Abstract
Previous studies on Ophiothrix in European waters demonstrated the existence of two distinct species, Ophiothrix fragilis and Ophiothrix sp. II. Using phylogenetic and species delimitation techniques based on two mitochondrial genes (cytochrome c oxidase I and 16S rRNA) we prove the existence of a new congeneric species (Ophiothrix sp. III), occurring in the deep Atlantic coast of the Iberian Peninsula and the Alboran Sea. We compared phylogeographic patterns of these three Ophiothrix species to test whether closely related species are differentially affected by past demographic events and current oceanographic barriers. We used 432 sequences (137 of O. fragilis, 215 of Ophiothrix sp. II, and 80 of Ophiothrix sp. III) of the 16S rRNA from 23 Atlantic-Mediterranean locations for the analyses. We observed different geographic and bathymetric distributions, and contrasted phylogeography among species. Ophiothrix fragilis appeared genetically isolated between the Atlantic and Mediterranean basins, attributed to past vicariance during Pleistocene glaciations and a secondary contact associated to demographic expansion. This contrasts with the panmixia observed in Ophiothrix sp. II across the Atlantic-Mediterranean area. Results were not conclusive for Ophiothrix sp. III due to the lack of a more complete sampling within the Mediterranean Sea.
Within the past few decades, phylogeographic patterns in marine benthic invertebrates with Atlantic-Mediterranean distribution have been documented in a plethora of species belonging to a range of phyla e.g. refs 1, 2, 3, 4, 5, 6, 7, 8, 9. The relatively well documented geological history of the Mediterranean Sea and its connection with the Atlantic Ocean has allowed in most cases relating historical processes (e.g. the Messinian Salinity Crisis –MSC– during Mio-Pliocene transition, and more recently, the Pleistocene glacial and interglacial periods during the Quaternary), to the current population connectivity patterns observed for different species e.g. refs 3, 10, 11, 12, 13, 14, 15, 16. Apart from these historical processes, the current hydrography, including major circulation patterns and local eddies, and geographical processes differentially affect the littoral areas from both the Atlantic and Mediterranean basins. These geographical, historical and oceanographic processes, altogether with the inherent biological characteristics of the species, are among the responsible processes for the present-day intraspecific distribution of genetic diversity in littoral species1,17. Nevertheless, the stochasticity of the recruitment processes and availability of substrate can also play an important role shaping the current genetic structure of benthic taxa18.
The Strait of Gibraltar represents a natural connection, and at the same time a transition area, between the different hydrographical conditions that go from the relatively cold waters of the North Atlantic to the warmer, saltier, and markedly seasonal waters of the Mediterranean19. However, the Alboran Sea and more specifically the Almeria-Oran Front (AOF), situated east of the Strait of Gibraltar, is considered as the real biogeographical break between these two basins. A number of marine species show genetic transition between Atlantic and Mediterranean populations at the AOF, which acts as a migration barrier preventing or significantly reducing genetic flow between these two areas (see a review in Patarnello et al.1, but see García-Merchán et al.7). Genetic connectivity between populations of these two basins including the AOF as a target area has been addressed for several benthic invertebrates, reaching in some cases different conclusions even when considering species with similar dispersal abilities e.g. refs 7, 8, 12 and 20, which may be explained by the effect of past historical processes differentially affecting the genetic diversity of these organisms. In this sense, the comprehensive study of congeneric species with a common evolutionary origin (i.e. sharing similar biological traits) and similar geographic distribution (i.e. being affected by similar hydrological and geological processes), has been proposed as a useful approach to identify phylogeographic signals and real permeability of major marine fronts1,7,21.
Due to their wide geographic distribution, extending from the intertidal to the deep subtidal22, and their ecological relevance, brittle stars of the genus Ophiothrix appear as an interesting study case to address questions about connectivity among populations occurring in both the Atlantic and Mediterranean basins, as well as potential processes of cryptic speciation and multi-specific complexes. In an investigation of Ophiothrix fragilis on different intertidal and subtidal Atlantic populations, also including a few Western Mediterranean specimens, Muths et al.23 identified two distinct lineages that might potentially correspond to cryptic species. One of these lineages corresponded to the northern European populations, and another one encompassed specimens from two sites of the south European coast: one at the Atlantic coast of the Iberian Peninsula, in the intertidal of Galicia, and another from a subtidal site at the French Coast of Banyuls-sur-mer (NW-Mediterranean). Later, Pérez-Portela et al.15 conducted an extensive study combining molecular and morphological analyses using populations from both the Atlantic and the Mediterranean basins, and concluded that the two different lineages identified by Muths et al.23 in fact corresponded to two different morphological species (Lineage I: O. fragilis, and Lineage II: Ophiothrix sp. II). One of the species identified, O. fragilis, inhabits the intertidal and shallow subtidal (0–50 m) in the North Atlantic, where it occurs in highly dense assemblages22, but it also occurs in the Mediterranean deep subtidal (>50 m), where it was previously described as O. quinquemaculata (see ref. 15). Unfortunately, the study by Pérez-Portela et al.15 only included two deep subtidal Mediterranean samples (~50–60 m), and sequences of the so-called O. quinquemaculata from Genbank; therefore conclusions about the distribution of O. fragilis along the Mediterranean area were very limited.
In the study by Pérez-Portela et al.15, shallow-water populations of Ophiothrix sp. II across the Atlantic and Mediterranean showed apparent panmixia with no gene-flow barriers across the study area. This pattern was explained by a combination of processes including a wide dispersal potential, large effective population size and a recent demographic expansion. On the other hand, information on the genetic structure between populations of O. fragilis in the Atlantic area, along the coast of France and England, showed a chaotic structure in this species23. Genetic connectivity between Atlantic individuals and the only individual analysed from the Western Mediterranean, though, were not conclusive and pointed to future studies to unravel the genetic connectivity of this species.
Here, we present a comprehensive genetic study in Ophiothrix spp. occurring in the Atlantic and Mediterranean basins covering from the intertidal to the shallow and deep subtidal. The aims of this work were: (i) to describe the distribution of Ophiothrix spp. across both space and depth, and to detect the possible occurrence of cryptic speciation; and (ii) to compare phylogeographic patterns based on mitochondrial DNA among congeneric species to determine whether present and past marine biogeographic barriers and demographic events may have differentially affected the distribution of genetic diversity among species, paying special attention to potential differentiation between the Atlantic and Mediterranean basins. To achieve our objectives we used two mitochondrial gene fragments, the 16S rRNA (hereafter 16S), which has been demonstrated to provide suitable resolution in population genetic studies of brittle stars15,24, and the cytochrome c oxidase subunit I (hereafter COI).
Results
Phylogeny and species delimitation
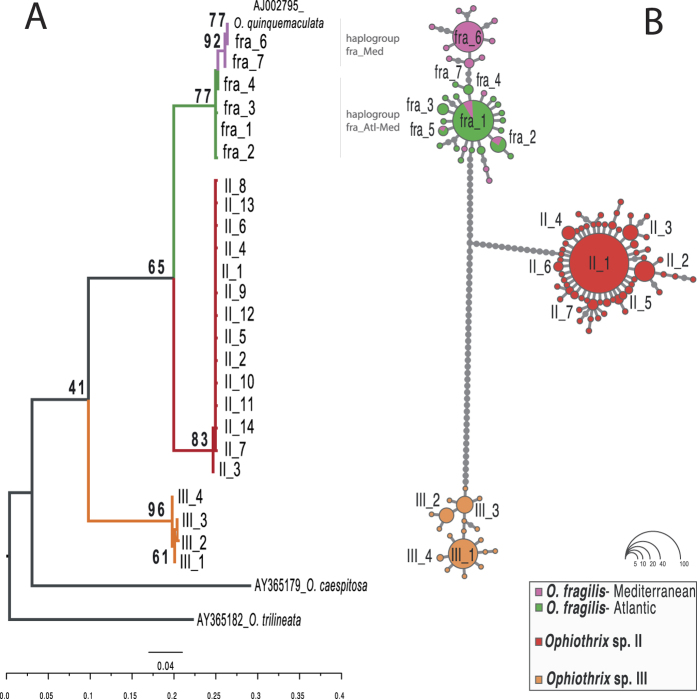
A total of 310 bp were obtained for the 16S fragment for the three species of Ophiothrix considered here. For the COI marker only three sequences (797 bp) of Ophiothrix sp. III from the Cantabrian Sea (two from station DEM91 and one from DEM45; see Table 1 and Fig. 1) were obtained, which were added to a previous dataset of 148 sequences of O. fragilis and Ophiothrix sp. II by Pérez-Portela et al.15. ML phylogenetic trees based on both the most common haplotypes of 16S and the total number of haplotypes of the 16S and COI showed three moderately supported clades, namely O. fragilis, Ophiothrix sp. II, and Ophiothrix sp. III. Although the three species can unambiguously be considered as different species due to the genetic distance between the three mitochondrial lineages, morphology and bathymetric segregation (see Results below), their relative phylogenetic relationships within the genus could not be resolved due to the low support of the nodes (Fig. 2A, supplementary Figs S2 and S3). Two different haplogroups were recovered for the 16S in O. fragilis corresponding to samples from the Mediterranean and from a mixture of samples from the Atlantic and Mediterranean (Fig. 2; fra_Med and fra_Atl-Med); these haplogroups, though, were not recovered in the COI phylogenetic tree because samples of this species from the Mediterranean were not available. Difficulties to obtain good quality sequences from additional genes, such as the COI from Mediterranean samples of O. fragilis, and the nuclear Histone 3, Internal Transcribed Spacers 1 and 2, prevented to generate better supported phylogenies to clarify the evolutionary relationships among lineages.
