Abstract
Chloroplast ATP synthase (cpATPase) is an importance thylakoid membrane-associated photosynthetic complex involved in the light-dependent reactions of photosynthesis. In this study, we isolated and characterized a rice (Oryza sativa) mutant yellow leaf 1 (yl1), which exhibits chlorotic leaves throughout developmental stages. The YL1 mutation showed reduced chlorophyll contents, abnormal chloroplast morphology, and decreased photochemical efficiency. Moreover, YL1 deficiency disrupts the expression of genes associated with chloroplast development and photosynthesis. Molecular and genetic analyses revealed that YL1 is a nucleus-encoded protein with a predicted transmembrane domain in its carboxyl-terminus that is conserved in the higher plant kingdom. YL1 localizes to chloroplasts and is preferentially expressed in green tissues containing chloroplasts. Immunoblot analyses showed that inactivation of YL1 leads to drastically reduced accumulation of AtpA (α) and AtpB (β), two core subunits of CF1αβ subcomplex of cpATPase, meanwhile, a severe decrease (ca. 41.7%) in cpATPase activity was observed in the yl1-1 mutant compared with the wild type. Furthermore, yeast two-hybrid and bimolecular fluorescence complementation assays revealed a specific interaction between YL1 and AtpB subunit of cpATPase. Taken together, our results suggest that YL1 is a plant lineage-specific auxiliary factor involved in the biogenesis of the cpATPase complex, possibly via interacting with the β-subunit.
Higher plant chloroplasts are semiautonomous organelles derived through endosymbiosis from a relative of present-day cyanobacteria1. Chloroplasts are responsible not only for the photosynthetic conversion of CO2 to carbohydrates, but also for the production of metabolites and phytohormones2,3. Therefore, the establishment of functional chloroplasts is undoubtedly one of the most important processes associated with growth and yield in the vast majority of crops. Much attention has focused on the biogenesis and homeostasis of chloroplasts, which has led to some important findings3,4,5,6. Despite maintaining their own genomes, chloroplasts possess relatively few of the genes responsible for their own biosynthesis, whereas numerous nuclear genes (~2,000–3,000) encode chloroplast-localized proteins involved in chloroplast development1,7. Some of these genes have been identified by screening for abnormal coloration or photosynthetic deficient mutants in plants including rice, maize, wheat, barley, and Arabidopsis thaliana4,8. Great strides have been made towards understanding the complex regulatory mechanisms for the biogenesis and development of plant chloroplasts6.
Chloroplast F1F0-ATP synthase (cpATPase) is an important thylakoid membrane-associated protein complex involved in the light-dependent reactions of photosynthesis9. This complex utilizes the proton motive force (pmf) across the thylakoid membrane to drive ATP biosynthesis from ADP and inorganic phosphate10. Like the bacterial F0F1-ATP synthases, active plant cpATPase complexes also comprise a rotary motor composed of two subcomplexes, the partially membrane-intrinsic, hydrophobic subcomplex CF0 and the membrane-extrinsic, soluble subcomplex CF1. The CF0 moiety (subunits ab2c10–15) consists of a membrane-intrinsic c-ring (also known as III or AtpH) and three membrane-intrinsic subunits, including b (I or AtpF), b’ (II or AtpG), and a (IV or AtpI). The CF0 moiety mainly functions in proton translocation across the membrane. The CF1 moiety is composed of five subunits, α (AtpA), β (AtpB), γ (AtpC), δ (AtpD), and ε (AtpE) in the stoichiometric ratio α3β3γδε. Subunits γ and ε form a central stalk that cooperates with a peripheral region comprising subunits δ, a, and b to pull the two motors together. In addition, the α3β3 subcomplex forms a hexamer with three catalytic nucleotide binding sites, each located at the αβ interfaces, which are involved in the catalytic reaction of reversible ATP biosynthesis11,12,13. Although the structure and function of F1F0-ATP synthase have been preliminarily determined, little is known about the biogenesis of this complex, especially chloroplast ATP synthase. Like most other photosynthetic complexes, the cpATPase subunits are encoded by both the nuclear (subunits b’, γ, and δ) and organellar (subunits a, b’, c, α, β, and ε) genomes11. Moreover, many nucleus-encoded cofactors are involved in the biosynthesis and assembly of the cpATPase complex14. Therefore, tight regulation of gene expression in both the organellar and nuclear genomes is required to ensure that the biosynthesis and assembly processes are coordinated, including the handling of the various subunits and many other cofactors11.
The biosynthesis and sequential assembly of ATP synthase is a multistep process guided by a series of specific auxiliary factors12. Over the years, several auxiliary proteins that support the biogenesis of mitochondrial ATP synthase have been identified in Saccharomyces cerevisiae by screening for respiration-defective yeast mutants15,16,17,18,19. However, many of these proteins do not have obvious homologs in chloroplasts14, and it is likely that plant-specific assembly factors are required for the biosynthesis and assembly of cpATPase20. Currently, three auxiliary factors, include ALB4 (the ALBINO3 homolog), CGL160 (CONSERVED ONLY IN THE GREEN LINEAGE160), and PAB (PROTEIN IN CHLOROPLAST ATPASE BIOGENESIS), have been shown to play important roles in the assembly of cpATPase in plants21,22,23. ALB4 is orthologous to the E. coli protein YidC, a thylakoid membrane insertase, and is strictly required for membrane insertion of the a and c subunits24,25. In Arabidopsis, ALB4 appears to act as a specific auxiliary protein, not only for facilitating the assembly of the c-ring structure, but also for stabilizing or promoting the assembly of the CF1 moiety during its attachment to the CF0 moiety21. CGL160 is a thylakoid-membrane protein in Arabidopsis with a conserved carboxyl-terminus that is distantly related to prokaryotic ATP SYNTHASEPROTEIN1 (Atp1/UncI) proteins, which are thought to function in CF0 assembly22,26,27. Like UncI in most bacteria, AtCGL160 is not essential for ATP synthase biosynthesis in Arabidopsis but is required for the assembly of c-rings. Consequently, the atcgl160 mutant exhibits strong defects in cpATPase accumulation and photosynthetic efficiency12,22. In addition, PAB is a recently identified plant-specific assembly chaperone of cpATPase. In Arabidopsis, AtPAB directly interacts with the nucleus-encoded γ subunit and functions downstream of chaperonin 60 (Cpn60)-mediated CF1γ subunit folding to promote its assembly into the active CF1 core23. The characterization of these auxiliary factors has opened up the possibility of gaining insights into the mechanism underlying the biogenesis of this complex. However, compared with our knowledge of the biogenesis of other thylakoid membrane-associated protein complexes, such as PSI, PSII, and Cyt b6/f 28,29,30,31, little is known about the assembly of the photosynthetic complex cpATPase.
Here, we report the characterization of a leaf-color mutant in rice, yellow leaf 1 (yl1), which exhibits a chlorotic leaf phenotype, with reduced chlorophyll levels throughout plant development. Map-based cloning of the responsible gene resulted in the identification of a single site mutation in the fourth exon of YL1, which encodes a chloroplast-localized protein that is predicted to contain a transit peptide and a transmembrane domain but lacks any other recognizable motifs or domains. Plants without YL1 display a yellow leaf phenotype and severe defects in the accumulation of the AtpA/AtpB subunits of cpATPase. Moreover, we show that YL1 physically interacts with AtpB, a plastid-encoded β subunit of cpATPase, indicating its possible role in efficient biogenesis of the cpATPase in plant chloroplasts.
