Abstract
The modulation of chromatin structure is a key step in transcription regulation in mammalian cells and eventually determines lineage commitment and differentiation. USF1/2, Setd1a and NURF complexes interact to regulate chromatin architecture in erythropoiesis, but the mechanistic basis for this regulation is hitherto unknown. Here we showed that Setd1a and NURF complexes bind to promoters to control chromatin structural alterations and gene activation in a cell context dependent manner. In human primary erythroid cells USF1/2, H3K4me3 and the NURF complex were significantly co-enriched at transcription start sites of erythroid genes, and their binding was associated with promoter/enhancer accessibility that resulted from nucleosome repositioning. Mice deficient for Setd1a, an H3K4 trimethylase, in the erythroid compartment exhibited reduced Ter119/CD71 positive erythroblasts, peripheral blood RBCs and hemoglobin levels. Loss of Setd1a led to a reduction of promoter-associated H3K4 methylation, inhibition of gene transcription and blockade of erythroid differentiation. This was associated with alterations in NURF complex occupancy at erythroid gene promoters and reduced chromatin accessibility. Setd1a deficiency caused decreased associations between enhancer and promoter looped interactions as well as reduced expression of erythroid genes such as the adult β-globin gene. These data indicate that Setd1a and NURF complexes are specifically targeted to and coordinately regulate erythroid promoter chromatin dynamics during erythroid lineage differentiation.
INTRODUCTION
Lineage differentiation is a highly regulated process involving commitment and differentiation of stem cells or progenitors into mature differentiated lineages such as red blood cells (RBCs) (1,2). During erythropoiesis, erythroid transcription programs are coordinated by erythroid-specific and ubiquitous transcription factors (TFs), chromatin insulators and histone modifying and remodeling factors. These activities cause dramatic changes in chromatin structure and gene expression patterns during erythropoiesis. Perturbation of these activities may lead to various forms of anemia (3,4). Recent global transcriptome analyses revealed that each erythroid differentiation stage exhibits unique transcription profiles that are temporally regulated (5). It remains unclear how gene expression patterns are switched from stem cell specific expression to lineage restricted expression and whether epigenetic modifier mediated chromatin dynamics underlies differentiation dependent transcription changes. During terminal erythroid differentiation, upstream stimulatory factor 1 (USF1) protects erythroid genes from encroachment of heterochromatin by interacting with SET domain containing 1A (Setd1a) and nucleosome remodeling factor (NURF) complexes (6). However, how these complexes are targeted to lineage specific genes and cooperate to regulate erythroid-specific chromatin structure and gene expression remains poorly understood.
USF1 and USF2 are ubiquitously expressed TFs that form heterodimers to bind to E-box elements (CANNTG). USF and associated cofactors act within domains that are rendered accessible by tissue-specific TFs during differentiation, providing tissue-specific activities. In erythroid cells, USFs are critical for the expression of erythroid-specific genes, including those encoding erythroid TFs and the globin genes. During erythropoiesis, GATA binding factor 1/globin transcription factor 1 (GATA1) and Krupple-like factor 1 (KLF1) have been implicated in establishing accessible chromatin domains. USFs may act within these accessible domains to remodel nucleosomes for recruiting transcription preinitiation complexes (PIC) to erythroid promoters.
In mammalian cells, conserved SET domain-containing SET1/MLL histone methyltransferase (HMT) complexes specifically methylate H3K4 (7). SET1/MLL complexes contain shared subunits, WDR5 (WD repeat domain 5), RBBP5 (Retinoblastoma-binding protein 5), ASH2L ((Absent, small or homeotic)-like) and HCF1 (host cell factor 1) that are required for enzymatic activity (8,9). In addition to the shared subunits, the complexes contain distinct enzymatic subunits (MLL1-4, SETD1A or SETD1B). MLL1 (mixed lineage leukemia 1) is required for definitive hematopoiesis (10), and the loss of Mll1 reduces H3K4 methylation at the HoxC locus (11). In contrast, MLL3/4 has been linked to adipogenesis (12).
Ablation of SETD1A, but not MLLs, has the most dramatic effect on global H3K4 trimethylation (H3K4me3) and gene expression (13,14). H3K4me3, one of the most studied histone marks, is highly enriched at transcription start sites (TSS) of active genes and controls gene transcription (15–18). Setd1a knock-out (KO) mice die at an early embryonic stage (19), while hematopoietic-specific Setd1a KO blocks progenitor-B to precursor-B cell development by inhibiting H3K4me3 levels and the immunoglobulin heavy chain (IgH) rearrangement (20). Thus, each SET1/MLL complex appears to have distinct context-dependent function in different tissues or cell lineages.