Table 1. Summary of the information related to the different localities surveyed and genetic variability for the 16S for the three different Ophiothrix species.
| Population | Code | Depth (m) | Coordinates | Geographical area | N | H | p | Hd | π | R2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Ophiothrix fragilis | ||||||||||
| Kristineberg† | KRI | Intertidal | 58°14′27.32“N 11°25′55.14“E | NE Atlantic- North Sea | 22 | 9 | 5 | 0.749 ± 0.094 | 0.00368 ± 0.0007 | 0.1467** |
| Roscoff | ROS | Intertidal | 48°45′9.55“N 3°58′38.10“W | NE Atlantic- English Channel | 19 | 6 | 3 | 0.468 ± 0.140 | 0.00204 ± 0.0007 | 0.1667* |
| Ferrol† | FER | 8–10 | 43°27′25.27“N 8°20′9.07“W | NE Atlantic | 16 | 7 | 4 | 0.775 ± 0.088 | 0.00391 ± 0.0008 | 0.1650** |
| Vigo-Cornide† | COR | 30–60 | 42°13′3.81“N 8°49′41.24“W | NE Atlantic | 22 | 7 | 4 | 0.481 ± 0.131 | 0.00206 ± 0.0007 | 0.1618** |
| Atlantic basin | 79 | 21 | 16 | 0.625 ± 0.063 | 0.00294 ± 0.0004 | 0.1040* | ||||
| Blanes† | ABL | 90 | 41°40′2.19“N 2°52′30.44“E | NW Mediterranean | 9 | 4 | 2 | 0.750 ± 0.139 | 0.00821 ± 0.0033 | 0.2036 |
| Indemares† | IMA | 123–139 | 41° 18,5752′ N 2°25,6324′ E | NW Mediterranean | 5 | 2 | 1 | 0.400 ± 0.237 | 0.00388 ± 0.0023 | 0.3000 |
| Planassa† | PLA | 80–100 | 41°38′47.92“N 2°54′40.61“E | NW Mediterranean | 21 | 12 | 8 | 0.824 ± 0.084 | 0.01235 ± 0.0022 | 0.1345 |
| Port de la Selva† | PSE | 80–100 | 42°21′59.49“N 3°13′26.37“E | NW Mediterranean | 23 | 4 | 1 | 0.577 ± 0.090 | 0.00849 ± 0.0012 | 0.1341** |
| Mediterranean basin | 58 | 17 | 12 | 0.698 ± 0.065 | 0.00960 ± 0.0012 | 0.1037* | ||||
| TOTAL | 137 | 34 | 28 | 0.777 ± 0.029 | 0.00979 ± 0.0005 | 0.0088* | ||||
| Ophiothrixsp. II | ||||||||||
| Ferrol† | FER | 8–10 | 43°27′25.27“N 8°20′9.07“W | NE Atlantic | 33 | 12 | 7 | 0.758 ± 0.122 | 0.00408 ± 0.0011 | 0.1112* |
| Cascais | CAS | Intertidal | 38°42′25.77“N 9°29′15.41“W | NE Atlantic | 18 | 11 | 5 | 0.810 ± 0.093 | 0.00409 ± 0.0008 | 0.0539** |
| Armação de Pera | APA | 10–15 | 37°5′41.52“N 8°21′37.31“W | NE Atlantic | 20 | 9 | 3 | 0.758 ± 0.101 | 0.00394 ± 0.0009 | 0.0541** |
| Ceuta | CEB | 18–25 | 35°52′52.19“N 5°19′10.20“W | Gibraltar Strait | 19 | 8 | 3 | 0.672 ± 0.119 | 0.00321 ± 0.0009 | 0.0857** |
| La Herradura | LH | 15–18 | 36°22′4.36“N 5°12′51.68“W | SW Mediterranean | 20 | 6 | 2 | 0.447 ± 0.137 | 0.00241 ± 0.0009 | 0.0921* |
| Xábea | XB | 10–20 | 38°47′39.48“N 0°12′18.10“E | SW Mediterranean | 20 | 7 | 3 | 0.521 ± 0.135 | 0.00219 ± 0.0007 | 0.0818** |
| Roses | RS | 10–20 | 42°14′11.17“N 3°15′45.80“E | NW Mediterranean | 14 | 4 | 1 | 0.494 ± 0.151 | 0.00172 ± 0.0006 | 0.1246* |
| Cadaqués | CAD | 2–15 | 42°17′27.85“N 3°17′43.95“E | NW Mediterranean | 19 | 9 | 5 | 0.772 ± 0.094 | 0.00407 ± 0.0009 | 0.0700** |
| Alcudia | AL | 15–18 | 39°52′1.28“N 3°11′39.87“E | W Mediterranean-Balearic Is. | 26 | 13 | 5 | 0.720 ± 0.099 | 0.00359 ± 0.0008 | 0.0412** |
| Ladiko | LK | 2–10 | 36°19′22.22“N 28°13′18.81“E | E Mediterranean | 16 | 9 | 5 | 0.817 ± 0.095 | 0.00459 ± 0.0013 | 0.0745** |
| Kalytea Bay | KB | 2–5 | 35°9′58.03“N 24°24′38.20“E | E Mediterranean | 9 | 6 | 3 | 0.833 ± 0.126 | 0.00418 ± 0.0012 | 0.1111** |
| TOTAL | 215 | 58 | 42 | 0.679 ± 0.0390 | 0.00345 ± 0.00031 | 0.0117** | ||||
| Ophiothrixsp. III | ||||||||||
| Demersales-45† | DEM45 | 153 | 43°36′44.7“N 8°29′49.5“W | NE Atlantic- Cantabrian Sea | 20 | 6 | 3 | 0.679 ± 0.101 | 0.0040 ± 0.0009 | 0.098* |
| Demersales-91† | DEM91 | 310 | 43°53′27.24“N 5°34′53.77“W | NE Atlantic- Cantabrian Sea | 22 | 5 | 5 | 0.745 ± 0.093 | 0.0039 ± 0.0007 | 0.1486** |
| Demersales-131† | DEM131 | 131 | 43°30′57.0“N 2°53′32.4“W | NE Atlantic- Cantabrian Sea | 23 | 8 | 5 | 0.744 ± 0.080 | 0.0039 ± 0.0006 | 0.1472** |
| Peniche† | POR | 100–150 | 39°27′53“N 9°12′48.33“W | NE Atlantic | 7 | 4 | 1 | 0.714 ± 0.181 | 0.0054 ± 0.0018 | 0.2417 |
| PINDALBV17† | PINDAL | 169 | 36°08′0“N 30°2′0”E | SW Mediterranean | 8 | 4 | 1 | 0.750 ± 0.139 | 0.0036 ± 0.0008 | 0.2311** |
| TOTAL | 80 | 19 | 15 | 0.714 ± 0.047 | 0.00407 ± 0.0004 | 0.100** | ||||
N number of individuals, H number of haplotypes, p private haplotypes, Hd haplotype diversity, π nucleotide diversity, R2 Ramos-Onsins & Rozas statistic.
For Ophiothrix sp. II and Ophiothrix sp. III information of the Atlantic and Mediterranean basins is not presented since these two species did not displayed significant differences between geographical areas. †New samples sequenced in this study. *p < 0.05 and **p < 0.01.
Figure 1.