Results
Isolation and characterization of the yl1-1 mutant
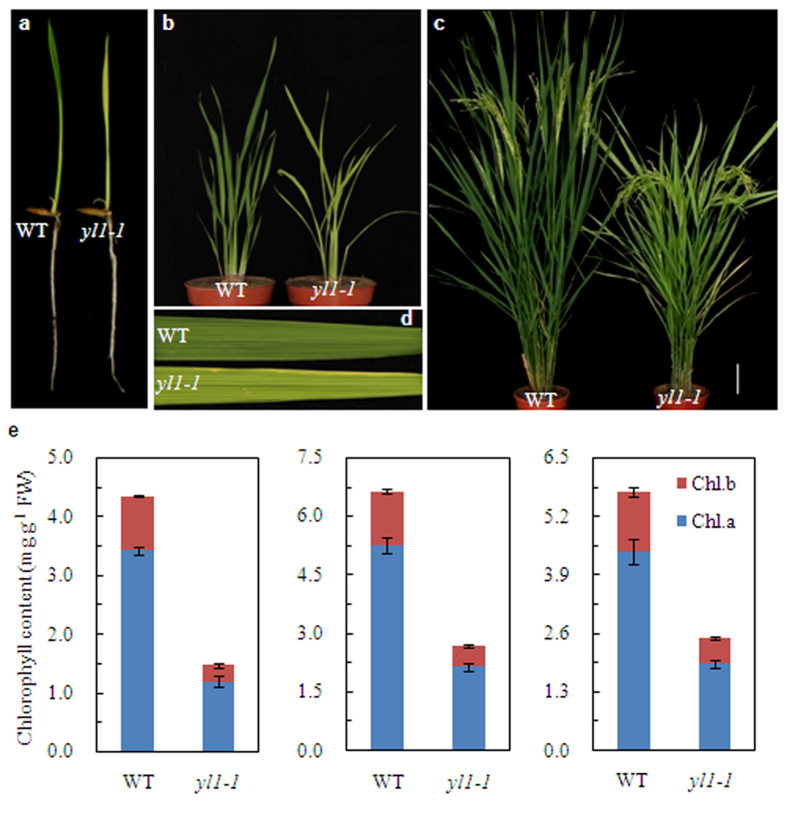
The rice yellow leaf mutant yl1-1 was identified from an ethyl methanesulfonate-mutagenized rice (indica cultivar Shuhui 527) population. Phenotypic analysis revealed that the yl1-1 mutant exhibits a yellow leaf phenotype throughout all developmental stages (Fig. 1a–d). We photographed the chlorotic phenotype of yl1-1 on day 10 (Fig. 1a), day 40 (Fig. 1b), and day 80 (Fig. 1c,d) after germination (DAG). In parallel, we measured the leaf chlorophyll contents of the yl1-1 mutant and wild-type plants at these three developmental stages. Significantly reduced levels of chlorophylls (Chl a + b) were detected in yl1-1 at all three stages, with 65.2%, 59.3%, and 56.6% reductions in Chl a levels and 70.3%, 60.3%, and 56.2% reductions Chl b levels compared to wild type at 10 DAG, 40 DAG, and 80 DAG, respectively (Fig. 1e). In addition, the yl1-1 mutant exhibited significantly earlier flowering and senescence than wild type, and as well as some altered agronomic traits. Compared with wild type, the yl1-1 mutant exhibited slightly reduced plant height, tiller number, and 1,000-grain weight (Supplemental Fig. S1).
Figure 1. Phenotypic characterization of wild type (WT) and yl1-1 mutants.
(a) Phenotypes of 7-day-old wild type and yl1-1 seedlings cultured in nutrition solution. (b) Phenotypes of 40-day-old wild type and yl1-1 seedlings under field condition. (c) Phenotypes of wild-type and yl1-1 plants at the booting stage. (d) Enlarged views of the leaves from (c). (e) Chlorophyll content of leaves in wild type and yl1-1 mutants at three different developmental stages: 10 (left), 40 (middle) and 80 (right) days after germination (DAG). Data are means ± SD (n = 5). Chl.a, Chlorophyll a; Chl.b, chlorophyll b; FW, fresh weight.
The yl1-1 mutant has impaired chloroplast development and photosynthesis
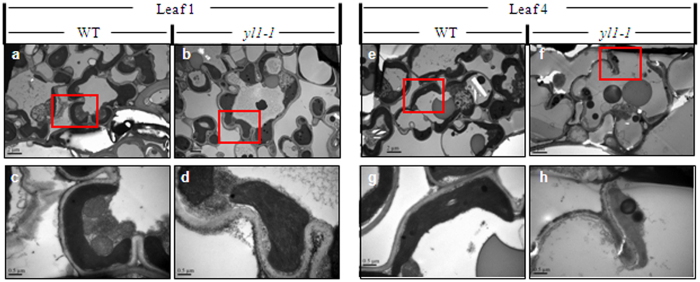
To investigate whether the lack of photosynthetic chlorophyll in the yl1-1 mutant is accompanied by defective chloroplast development, we examined ultrastructural changes in the chloroplasts of different-aged leaves from 40-day-old wild-type and yl1-1 plants. Transmission electron microscopy (TEM) revealed that chloroplasts in the leaves of wild-type plants displayed well-developed membrane structures, with dense thylakoids arranged in the grana and in membranes interconnecting the grana; no significant differences in chloroplast ultrastructure were observed between young and old leaves (Fig. 2a–d). However, chloroplasts from the leaves of mutant plants exhibited abnormal morphology, with loose thylakoid membranes and less dense grana stacks, and they also exhibited markedly accelerated degradation compared to those of age-matched wild-type plants (Fig. 2e–h; Supplemental Fig. S2). These observations indicate that the mutation of YL1 impairs chloroplast development.
Figure 2.
Transmission electron microscopy (TEM) of chloroplasts from Leaf 1 (a–d) and Leaf 4 (e–h) of the wild type (WT, left) and yl1-1 mutant (right) seedlings at 40-day-old. Leaf 1 to Leaf 4 represent leaves from the youngest to the oldest ones. (c,d,g,h) Enlarged images of chloroplasts shown in (a,b,e,f), respectively. Bars = 2.0 μm in (a,b,e,f); and 0.5 μm in (c,d,g,h).
We then examined the changes in photosynthetic capacity in the yl1-1 mutant. We measured photosynthetic parameters in 80-day-old seedlings of yl1-1 and wild-type plants grown in the field. As shown in Table 1, significantly reduced net photosynthetic rate (Pn), stomatal conductance (Gs), and transpiration rate (Tr) were detected in yl1-1 compared to wild-type plants. In addition, light-induced chlorophyll fluorescence measurements also showed that both the maximal efficiency of PSII photochemistry (Fv/Fm) and the effective quantum yield of PSII (ΦII) were slightly but significantly reduced in the yl1-1 mutant (Table 1). These results indicate that the YL1 mutation leads to reduced photosynthetic capacity.
Table 1. Chlorophyll Contents, photosynthetic and chlorophyll fluorescence parameters in wild-type and yl1-1 mutant.
| Pn (μmol CO2·m−2·s−1) | Gs (mol CO2·m−2·s−1) | Tr (mol CO2·m−2·s−1) | Fv/Fm | ΦII | |
|---|---|---|---|---|---|
| Wild type | 20.6 ± 0.41 | 0.79 ± 0.05 | 8.17 ± 0.28 | 0.78 ± 0.01 | 0.65 ± 0.01 |
| yl1-1 | 17.3 ± 0.22** | 0.62 ± 0.08** | 6.99 ± 0.25** | 0.75 ± 0.01** | 0.60 ± 0.02** |
Data are presented as means ± SD (n = 6). The asterisk indicates significant difference between the wild type and yl1-1 mutant (Student’s t-test, *p < 0.05; **p < 0.01). Chl, chlorophyll; Pn, net photosynthetic rate; Gs, stomatal conductance; Tr, transpiration rate; Fv/Fm, maximum quantum yield of P SII; ΦII, effective quantum yield of PSII.
Cloning and characterization of YL1
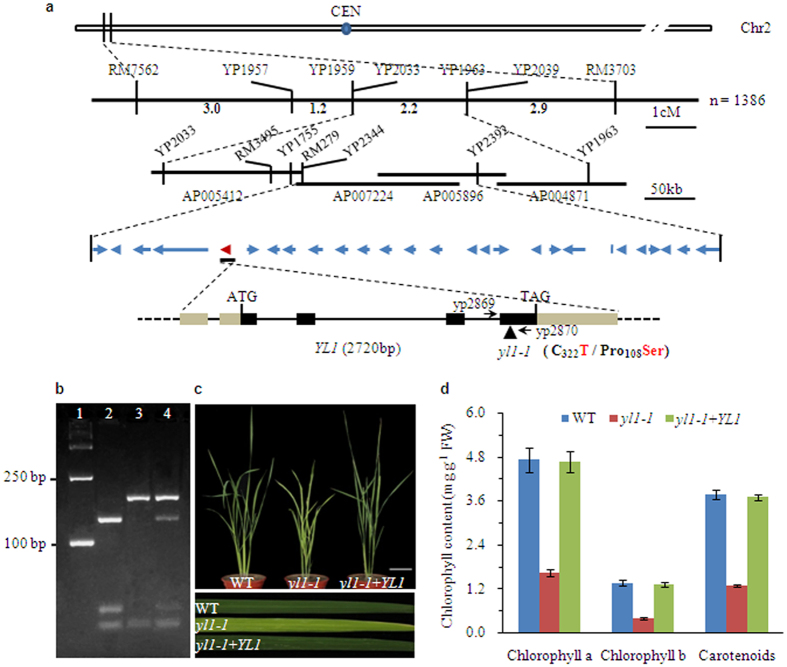
Using an F2 population generated from a cross between yl1-1 and japonica rice cultivar Nipponbare, the YL1 locus was initially mapped to an interval between simple sequence repeat (SSR) markers RM7562 and RM3703 at the top of rice chromosome 2 (Fig. 3a). To perform fine mapping of YL1, we utilized nine newly developed sequence-tagged site (STS) markers located between RM7562 and RM3703, ultimately localizing the YL1 locus to a 199-kb interval between markers YP2344 and YP2392 on BAC (bacterial artificial chromosome) clones AP007224 and AP005896 (Fig. 3a). This region contains 27 annotated genes (TIGR Rice Genome Annotation Database). Sequencing of these genes from wild type and yl1-1 revealed a single nucleotide substitution (C-to-T) in exon 4 of LOC_Os02g05890, leading to the transition of an amino acid residue from proline (Pro) in wild type to serine (Ser) in the yl1-1 mutant.
