Abstract
Acute allograft rejection is a serious and life-threatening complication of organ transplantation. Th17 cells induced inflammation has been described to play an important role in allograft rejection. Since there is a plenty of evidence indicating that transcriptional factor BATF regulates the differentiation of Th17 and follicular T helper cells both in vitro and in vivo, we investigated whether is BATF involved in acute rejection and allograft survival by injecting lentivirus containing BATF shRNA through tail vein before the cardiac transplantation operation. We found that the allograft survival time of the mice treated with BATF shRNA was significantly prolonged compared with that of negative shRNA treated group and the control group. Further pathological analysis revealed that the BATF shRNA treatment group had significantly lower rejection degree than the negative shRNA group, while there was no significant difference between the negative shRNA group and the control group. Furthermore, flow cytometry analysis and quantitative polymerase chain reaction and enzyme-linked immuno sorbent assay were used to determine the proportion of T helper cells, the expression of specific transcription factor and the inflammatory cytokines respectively. Data showed that BATF regulated Th17 and Treg responses during allograft rejection. And BATF inhibition led to reduction of the expression level of Rorγ-t and enhancement of the Foxp-3. In addition, cytokines IL-17A and IL-4 were found decreased. This may indicate BATF as a novel therapy target for treatment of acute allograft rejection.
Keywords: Acute allograft rejection, Th17, BATF, shRNA, heterotopic cardiac transplantation, lentiviral vectors
Introduction
Acute rejection is the most important factor to cause allograft loss, which is still a challenge for patients receiving organ transplantation. Several studies have demonstrated that the development of acute allograft rejection is associated with poor prognosis [1,2]. Currently, the prevention and treatment of this rejection is mainly dependent on immunosuppressant, and its diagnosis relies on pathological examination of the allografts. Although some immunosuppressants have been proved to efficiently inhibit the acute rejection following transplantation, this treatment often fails to provide satisfactory results for numerous side effects such as renal toxicity and neurotoxicity [3,4]. Thus, great efforts have been made to explore novel targets and safe methods to prevent acute rejection.
Despite medical and scientific efforts made over the past decades, the mechanisms of allograft rejection remain unclear. Generally, Th1 mediated immune response along with activation of macrophages are considered to be responsible for allograft rejection, while Th2 response is thought to be beneficial to long-term allograft survival [5,6]. Although previous extensive studies indicated Th1 and Th2 responses play a critical role in allograft rejection, increasing evidence has challenged the this theory. A study reported that cardiac grafts infiltrated with Th17 cells undergo accelerated vascular rejection in Tbet-/- mice model, which is deficient in Th1 induced inflammation [7]. Th17 cells originate from a common naive T-cell precursor and are characterized by their ability to produce the pro-inflammatory cytokines IL-17A and IL-17F [8,9]. There is plenty of evidence indicating Th17 response is involved in the pathogenesis of acute allograft rejection in humans and in other animal models, which was also in accordance with our previous study. Our previous works had indicated that inhibition of IL-6, which interferes Th17 differentiation, could effectively reduce allograft rejection [10]. The role of Th17 and relative regulatory mechanisms that control the different Th cell subsets during allograft rejection are still unknown. Therefore, a series of experiments have been undertaken to evaluate the molecular mechanisms of Th17 cells underlying allograft rejection.
Basic leucine zipper transcription factor ATF-like (BATF) is a member of the AP-1/ATF family of transcription factors, which is expressed predominantly by the immune cells and plays an important role in its development and function. Recently, the effect of regulating BATF expression in both vivo and vitro has been examined by several groups. BATF knockout mice are deficient in both CD4+ Th17 cells and T follicular helper cells and also possess an intrinsic B cell defect [11]. The ability of BATF to promote the differentiation of naive CD4+ T cells to the Th17 lineage relies on the formation of IRF-4/BATF protein complexes that bind to and transactivate a number of genes, including IL-17A/F [12]. Additionally, it has been reported that BATF can suppress Sirt1 expression and control the ATP level and effector function of CD8+ T cells [13].
Thereupon, we hypothesized that the transcription factor BATF may play an important role in the development of acute allograft rejection that is likely related to Th17 cells. In order to answer these questions, we established the acute rejection models of heterotopic cardiac transplantation in mice according to the protocol described by Corry et al. [14] and detected the expression of BATF mRNA in cardiac graft. Furthermore, we investigated whether BATF inhibition could lengthen allograft survival time and lower acute rejection degree by RNAi technology before the transplantation operation.
Materials and methods
Animals
This study was approved by the Institutional Animal Care and Use Committee of Tongji Medical College. Male BALB/C (H2d) and C57BL/6 (B6, H2b) mice at age of 4-8 weeks were maintained at the Animal Facility of Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China, under controlled conditions (specific pathogen-free, 22°C, 55% humidity and 12 h day/night).
Experimental models
For acute rejection model of allogeneic cardiac transplantation, C57BL/6 mice were used as recipients, whereas BALB/C mice were used as donors. For negative control model of syngeneic cardiac transplantation, C57BL/6 mice were used as both recipients and donors. The mouse abdominal cardiac transplantation was performed using standard microsurgical techniques according to the protocol described before [14]. The day of operation was recorded as POD (postoperative day) 0. Graft beating was monitored by daily palpation. The day of rejection was defined as the last day of a detectable heartbeat in the graft, which was confirmed by direct palpation and observation after abdominal cavity opened. Allograft and spleen were collected and divided into equal sections for histology, real-time polymerase chain reaction assay and flow cytometry. And the blood samples were collected from recipients after eyeball removal for enzyme-linked immuno sorbent assay.