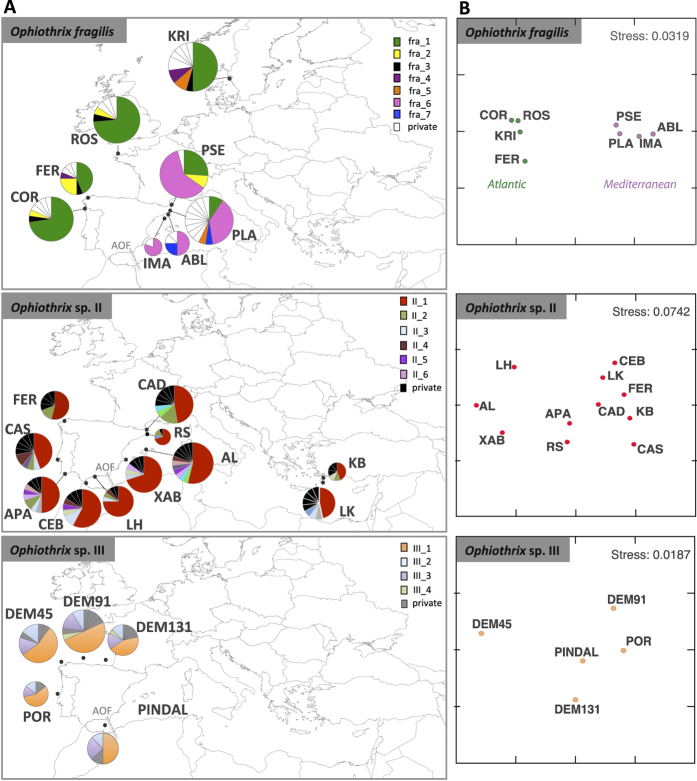
(A) Map of the Atlantic-Mediterranean area showing the locations where samples of Ophiothrix fragilis, Ophiothrix sp. II, and Ophiothrix sp. III were collected. See Table 1 for further details about locations. Circles represent the 16S haplotype diversity within each location and their size is proportional to the number of individuals per site. Partitions inside the circles represent the relative proportion of each haplotype within each location. The dashed-grey line represents the Almeria-Oran Front (AOF). See Fig. 2 for details about haplotypes for each species. (B) MDS representation of the ФST values at each location for each of the three species. Figures were created with the free software QGIS (http://qgis.osgeo.org/es/site/), and edited in Adobe Illustrator CS5.1 (http://www.adobe.com) for this study.
Figure 2.
(A) Maximum Likelihood phylogenetic tree of the most frequent haplotypes of three species of Ophiothrix resulting from the 16S marker. Only bootstrap support values >40 are indicated in the main nodes. (B) Haplotype network for the three Ophiothrix species based on the complete 16S alignment. Circles are proportional to the number of individuals per haplotype. Grey dots correspond to mutational steps.
PTP analyses based on both the 16S and COI markers corroborated the existence of three species corresponding to the three main clades identified in the phylogenetic trees (supplementary Table S1, Figs S4 and S5). Inter-specific genetic distances (K2p) between the three species ranged from 11–20% for the 16S, and from 19–22% for the COI (Table 2); these values were lower than those between these three species and other congeneric species (see supplementary Table S2 for a complete list of divergence values of the COI). Intra-specific values (K2p) were ca. ten fold lower than inter-specific ones and ranged from 0.9–1.6% for 16S, and between 0.11–0.17% for COI (Table 2).
Table 2. Genetic distances (±standard error) between Ophiothrix spp. based on a Kimura 2-Parameters model for 16S and COI markers.
| 16S | O. fragilis | Ophiothrix sp. II | Ophiothrix sp. III | O. trilineata | O. caespitosa |
|---|---|---|---|---|---|
| O. fragilis | (0.0158 ± 0.003) | ||||
| Ophiothrix sp. II | 0.111 ± 0.018 | (0.0091 ± 0.001) | |||
| Ophiothrix sp. III | 0.195 ± 0.027 | 0.203 ± 0.029 | (0.0089 ± 0.002) | ||
| O. trilineata | 0.308 ± 0.036 | 0.324 ± 0.039 | 0.318 ± 0.038 | — | |
| O. caespitosa | 0.262 ± 0.031 | 0.338 ± 0.039 | 0.295 ± 0.035 | 0.385 ± 0.043 | — |
| COI | O. fragilis | Ophiothrix sp. II | Ophiothrix sp. III | O. trilineata | O. caespitosa |
| O. fragilis | (0.0175 ± 0.003) | ||||
| Ophiothrix sp. II | 0.189 ± 0.022 | (0.0117 ± 0.001) | |||
| Ophiothrix sp. III | 0.202 ± 0.023 | 0.220 ± 0.024 | (0.0112 ± 0.004) | ||
| O. trilineata | 0.237 ± 0.026 | 0.280 ± 0.029 | 0.264 ± 0.027 | — | |
| O. caespitosa | 0.267 ± 0.029 | 0.269 ± 0.029 | 0.253 ± 0.026 | 0.293 ± 0.032 | — |
Inter-specific distance values are presented below the diagonal. Numbers along the diagonal in brackets represent intra-specific variation for the three species analysed in this study.
Genetic diversity and population connectivity
A total of 237 new individuals were sequenced for the 16S, corresponding to 137 from O. fragilis (34 haplotypes), 80 from Ophiothrix sp. III (19 haplotypes), and 20 from Ophiothrix sp. II, the latter collected at FER; this dataset was analysed together with the 215 individuals of Ophiothrix sp. II sequenced by Pérez-Portela et al.15 (Table 1). The three different species did not occur in the same locality except for FER, where both O. fragilis (16 individuals) and Ophiothrix sp. II (33 individuals) were found in sympatry. Out of the total 111 haplotypes found for all species, 85 (76.6%) were private. Values of genetic diversity are detailed in Table 1.
The haplotype network based on the joined 16S alignment of the three species was structured in three different main clades (species) lacking intermediate haplotypes (Fig. 2B). Divergence among clades was caused by 27 mutations between O. fragilis and Ophiothrix sp. II, and 51 between Ophiothrix sp. III and the other two species (Fig. 2B). The O. fragilis network exhibited two star-like patterns (haplogroups: fra_Md and fra_Atl-Med) connected by two mutational steps. In both cases, two dominant haplotypes were located at the center of the haplogroups: fra_6, appearing only at Mediterranean locations; and fra_1, broadly distributed across the Atlantic and Mediterranean basins, being the most common haplotype in the Atlantic basin. Apart from fra_1 (present in the Atlantic and in the Mediterranean populations of PSE and PLA), only two other haplotypes were shared between the two basins: fra_2, in three Atlantic populations (FER, COR, and ROS) and the Mediterranean PSE; and fra_5, which appeared in the Atlantic KRI and the Mediterranean PLA (Figs 1A and 2B). Ophiothrix sp. II showed a clear star-like pattern with II_1 as the ancestral haplotype, present in a similar proportion in all populations in both basins, surrounded by low-frequency (II_2 to II_7) and private haplotypes (Figs 1A and 2B). Ophiothrix sp. III revealed a network with a few dominant haplotypes (III_1 to III_4) surrounded by private haplotypes (Fig. 2B). Similar contributions of III_1, III_2 and III_3, the most common haplotypes in Ophiothrix sp. III, were observed in all populations (Fig. 1A).
AMOVA results showed no significant difference in the genetic structure between the Atlantic and Mediterranean basins and among sites for Ophiothrix sp. II and Ophiothrix sp. III, which retained 100% of their genetic variability within sites (Table 3). In contrast, O. fragilis showed significant differences between the two basins (58% of total genetic variation), and within sites (41%) (Table 3).
Table 3. Results of the AMOVA analyses for each of the three Ophiothrix species based on the 16S.
| Source of variation | d.f. | Sum of squares | Fixation index | % of variation | p-value |
|---|---|---|---|---|---|
| Ophiothrix fragilis | |||||
| Among basins | 1 | 84.1 | FCT = 0.580 | 58.02 | 0.028* |
| Among sites within basins | 6 | 6.7 | FSC = 0.015 | 0.65 | 0.293 |
| Within sites | 128 | 114.2 | FST = 0.587 | 41.3 | 0.000** |
| Ophiothrixsp. II | |||||
| Among basins | 1 | 0.512 | FCT = −0.0007 | 0.000 | 0.483 |
| Among sites within basins | 9 | 4.854 | FSC = −0.0007 | 0.000 | 0.487 |
| Within sites | 182 | 99.323 | FST = −0.0013 | 100 | 0.535 |
| Ophiothrixsp. III | |||||
| Among basins | 1 | 0.315 | FCT = −0.0158 | 0.000 | 1.000 |
| Among sites within basins | 3 | 0.933 | FSC = −0.0295 | 0.000 | 0.944 |
| Within sites | 74 | 44.930 | FST = −0.0458 | 100 | 0.966 |
ФST values confirmed AMOVA results, showing only significant differences between pairwise comparisons among populations of O. fragilis corresponding to different basins (Fig. 1B; Table 4, supplementary Tables S3 and S4). These results indicated panmixia across the geographical study area in Ophiothrix sp. II and III, while a strong phylogeographic break was observed between basins in O. fragilis (Fig. 1B). Genetic differentiation was weak although significantly correlated to geographical distances when the complete O. fragilis dataset was considered (Mantel test: r = 0.0004; p = 0.043), but this pattern of isolation by distance was not significant when basins were analysed separately (Atlantic: r = 0.0000; p = 0.588; and Mediterranean: r = −0.0002, p = 0.547).
Table 4. ФST values between pairs of populations for Ophiothrix fragilis based on the 16S.
| Ophiothrix fragilis | |||||||
|---|---|---|---|---|---|---|---|
| ABL | PLA | IMA | PSE | ROS | FER | KRI | |
| PLA | −0.02310 | — | |||||
| IMA | −0.04994 | 0.01724 | — | ||||
| PSE | 0.01959 | −0.01746 | 0.11168 | — | |||
| ROS | 0.74571** | 0.49423** | 0.86336** | 0.54393** | — | ||
| FER | 0.67420** | 0.45448** | 0.78977** | 0.49536** | 0.04975 | — | |
| KRI | 0.68246** | 0.46144** | 0.79129** | 0.50192** | 0.00596 | 0.04456 | — |
| COR | 0.75560** | 0.50949** | 0.86475** | 0.55582** | −0.01535 | 0.05551 | 0.01485 |
**Significant values after applying the false discovery rate method.
LAMARC results for O. fragilis showed an almost unidirectional migration for mitochondrial DNA from the Atlantic Ocean to the Mediterranean Sea according to the posterior likelihood values obtained (Fig. 3).