Figure 3. Map-based cloning of the YL1 gene.
(a) Schematic diagram of the YL1 gene inferred by DNA sequence analysis. YL1 was mapped primarily to the top of rice chromosome 2 between markers RM7562 and RM3703 and then narrowed to a 199-kb region using an enlarged F2 mapping population. Amplification of relevant DNA fragments and sequence comparison revealed that the yl1 alleles resulted from a single base substitution (C to T) in the fourth exon of YL1 gene leading to the change of amino acid residue from Pro to Ser in residue 108. The blue arrows denote the 27 putative ORFs in the 199-kb genomic region. YL1 (LOC_Os02g05890) is shown as the red arrow. Exons (black boxes), introns (black lines) and UTR region (brown boxes) are indicated. ATG start codon and TGA stop codon are shown. (b) The presence of normal transcripts of YL1 in transgenic line of yl1-1 with complemented expression of YL1 was confirmed by PCR followed by restriction enzyme digestion. A 207-bp DNA fragment around the mutation is amplified with the specific primers (YP2869 and YP2870) and then digested with the restriction enzyme AciI. For wild type, the DNA is cleaved at position 129 and 172 and three fragments are generated. There is only one AciI site (at position 172) in the corresponding DNA of yl1-1 mutant due to a single base substitution, and therefore its DNA is cut to yield two fragments. Line 1, molecular weight (D2000 DNA marker); line 2, wild type; line 3, yl1-1 mutant; line 4, transgenic line of yl1-1 with complemented expression of YL1. (c) Phenotypic complementation of the yl1-1 mutant by introduction of the YL1 gene. Left, wild type; center, yl1-1 mutant; right, transgenic line of yl1-1 with complemented expression of YL1 (40-day-old plants, bar = 10 cm). Flag leaves of each line are enlarged at the bottom section of (c) to highlight the leaf color. (d) Chlorophylls contents of 40-day-old leaves in wild-type, yl1-1 mutant and transgenic line of yl1-1 with complemented expression of YL1. Mean values were obtained from five independent experiments and error bars indicate SD.
To help confirm that LOC_Os02g05890 is the corresponding YL1 gene, we introduced a wild-type genomic DNA fragment 5.38 kb in size, including the entire coding region of the candidate gene, the 2,099-bp 5′-upstream sequence, and the 1,382-bp 3′-downstream, into yl1-1 by Agrobacterium-mediated transformation (Supplemental Fig. S3a). The authenticity of independent transgenic plants was verified by restriction enzyme digestion analysis, because an AciI restriction site was abolished by the C-to-T substitution in the mutant sequence (Fig. 3b; Supplemental Fig. S3b). Of the 28 plants generated, 24 independent transgenic-positive plants phenocopied the wild type (Fig. 3c). In addition, the levels of Chl a, Chl b, and carotenoids in transgenic yl1-1 lines with complemented expression of YL1 were also restored to wild-type levels (Fig. 3d). These results indicate that the yellow-leaf phenotype indeed resulted from the mutation of LOC_Os02g05890.
To further confirm that the mutated gene is responsible for the observed phenotype, we obtained an additional independent homozygous T-DNA insertion mutant of LOC_Os02g05890 (RMD_03Z11BQ88) from the Rice Mutant Database, which was subsequently designated yl1-2. Analysis of the flanking sequence revealed that the mutant carries a T-DNA insertion in the second exon of LOC_Os02g05890. Homozygous mutant plants with the T-DNA insertion were confirmed by PCR using gene-specific primers (YP3180 and YP3181) and a T-DNA border primer (LBT2; Supplemental Fig. S4a,b). RT and qRT-PCR analysis showed that YL1 transcripts were not present in homozygous mutant plants (Supplemental Fig. 4c,d), indicating that yl1-2 is a loss-of-function mutant. Leaves of the homozygous yl1-2 mutant exhibited an identical yellow phenotype to that of the yl1-1 mutant (Supplemental Fig. 4e). We also observed chlorophyll deficiency and photosynthetic defects in yl1-2 mutant plants (Supplemental Fig. 4f; Supplemental Table S1). These results indicate that inactivated YL1 is responsible for the observed yl1-2 phenotype and that LOC_Os02g25890 is indeed YL1.
Sequence analysis revealed that YL1 encodes a protein of 165 amino acids with a putative chloroplast transit peptide of 51 amino acids at the N-terminus and one transmembrane region between residues 116 and 138 near the C-terminus of the sequence but with no other recognizable domains or motifs (http://www.cbs.dtu.dk/services/ChloroP/; http://smart.embl-heidelberg.de/smart/; Supplemental Fig. S5). Analysis of the rice genome annotation databases (http://rice.plantbiology.msu.edu/) revealed two other homologs in the rice genome (LOC_Os02g17380 and LOC_Os06g22660) sharing 49% and 51% identity with YL1, respectively. BLASTP searches (http://blast.ncbi.nlm.nih.gov/) also revealed homologous proteins in many other higher plants, including Aegilops tauschii, Arabidopsis lyrata subsp. lyrata, Arabidopsis thaliana, Amborella trichopoda, Brachypodium distachyon, Capsella rubella, Cucumis sativus, Fragaria vesca subsp. vesca, Glycine max, Medicago truncatula, Setaria italica, Solanum lycopersicum, Sorghum bicolor, Theobroma cacao, Triticum urartu, Vitis vinifera, and Zea mays. These proteins share 38–85% amino acid sequence identity, with a highly conserved C-terminus (Supplemental Fig. S5). Interestingly, the mutation in yl1-1 only affects this conserved region (proline [P] at position 108 of YL1), indicating the importance of this region for the regulatory function of YL1. Phylogenetic analysis showed that the YL1 homologs clearly clustered into two major groups, monocotyledons and dicotyledons, revealing the evolution of two distinct clades of YL1 genes in dicotyledons and monocotyledons (Supplemental Fig. S5b). However, the functions of these homologous genes are largely unknown.
YL1 is targeted to the chloroplast
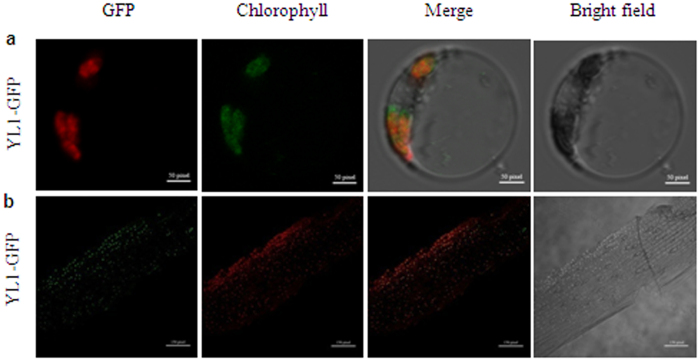
To determine the intracellular localization of YL1, we fused the full-length YL1 gene to the green fluorescent protein (GFP) gene driven by the cauliflower mosaic virus (CaMV) 35S promoter (35S::YL1::GFP) and transformed this construct into rice protoplasts by transient transformation. Confocal Laser Scanning Microscopy (CLSM) observation showed that the green fluorescence of YL1::GFP fusion protein was exclusively colocalized with the red autofluorescence of chloroplastic chlorophyll (Fig. 4a). Moreover, we observed a consistent localization pattern of YL1-GFP in transgenic rice plants containing the 35S::YL1::GFP cassette: the fluorescence signals of the YL1-GFP fusion protein were completely overlapping with chloroplast autofluorescence in the living cells of leaf epidermis from transgenic plants (Fig. 4b). These observations confirm that YL1 is a chloroplast-localized protein.
Figure 4. Subcellular localization of the YL1 protein.
(a) In vivo targeting of YL1-GFP in rice protoplast cells, bar = 50 pixel. (b) fluorescence signals in the living cells of leaf epidermis from YL1-GFP transgenic seedlings, bar = 150 pixel. From left to right, GFP fluorescence, chloroplast’s autofluorescence, merge images, and bright field images.