In this study, C57BL/6 mice (4-5 weeks) were also randomly divided into three groups, namely control group, negative shRNA group and the BATF shRNA group. In each group, animals were injected with 0.20 ml NS solution, 0.20 ml NS solution containing 2×107 TU scrambled shRNA lentivirus, or 0.20 ml NS solution containing 2×107 TU shBATF-3 lentivirus respectively through the tail vein [15]. And 14 days after transduction, all animals received an allogeneic cardiac transplantation operation.
Construction of BATF-shRNA expression plasmid and lentivirus production
The mice BATF was treated as the target gene and three pairs of shRNA oligonucleotides were designed and synthesized (Cyagen Biosciences, United States). A pair of random scrambled sequences as negative shRNA, which have no homologous relation with any mice gene sequences and whose shRNAs would not interfere with mice mRNA was also designed (Table 1). The plasmid used in this study was provided and sequenced by Cyagen Biosciences Inc. The BATF-shRNA recombinant lentivirus was produced by co-transfecting 293T cells with the help of Lipofectamine 2000 (Invitrogen, United States) according to standard protocols. Viral supernatant was harvested 48 h after transduction and the titer was detected.
Table 1.
Short hairpin RNA oligo sequences
| Oligo name | Sequence 5’ to 3’ |
|---|---|
| shBATF-1 | GGTGGTATACAGTGCCCAT |
| anti-shBATF-1 | ATGGGCACTGTATACCACC |
| shBATF-2 | GAGCTCAAGTACTTCACAT |
| anti-shBATF-2 | ATGTGAAGTACTTGAGCTC |
| shBATF-3 | GGACTCATCTGATGATGTG |
| anti-shBATF-3 | CACATCATCAGATGAGTCC |
| Scramble shRNA | CCTAAGGTTAAGTCGCCCTCG |
| anti-Scramble shRNA | CGAGGGCGACTTAACCTTAGG |
Cell culture and transfection
The Jurkat cells (ATCC, United States) were grown in RPMI 1640 medium containing 10% fetal bovine serum and induced with CD3 and CD28 antibodies. Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2. For transfection, the cells were pre-seeded into 24-well plates at 1×104 cells per well according to the manufacturer’s instructions. When cells reached the confluent rate of about 50% in complete medium, they were infected with the lentiviral constructs at different MOIs (multiplicity of infection, or infectious units ratio). Transfection efficiency was observed daily with an inverted fluorescence microscope (DP72, Olympus, Japan). BATF interference expression levels in transfected cell lines were assayed by RT-PCR and western-blot 48 hours after transfection.
Real-time quantitative PCR
Total mRNA was extracted and reverse transcription was performed using a mix kit (total RNA extraction reagent) (Takara, Japan) according to the manufacturer’s protocol. RT-PCR was performed with a SYBR master mix kit (Takara, Japan) on a Roche LC480 (Roche, Switzerland) RT-PCR System. Primers used for real-time quantitative PCR are shown in Table 2. The initial denaturation step was 95°C for 15 seconds, followed by 40 cycles of amplification at 95°C for 10 seconds and 55°C for 30 seconds. Each sample was measured in triplicate. A mean value was used to determine mRNA levels using the comparative Ct method, using the formula 2-ΔΔCT and the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene as the reference.
Table 2.
Primer sequences were as follows
| Gene | Primer sequences 5’ to 3’ | Length (bp) |
|---|---|---|
| RORγ-t | F-GAGCAATGGAAGTCGTCCTAGTCAG | 190 |
| R-AGGGCAATCTCATCCTCAGAAAA | ||
| Foxp-3 | F-TCACCTATGCCACCCTTATCC | 236 |
| R-GCTCCTCTTCTTGCGAAACTC | ||
| IFN-γ | F-GCTCTGAGACAATGAACGCTACA | 150 |
| R-TTTCTTCCACATCTATGCCACTT | ||
| IL-4 | F-TCCTGCTCTTCTTTCTCG | 101 |
| R-TTCTCCTGTGACCTCGTT | ||
| BATF | F-TGCCCTCATACCTCTACCCA | 151 |
| R-TCCAGTACATTGGCTCGAC | ||
| GAPDH | F-GACAAAATGGTGAAGGTCGGT | 120 |
| R-GAGGTCAATGAAGGGGTCG |
Western blotting
The protein levels were quantified by standard western blotting procedures. Protein extracted from cells or tissue was separated on 12% SDS-polyacrylamide electrophoresis gels and transferred to nitrocellulose membranes. After being blocked with 5% non-fat milk in TBS for 3 hours, the membranes were incubated with indicated primary antibodies (Cell Signaling, United States; dilution 1:1000) at 4°C overnight, followed by incubation with HRP-conjugated secondary antibody for 3 hours. GAPDH (Sigma, United States) was used as a loading control for comparison between samples.