Figure 3. Results of migration analyses between the Atlantic and Mediterranean basins in Ophiothrix fragilis based on 16S sequences.
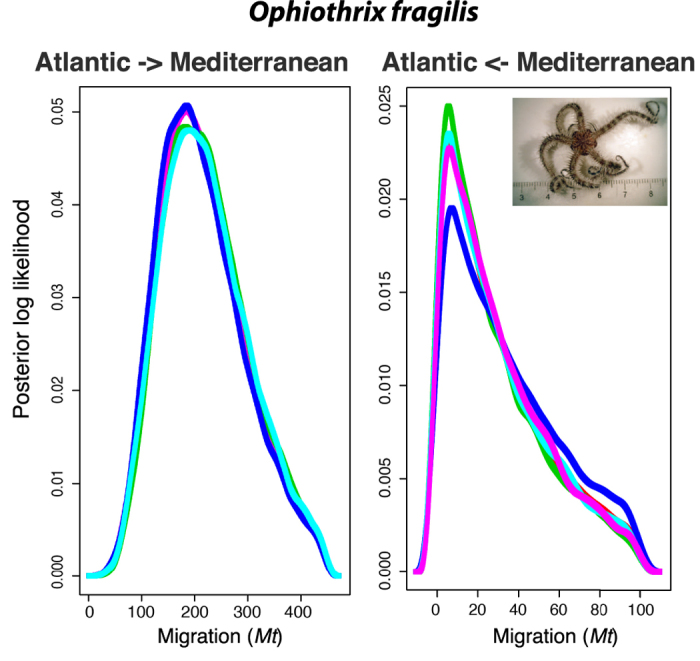
The two graphs represented the final results of LAMARC for 5 different replicates (left graph: Migration from the Atlantic basin to the Mediterranean basin; and right graph: Migration from the Mediterranean basin to the Atlantic basin).
Demography
The neutrality Rozas’ R2 test was significant for the three Ophiothrix species, which can be interpreted as an evidence of past demographic expansion (see 16S results in Table 1). The mismatch distribution based also on the 16S marker showed a unimodal distribution in Ophiothrix sp. II and III, also attributable to demographic expansion (supplementary Fig. S6). In the case of O. fragilis, though, despite showing a clear bimodal distribution when all populations were analysed together, a unimodal distribution, likely due to a past demographic expansion, was detected for Atlantic samples separately analysed, a pattern not observed for the Mediterranean dataset (supplementary Fig. S6). BSP results further confirmed demographic expansion in the three species. According to our analyses based on the 16S fragment, demographic expansion for the three species happened more than 50,000 years ago, remaining stable for approximately the last 40,000 years (Fig. 4). These analyses also suggest differences in female effective population sizes among species, Ophiothrix sp. II displaying the largest effective population size (Fig. 4). BSP results based on the COI of Ophiothrix sp. II also detected a demographic expansion but much earlier, ca. 100,000–180,000 years ago (see supplementary Fig. S7), than for the 16S marker. These differences may be related with the fact that there is not a well-calibrated molecular clock for these markers in ophiuroids, and the values obtained need to be taken as an approximation. For O. fragilis, estimations based on COI could not be performed due to the absence of coalescence in the dataset.
Figure 4. Demographic analyses of the three Ophiothrix species based on Bayesian Skyline plots of the 16S marker.
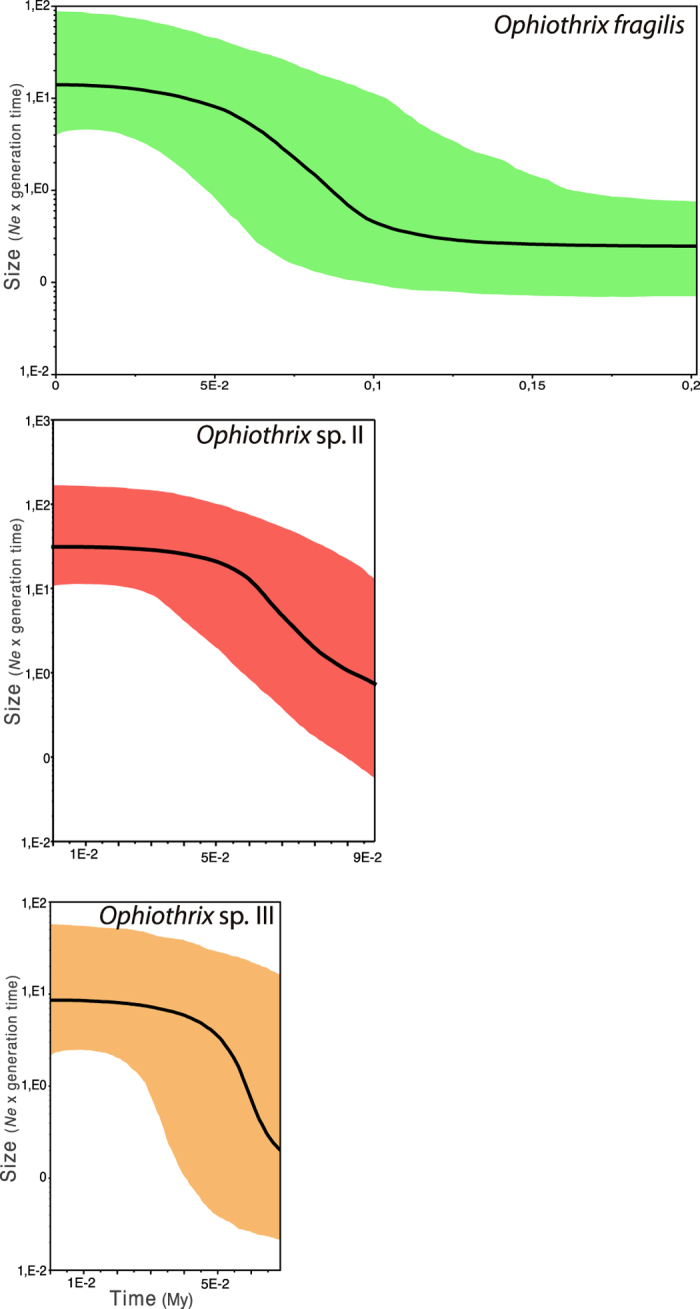
Time is measured in million years (Mya). Black lines illustrate mean size estimations, and colour shadows show 95% confidence interval.
Discussion
Our study revealed unexpected biodiversity in Ophiothrix along the Atlantic-Mediterranean area, with the occurrence of three different lineages that may be considered as species. Despite the closely evolutionary relationships among them, they displayed different geographic and bathymetric distributions, and contrasted phylogeographic patterns for the mitochondrial gene 16S, a fact that should be contrasted in the future with the analysis of nuclear markers to infer if they are congruent with mitochondrial data. Our study represents an important contribution in the context of phylogeography of ophiuroids, since this has only been addressed for a few taxa worldwide15,23,24,25,26,27,28,29,30.
Two of the species found in this study, O. fragilis and Ophiothrix sp. II, were distinguished in great detail in a previous study15, while Ophiothrix sp. III is reported here for the first time. Phylogenetic inference using 16S and COI combined with species delimitation analyses unambiguously confirmed that these three organisms are in fact three closely related congeneric species (Fig. 2, supplementary Figs S2–S5). In addition, no haplotype was shared among them and levels of inter-specific genetic divergence in 16S and COI were high and larger that those measured at the intra-specific level. In this sense, divergence between Ophiothrix in our study was an order of magnitude greater (19–22% for COI) than the limit commonly accepted to discriminate species in echinoderms (0.9–1% for COI31). Importantly, remarkable morphological and ecological (bathymetric distribution) differences supporting the genetic divergence exist (supplementary Fig. S1; see Methods section), although they were not fully discussed in this work. Forthcoming and on-going morphological studies will lead to the formal description of both Ophiothrix sp. II and Ophiothrix sp. III (Manjón E. et al. in preparation).