YL1 is expressed in green tissues
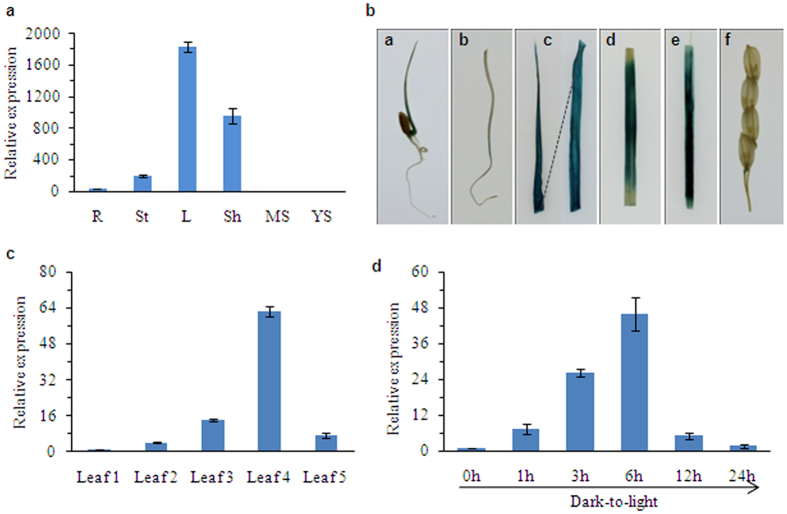
To investigate the tissue-specific expression pattern of YL1, we analyzed RNA from roots, stems, leaves, leaf sheaths, and young spikes of wild-type plants by quantitative reverse-transcription PCR (qRT-PCR). Our results show that YL1 is predominantly expressed in green organs, with the highest transcript levels in leaves, followed by leaf sheaths and stems, but with very low expression levels in roots and young spikes (Fig. 5a). We performed histochemical staining of transgenic rice plants harboring the pYL1::GUS cassette to further evaluate YL1 expression in vivo. GUS activity was strongly detected in leaves, leaf sheaths, and stems, but not in roots or young spikes (Fig. 5b), which is consistent with the results obtained from qRT-PCR analysis. These results suggest that YL1 might be responsible for the development of green tissues.
Figure 5. YL1 Expression Pattern Analysis.
(a) qRT-PCR analysis of YL1 expression in various tissues, including roots (R), stem (S), leaves (L), sheath(Sh), spike for mature stage (MS) and young spike (YS) were detected. The YL1 transcript level was normalized to the Actin gene transcript (LOC_Os03g50885). (b) pYL1::GUS expression patterns in transgenic rice plants. From left to right: young buds, root, leaves, stems, leaf sheath and young spike. (c) qRT-PCR analysis of the YL1 expression in different-aged leaves. Leaf1 to Leaf5 represent leaves from the youngest to the oldest ones in 80-day-old wild-type plants grown under field condition (see Supplemental Fig. S6). (d) qRT-PCR analysis of the YL1 expression during greening of etiolated seedlings. Wild type rice seeds were germinated and grown in darkness for 7 d, etiolated seedlings were then illuminated for 0 (control), 1, 3, 6, 12 and 24 h under normal light condition. All the data are means ± SD (n = 3).
To further investigate the possible involvement of YL1 in leaf development, we examined the expression patterns of YL1 during leaf ontogenesis. We investigated YL1 mRNA expression patterns in a series of leaves harvested from 80-day-old wild-type plants by qRT-PCR. In these plants, leaf number 1 represents the youngest leaves and leaf number 5 represents the oldest leaves (Supplemental Fig. S6a). An age-dependent increase in YL1 expression was observed from leaf 1 to leaf 4, but YL1 expression decreased in leaf 5 (Fig. 5c). Interestingly, this expression pattern is similar to the variation in chlorophyll contents between young and old leaves (Supplemental Fig. S6b). These results help confirm that YL1 plays an important role in chlorophyll biosynthesis during leaf development.
Light is an important element during periods of rapid chlorophyll production. We therefore investigated whether the expression of YL1 is induced by light stimulation. We analyzed the expression levels of YL1 in shoots of seven-day-old wild-type etiolated seedlings by qRT-PCR at different times after illumination. Relatively low levels of YL1 transcript were detected in etiolated leaves, but its expression rapidly increased within 6 hours of illumination and decreased gradually over time (Fig. 5d), indicating that YL1 is a light-induced gene that is likely involved in light-regulated chlorophyll production and chloroplast development.
Expression profiles of genes involved in chloroplast development and photosynthesis
We then examined the expression of genes involved in chloroplast development in both yl1-1 and wild-type plants. Eight genes previously reported to function in chloroplast biogenesis and development were selected for expression analysis by qRT-PCR, including genes encoding Virescent1 (V1, nuclear undecaprenyl pyrophosphate synthase 1, NUS1), V2 (a guanylate kinase), V3 (a large subunit of ribonucleotide reductase, RNR), VYL (a plastidic caseinolytic protease, OsClpP6), RpoTP (a nucleus-encoded RNA polymerase), RpoA (a plastid-encoded RNA polymerase), Rps15 (Ribosomal Protein S15), and OsSig2A (a nucleus-encoded chloroplast sigma factor). The results show that almost all of these genes were significantly upregulated, except for RpoTP (which appeared to be upregulated but not significantly so), in the yl1-1 mutant compared to wild type (Fig. 6). Moreover, we also investigated the transcriptional levels of genes associated with photosynthesis. The mRNA levels of several specific photosynthesis-related genes encoding the reaction-center proteins of photosystem I/II (PsaA, PsaD, PsaE, PsaF, PabA, PsbB, and PsbO), the large subunit of Rubisco (RbcL), light-harvesting complex protein Lhcp2, cytochrome b6/f protein (Cyt f), and light harvesting Chla/b binding protein 1 (Cab1), were significantly reduced in the yl1-1 mutant compared to wild type (Supplemental Fig. S7). These results suggest that YL1 participates in regulating the expression of genes associated with chloroplast development and photosynthesis.
Figure 6. Expression analysis of genes involved in chloroplast development and plastidic transcription apparatus in leaves of wild type (WT) and yl1-1 mutants.
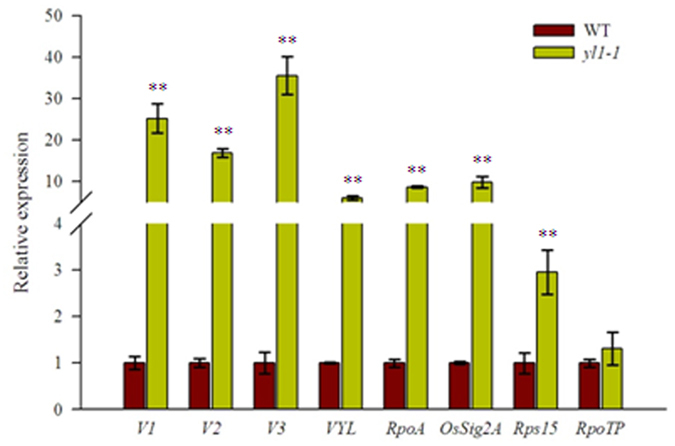
The relative expression level of each gene were analyzed by qRT-PCR and normalized using the Actin gene (LOC_Os03g50885) as an internal control (mean ± SD, n = 3). The asterisk indicates significant difference between the wild type and yl1-1 mutant (Student’s t-test, **p < 0.01).
Accumulation of photosynthetic complexes is impaired in yl1-1
The obvious difference in chlorophyll content and photosynthesis related genes expression between wide type and yl1-1 mutant prompted us to assess the changes in levels of photosynthetic complexes in yl1-1. We extracted thylakoid membranes proteins from wild-type and yl1-1 leaves and performed an immunoblot analysis using specific antibodies against several representative subunits of the thylakoid protein complexes (i.e. PSI, PSII, Cyt b6f, LHC, RbcL and ATP synthase). The results showed that, on an equal fresh weight basis, the levels of the PSI core subunit, PsaA, was decreased to ~30% of wild-type levels and that the amounts of the PSII proteins, D1 and PsbO, were reduced to 60% to 70% of wild-type levels. The levels of the cpATPase subunits, AtpA and AtpB, were also reduced to approximately 50–60% of wild-type levels (Fig. 7). Moreover, marked reductions in the levels of Cyt f, LHcb and RbcL were also detected in the yl1-1 mutants, to about ~56%, ~44% and ~75% of those seen in the wild type, respectively (Fig. 7).
Figure 7. Immunoblot analysis of thylakoid membrane proteins from wild type (WT) and yl1-1 mutant plants.
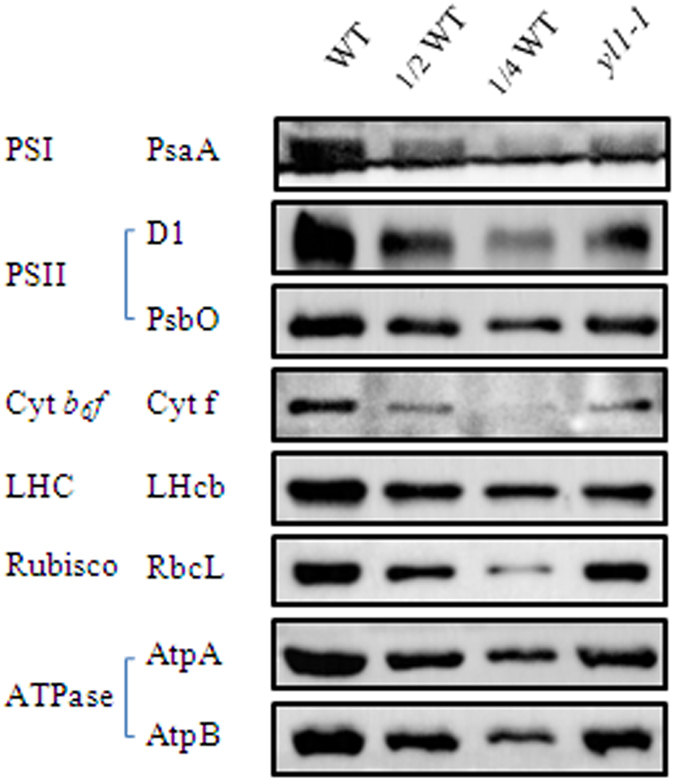
Total proteins were extracted from leaves of 4-week-old wild type and yl1-1 mutant plants and separated by 10% SDS-PAGE on the basis of equal fresh weight. The blots were probed with specific anti-PsaA, anti-PsbA (D1), anti-PsbO, anti-Cyt f, anti-LHCII, anti-RbcL, anti-AtpA and anti-AtpB antibodies.
