol. 2004;335(4):1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45(25):7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ota N, Agard DA. Intramolecular signaling pathways revealed by modeling anisotropic thermal diffusion. J Mol Biol. 2005;351(2):345–354. doi: 10.1016/j.jmb.2005.05.043. [DOI] [PubMed] [Google Scholar]
- 43.Ke HM, Lipscomb WN, Cho YJ, Honzatko RB. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J Mol Biol. 1988;204(3):725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- 44.Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991;218(1):233–260. doi: 10.1016/0022-2836(91)90887-c. [DOI] [PubMed] [Google Scholar]
- 45.Thompson JR, Bell JK, Bratt J, Grant GA, Banaszak LJ. Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry. 2005;44(15):5763–5773. doi: 10.1021/bi047944b. [DOI] [PubMed] [Google Scholar]
- 46.Monod J, Wyman J, Changeux JP. On The Nature Of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 47.Chennubhotla C, Bahar I. Markov propagation of allosteric effects in biomolecular systems: application to GroEL-GroES. Mol Syst Biol. 2006;2:36. doi: 10.1038/msb4100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eyal E, Gerzon S, Potapov V, Edelman M, Sobolev V. The limit of accuracy of protein modeling: influence of crystal packing on protein structure. J Mol Biol. 2005;351(2):431–442. doi: 10.1016/j.jmb.2005.05.066. [DOI] [PubMed] [Google Scholar]
- 49.Cormen TH, Cormen TH. Introduction to algorithms. Cambridge, Mass: MIT Press; 2001. [Google Scholar]
- 50.Beauchamp MA. An Improved Index of Centrality. Behav Sci. 1965;10:161–163. doi: 10.1002/bs.3830100205. [DOI] [PubMed] [Google Scholar]
- 51.Sabidussi G. The centrality of a graph. Psychometrika. 1966;31(4):581–603. doi: 10.1007/BF02289527. [DOI] [PubMed] [Google Scholar]
- 52.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34(Database issue):D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19(3):427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 55.Samuels ML, Witmer JA. Statistics for the life sciences. Upper Saddle River, NJ: Prentice Hall; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.