The three Ophiothrix species studied here overlapped at least partially across their geographical distribution range, although they were in general bathymetrically segregated (Fig. 1A; Table 1). Ophiothrix sp. II is an intertidal and shallow subtidal species (0–60 m)15, whereas O. fragilis commonly occurs from the intertidal to the deep subtidal in the NE Atlantic, and from ~60 m to 100–130 m depth in the NW Mediterranean, being absent from shallower waters in the Mediterranean area. Such a bathymetric segregation between closely related ophiuroids is not the first time to be noticed. Muths et al.27 also reported the occurrence of two brittle stars of the genus Acrocnida that appeared in the intertidal (Acrocnida spatulispina) and the subtidal (Acrocnida brachiata) along the English Channel and the coast of Brittany, that had little overlapping in their distribution. Similarly, the widely distributed Atlantic-Mediterranean O. fragilis and Ophiothrix sp. II, only occurred in sympatry in the intertidal and shallow subtidal of the Atlantic coast of Ferrol and at ~50–60 m in the NW Mediterranean. Interestingly, preliminary results of in vitro cross-fertilization experiments indicated fertilization success in both intra- and inter-specific trials (the latter with less success) using a higher concentration of sperm than in natural conditions (authors’ unpublished data), which may suggest that these species are able to hybridize despite their relatively high genetic divergence (11% for 16S, 19% for COI; Table 2). These preliminary results are in agreement with the ones reported for the above-mentioned Acrocnida ophiuroids, where it was proved that hybrids occurred despite the two species showing a divergence of about 20% for COI27,28. Prezygotic barriers, such as reproductive asynchrony and/or ecological preferences (i.e. bathymetric segregation), were suggested to be limiting hybridization in Acrocnida28; these barriers might also be preventing the opportunity to hybridize between O. fragilis and Ophiothrix sp. II, since the principal annual recruitments for these species do not overlap22,23 and their habitat preferences differ. Prezygotic barriers have been widely documented in other echinoderms, especially for echinoids, including examples on habitat separation between species but also on gametic isolation due to the evolution of molecules involved in gamete recognition e.g. refs 32 and 33.
A possible explanation for the contrasting distribution in O. fragilis and Ophiothrix sp. II was already anticipated by Pérez-Portela et al.15, who suggested that their speciation processes may have taken place during the Mio-Pliocene transition, a period coincident with the isolation of the North Atlantic area into two main basins. Briefly, after the opening of the English Channel and the subsequent contact of the two North Atlantic basins previously isolated, Ophiothrix sp. II, originally preadapted to the warmer and shallower conditions of the southern basin of the North Atlantic, colonized the shallow intertidal-subtidal across the South European coast and the Mediterranean Sea after the opening of Gibraltar Strait following the Messinian Salinity Crisis. On the other hand, O. fragilis, preadapted to colder temperatures and deeper waters from the northern basin of the North Atlantic, subsequently colonized the deeper Mediterranean subtidal, where conditions are more similar in temperature to those currently found in the shallow Atlantic subtidal15. Thus, the distribution of these two Ophiothrix spp. found in our study confirms the hypothesis already presented by Pérez-Portela et al.15. Interestingly, a similar hypothesis was suggested to explain the current ecological segregation observed for the two Acrocnida species mentioned earlier28.
Ophiothrix sp. III was exclusively found in deep subital waters (>100 m) of the Atlantic Cantabrian Sea and Portugal shore, as well as also in the Mediterranean Alboran Sea, which lies just behind the AOF, typically considered as one of the most important biogeographic barriers separating the Atlantic from Mediterranean basins1. Hence, our observations for Ophiothrix sp. III do not allow us venturing any hypothesis about the current distribution of this species within the Mediterranean, although its absence in NW Mediterranean deep subtidal areas, where O. fragilis is commonly found, might indicate that its distribution limit in the Mediterranean is at the AOF, as observed in other marine invertebrates e.g. refs 9, 13 and 34. Nevertheless, further sampling in deep waters of the SW Mediterranean and Eastern Mediterranean sub-basin would be necessary to have a complete picture of the geographical distribution of this species.
Overall, the three Ophiothrix species showed high levels of genetic diversity comparable to those reported in other brittle stars e.g. refs 24 and 29, although they showed different phylogeographic patterns. Panmixia in the shallow-water Ophiothrix sp. II, with no barrier preventing gene flow between Atlantic and Mediterranean populations15, contrasts with the genetic structure between basins observed in O. fragilis, supported by the significant values of AMOVA and ФST (Fig. 1B; Table 3 and supplementary Tables S3 and S4). Examples of marine species displaying similar dispersal capabilities but contrasted patterns of genetic structure are widely documented in the Atlantic-Mediterranean area (see Patarnello et al.1 for a review). Among these, the studies on the broadcast spawning invertebrates Nephrops norvegicus (Norwegian lobster) and the Homarus gammarus (European lobster) showed no clear differentiation pattern between basins in the former while well differentiated groups occurring in the different basins were observed for the latter12,20. More recently, Fernández et al.8 described admixture with high gene flow for Chiton olivaceous, and a ‘chaotic patchiness’ pattern with high genetic variability and private haplotypes in all sites for Lepidopleurus cajetanus, two closely related chiton molluscs occurring in sympatry with presumably limited dispersal ability due to their lecithotrophic larva.
Large population size, high dispersal potential due to a long-lasting larval phase and shallow-water near shore currents along the shore, and a past demographic expansion event were used as possible reasons to explain the genetic homogeneity observed in Ophiothrix sp. II15. Effective population size estimations using the BSP confirmed the existence of larger population size in Ophiothrix sp. II when compared to the other two species (Fig. 4). Ophiothrix fragilis with smaller effective population size might be more vulnerable to the effect of genetic drift, a process that changes allele frequencies promoting genetic divergence between populations over short periods of time35.
Populations showing panmixia, as in Ophiothrix sp. II, have been reported for many echinoderms with planktotrophic larvae. In the holothurian Holothuria mammata genetically homogeneous populations were reported along the Macaronesian Islands, Algarve, and Western Mediterranean, although a significant break in genetic structure was detected in the Aegean Sea16. In other latitudes, the congeneric brittle star Ophiothrix suensonii also displayed lack of genetic differentiation across Florida and the Caribbean up to 1,700 km apart, combined with significant differences in genetic structure for some populations29.
Ophiothrix fragilis likely displayed genetic homogeneity within basins due to a long-lasting planktotrophic larva that promotes connectivity between distant populations, but clear divergence between basins. Examples of divergence between basins are not scarce in the literature, and have been reported for a diversity of marine invertebrates e.g. refs 2, 4, 11, 13 and 36. Two of the most common sea urchins across the Atlantic-Mediterranean area, Paracentrotus lividus and Arbacia lixula, showed no genetic differentiation over distances of thousands of km but divergence between basins despite having presumable high dispersal ability due to a long-lasting planktotrophic larva, demonstrating the strong disruptive effect of the Gibraltar Strait and/or Alboran Front3,6,11. The sea star Astropecten auranciacus also showed some significant differentiation of Atlantic versus Mediterranean populations and Western versus Eastern Mediterranean sub-basins that could not be attributed to any of the specific marine barriers targeted in their study (Strait of Gibraltar, AOF, Siculo-Tunisian Strait), but instead of isolation by geographical distance5. In contrast, divergence in O. fragilis did not follow a strong pattern by isolation by distance. The weak correlation detected with the Mantel test (r = 0.0004, p = 0.043) for the whole species dataset was magnified by the fact that all Mediterranean populations (very divergent from the Atlantic ones) were geographically close to each other, which may explain why this pattern of isolation by distance was not maintained within basins. Hence, we hypothesize the disruptive effect of the Gibraltar Strait and/or the AOF to be the most plausible genetic barrier explaining the divergence detected between basins in O. fragilis.
Nevertheless, the pattern observed for O. fragilis is complex and cannot be explained by a simple process of divergence between basins due to current genetic isolation. The two main haplogroups identified represent phylogeographical discontinuities that typically evolved in response to long-term extrinsic barriers to gene flow, although the present day structure may be affected by a redistribution of genetic diversity by contemporary contact. The asymmetrical distribution of haplogroups is congruent with a process of past vicariance due to allopatry between basins followed by secondary contact within the Mediterranean37, which is supported by the bimodal mismatch distribution (supplementary Fig. S6). This pattern in O. fragilis resembles that in Marthasterias glacialis, a widely distributed sea star in the Atlantic-Mediterranean region, with two distant lineages occurring in the Mediterranean Sea, but only one of them endemic from the Mediterranean14. Lineage splitting in M. glacialis was attributed to vicariance and secondary contact during Pleistocene glaciations (800–12 Kya) that caused sea level drops off up to 150 m with dramatic range shifts along the European coastline, ultimately interrupting circulation of species across the most important marine European corridors38. Hence, most of the present-day genetic patterns of marine coastal populations across the Atlantic-Mediterranean arch are the result of periodical limitations of larval interchange and migration across the Gibraltar Strait1,10. Our results for O. fragilis suggest a similar pattern to that observed in M. glacialis, with a recent demographic expansion during the last 50,000 years (Fig. 4), which could have followed a subsequent geographical expansion from the Atlantic to the Mediterranean. This is supported by the higher haplotype and nucleotide diversity in the Mediterranean (Table 1) due to the admixture of haplotypes from the two haplogroups in this basin, and by the unidirectional gene flow (migration) from the Atlantic to the Mediterranean detected in LAMARC (Fig. 3). However, the lack of a specific molecular clock for 16S and COI in ophiuroids make that estimations of demographic expansions need to be taken with caution and can be only used for comparison among species. At present, the predominant gene flow from the Atlantic to the Mediterranean might be maintained by major circulation currents across the Gibraltar Strait, due to a strong surface inflow from the Atlantic to the Mediterranean and water stratification39. The reverse current slides down 400–800 m depth, environmental conditions under which larvae of some echinoderms cannot survive40, might prevent the endemic Mediterranean haplogroup to disperse further in the Atlantic. Despite the disruptive effect of marine circulation across the Gibraltar Strait, the bathymetric fractioning between Atlantic (intertidal and shallow subtidal down to 60 m) and Mediterranean populations (deep subtidal, >60 m) in O. fragilis might be reinforcing the genetic divergence observed between basins by limiting the connectivity among populations41. The absence of the Mediterranean haplogroup (fra_Med) in Atlantic populations could be also related to the adaptation of lineages to different environmental conditions between habitats (shallow vs deep water) or basins, a hypothesis that cannot be completely ruled out. For instance, in the European anchovy water temperature shapes the contemporary distribution of mitochondrial DNA lineage frequencies due to strong selection on some codons42.