Flow cytometry analysis
The spleen lymphocytes from recipients were isolated on POD 7 for flow cytometry analysis. Briefly, the cells were prepared as a single-cell suspension and cultured with 1×Cell Stimulation Cocktail (eBioscience, United States) for 16 hours used as the activation and blocking agent. For Th17 analysis, FITC-conjugated anti-CD4 mAb was added to the cell suspension simultaneously and incubated at 4°C for 30 min. After fixation and permeabilization, the cell suspensions were stained with anti-IL-17A PE mAb (eBioscience). Similarly, the cell suspensions were stained with anti-IFN-γ APC mAb to analyze Th1 cells and anti-Foxp3 APC mAb (eBioscience) for Treg cells. After washing twice, the stained cells were re-suspended in 200 μl of cold staining buffer and analyzed by fluorescence activated cell sorter (FACS, BD Biosciences, United States).
ELISA measurement
The inflammatory cytokines levels of IFN-γ, IL-4, IL-10 and IL-17A in the serum of recipients were measured using an ELIAS kits (NeoBioscience, China). The procedures were performed in accordance with the manufacturer’s protocol. Absorbance values (at 450) by duplicate were plotted against dilutions and expressed as pg/mL.
Pathological examination
Seven days after transplantation, six mice of each group were randomly selected to do biopsy of allograft heart for the detection of rejection. The hearts were obtained and fixed in formaldehyde. Then, heart tissues were embedded in paraffin, cut into 4-μm sections, dehydrated with graded alcohol, transparentized with xylene, and stained with H&E.
Statistical analysis
Allograft survival curves were generated by the Kaplan and Meier method. The difference in allograft survival between multiple groups was determined using the log-rank (Mantel-Cox) test. Difference in data from flow cytometry, RT-PCR and ELISA assay was calculated by one-way ANOVA test using the GraphPad Prism statistical program (GraphPad Software, United States). Data were expressed as mean ± SD. A value of P<0.05 was treated as statistically significant.
Results
BATF expression in acute allografts rejection
We successfully performed the acute rejection model of heterotopic cardiac transplantation in mice (Supplementary Figure 1), the histological analysis of cardiac grafts harvested on POD 7 after allogeneic cardiac transplantation revealed severe pathological feature of acute rejection by comparing with those mice treated with syngeneic transplantation and untreated. The cardiac grafts in the rejection group showed rigorous infiltration of inflammatory cells and allograft destruction (Figure 1A-C). The BATF mRNA levels were significantly higher in the rejection group compared with other groups (Supplementary Figure 2). Furthermore, the proportion of IL-17A secreting cells from splenocytes of recipients was analyzed by flow cytometry. We found that CD4+IL-17A+ cells in the recipients of rejection group increased significantly as compared with those mice in negative control and control group (Figure 1D), which was in accordance with our previous report [10].
Figure 1.
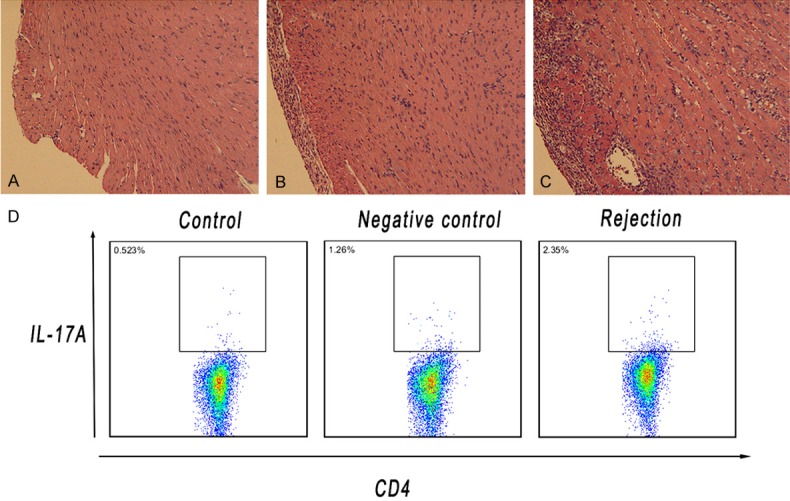
A-C. Representative histological findings of cardiac grafts from normal mice (Control), syngeneic (Negative control) and allogeneic cardiac transplantation mice (Rejection). (20×). Relatively intact myocardium accompanied by markedly increased infiltration of inflammatory cells was observed in allografts of recipients in acute rejection group. D. CD4+IL-17A+ T cells were analyzed in splenocytes of each study group. The specimens of recipients were collected on postoperative day (POD) 7.
Down-regulation of BATF expression in vitro and in vivo
To further confirm the role of BATF in development of acute rejection, shRNA was used to knockdown expression of BATF both in vitro and in vivo. The in vitro transfection efficiency and related gene expression as well as protein expression are shown in Figure 2. There is an approximately 80% decrease of BATF gene expression as well as BATF protein expression in shBATF-3 transfected cells compared with the negative shRNA transfected and untransfected cells, which suggests that the shBATF-3 is the most effective shRNA among the three (Figure 2D and 2E). Therefore, shBATF-3 was chosen as the shRNA construct for generation of lentivirus for following in vivo studies. The in vivo transfection and silencing efficiency of shBATF-3 could reach almost 70% (Figure 2C and 2F).
Figure 2.