We next performed Blue-Native PAGE (BN-PAGE) analysis to investigate the possible changes in the structure of photosynthetic complexes in the yl1-1 mutant. Thylakoid membranes were solubilized with dodecyl-b-D-maltopyranoside (DM) and separated by blue native PAGE (BN-PAGE) based on an equal chlorophyll content. After the first-dimensional separation, six major bands were observed, representing PSI-PSII supercomplex (band I), PSI-PSII dimer (band II), PSI monomer (band III), CP43-less PSII core monomer (band IV), LHCII dimer (band V), and LHCII monomer (band VI) according to previous reports32. Notably, the relative level of PSI and PSII (band I and band III) per unit of chlorophyll was significantly reduced in the thylakoid membranes of yl1-1 compared to wild type (Supplemental Fig. S8). The protein complexes resolved by BN-PAGE were then separated into their subunits by SDS-urea-PAGE in the second dimension. The results confirm that the amount of the PSI subunits PsaA/PsaB was considerably reduced in the yl1-1 mutant compared with wild type, whereas no significant differences in the levels of PSII core subunits, ATPase or light-harvesting complex II (LHCII) were observed between wild-type and yl1-1 mutant plants (Supplemental Fig. S8). Taken together, these results suggest that the relative amount of protein subunits of photosynthetic complexes is disturbed in the mutant, especially the PSI core subunits.
YL1 interacts with the β-subunit of chloroplast ATP synthase
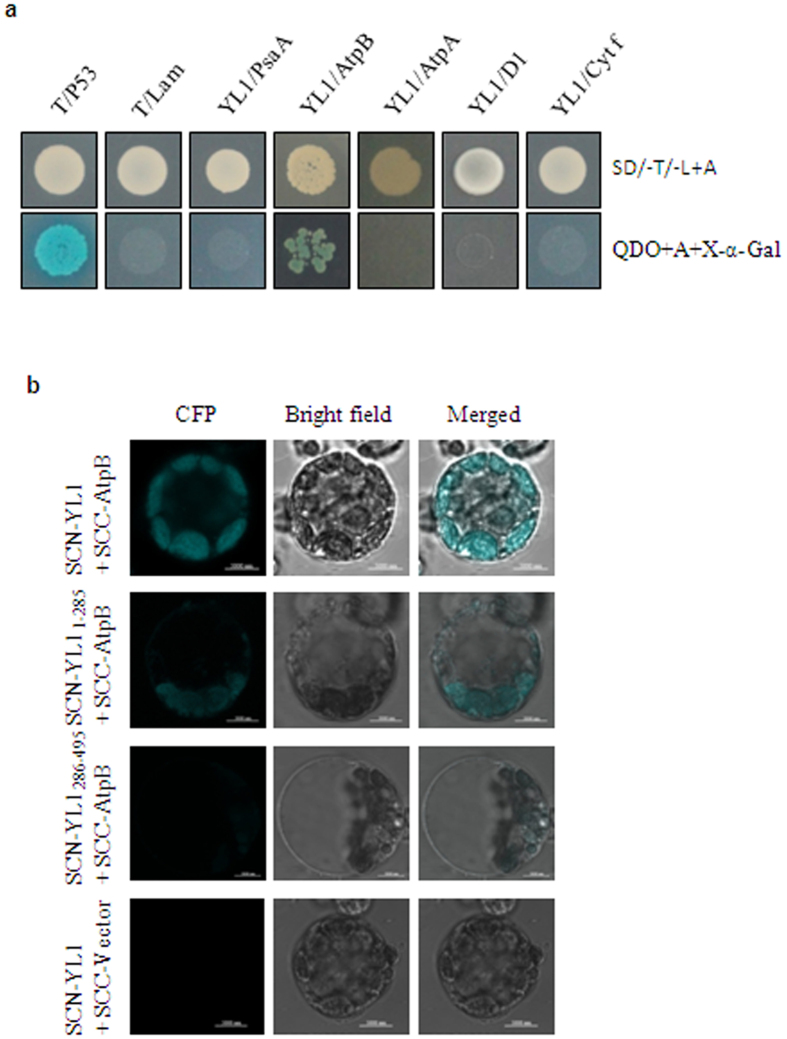
Since YL1 is involved in the biogenesis of photosynthetic complexes, we reasoned that YL1 might directly interact with specific subunits of these complexes. To test this possibility, we performed yeast two-hybrid assays to identify potential interactions between YL1 and several representative subunits of the PSI, PSII, cpATPase, and Cyt b6/f complexes. Notably, only yeast cells co-transformed with the BD-YL1 prey construct and AD-AtpB bait vector grew on SD/-Ade/-His/-Leu/-Trp medium containing X-α-gal (Fig. 8a). These results suggest that YL1 directly interacts with AtpB but not with AtpA or PsaA of the PSI complex, D1 of the PSII complex or Cyt f of the Cyt b6/f complex. In order to confirm the interaction between YL1 and AtpB protein occurs in vivo, bimolecular fluorescence complementation (BiFC) analysis was further conducted. Cyan fluorescence protein (CFP) fluorescence was reconstituted when the full-length YL1 and AtpB proteins were co-expressed in rice protoplasts (Fig. 8b), showing that they physically interact with each other in vivo. Moreover, deletion derivatives of YL1 were further analyzed for interaction to determine which domains in YL1 are involved in the interaction with AtpB, and results showed that construct with only the C-ternimal failed to interact with AtpB (Fig. 8b). Based on these results, we concluded that YL1 physically interacts with AtpB and the N-ternimal is essential for the interaction of YL1 with AtpB.
Figure 8. Interaction of YL1 with AtpB.
(a) Yeast two-hybrid analysis of the interaction between YL1 and several subunits of thylakoid membrane complexes. The full-length coding sequence of YL1 was cloned into pGBKT7 (bait vector), and the PsaA, AtpA, AtpB and Cyt f genes were individually cloned into pGADT7 (prey vector). The prey and bait constructs were cotransformed into S. cerevisiae strain AH109. “T/P53” and “T/Lam” represent positive and negative control, respectively. (b) BiFC assays showing the interaction between YL1 and AtpB in the chlorolasts of rice protoplasts. As negative controls, SCN-YL1 and empty vector of SCC (SCC-Vector) were cotransfected into protoplasts. CFP, cyan fluorescence protein. Bars = 5 μm.
ATPase activity is markedly reduced in chloroplasts of the yl1-1 mutant
To determine whether the chloroplast ATPase activity is affected in yl1-1 mutant, intact chloroplasts were isolated from leaves of wild type and yl1-1 mutant plants, and the ATPase activity in isolated chloroplasts was measured on the basis of equal fresh weight. As shown in Supplemental Fig. S9, the chloroplast ATPase activity in yl1-1 mutant was markedly reduced to 58.3% of the wide type levels, indicating that the accumulation of functionally active ATPase complex in chloroplasts of yl1-1 mutant was significant affected.
Discussion
The biogenesis of chloroplast ATP synthase is a complicated and highly regulated process that requires coordination between the nuclear and plastid genome and also depends on the action of various nucleus-encoded auxiliary factors involved in transcription, translation, import, protein turnover, and complex assembly33. Identifying and characterizing these assembly factors will provide meaningful insights into the mechanism underlying cpATPase biogenesis. However, compared to the number of auxiliary proteins found to function in the assembly of mitochondrial ATP synthase in yeast, surprisingly few auxiliary proteins involved in cpATPase assembly have been characterized to date13. Therefore, additional cpATPase assembly factors likely remain to be identified. In this study, we report the identification of rice protein YL1, a nucleus-encoded chloroplast protein, which appears to be involved in the biogenesis of the chloroplast ATPase complex, possibly through interaction with the AtpB subunit.