In conclusion, we present here important insights in Ophiothrix diversity, geographic and bathymetric distribution, and most relevant processes shaping intraspecific diversity of three Atlantic-Mediterranean species. It is a remarkable example of how closely related species, with similar biological features, display divergent phylogeographic patterns inferred from the mitochondrial markers due to the different effect of both historical and/or contemporary events. Future studies should be directed to determine the distribution of O. fragilis and the new species Ophiothrix sp. III across the whole Mediterranean basin by surveying under-sampled areas, with an especial emphasis in the deep subtidal. Also, efforts should be directed to investigate the genetic patterns across transition areas and biogeographic barriers such as the AOF, as well as the influence of other oceanographic fronts across the Mediterranean21. Importantly, further studies should develop and target nuclear markers for these three species since gene genealogies obtained for the mitochondrial markers, 16S and COI, can be different from the complete history of species. Additional nuclear markers could refine connectivity estimates, as well as the potential for hybridization between species, mitochondrial introgression and effective population size43,44.
Methods
Sample collection
Samples of Ophiothrix fragilis were collected between 2005 and 2012 from eight shallow and deep sites (Fig. 1A and Table 1), including four Atlantic locations [Kristineberg (KRI), Roscoff (ROS), Ferrol (FER), and Vigo collected on board the R/V Cornide de Saavedra (COR)], and four Mediterranean ones [Blanes (ABL), Barcelona-INDEMARES project (IMA), Planassa area (PLA), and Port de la Selva (PSE)]. KRI, ROS, and FER samples were collected by scuba-diving (8–10 m depth) and by hand in the intertidal; the rest of samples were collected by trawling on the R/V Cornide de Saavedra and on fishing trawlers (30 to 139 m depth). Samples of a species morphologically similar to O. fragilis and Ophiothrix sp. II (see Results section), were collected in 2013 from five deep subtidal sites (100–310 m depth; Table 1), four of them located in the Atlantic basin [by trawling during the DEMERSALES cruise (DEM45, DEM91, and DEM131) on board the R/V Miguel Oliver- ERDEM3 project, and the ones from Portugal (POR) by a fishing trawler], while the Mediterranean population (PINDAL) was collected during the INDEXARES cruise on board the ship Isla de Alborán (Fig. 1A; Table 1). Complete specimens were preserved in absolute ethanol, and stored at −20 °C until processed.
Samples of Ophiothrix sp. II analysed here for comparative purposes correspond to the collection of Pérez-Portela et al.15 plus 20 extra individuals from FER (Table 1). These samples were collected by scuba-diving or in the intertidal from 11 different shallow-water locations (Fig. 1A; Table 1), including three Atlantic locations [Ferrol (FER), Cascais (CAS), and Armação de Pera (APA)] and eight Mediterranean ones [Ceuta (CEB), La Herradura (LH), Xábea (XB), Roses (RS), Cadaqués (CAD), Alcudia (AL), Ladiko (LK), and Kalytea Bay (KB)].
Morphology and Biology
Specimens of Ophiothrix spp. used for DNA sequencing were morphologically inspected, although we do not provide here detailed morphological descriptions. Specimens of O. fragilis displayed typical characters of the species: rounded disc with diameter between 7–10.6 mm, wide and naked radial shields covering approximately half of the aboral disc area. They also had the typical slender spines in the central area of the aboral side of the disc and in the inter-radial fields, and pale colours from brown to whitish15 (supplementary Fig. S1 and Table S1). Specimens of Ophiothrix sp. II showed strikingly colour plasticity and were within the range of size described by Pérez-Portela et al.15, with disc diameter between 6.5–10 mm, small radial shields on the aboral side covering 1/3–1/4 of its surface, and partially covered by small spines and tubercles. The central and inter-radial fields of the aboral side was homogeneously covered by small spines. Specimens of Ophiothrix sp. III were in general larger than the other two species displaying a rounded disc reaching up to 18 mm in diameter, although disc diameter of specimens largely varied among sites. Their wide radial shields covered approximately half of the aboral side of the disc as in O. fragilis, but they presented tubercles and small spines on the shields as in Ophiothrix sp. II (supplementary Fig. S1 and Table S1).
The only information about the reproductive patterns of O. fragilis comes from studies on Atlantic individuals, and concluded that the reproductive dynamics of O. fragilis has four annual recruitments, the principal of which taking place between September-October, with a planktotrophic larvae with a moderately long lifespan often recruiting directly onto the discs and arms of large adults45. In fact, several young recruits were found on the disc of adult specimens of O. fragilis collected in Kristineberg (KRI) during morphological analysis of the samples. In contrast to that reported for O. fragilis, Ophiothrix sp. II has a main annual reproductive period (late spring-early summer), through a planktotrophic larva and with juveniles settling on top of sponges and adults living in small numbers between algae and under rocks46.
DNA sequencing
Total DNA was extracted from tube feet using the REDExtract-N-Amp kit (www.sigma.com) following the manufacturer’s protocol. Fragments of the 16S were amplified and sequenced using the primers designed by Pérez-Portela et al.15 for all new samples (Table 1). In addition, the mitochondrial COI was sequenced for species delimitation purposes for a subset of samples of Ophiothrix sp. III. Fragments of the COI were amplified and sequenced using the primers polyLCO/polyHCO46. PCR amplification reactions were performed in a 20-μL total reaction with 10 μL of REDEXtract-N-ampl PCR reaction mix, 0.8 μL of each primer (10 μM), 7.4 μL of ultrapure water, and 1 μL of DNA template. PCR temperature profile was 95 °C/5 min-(94 °C/1 min–46 °C/30 s–72 °C/2 min)*37 cycles–72 °C/8 min for 16S and 94 °C/5 min-(94 °C/1 min–55 °C/1 min–72 °C/1 min)*35 cycles–72 °C/5 min for COI. PCR products were purified and sequenced by Macrogen, Inc. (Seoul, Korea) with the same primers used for amplification. New sequences have been deposited in GenBank (accession numbers KX577803–KX578016).
Phylogeny and species delimitation
Due to the presence of individuals with different morphology, we first applied phylogenetic and species delimitation approaches to avoid mixing different species for further analyses.
Sequences were edited using Geneious vs. R8 and aligned with the Q-INS-I option of MAFFT. To detect different evolutionary units, 16S and COI sequences were collapsed into haplotypes using DnaSP vs. 5.10.147, and used for phylogenetic analyses, including as outgroups sequences of congeneric Ophiothrix species and other phylogenetically close ophiuroids. Outgroups varied between markers depending on the sequences available from Genbank (Fig. 2, and supplementary Figs 2S and 3S). The most appropriate evolutionary model for 16S (HKY + I) and COI (GTR + I + G) were inferred using jModelTest48 via the Akaike Information Criterion (AIC). Maximum Likelihood (ML) phylogeny trees were estimated separately for 16S and COI using PhyML as implemented in Seaview49, with 1,000 bootstrap replicates, and an optimised tree searching (Best of NNI and SPR). The resulting trees were edited in FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). Phylogenetic reconstruction was applied for the whole dataset of haplotypes of the 16S and COI alignments, but also for the most frequent haplotypes of the 16S, to simplify the interpretation of the intra-specific data.
The Poisson Tree Processes (PTP) model was applied to infer putative species boundaries among samples. This method is a powerful tool developed to identify cryptic species and to solve taxonomic ambiguities50,51. Based on the rooted ML phylogenetic trees of COI and 16S sequences the PTP model was implemented in the webserver (http://species.h-its.org)52 using 100,000 MCMC generations, thinning of 100, and a burning of 0.1. Levels of genetic divergence between and within species of Ophiothrix detected from PTP were calculated based on the uncorrected Kimura 2 parameters (K2p) model using MEGA vs. 5.0553, with deviation estimated after 1,000 bootstrap replicates. The K2p model was applied to compare with previous studies in echinoderms.
Genetic structure
We only used 16S alignments due to its better resolution for populations’ genetic structure than COI as observed in previous studies with Ophiothrix and other echinoderms3,15. An unrooted haplotype network for the different species was constructed based on a ML tree with the program Haploviewer (www.cibiv.at/~greg/haploviewer).