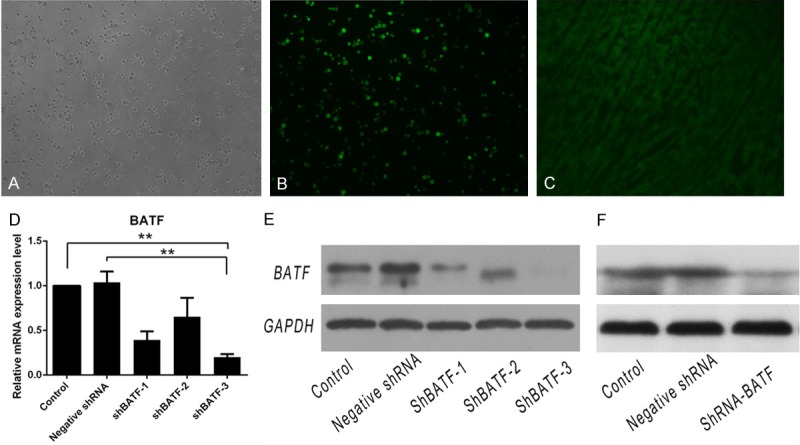
A and B. The bright and dark field images of Jurkat cells after shBATF stable transfection by fluorescence microscopy respectively. C. The mice hearts were collected and directly fluorescent photographing at 14 days after lentivirus injection through the tail vein. D. The interference efficiency of BATF in shBATF transfected cells using real-time PCR. The results are presented as mean ± SD. **p<0.01. E and F. The down-regulation levels of BATF were evaluated by western blot in vitro and vivo (spleen tissues). Sample size of each group was 3.
BATF shRNA transfer prolongs allograft survival and reduces acute rejection degree
C57BL/6 mice recipients of fully allogeneic BALB/C cardiac grafts injected with BATF shRNA before transplantation shows significant prolongation of allograft survival from 7.0 days median graft survival time (MST) for recipients of rejection group to 10.5 days MST (Figure 3A). However, there is no obvious prolongation of allograft MST observed in negative shRNA group compare with rejection group, indicating the improvement of survival time might be specific for knockdown of BATF.
Figure 3.
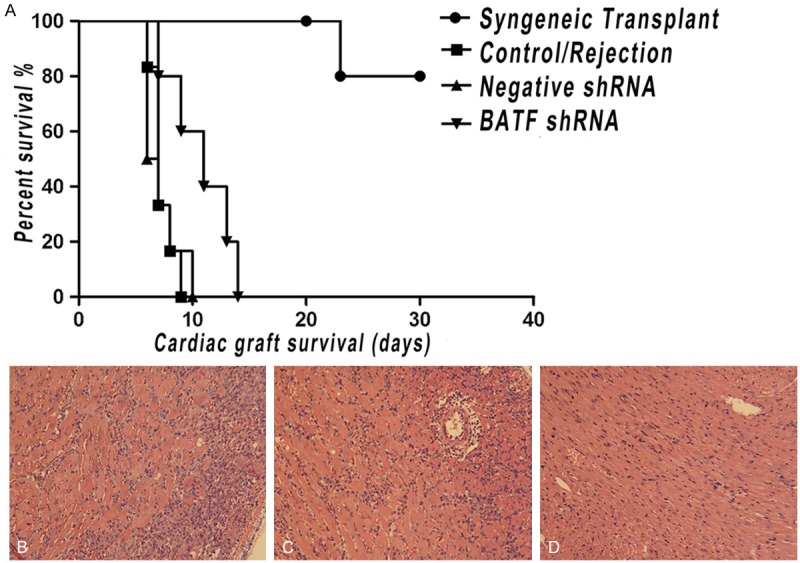
A. The recipients were treated with BATF shRNA lentivirus through the tail vein injection before transplantation prolonged allograft survival. The cardiac allograft survival was examined over time. (n = 6 mice/group). B-D. On day 7 post-transplant, cardiac grafts from C57BL/6 mice treated with BATF shRNA, negative shRNA and untreated mice were collected and histological assayed. (20×).
Routine histology demonstrates that cardiac grafts seven days after transplantation develop a similar minimal cell infiltration injected with negative shRNA and control mice, which indicates that such dose of shRNA is not toxic and it won’t cause cytopathic effects in the allograft. By contrast, relatively decreased myocardial damage and remarkable reduced infiltration of inflammatory cells are observed in allografts of BATF shRNA group (Figure 3B-D). The level of allograft rejection on POD 7 is shown in Table 3 (Supplementary Figure 3). Thus, it is possible that the BATF shRNA therapy could not only prolong allograft survival, but also reduce acute rejection degree with a moderate decrease of inflammatory cell infiltration and less tissue damage.
Table 3.