Database searches revealed that YL1 and its putative homologs are only present in green land plants (Supplemental Fig. S5). These proteins contain a conserved C-terminus but lack any functional domain or motif. However, a specific function for these proteins has not yet been experimentally demonstrated. In the present study, we found that the rice yl1-1 mutant is characterized by a yellow leaf phenotype throughout development, which is compatible with its reduced chlorophyll content, abnormal chloroplast morphology, and reduced photochemical efficiency (Figs 1 and 2; Table 1). In vivo transformation of the YL1::GFP fusion protein construct into rice protoplasts revealed its intracellular targeting to the chloroplast (Fig. 4). Moreover, similar to other rice mutants impaired in chloroplast development34, several well-known genes involved in chloroplast development and the plastidic transcription apparatus were highly upregulated in the yl1-1 mutant, whereas chlorophyll biosynthesis- and photosynthesis-related genes were dramatically downregulated (Fig. 6; Supplemental Fig. S7). These results suggest that YL1 functions in chloroplast development. In Arabidopsis, the YL1 homolog At1g56200 (EMB1303), which shares 45% amino acid sequence identity with rice YL1, also plays an essential role in chloroplast development35. However, the molecular nature of YL1 might differ from that of Arabidopsis EMB1303. First, the rice yl1 mutant (both yl1-1 and yl1-2 alleles) phenotype is less severe than that of emb1301, which exhibits an albino seedling-lethal phenotype, even when grown on medium containing sucrose35. Second, YL1 is preferentially expressed in green tissues containing chloroplasts, such as leaves, sheaths, and stems (Fig. 5a,b). By contrast, AtEMB1303 is constitutively expressed in various tissues, including young leaves, roots, and flowers35. Third, phylogenetic analysis revealed that the YL1 homologs were consistently subdivided into two major groups, dicotyledons and monocotyledons (Supplemental Fig. S5), suggesting the functional specialization of YL1 proteins between monocot and dicot plants. Taken together, these results suggest that YL1 is a plant lineage-specific protein that functions in chloroplast development in monocotyledonous plants.
Plants lacking YL1 exhibited abnormal chloroplast morphology, with loose thylakoid membranes and less dense grana stacks (Fig. 2; Supplemental Fig. S2), which is compatible with the reduced photosynthetic capacity revealed by the obvious reduction in Pn, Gs, and Tr levels (Table 1; Supplemental Table S1). Meanwhile, our immunoblot analyses revealed that the abundance of several representative subunits of the thylakoid protein complexes, such as subunits of PSI (PsaA), PSII (D1, PsbO) and cpATPase (AtpA, AtpB), was disturbed in the yl1-1 mutant, especially the accumulation of PSI core subunit PsaA, which reduced to ~30% of wild-type levels (Fig. 7). These results indicate that the accumulation of thylakoid membrane proteins was disturbed in the yl1-1 mutant, which might be effected the formation of photosynthetic complex. However, chlorophyll fluorescence measurements showed that mutation of YL1 only caused a slight reduction in PSII efficiency (Table 1), indicating that the PSII complex was accumulate in a stable manner in yl1-1 mutant and YL1 is unlikely to be involved in the formation of the PSII complex. Notably, a severe decrease (ca. 41.7%) in chloroplast ATPase activity was observed in the yl1-1 mutant compared with the wild type (Supplemental Fig. 9), suggesting that YL1 might be responsible for the formation of functionally active ATPase in chloroplasts. In addition, relatively low accumulation of PSI proteins has also been observed in several cpATPase mutants, suggesting that the reduced PSI protein level in the mutant is mostly a secondary effect of the lack of cpATPase22,36. Perhaps this secondary effect involves the strong lumenal overacidification caused by the repression of cpATPase36. We therefore speculate that YL1 might be involved in the biogenesis of cpATPase complexes.
This hypothesis was confirmed by yeast two-hybrid analysis: the results show that YL1 interacts directly and specifically with AtpB in cpATPase but not with AtpA or PsaA of the PSI complex, D1 of the PSII complex or with Cyt f of the Cyt b6/f complex (Fig. 8). Therefore, YL1 is most likely involved in the assembly of the CF1 subcomplex during cpATPase biogenesis. Previous studies have demonstrated that assembly of the CF1 complex is accomplished in a step-by-step manner in the chloroplast. After sequential formation of the subunits, the α and β subunits are folded and assembled into a dimer in a chaperone-dependent process. Subsequently, three such dimers are further assembled into a hexamer20. However, little is known about this process at the molecular level13. In yeast mitochondria, CF1-α3β3 hexamer assembly requires two chaperone proteins, Atp11p and Atp12p, which bind to the α- and β-subunits respectively37,38,39. However, homologs of these two chaperones are absent in chloroplasts. Thus, an entirely different assembly mechanism was postulated for CF1 subcomplex assembly in the chloroplast12. In this study, the specific interaction between YL1 and the β subunit (AtpB) was confirmed by in vivo and in vitro tests (Fig. 8), and a significantly reduction in cpATPase activity was detected in the mutant, suggesting that YL1 might be serve as an auxiliary protein that is required for biogenesis of the cpATPase catalytic center CF1-α3β3 hexamer in chloroplasts. However, the precise role of YL1 in α3β3 hexamer fomation requires further investigation.
Notably, rice plants lacking YL1 retain approximately 58.3% of normal levels of the ATPase activity (Supplemental Fig. 9), which contributes to the mild phenotype of these mutant plants (Fig. 1). This finding suggests that relatively low but still significant levels of active cpATPase are present in the yl1-1 mutant. A similar phenomenon has been also observed in the Arabidopsis atcgl160-1 (AtCGL160 is required for c-ring assembly) and atpab (AtPAB functions in the assembly of CF1γ into the CF1 core) mutants, in which the assembly of cpATPase is affected22,23. Therefore, the mild phenotype of the yl1 mutant might be due to the presence of residual protein in this mutant, which might retain some normal functions, as is true for the atpab mutant in Arabidopsis23. It is also possible that YL1-independent assembly of functional ATP synthase occurs in the yl1-1 mutant, suggesting functional redundancy. Such functional redundancy has been experimentally demonstrated in the PSII repair and assembly process40,41. Therefore, additional auxiliary protein(s) might be involved in the accumulation of the CF1-α3β3 hexamer in chloroplasts. Further studies of YL1 and its interaction partners will provide a more detailed understanding of cpATPase biogenesis in higher plants.
Materials and Methods
Plant materials and growth conditions
The rice (Oryza sativa) mutant yl1-1 was identified from a mutagenized population of rice ssp. indica cv. Shuhui 527 treated with ethyl methanesulfonate (EMS). F2 mapping populations were generated from a cross between the yl1-1 mutant and japonica rice cultivar Nipponbare. The T-DNA insertion mutant line yl1-2 (RMD_03Z11BQ88) in a rice ssp. japonica cv. Zhonghua 11 background was obtained from the Rice Mutant Database (http://rmd.ncpgr.cn)42,43. Shuhui 527 and Zhonghua 11 represent the wild-type (WT) controls for yl1-1 and yl1-2, respectively. Rice plants were grown in paddy fields under natural conditions or in a growth chamber under a 14-h-light (30 °C)/10-h-dark (24 °C) cycle at the Hangzhou Normal University and the China National Rice Research Institute in Hangzhou, China (latitude 30° 26N; longitude 120° 19E).
Map-based cloning of YL1
The YL1 locus was mapped and cloned using 1,386 individual F2 mutant plants screened from a population of yl1-1 and Nipponbare. YL1 was preliminarily mapped to the top of rice chromosome 2 using SSR (simple sequence repeat) markers and STS (sequence-tagged site) markers that are evenly distributed on the 12 rice chromosomes (Supplemental Table S2). New STS markers for fine mapping were developed based on genome polymorphisms between Nipponbare and 93-11 (ssp. indica) around the yl1 locus44, and YL1 was ultimately mapped to a 199-kb region on chromosome 2 between the two new STS markers YP2344 and YP2392. DNA fragments corresponding to the 27 candidate genes in this region were amplified by PCR from wild-type and mutant plants and sequenced to identify the yl1-1 mutation. Molecular markers used in this study are described in Supplemental Table S2.
Complementation of the yl1-1 mutant
For yl1-1 mutant complementation, a 5.38-kb genomic DNA fragment containing the entire YL1 coding region, a 2,099-bp promoter region, and a 1,382-bp downstream sequence (Supplemental Fig. S3a) was amplified from Nipponbare using primers pCYL1-F and pCYL1-R (Supplemental Table S2). The PCR product was fully sequenced and cloned into binary vector pCAMBIA1301 using the XbaI and SalI restriction sites to generate the transformation plasmid. Subsequently, the binary construct was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into yl1-1 mutant plants via Agrobacterium-mediated transformation as described45,46.
Bioinformatics analysis
Putative chloroplast transit peptides and transmembrane domains were predicted using the online tools ChloroP (http://www.cbs.dtu.dk/services/ChloroP/)47 and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/)48, respectively. BLASTp analysis was performed using the NCBI database to search for YL1 homologs (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment of YL1 and its homologs was performed with the ClustalW program (www.ebi.ac.uk/clustalw/)49. The phylogenetic tree was generated with MAGE software version 5.0550 using the neighbor-joining method and the bootstrap method with 1,000 bootstrap replications.
Measurement of chlorophylls contents and photosynthetic characteristics
Chlorophylls and total carotenoids were measures in fresh leaves collected from wild-type and yl1-1 plants at three different developmental stages (10, 40, and 80 days after germination), two-week-old wild-type and yl1-2 plants, and 40-day-old wild-type, yl1-1 mutant, and transgenic yl1-1 plants with complemented expression of YL1. Leaf samples (~50 mg) were immersed in 10 ml extract solution (45% ethanol + 45% acetone + 10% water) for 16 h in the dark and examined spectrophotometrically at 663, 645, and 470 nm following the method described by Lichtenthaler51.