All 16S sequences were used to calculate number of haplotypes (H), number of private haplotypes (Hp), haplotype diversity (Hd), and nucleotide diversity (π) with DnaSP.
Differences in the genetic structure of populations were assessed by computing pairwise ФST statistics in ARLEQUIN v 3.554 between sites within each species. The corresponding p-values were evaluated by 10,000 permutations, and adjusted by a false discovery rate method55. The matrixes of the ФST values were plotted graphically in a Multidimensional Scaling analysis (MDS). The fit of genetic differentiation between sites in O. fragilis (the only species displaying divergence between sites; see Results section) to a pattern of isolation by geographical distance was tested in ARLEQUIN, using the Mantel test procedure with 16,000 permutations. Geographical distances were calculated as the minimum distance in Km between sites over the sea.
Differentiation between the Atlantic and Mediterranean basins was assessed separately for each species by conducting hierarchical analyses of molecular variance (AMOVA) using genetic distances. Their significance was tested running 16,000 permutations in ARLEQUIN. Different components of the genetic variance were quantified per each species as follows: between basins (Atlantic vs Mediterranean), among sites within basins, and within sites. Due to the existence of genetic divergence between basins in O. fragilis (see Results), a Bayesian analyses was performed to evaluate migration rates between basins in LAMARC v 2.1.956. The best evolutionary method for O. fragilis as inferred with jModelTest 2 (JC) was implemented. Three initial runs were performed to estimate the most likely priors for our dataset. The first run was performed with default parameters, and two additional runs were computed with most accurate parameters. Once accurate priors were obtained, they were implemented in a final run, with variation values of migration between 0 and 500, from the Atlantic to the Mediterranean basin, and 0 and 150 on the reverse way. The final run was based on five different replicates with 40 initial chains of 5,000 MCMC each, burning period of 500, and 5 final chains of 40,000 MCMC each with a burning period of 1,000. Two simultaneous heating searches (1 and 1.5) were performed per replicate. To visualise whether the run was long enough reaching a plateaus of probability, and to confirm the existence of at least 200 independent simulations (Effective Sample Size-ESS) for each parameter, results were summarised in Tracer v 1.5 (http://beast.bio.ed.ac.uk/Tracer). Migration rates (Mt) were expressed as the number of migrants per generation Mt = n/u, where n is the immigration rate per generation, and u the substitution rate.
Demography
To test departures from a constant population size we used different approaches for the 16S. First we estimated neutrality Rozas’ R2 statistic57 with DnaSP per population. Then the history of effective population size was assessed by: a) the classical pairwise mismatch distribution following the model of Rogers & Harpending58 in ARLEQUIN, in which populations tend to display unimodal and smoother distributions while stationary populations commonly show multimodal distributions; and b) the coalescent-based Bayesian skyline plots (BSP) using BEAUti v 2 and BEAST v 2.1.259. For BSP priors included the implementation of the substitution models defined by jModelTest (JC, K80, and HKY for O. fragilis, Ophiothrix sp. II, and Ophiothrix sp. III, respectively), a strict clock model, and the constant skyline model. As no molecular clock has been calibrated for ophiuroids, a mutation rate of 1.25% per nucleotide per million years was used for the 16S, following other studies in echinoderms60. Between 50 and 500 million MCMC generations were run per species, sampled every 5,000th–50,000th step, and a burnin of 1,000–10,000. As explained for LAMARC, we ensured that the runs were long enough and above 200 ESS by visualizing the result outputs in Tracer. Tracer was also used to generate the evolution of effective population size under the skyline plot model, expressed as NeT (T = generation time) over time. BSP analyses were additionally run for the COI sequences available for O. fragilis and Ophiothrix sp. II. It was not performed for Ophiothrix sp. III because only three sequences were available for the species. A mutation rate of 2.48% per nucleotide per million years was used for the COI, following other studies in echinoderms61. Between 50 million and one billion MCMC generations were run per species, sampled every 5,000th–100,000th step, and a burnin of 1,000–100,000 with the corresponding substitution model (GTR).
Additional Information
How to cite this article: Taboada, S. and Pérez-Portela, R. Contrasted phylogeographic patterns on mitochondrial DNA of shallow and deep brittle stars across the Atlantic-Mediterranean area. Sci. Rep. 6, 32425; doi: 10.1038/srep32425 (2016).
Supplementary Material
Acknowledgments
Thanks are due to E. Velasco, J. Cristobo, E. Manjón, J.M. Missé, N. Rodriguez, C. Palacín, O. Wangensteen, E. Mateos, J. Moles, X. Turon, D. Costalago, A. Riesgo, Estelada crew, and INDEMARES cruise crew, who kindly provided the material from O. fragilis and Ophiothrix sp. III. This research was funded by a research project from the Linnean Society of London & The Systematics Association to R.P.-P. (Systematics Research Funds 2015), the Spanish Ministry of Science (projects CTM2010-22218 and CTM2013-48163) and a “Juan de la Cierva” contract to R.P.-P. This paper is a contribution of the Consolidated Research Group 2014SRG336 of the Generalitat of Catalunya.
Footnotes
Author Contributions S.T. and R.P.-P. designed the study, collected most of the specimens and identified them, sequenced all the samples, performed the molecular analyses, and wrote the paper.
References
- Patarnello T., Volckaert F. A. & Castilho R. Pillars of Hercules: is the Atlantic–Mediterranean transition a phylogeographical break? Mol. Ecol. 16, 4426–4444 (2007). [DOI] [PubMed] [Google Scholar]
- Duran S., Giribet G. & Turon X. Phylogeographical history of the sponge Crambe crambe (Porifera, Poecilosclerida): range expansion and recent invasion of the Macaronesian islands from the Mediterranean Sea. Mol. Ecol. 13, 109–122 (2004a). [DOI] [PubMed] [Google Scholar]
- Calderón I., Giribet G. & Turon X. Two markers and one history: phylogeography of the edible common sea urchin Paracentrotus lividus in the Lusitanian region. Mar. Biol. 154, 137–151 (2008). [Google Scholar]
- Pérez-Portela R. & Turon X. Cryptic divergence and strong population structure in the colonial invertebrate Pycnoclavella communis (Ascidiacea) inferred from molecular data. Zoology, 111, 163–178 (2008). [DOI] [PubMed] [Google Scholar]
- Zulliger D. E., Tanner S., Ruch M. & Ribi G. Genetic structure of the high dispersal Atlanto-Mediterreanean sea star Astropecten aranciacus revealed by mitochondrial DNA sequences and microsatellite loci. Mar. Biol. 156, 597–610 (2009). [Google Scholar]
- Wangensteen O. S., Turon X., Pérez-Portela R. & Palacín C. Natural or naturalized? Phylogeography suggests that the abundant sea urchin Arbacia lixula is a recent colonizer of the Mediterranean. PLoS One 7, e45067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Merchán V. H. et al. Phylogeographic patterns of decapod crustaceans at the Atlantic–Mediterranean transition. Mol. Phylogenet. Evol. 62, 664–672 (2012). [DOI] [PubMed] [Google Scholar]
- Fernández R., Lemer S., McIntyre E. & Giribet G. Comparative phylogeography and population genetic structure of three widespread mollusc species in the Mediterranean and near Atlantic. Mar. Ecol. 36, 701–715 (2015). [Google Scholar]
- De Luca D., Catanese G., Procaccini G. & Fiorito G. Octopus vulgaris (Cuvier, 1797) in the Mediterranean Sea: Genetic Diversity and Population Structure. PLoS One 11, e0149496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggs C. A. et al. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89, S108–S122 (2008). [DOI] [PubMed] [Google Scholar]
- Duran S., Palacín C., Becerro M. A., Turon X. & Giribet G. Genetic diversity and population structure of the commercially harvested sea urchin Paracentrotus lividus (Echinodermata, Echinoidea). Mol. Ecol. 13, 3317–3328 (2004b). [DOI] [PubMed] [Google Scholar]
- Triantafyllidis A. et al. Mitochondrial DNA variation in the European lobster (Homarus gammarus) throughout the range. Mar. Biol. 146, 223–235 (2005). [Google Scholar]
- Pérez-Losada M., Nolte M. J., Crandall K. A. & Shaw P. W. Testing hypotheses of population structuring in the Northeast Atlantic Ocean and Mediterranean Sea using the common cuttlefish Sepia officinalis. Mol. Ecol. 16, 2667–2679 (2007). [DOI] [PubMed] [Google Scholar]
- Pérez-Portela R., Villamor A. & Almada V. Phylogeography of the sea star Marthasterias glacialis (Asteroidea, Echinodermata): deep genetic divergence between mitochondrial lineages in the North-Western Mediterranean. Mar. Biol. 157, 2015–2028 (2010). [Google Scholar]