Score of the graft rejection among groups
| Groups | Score of the graft rejection | ||||
|---|---|---|---|---|---|
|
| |||||
| 0 | 1 | 2 | 3 | 4 | |
| Control | 0 | 0 | 0 | 2 | 4 |
| Negative shRNA | 0 | 0 | 0 | 3 | 3 |
| BATF shRNA | 0 | 1 | 3 | 2 | 0 |
**P<0.01.
BATF inhibition interferes the Th17/Treg responses and IL-4 induction
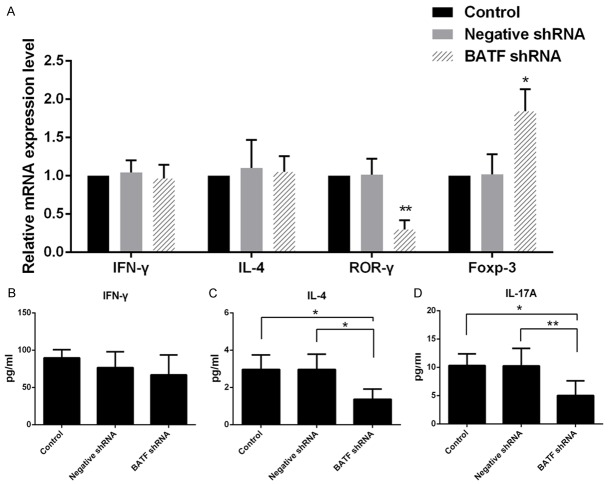
In order to determine the molecular mechanisms underlying BATF shRNA mediated allograft protection, the proportions of the inflammatory T helper subset cells in splenocytes of recipients from each group on POD 7 were analyzed using flow cytometey. The proportion of CD4+IL-17A+ cells in the recipients of BATF shRNA treated group decreased significantly compared with the negative shRNA group and the control group, while the CD4+Foxp-3+ cells proportion altered contrarily. As is shown, BATF shRNA treatment suppresses the differentiation of Th17 cells and induced Treg cells. However, the proportion of CD4+IFN-γ+ cells was not significantly altered among the different groups (Figure 4 and Supplementary Figure 4). It is observed that the response of Th17 and Treg in allograft rejection was regulated by BATF.
Figure 4.
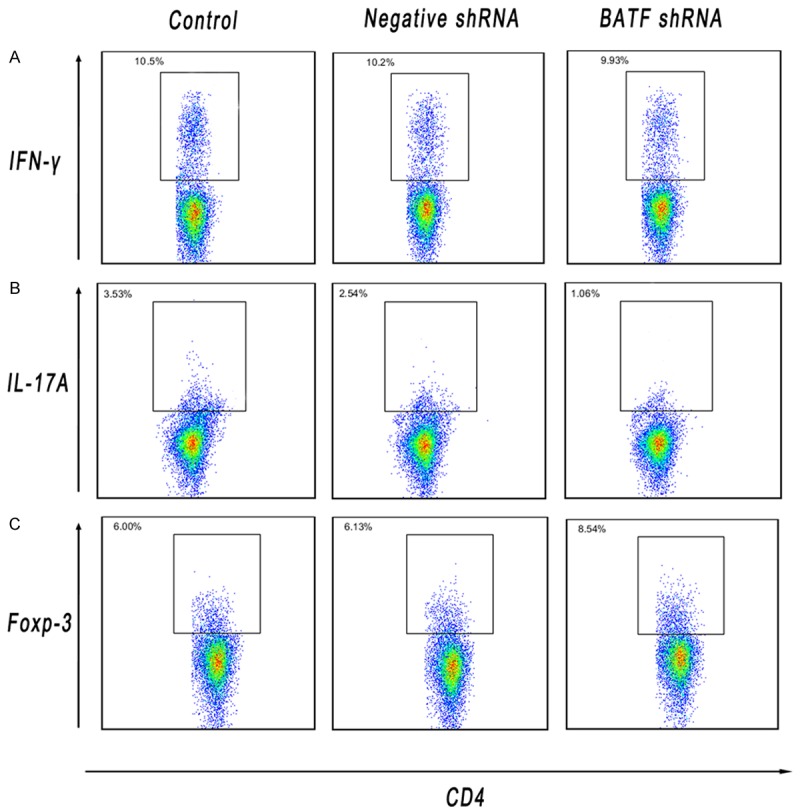
A-C. Splenic T cells were obtained from allograft recipients in each control, negative shRNA and BATF shRNA group and stained on POD 7. The percentages of CD4+ cells that were positive for IFN-γ, IL-17A and Foxp-3 were determined using flow cytometric analysis.
The specific transcription factors and inflammatory cytokines in the allografts and blood were further detected. The transcript levels for IFN-γ, IL-4, Rorγ-t and Foxp-3 in allograft of each study group on POD 7 were determined by real-time PCR, respectively (Figure 5A). The Rorγ-t expression in cardiac allograft decreases significantly in BATF shRNA treated mice. By contrast, BATF shRNA treatment increases the mRNA expression of Foxp-3 dramatically on the same day. However, the IFN-γ and IL-4 expression levels in BATF shRNA treated allografts are not significantly different from the negative shRNA and the control group. In addition, the inflammatory cytokines levels in recipients’ peripheral blood serum shows similar results except for the induction level of IL-4 (Figure 5B-D). Notably, the serum IL-4 level is significantly decreased in mice with BATF shRNA compared with negative shRNA and control mice.
Figure 5.
A. RT-PCR analysis of IFN-γ, IL-4, Foxp-3 and Rorγ-t expression in cardiac grafts by BATF shRNA treatedment compared with those treated with negative shRNA and control. B-D. The associated cytokines IFN-γ, IL-4 and IL-17A in serum were detected by ELISA. Data were presented as mean ± SD, and experiments were performed in duplicates. *p<0.05, **p<0.01.