Photosynthetic parameters including net photosynthetic rate (Pn), stomatal conductance (Gs), transpiration rate (Tr), and intracellular CO2 concentration (Ci) were measured with an LI-6400 portable photosynthesis system (LI-COR, Lincoln, USA). Chlorophyll fluorescence parameters (Fv/Fm and ΦII) were determined using a chlorophyll fluorescence system (PAM-2500, Heinz-Walz Instruments, Germany), as described by Liu et al.52. The flag leaves of wild-type and mutant plants at booting stage were used for measurement.
Transmission electron microscopy
Chloroplast ultrastructure in different-aged leaves from 40-day-old wild-type and yl1-1 mutant plants was examined by transmission electron microscopy (TEM). Leaf samples for TEM were prepared as previously described53. TEM observation was performed under a transmission electron microscope (JEOL JEM-1230 EX, Japan).
RT-PCR and quantitative RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) and reverse transcribed into first-strand cDNA using ReverTra Ace qPCR RT Master Mix with gDNA remover (TOYOBO). The qRT-PCR was performed on a CFX96 instrument (Bio-Rad) using SYBR Green Supermix (Bio-Rad) following the manufacturer’s instructions. The rice ACTIN gene (LOC_Os03g50885) was used as an internal control for RT-PCR and qRT-PCR analyses. Primer sequences used for RT-PCR and qRT-PCR are listed in Supplemental Table S2.
GUS staining
To generate the pYL1::GUS construct for plant transformation, a 2,099 bp promoter fragment immediately before the start codon of YL1 was amplified using the primers pGUS-F and pGUS-R (Supplemental Table S2). The PCR product was cloned into the binary vector pCAMBIA1301 containing the GUS reporter gene using the KpnI and NcoI restriction sites. The plasmid pYL1::GUS was then introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Nipponbare plants. GUS activity in young buds, roots, leaves, stems, leaf sheaths, and young spikes of T2 transgenic plants containing pYL1::GUS was detected as described previously54.
Subcellular localization of YL1
To determine the subcellular localization of YL1, the coding sequence (CDS) of YL1 was amplified using the primers YL1-GFP-F and YL1-GFP-R (Supplemental Table S2) and ligated into a modified pCAMBIA1300 vector containing a CaMV 35S::GFP cassette using the SacI and SalI restriction sites to generate pCaMV 35S::YL1-GFP. The resulting construct was transferred into rice protoplasts that were freshly isolated from young stem tissue of Nipponbare based on the method described by Zhang et al.55. The transformed protoplasts were observed under a confocal laser-scanning microscope (LSM710, Zeiss, Germany).
The binary plasmid was also introduced into yl1-1 mutant plants to generate YL1-GFP transgenic plants by Agrobacterium-mediated transformation. GFP florescence was examined in the living cells of leaf epidermis from YL1-GFP transgenic seedlings by confocal microscopy as described above (LSM710).
Thylakoid membranes isolation
Thylakoid membrane proteins were isolated from four-week-old rice leaves as previously described56. Briefly, 20 g of leaf samples was homogenized in 30 ml ice-cold isolation buffer (0.33 M sorbitol, 2.5 mM EGTA, 5 mM EDTA, 10 mM NaHCO3, 20 mM HEPES/KOH, pH 8.0) and filtered through a double layer of Miracloth. The filtrate was centrifuged at 4,200× g for 5 min at 4 °C, and the pellet was resuspended in cold isolation buffer (without sorbitol) for 10 min to lyse the chloroplasts. After re-centrifugation at 8,000× g for 2 min, the pellet was resuspended in 300–500 μl of the same buffer, and the chlorophyll content was determined spectrophotometrically as described57.
2D-BN/SDS-PAGE and immunoblot analyses
For BN-PAGE analysis, fresh isolated thylakoid membrane proteins were pre-treated as described by Peng et al.32 and separated in a NativePAGE Novex 4–16% Bis-Tris Gels System (Invitrogen). Samples were loaded on an equivalent chlorophyll content basis and electrophoresis was performed at 4 °C. The second-dimension separation was performed as described by Ma et al.58. After electrophoresis, the proteins were visualized by Coomassie Blue staining.
For immunoblot analyses, thylakoid membrane samples were boiled in 2 × SDS loading buffer (125 mM Tris-HCl [pH 6.8], 2% SDS, 20% glycerol, 0.02% bromophenol blue, and 5% β-mercaptoethanol). Samples of thylakoid membranes were loaded on an equal-fresh-weight basis and resolved by 10% SDS-PAGE. After electrophoresis, the proteins were transferred to PVDF membranes (Millipore) and incubated with antibodies specific for PsaA, PsbA (D1), PsbO, Cytb 6f, LHCII, RbcL, AtpA, and AtpB. Signals were detected with an enhanced chemiluminescence kit (Novex ECL Chemiluminescent Substrate Reagent Kit, Invitrogen).
ATPase activity assay
Intact chloroplasts were isolated from 4-week-old plants according to Mao et al.23. The ATPase activity was determined by measuring by the amount of inorganic phosphate (Pi) in the reaction using an ATPase Activity Assay Kit (Sigma–Aldrich). Isolated chloroplast suspension (10 ul) was incubated for 30 min at room temperature in 30 ul assay buffer containing 40 mM Tis-HCl (pH 7.5), 80 mM NaCl, 8 mM MgAc2, 1 mM EDTA and 1 mM ATP. The reaction was stopped by adding 200 ul Reagent and the released Pi was monitored by a microcolorimetric method after incubated for additional 30 min at room temperature.
Yeast two-hybrid assays
The yeast two-hybrid assays were performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech). The full-length coding sequence of YL1 and several genes encoding core subunits of photosynthetic complexes (PsaA in PSI, AtpA and AtpB in cpATPase, and Cyt f in Cytochrome b6/f) were amplified using gene-specific primers (Supplemental Table S2). YL1 was cloned into pGBKT7 (bait vector), and PsaA, AtpA, AtpB, Cyt f and D1 were individually cloned into pGADT7 (prey vector). The prey and bait constructs were cotransformed into S. cerevisiae strain AH109. The yeast transformants were cultured in synthetic dropout (SD) medium lacking Leu and Trp (SD/-Leu/-Trp), followed by screening on SD/-Ade/-His/-Leu/-Trp medium containing X-α-gal.
Bimolecular Fluorescence Complementation Assays
BiFC assays were performed according to the methods of our previous study59. For generation of BiFC vectors, cDNA encoding full-length YL1, YL-N (CDS1–285) and YL1-C (CDS286–495) were amplified by primer pairs (Supplementary Table S2) respectively and were individually cloned at BamhI - SalI sites in pSCYNE. The CDS of AtpB was amplified and cloned into BamhI -KpnI sites of pSCYCE (R). Rice protoplasts were prepared as described above and were cotransformed with combinations of constructs. CFP fluorescence was imaged using a confocal laser scanning microscope (LSM710, Zeiss, Germany).
Statistical analysis
The data presented are the averages of at least three independent replicates. Statistical analyses were performed with the Data Processing System (DPS) statistical software package60 using ANOVA followed by the Duncan’s multiple range test (SSR) to evaluate significant effects of the treatments at a significance level of P ≤ 0.05.
Additional Information
Accession codes: Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: rice YL1 (Os02g0152900), Arabidopsis EMB1303 (At1g56200). A. thaliana (At1g30475), A. lyrata subsp. lyrata (ARALYDRAFT_473274), A. tauschii (F775_14531), A. trichopoda (AMTR_s00007p00184720), B. distachyon (LOC100823286), C. rubella (CARUB_v10010523mg), C. sativus (LOC101207190), F. vesca (LOC101302917), G. max (LOC100817121), M. truncatula (ACJ85874), S. lycopersicum (LOC101255489), S. bicolor (SORBIDRAFT_04g003770), S. italica (LOC101773543), T. cacao (Embryo defective 1303), T. urartu (TRIUR3_32364), V. vinifera (CAO45180), and Z. mays (LOC100274890).
How to cite this article: Chen, F. et al. A Nucleus-Encoded Chloroplast Protein YL1 Is Involved in Chloroplast Development and Efficient Biogenesis of Chloroplast ATP Synthase in Rice. Sci. Rep. 6, 32295; doi: 10.1038/srep32295 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Lixin Zhang and Dr. Lianwei Peng (The Chinese Academy of Sciences) for kindly gift of all of the photosynthetic antibodies and assistance for BN/SDS-PAGE. We thank Ms. Junying Li, the technician of 985-Institute of Agrobiology and Environmental Sciences of Zhejiang University, for her assistance in using the transmission electron microscope. This work was supported by Funds from the National Natural Science Foundation of China (91335103, 31271696, 31170346 and 31501282), National Key R&D program of China (No. 2016YFD0101801), Hangzhou Overseas Students Merit Funded Projects (20140005), the Outstanding Researcher Program of Hangzhou Normal University (20141215), and the Startup funding of Hangzhou Normal University (20110004).