Mammalian genomes encode two ISWI genes, SNF2H (SNF2 homolog) and SNF2L (SNF2-like). SNF2H catalyzes the formation of regularly ordered nucleosome arrays and facilitates heterochromatin formation. In contrast, the SNF2L-containing NURF complex is involved in transcriptional activation by creating nucleosome-free regions at promoters via sliding nucleosomes on DNA templates (21). Mechanistically, nucleosome sliding could be directly coupled to H3K4me3 because NURF recognizes the H3K4me3 marks (22). NURF is required for USF1-mediated barrier insulator activity to protect erythroid genes from silencing (6). Interestingly, recent genome-wide studies indicated that USF1 predominantly occupied proximal promoters in human erythroleukemia K562 cells (23). This raises the possibility that USF1 may collaborate with Setd1a and NURF complexes to create a promoter-accessible region allowing recruitment of PIC.
To understand how USF1, SETD1A and NURF interact to regulate erythroid chromatin dynamics and gene transcription, we performed ChIP (chromatin immunoprecipitation)-seq in human primary erythroid cells and compared these results to MNase (micrococcal nuclease)-seq data (24). USF1, BPTF, a component of the NURF complex and SETD1A co-occupied the promoters of active erythroid genes and this correlated with changes in promoter-H3K4me3 levels and promoter accessibility. Depletion of Setd1a led to reduced transcription of erythroid genes accompanied by decreases in H3K4me3 levels, NURF occupancy and chromatin accessibility at erythroid promoters. Finally, erythroid-specific Setd1a KO mice revealed a reduced number of splenic CD71+/Ter119+ erythroblasts, reduced peripheral blood RBC counts and hemoglobin (HGB) levels. Together, these data revealed a collaborative role of Setd1a HMT complexes and NURF chromatin-remodeling complexes on erythroid-specific chromatin structure alterations at lineage specific promoters during differentiation.
MATERIALS AND METHODS
Constructs, tissue culture and cell transfections
Setd1a shRNAs were cloned into the pSuper.retro.puro vector (Oligoengine) or pTRIZP lentiviral inducible vector (Open Biosystems) following the manufacturer's instructions. Infectious viruses were produced in packaged cells and used to infect embryonic stem cells (ESCs). The infected cells were selected with 1 μg/ml puromycin as described previously (14). Primary human CD34+ hematopoietic stem cells (HSCs) were isolated and differentiated to CD36+ or R3/R4 erythroblast cells, which were purified as described (6,25). Briefly, human peripheral/core blood derived CD34+ cells were enriched through positive immune selection by flow cytometry. The CD34-selected stem and progenitor cells were cultured in StemSpan SF expansion media (StemSpan, 09650) with estradiol (100 ng/ml), dexamethasone (10 ng/ml), human transferrin (200 ng/ml), insulin (10 ng/ml), Flt3 ligand (100 ng/ml), stem cell factor (100 ng/ml), IL3 (50 ng/ml), IL6 (20 ng/ml), insulin like growth factor-1 (50 ng/ml) and erythropoietin (EPO) (3 U/ml) for 9–14 days. During the course of erythroid differentiation, CD36+ erythroid progenitors could be purified by flow cytometry. The CD36+ cells continue to differentiate and were used to isolate cells expressing CD71 (transferrin receptor) and CD235a (glycophorin A), defined as the R3/R4 population of differentiating erythroid cells (6). Murine erythroleukemia (MEL) cells were cultured in Dulbecco's modified Eagle's medium media supplemented with 10% fetal bovine serum (FBS) and induced to differentiate with 1.8% Dimethyl Sulphoxide (DMSO) for 5 days. In vitro ES cell–derived erythroid embryoid body (EB) differentiation was performed as described (26) with modifications (Supplementary Figure S3A). To induce EB formation, 5 × 105 R1E ES cells/ml were plated into non-treated 10 cm petri dishes containing ES cell culture medium without leukemia inhibitory factor (LIF). At day 4 of differentiation, EBs were harvested and replated into fresh EB differentiation medium (IMDM, 1% penicillin/streptomycin, 15% FBS) plus 20 ng/ml mouse SCF (Peprotech). At day 7, EBs were replated into fresh EB differentiation medium plus 20 ng/ml mouse SCF, 10 ng/ml mouse IL-3, 10 ng/ml mouse IL-6 and 2 U/ml EPO. At day 10, EBs were replated into fresh EB differentiation medium plus 10 ng/ml mouse IL-3, 10 ng/ml mouse IL-6, 2 U/ml EPO and extra 5 µg/ml doxycycline for inducible Setd1a KD R1E ES cells. From day 13 on, EBs were replated every 3 days into fresh EB differentiation medium only containing 2 U/ml EPO and 5 µg/ml doxycycline. At day 19 of differentiation, EBs were harvested and appeared pink in color.