Discussion
Cardiac transplantation is an effective treatment for congestive cardiac failure, particularly for those patients who are resistant to aggressive medical therapy. However, acute allograft rejection is still a challenge for patients receiving transplantation. Th1 and Th2 response induced inflammations have been considered to be responsible for allograft rejection and long-term allograft survival in past decades. A number of immunosuppressants have been developed to control this process [16,17]. However, rejection is still the one of the most common complications in organ transplantation.
It is well known that acute allograft rejection is dependent on T cells, including a variety of cell subsets of CD4+ and CD8+ T cells. Recently, a large amount of evidence has demonstrated Th17 cells, a novel pleiotropic pro-inflammatory T helper subset, have been shown to involve in a number of immune responses [18,19]. There is some studies supporting the notion that Th17 cells contribute to chronic destructive disorder and inflammation in rheumatoid arthritis (RA) patients and the pathogenesis of inflammatory bowel disease (IBD) [20,21]. Moreover, recent clinical and experimental transplantation studies have suggested the involvement of Th17 cells in allograft rejection. In liver transplantation, serum IL-23 and IL-17 levels are elevated during acute hepatic rejection, and IL-17 production is implicated in graft versus host disease [22]. In lung transplant recipients, studies have shown that the IL-23/IL-17 axis is involved in patients with bronchiolitis obliterans syndrome (BOS) [23]. These results have important implication that Th17 cells mediate an aggressive pro-inflammatory response culminating in severe accelerated allograft rejection [24,25]. However, Th17 cells are distinct from Th1 and Th2 cells. They produce IL-17A, IL-17F, IL-21 and IL-22. Orphan nuclear receptor RORγ-t is suggested to play a crucial role as a key transcription factor that orchestrates differentiation of the Th17 lineage [26]. Hence, the Th17-mediated alloimmune response is an important field of research regarding solid organ transplantation, which requires extensive studies.
BATF plays an important role in regulating differentiation and function of many lymphocyte lineages. A previous study showed the differentiation and function of CD4+ T cells rely mainly on the binding of BATF to AP-1-IRF-4 composite elements (AICEs) [27]. Furthermore, other findings have revealed that the development of murine Th17 cells is critically dependent on the expression of the lineage-specifying transcription factors BATF [28]. Researchers have already started to analyze whether BATF readjust the autoimmune diseases through regulating the secretion of Th17 cells. Some of them speculate the regulatory function may take effect through synergy with RORγ-t. In particular, Gong et al. revealed that D1-like-R signaling enhances BATF activity, which then transcribes the expression of RORγ-t; accordingly, D1-like-R signaling regulates Th17 differentiation to promote the development of allergic asthma [29]. Moreover, another group of researchers, Xu et al., assert that BATF plays a critical role in the pathogenesis of anti-major histocompatibility complex-induced obliterative airway disease. Antibodies against major histocompatibility complex (MHC) results in Th17 cells mediated immunity [30]. Thus, we proposed that the transcription factor BATF might also control acute allograft rejection by regulating Th17 differentiation. Recently, a similar research shows that tacrolimus could control interferon regulatory factor 4 (IRF-4) to attenuate acute rejection responses after liver transplantation. IRF-4 is a crucial transcription factor as same as BATF, which might be related to the function of T helper subsets (Treg and Th17 cells) [31]. To our knowledge, our study will be a novel finding as the transcription factor BATF interaction with organ transplantation has never before been explored, particularly regarding acute allograft rejection.
Our findings are in consistent with many others’, which show that BATF inhibition could diminish the inflammatory response mainly through control Th17 cell differentiation during the development of acute allograft rejection. Furthermore, we revealed that that BATF not only could control Th17 cell differentiation but also impact Treg cell through detecting their corresponding transcription factors and inflammatory cytokines. Our research group considered that BATF might not directly regulate the Treg cell function. Since Th17 and Treg share reciprocal development pathways, certain inflammatory milieu could convert differentiated Tregs into Th17 cells [32,33]. Thus, BATF could be another critical factor for the balance between Th17 and Treg. In addition, BATF could regulate the inflammatory cytokine IL-4 which has been strongly implicated in a number of inflammation and autoimmune diseases, such as atopic and allergic diseases. The IL-4 is originally identified as a B cell-stimulating factor critical for class-switch recombination of B cells to IgG1- and IgE-producing cells. Last year, the specific relationship between BATF and IL-4 has been found by the group of researchers, Anupama Sahoo et al., BATF could cooperate with IRF-4 along with Stat3 and Stat6 trigger IL-4 production in Tfh cells by directly binding to and activation of the CNS2 region in the IL-4 locus [34]. Taken together, we concluded that BATF inhibition could mainly impact Th17 and Treg cells balance in recipient mice after allogeneic cardiac transplantation during the development of acute rejection.