- Pérez‐Portela R., Almada V. & Turon X. Cryptic speciation and genetic structure of widely distributed brittle stars (Ophiuroidea) in Europe. Zool. Scr. 42, 151–169 (2013). [Google Scholar]
- Borrero-Pérez G. H., González-Wangüemert M., Marcos C. & Pérez-Ruzafa A. Phylogeography of the Atlanto‐Mediterranean sea cucumber Holothuria (Holothuria) mammata: the combined effects of historical processes and current oceanographical pattern. Mol. Ecol. 20, 1964–1975 (2011). [DOI] [PubMed] [Google Scholar]
- Avise J. C. et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 489–522 (1987). [Google Scholar]
- Siegel D. A. et al. The stochastic nature of larval connectivity among nearshore marine populations. PNAS 105, 8974–8979 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Geziry T. M. & Bryden I. G. The circulation pattern in the Mediterranean Sea: issues for modeller consideration. J. Oper. Oceanogr. 3, 39–46 (2010). [Google Scholar]
- Stamatis C., Triantafyllidis A., Moutou K. A. & Mamuris Z. Mitochondrial DNA variation in Northeast Atlantic and Mediterranean populations of Norway lobster. Nephrops norvegicus. Mol. Ecol. 13, 1377–1390 (2004). [DOI] [PubMed] [Google Scholar]
- Villamor A., Costantini F. & Abbiati M. Genetic structuring across marine biogeographic boundaries in rocky shore invertebrates. PLoS One 9, e101135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoult D. Structure démographique et production de la population d’Ophiothrix fragilis (Abildgaard) du détroit du Pas de Calais (France). Vie Marine 10, 116–127 (1989). [Google Scholar]
- Muths D., Jollivet D., Gentil F. & Davoult D. Large-scale genetic patchiness among NE Atlantic populations of the brittle star Ophiothrix fragilis. Aquat. Biol. 5, 117–132 (2009). [Google Scholar]
- Hunter R. L. & Halanych K. M. Phylogeography of the Antarctic planktotrophic brittle star Ophionotus victoriae reveals genetic structure inconsistent with early life history. Mar. Biol. 157, 1693–1704 (2010). [Google Scholar]
- Boissin E., Feral J. P. & Chenuil A. Defining reproductively isolated units in a cryptic and syntopic species complex using mitochondrial and nuclear markers: the brooding brittle star, Amphipholis squamata (Ophiuroidea). Mol. Ecol. 17, 1732–1744 (2008). [DOI] [PubMed] [Google Scholar]
- Boissin E., Stöhr S. & Chenuil A. Did vicariance and adaptation drive cryptic speciation and evolution of brooding in Ophioderma longicauda (Echinodermata: Ophiuroidea), a common Atlanto‐Mediterranean ophiuroid? Mol. Ecol. 20, 4737–4755 (2011). [DOI] [PubMed] [Google Scholar]
- Muths D., Davoult D., Gentil F. & Jollivet D. Incomplete cryptic speciation between intertidal and subtidal morphs of Acrocnida brachiata (Echinodermata: Ophiuroidea) in the Northeast Atlantic. Mol. Ecol. 15, 3303–3318 (2006). [DOI] [PubMed] [Google Scholar]
- Muths D., Davoult D., Jolly M. T., Gentil F. & Jollivet D. Pre-zygotic factors best explain reproductive isolation between the hybridizing species of brittle-stars Acrocnida brachiata and A. spatulispina (Echinodermata: Ophiuroidea). Genetica 138, 667–679 (2010). [DOI] [PubMed] [Google Scholar]
- Richards V. P., DeBiasse M. B. & Shivji M. S. Genetic evidence supports larval retention in the Western Caribbean for an invertebrate with high dispersal capability (Ophiothrix suensonii: Echinodermata, Ophiuroidea). Coral Reefs 34, 313–325 (2015). [Google Scholar]
- Sands C. J., O’Hara T., Barnes D. K. & Martín-Ledo R. Against the flow: evidence of multiple recent invasions of warmer continental shelf waters by a Southern Ocean brittle star. Frontiers in Ecology and Evolution 3 (2015). [Google Scholar]
- Landry C., Geyer L. B., Arakaki Y., Uehara T. & Palumbi S. R. Recent speciation in the Indo-West Pacific: rapid evolution of gamete recognition and sperm morphology in cryptic species of the sea urchin. P. Roy. Soc. London 270, 1839–1847 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios H. A. & Cunningham C. W. Gametic incompatibility between species of the sea urchin Echinometra on the two sides of the Isthmus of Panama. Evolution 933–941 (1990). [DOI] [PubMed] [Google Scholar]
- Hart M. W., Sunday J. M., Popovic I., Learning K. J. & Konrad C. M. Incipient speciation of sea star populations by adaptive gamete recognition coevolution. Evolution 68, 1294–1305 (2014). [DOI] [PubMed] [Google Scholar]
- Dailianis T., Tsigenopoulos C. S., Dounas C. & Voultsiadou E. Genetic diversity of the imperilled bath sponge Spongia officinalis Linnaeus, 1759 across the Mediterranean Sea: patterns of population differentiation and implications for taxonomy and conservation. Mol. Ecol. 20, 3757–3772 (2011). [DOI] [PubMed] [Google Scholar]
- Allendorf F. W., Luikart G. & Aitken S. N. Conservation and the genetics of populations John Wiley & Sons, Chichester, UK (2012). [Google Scholar]
- Pérez‐Portela R., Noyer C. & Becerro M. A. Genetic structure and diversity of the endangered bath sponge Spongia lamella. Aquat. Conser 25, 365–379 (2014). [Google Scholar]
- Pellerito R., Arculeo M. & Bonhomme F. Recent expansion of Northeast Atlantic and Mediterranean populations of Melicertus (Penaeus) kerathurus (Crustacea: Decapoda). Fisheries Sci. 75, 1089–1095 (2009). [Google Scholar]
- Svendsen J. I. et al. Late Quaternary ice sheet history of northern Eurasia. Quaternary Sci. Rev. 23, 1229–1271 (2004). [Google Scholar]
- Candela J. The Gibraltar Strait and its role in the dynamics of the Mediterranean Sea. Dynam. Atmos. Oceans 15, 267–299 (1991). [Google Scholar]
- Villalobos F. B., Tyler P. A. & Young C. M. Temperature and pressure tolerance of embryos and larvae of the Atlantic seastars Asterias rubens and Marthasterias glacialis (Echinodermata: Asteroidea): potential for deep-sea invasion. Mar. Ecol. Prog. Ser. 314, 109–117 (2006). [Google Scholar]
- Brown A. & Thatje S. Explaining bathymetric diversity patterns in marine benthic invertebrates and demersal fishes: physiological contributions to adaptation of life at depth. Biol. Rev. 89, 406–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G., Lima F. P., Martel P. & Castilho R. Thermal adaptation and clinal mitochondrial DNA variation of European anchovy. P. Roy. Soc. B- Biol. Sci. 281, 20141093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard J. W. O. & Whitlock M. C. The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744 (2004). [DOI] [PubMed] [Google Scholar]
- Davoult D., Gounin F. & Richard A. Dynamique et reproduction de la population d’Ophiothrix fragilis (Abildgaard) du de’troit du Pas de Calais (Manche Orientale). J. Exp. Mar. Biol. Ecol. 138, 201–216 (1990). [Google Scholar]
- Turon X., Codina M., Tarjuelo I., Uriz M. J. & Becerro M. A. Mass recruitment of Ophiothrix fragilis (Ophiuroidea) on sponges: settlement patterns and post-settlement dynamics. Mar. Ecol. Prog. Ser. 200, 201–212 (2000). [Google Scholar]
- Carr C., Hardy S., Brown T., Macdonald T. & Hebert P. N. A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in Canadian polychaetes. PLoS One 6, e22232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Gouy M., Guindon S. & Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010). [DOI] [PubMed] [Google Scholar]
- Romano C., Voight J. R., Pérez-Portela R. & Martin D. Morphological and genetic diversity of the wood-boring Xylophaga (Mollusca, Bivalvia): new species and records from deep-sea Iberian canyons. PLoS One 9, e102887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas P. et al. Phylogenetic molecular species delimitations unravel potential new species in the pest genus Spodoptera Guenée, 1852 (Lepidoptera, Noctuidae). PLoS One 10, e0122407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kapli P., Pavlidis P. & Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L. & Lischer H. E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 1165–1188 (2001). [Google Scholar]
- Kuhner M. K. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22, 768–770 (2006). [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins S. E. & Rozas J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19, 2092–2100 (2002). [DOI] [PubMed] [Google Scholar]
- Rogers A. R. & Harpending H. Population-growth makes waves in the distribution of pairwise genetic-differences. Mol. Biol. Evol. 9, 552–569 (1992). [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Rambaut A., Shapiro B. & Pybus O. G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005). [DOI] [PubMed] [Google Scholar]
- Boissin E., Egea E., Féral J. P. & Chenuil A. Contrasting population genetic structures in Amphipholis squamata, a complex of brooding, self-reproducing sister species sharing life history traits. Mar. Ecol. Prog. Ser. 539, 165 (2015). [Google Scholar]
- Naughton K. M., O’Hara T. D., Appleton B. & Cisternas P. A. Antitropical distributions and species delimitation in a group of ophiocomid brittle stars (Echinodermata: Ophiuroidea: Ophiocomidae). Mol. Phylogenet. Evol. 78, 232–244 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.