Footnotes
Author Contributions Y.Y. and Q.Q. designed the research; F.C., G.D., L.W., F.W., X.Y., X.M., H.W., J.W. and Y.Z. performed the experiments; F.C., G.D., H.W., Y.Y. and Q.Q. analyzed the data; F.C. and Y.Y. wrote the manuscript.
References
- Sakamoto W., Miyagishima S. Y. & Jarvis P. Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. Arabidopsis Book 6, e0110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Finazzi G. & Wollman F. A. The dynamics of photosynthesis. Annu Rev Genet 42, 463–515 (2008). [DOI] [PubMed] [Google Scholar]
- Pogson B. J. & Albrecht V. Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155, 1545–1551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T. & Langdale J. A. The making of a chloroplast. EMBO J 28, 2861–2873 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Sun X. & Zhang L. Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64, 559–582 (2013). [DOI] [PubMed] [Google Scholar]
- Jarvis P. & Lopez-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14, 787–802 (2013). [DOI] [PubMed] [Google Scholar]
- Li H. M. & Chiu C. C. Protein Transport into Chloroplasts. Annual Review of Plant Biology, Vol 61 61, 157–180 (2010). [DOI] [PubMed] [Google Scholar]
- Pogson B. J., Ganguly D. & Albrecht-Borth V. Insights into chloroplast biogenesis and development. Biochimica Et Biophysica Acta-Bioenergetics 1847, 1017–1024 (2015). [DOI] [PubMed] [Google Scholar]
- Lyska D., Meierhoff K. & Westhoff P. How to build functional thylakoid membranes: from plastid transcription to protein complex assembly. Planta 237, 413–428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. & Ben-Shem A. The complex architecture of oxygenic photosynthesis. Nat Rev Mol Cell Biol 5, 971–982 (2004). [DOI] [PubMed] [Google Scholar]
- von Ballmoos C., Wiedenmann A. & Dimroth P. Essentials for ATP synthesis by F1F0 ATP synthases. Annu Rev Biochem 78, 649–672 (2009). [DOI] [PubMed] [Google Scholar]
- Ruhle T. & Leister D. Assembly of F1F0-ATP synthases. Biochim Biophys Acta 1847, 849–860 (2015). [DOI] [PubMed] [Google Scholar]
- Schottler M. A., Toth S. Z., Boulouis A. & Kahlau S. Photosynthetic complex stoichiometry dynamics in higher plants: biogenesis, function, and turnover of ATP synthase and the cytochrome b6f complex. J Exp Bot 66, 2373–2400 (2015). [DOI] [PubMed] [Google Scholar]
- Pickova A., Potocky M. & Houstek J. Assembly factors of F1FO-ATP synthase across genomes. Proteins 59, 393–402 (2005). [DOI] [PubMed] [Google Scholar]
- Tzagoloff A. & Dieckmann C. L. PET genes of Saccharomyces cerevisiae. Microbiol Rev 54, 211–225 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M., Zeng X., Briere J. J. & Tzagoloff A. Assembly of F0 in Saccharomyces cerevisiae. Biochim Biophys Acta 1793, 108–116 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M., Gokova S. & Tzagoloff A. Modular assembly of yeast mitochondrial ATP synthase. EMBO J 30, 920–930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D. Mitochondrial protein synthesis, import, and assembly. Genetics 192, 1203–1234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckers-Hebestreit G. Assembly of the Escherichia coli FoF1 ATP synthase involves distinct subcomplex formation. Biochem Soc Trans 41, 1288–1293 (2013). [DOI] [PubMed] [Google Scholar]
- Chen G. G. & Jagendorf A. T. Chloroplast molecular chaperone-assisted refolding and reconstitution of an active multisubunit coupling factor CF1 core. Proc Natl Acad Sci USA 91, 11497–11501 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz M. et al. Alb4 of Arabidopsis promotes assembly and stabilization of a non chlorophyll-binding photosynthetic complex, the CF1CF0-ATP synthase. Mol Plant 2, 1410–1424 (2009). [DOI] [PubMed] [Google Scholar]
- Ruhle T. et al. The Arabidopsis protein CONSERVED ONLY IN THE GREEN LINEAGE160 promotes the assembly of the membranous part of the chloroplast ATP synthase. Plant Physiol 165, 207–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. et al. PAB is an assembly chaperone that functions downstream of chaperonin 60 in the assembly of chloroplast ATP synthase coupling factor 1. Proc Natl Acad Sci USA 112, 4152–4157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L. et al. YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry 42, 10537–10544 (2003). [DOI] [PubMed] [Google Scholar]
- Wang P. & Dalbey R. E. Inserting membrane proteins: the YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim Biophys Acta 1808, 866–875 (2011). [DOI] [PubMed] [Google Scholar]
- Suzuki T., Ozaki Y., Sone N., Feniouk B. A. & Yoshida M. The product of uncI gene in F1Fo-ATP synthase operon plays a chaperone-like role to assist c-ring assembly. Proc Natl Acad Sci USA 104, 20776–20781 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R. et al. The thylakoid membrane protein CGL160 supports CF1CF0 ATP synthase accumulation in Arabidopsis thaliana. PLoS One 10, e0121658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottler M. A., Albus C. A. & Bock R. Photosystem I: its biogenesis and function in higher plants. J Plant Physiol 168, 1452–1461 (2011). [DOI] [PubMed] [Google Scholar]
- Chi W., Ma J. & Zhang L. Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos Trans R Soc Lond B Biol Sci 367, 3420–3429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J. & Rengstl B. Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 64, 609–635 (2013). [DOI] [PubMed] [Google Scholar]
- Pagliano C., Saracco G. & Barber J. Structural, functional and auxiliary proteins of photosystem II. Photosynth Res 116, 167–188 (2013). [DOI] [PubMed] [Google Scholar]
- Peng L. W. et al. LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18, 955–969 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann H., Shavit N. & Leu S. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas (eds Rochaix J. D., Goldschmidt-Clermont M. & Merchant S.) pp 477–500 (Springer Netherlands, 1998).
- Dong H. et al. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol 162, 1867–1880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Z., Zhang X. Y. & Yang S. H. A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Research 19, 1205–1216 (2009). [DOI] [PubMed] [Google Scholar]
- Rott M. et al. ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by overacidification of the thylakoid lumen. Plant Cell 23, 304–321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. G. & Ackerman S. H. The assembly factor Atp11p binds to the beta-subunit of the mitochondrial F(1)-ATPase. J Biol Chem 275, 5767–5772 (2000). [DOI] [PubMed] [Google Scholar]
- Wang Z. G., Sheluho D., Gatti D. L. & Ackerman S. H. The alpha-subunit of the mitochondrial F(1) ATPase interacts directly with the assembly factor Atp12p. EMBO J 19, 1486–1493 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre-Legendre L. et al. Failure to assemble the alpha 3 beta 3 subcomplex of the ATP synthase leads to accumulation of the alpha and beta subunits within inclusion bodies and the loss of mitochondrial cristae in Saccharomyces cerevisiae. J Biol Chem 280, 18386–18392 (2005). [DOI] [PubMed] [Google Scholar]
- Lu Y., Hall D. A. & Last R. L. A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23, 1861–1875 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. et al. Hypersensitive to high light1 interacts with LOW QUANTUM yield of photosystem II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell 26, 1213–1229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35, 418–427 (2003). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34, D745–D748 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. et al. Independent losses of function in a polyphenol oxidase in rice: differentiation in grain discoloration between subspecies and the role of positive selection under domestication. Plant Cell 20, 2946–2959 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T. & Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6, 271–282 (1994). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Control of tillering in rice. Nature 422, 618–621 (2003). [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H. & von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8, 978–984 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G. & Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- Chenna R. et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31, 3497–3500 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods in Enzymology 148, 350–382 (1987). [Google Scholar]
- Liu J. et al. PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24, 4992–5006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. et al. Genotype-Dependent Effect of Exogenous Nitric Oxide on Cd-induced Changes in Antioxidative Metabolism, Ultrastructure, and Photosynthetic Performance in Barley Seedlings (Hordeum vulgare). Journal of Plant Growth Regulation 29, 394–408 (2010). [Google Scholar]
- Xiong G. et al. The rice dynamin-related protein DRP2B mediates membrane trafficking, and thereby plays a critical role in secondary cell wall cellulose biosynthesis. Plant J 64, 56–70 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., van Wijk K. J. & Aro E. M. Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274, 16062–16067 (1999). [DOI] [PubMed] [Google Scholar]
- Porra R. J., Thompson W. A. & Kriedemann P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim Biophys Acta 975, 384–394 (1989). [Google Scholar]
- Ma J. F. et al. LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19, 1980–1993 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xu C. et al. Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering. Nat Commun 3, 750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q. Y. & Zhang C. X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci 20, 254–260 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.