Here, our study also provides evidence that lentivirus vectors expressing shRNA molecules could be used to investigate gene function in the immune system, although there is limited information about the toxicity of and immune reaction to lentiviral vectors [35]. RNA interference has the potential to silence any target gene, and to treat a variety of diseases, including but not limited to cancers, viral infections, and hereditary disorders. For example, a novel study showed the incorporation of siRNA into organ storage solution could be a feasible and effective method of attenuating ischemia and reperfusion injury, protecting cardiac function, and prolonging graft survival [36]. However, although the RNAi technology can knock down genes by targeting and cleaving complementary mRNA with high efficiency, the interference stability is not compared to the gene knockout mice. In addition, since the limited number of animals used in this study, more research is needed to define that relationship more clearly. In the near future, we will continue to clarify the precise molecular and cellular mechanism involved in this immunoregulation. Overall, the lentivirus-mediated gene therapy might become a promising mode of treatment for various clinical applications and the therapeutic approach to inhibit BATF could be a highly promising candidate for use in organ transplantation.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No. 81370581, 81470936) and the Natural Science Foundation of Hubei Province of China (No. 2015CFB573).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Kantrowitz A, Haller JD, Joos H, Cerruti MM, Carstensen HE. Transplantation of the heart in an infant and an adult. Am J Cardiol. 1968;22:782–790. [PubMed] [Google Scholar]
- 2.Lund LH, Edwards LB, Kucheryavaya AY, Dipchand AI, Benden C, Christie JD, Dobbels F, Kirk R, Rahmel AO, Yusen RD, Stehlik J International Society for Heart and Lung Transplantation. The Registry of the International Society for Heart and Lung Transplantation: Thirtieth Official Adult Heart Transplant Report-2013; Focus Theme: Age. J Heart Lung Transplant. 2013;32:951–964. doi: 10.1016/j.healun.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Pallardo Mateu LM, Sancho Calabuig A, Capdevila Plaza L, Franco Esteve A. Acute rejection and late renal transplant failure: risk factors and prognosis. Nephrol Dial Transplant. 2004;19(Suppl 3):iii38–42. doi: 10.1093/ndt/gfh1013. [DOI] [PubMed] [Google Scholar]
- 4.Kobashigawa JA, Kirklin JK, Naftel DC, Bourge RC, Ventura HO, Mohanty PK, Cintron GB, Bhat G. Pretransplantation risk factors for acute rejection after heart transplantation: a multiinstitutional study. The Transplant Cardiologists Research Database Group. J Heart Lung Transplant. 1993;12:355–366. [PubMed] [Google Scholar]
- 5.Barrat FJ, Cua DJ, Boonstra A, Richards DF, Crain C, Savelkoul HF, de Waal-Malefyt R, Coffman RL, Hawrylowicz CM, O’Garra A. In vitro generation of interleukin 10-producing regulatory CD4(+) T cells is induced by immunosuppressive drugs and inhibited by T helper type 1 (Th1)- and Th2-inducing cytokines. J Exp Med. 2002;195:603–616. doi: 10.1084/jem.20011629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariotti J, Foley J, Jung U, Borenstein T, Kantardzic N, Han S, Hanson JT, Wong E, Buxhoeveden N, Trepel JB, Fojo AT, Telford W, Fowler DH. Ex vivo rapamycin generates apoptosis-resistant donor Th2 cells that persist in vivo and prevent hemopoietic stem cell graft rejection. J Immunol. 2008;180:89–105. doi: 10.4049/jimmunol.180.1.89. [DOI] [PubMed] [Google Scholar]
- 7.Yuan X, Paez-Cortez J, Schmitt-Knosalla I, D’Addio F, Mfarrej B, Donnarumma M, Habicht A, Clarkson MR, Iacomini J, Glimcher LH, Sayegh MH, Ansari MJ. A novel role of CD4 Th17 cells in mediating cardiac allograft rejection and vasculopathy. J Exp Med. 2008;205:3133–3144. doi: 10.1084/jem.20081937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong C. Genetic controls of Th17 cell differentiation and plasticity. Exp Mol Med. 2011;43:1–6. doi: 10.3858/emm.2011.43.1.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockinger B, Veldhoen M. Differentiation and function of Th17 T cells. Curr Opin Immunol. 2007;19:281–286. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Lei J, He F, Wu M, Zheng X, Chen X, Chen Z. Administration of anti-interleukin-6 monoclonal antibody prolongs cardiac allograft survival. Transpl Int. 2010;23:1271–1281. doi: 10.1111/j.1432-2277.2010.01125.x. [DOI] [PubMed] [Google Scholar]
- 11.Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, Murphy KM. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, Wherry EJ, Haining WN. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014;15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corry RJ, Winn HJ, Russell PS. Primarily vascularized allografts of hearts in mice. The role of H-2D, H-2K, and non-H-2 antigens in rejection. Transplantation. 1973;16:343–350. doi: 10.1097/00007890-197310000-00010. [DOI] [PubMed] [Google Scholar]
- 15.Croyle MA, Callahan SM, Auricchio A, Schumer G, Linse KD, Wilson JM, Brunner LJ, Kobinger GP. PEGylation of a vesicular stomatitis virus G pseudotyped lentivirus vector prevents inactivation in serum. J Virol. 2004;78:912–921. doi: 10.1128/JVI.78.2.912-921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Textor SC, Wiesner R, Wilson DJ, Porayko M, Romero JC, Burnett JC Jr, Gores G, Hay E, Dickson ER, Krom RA. Systemic and renal hemodynamic differences between FK506 and cyclosporine in liver transplant recipients. Transplantation. 1993;55:1332–1339. doi: 10.1097/00007890-199306000-00023. [DOI] [PubMed] [Google Scholar]
- 17.Gardiner SM, March JE, Kemp PA, Fallgren B, Bennett T. Regional haemodynamic effects of cyclosporine A, tacrolimus and sirolimus in conscious rats. Br J Pharmacol. 2004;141:634–643. doi: 10.1038/sj.bjp.0705659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J Exp Med. 2006;203:1685–1691. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hamburg JP, Asmawidjaja PS, Davelaar N, Mus AM, Colin EM, Hazes JM, Dolhain RJ, Lubberts E. Th17 cells, but not Th1 cells, from patients with early rheumatoid arthritis are potent inducers of matrix metalloproteinases and proinflammatory cytokines upon synovial fibroblast interaction, including autocrine interleukin-17A production. Arthritis Rheum. 2011;63:73–83. doi: 10.1002/art.30093. [DOI] [PubMed] [Google Scholar]
- 21.Punkenburg E, Vogler T, Buttner M, Amann K, Waldner M, Atreya R, Abendroth B, Mudter J, Merkel S, Gallmeier E, Rose-John S, Neurath MF, Hildner K. Batf-dependent Th17 cells critically regulate IL-23 driven colitis-associated colon cancer. Gut. 2015 doi: 10.1136/gutjnl-2014-308227. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.Fabrega E, Lopez-Hoyos M, San Segundo D, Casafont F, Pons-Romero F. Changes in the serum levels of interleukin-17/interleukin-23 during acute rejection in liver transplantation. Liver Transpl. 2009;15:629–633. doi: 10.1002/lt.21724. [DOI] [PubMed] [Google Scholar]
- 23.Vanaudenaerde BM, De Vleeschauwer SI, Vos R, Meyts I, Bullens DM, Reynders V, Wuyts WA, Van Raemdonck DE, Dupont LJ, Verleden GM. The role of the IL23/IL17 axis in bronchiolitis obliterans syndrome after lung transplantation. Am J Transplant. 2008;8:1911–1920. doi: 10.1111/j.1600-6143.2008.02321.x. [DOI] [PubMed] [Google Scholar]
- 24.Heidt S, Segundo DS, Chadha R, Wood KJ. The impact of Th17 cells on transplant rejection and the induction of tolerance. Curr Opin Organ Transplant. 2010;15:456–461. doi: 10.1097/MOT.0b013e32833b9bfb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deteix C, Attuil-Audenis V, Duthey A, Patey N, McGregor B, Dubois V, Caligiuri G, Graff-Dubois S, Morelon E, Thaunat O. Intragraft Th17 infiltrate promotes lymphoid neogenesis and hastens clinical chronic rejection. J Immunol. 2010;184:5344–5351. doi: 10.4049/jimmunol.0902999. [DOI] [PubMed] [Google Scholar]
- 26.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 27.Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, Hackney J, Nurieva R, Escalante CR, Ouyang W, Littman DR, Murphy KM, Singh H. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A Validated Regulatory Network for Th17 Cell Specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong S, Li J, Ma L, Li K, Zhang L, Wang G, Liu Y, Ji X, Liu X, Chen P, Ouyang R, Zhang S, Zhou Z, Wang CY, Xiang X, Yang Y. Blockade of dopamine D1-like receptor signalling protects mice against OVA-induced acute asthma by inhibiting B-cell activating transcription factor signalling and Th17 function. FEBS J. 2013;280:6262–6273. doi: 10.1111/febs.12549. [DOI] [PubMed] [Google Scholar]
- 30.Xu Z, Ramachandran S, Gunasekaran M, Nayak D, Benshoff N, Hachem R, Gelman A, Mohanakumar T. B Cell-Activating Transcription Factor Plays a Critical Role in the Pathogenesis of Anti-Major Histocompatibility Complex-Induced Obliterative Airway Disease. Am J Transplant. 2016;16:1173–82. doi: 10.1111/ajt.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang T, Lu Q, Yang X, Liu X, Liao R, Zhang Y, Yang Z. Roles of the tacrolimus-dependent transcription factor IRF4 in acute rejection after liver transplantation. Int Immunopharmacol. 2015;28:257–263. doi: 10.1016/j.intimp.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Barbi J, Pardoll D, Pan F. Metabolic control of the Treg/Th17 axis. Immunol Rev. 2013;252:52–77. doi: 10.1111/imr.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 34.Sahoo A, Alekseev A, Tanaka K, Obertas L, Lerman B, Haymaker C, Clise-Dwyer K, McMurray JS, Nurieva R. Batf is important for IL-4 expression in T follicular helper cells. Nat Commun. 2015;6:7997. doi: 10.1038/ncomms8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Pettigrew GJ, Bolton EM, Murfitt CR, Carmichael A, Bradley JA, Lever AM. Lentivirus-mediated gene transfer of viral interleukin-10 delays but does not prevent cardiac allograft rejection. Gene Ther. 2005;12:1509–1516. doi: 10.1038/sj.gt.3302547. [DOI] [PubMed] [Google Scholar]
- 36.Zheng X, Lian D, Wong A, Bygrave M, Ichim TE, Khoshniat M, Zhang X, Sun H, De Zordo T, Lacefield JC, Garcia B, Jevnikar AM, Min WP. Novel small interfering RNA-containing solution protecting donor organs in heart transplantation. Circulation. 2009;120:1099–1107. doi: 10.1161/CIRCULATIONAHA.108.787390. 1091 p following 1107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.