Purification of USF1 complexes and co-immunoprecipitation
USF1 containing protein complexes were isolated from HeLaS3 cells stably transduced with a FLAG-Hemagglutinin (HA)-tagged USF1. Complexes were first purified with M2-anti-FLAG resin (Sigma) by incubating 100 μl packed resin with nuclear extracts for 4 h at 4°C, which were then washed three times with wash buffer (20 mM Tris, pH7.9, 0.1M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 1 mM DTT, protease inhibitors) in a disposable column followed by two elutions with 100 μl each of Wash buffer plus 0.16 μg/ml FLAG peptide. The eluted complexes were resolved in sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by western blot analysis. For co-immunoprecipitation assays, nuclear extracts were incubated with antibody immobilized on protein A beads, which were then washed briefly four times with RIPA buffer followed by a wash with phosphate buffered saline (PBS) for 10 min at 4°C with agitation. Immunoprecipitated proteins were fractionated by SDS-PAGE and electrotransferred to polyvinylidene difluoride membranes. Blots were incubated with the indicated primary antibodies, and then with HRP-conjugated secondary antibodies. Proteins were visualized by enhanced chemiluminescence (Pierce, Rockford, IL, USA).
Quantitative polymerase chain reaction (qPCR) and qRT-PCR
Total RNA was prepared using the RNeasy mini-isolation kit according to manufacturer's instructions (Qiagen, MD, USA). A total of 2 μg RNA was subjected to reverse-transcription with Superscript II reverse Transcriptase (Invitrogen) and analyzed by a CFX 96 real-time polymerase chain reaction (PCR) Detection System (Bio-Rad).
ChIP and sequential ChIP
ChIP, ChIP-Seq and sequential-ChIP assays were performed as described previously (6,14). Briefly, Nuclei were sonicated with the Bioruprtor™ UCD200. Chromatin samples prepared from 1 × 106 cells of Day 19 EBs or bone marrow (BM) cells were immunoprecipitated with antibodies specific for histone modifications or epigenetic regulators. The immunoprecipitated chromatin complexes were reverse cross-linked and purified. The recovered DNA was analyzed by qPCR. The relative enrichment was determined by the following equation: 2Ct(IP)-Ct(ref). The sequential ChIP assays were carried out essentially as described (14,27) with some modifications. Briefly, chromatin prepared from 5 × 106 cells was immunoprecipitated with antibodies specific for H3K4me3 or USF1. The H3K4me3 or USF1-selected chromatin complexes were eluted, dialyzed and subsequently immunoprecipitated with antibody specific to BPTF. The bound protein–DNA complexes were reverse cross-linked and purified. The recovered DNA was analyzed by qPCR.
Antibodies against H3K4me2 (07-730), H3K4me3 (04-745), SNF2L (05-698) and BPTF (ABE24) were purchased from Millipore, H3K4me1 (ab8895) was from Abcam, ASH2L (A300-489A) and Setd1a (A300-289A) were from Bethyl laboratories and USF1 (H-86) was from Santa Cruz Biotechnology.
ChIP-seq and MNase-seq
After chromatin immunoprecipitation, The ChIP DNA was ligated to Illumina ChIP-Seq adaptors, amplified using the Illumina primers and sequenced on the Illumina GAII platform. Sequence reads of 25-bp were obtained and mapped to the human genome (UCSC Genome Browser hg18). For BPTF, SETD1A and H3K4me3, the reads per sample were ranged from 28 to 34 million reads. The USF1 average reads were around 10.3 million and USF2 average reads were 8.3 million reads. The Model-based Alignment of ChIP-seq (MACS) program was used to identify and call peaks with a default cutoff P-value of <10e-5 (28). The USF1 motif was predicted by MEME SUITE program (29) and the annotation of protein binding sites was done by using CEAS (30) and HOMER 4.7 (31) with default settings. The overlapping of USF1 and USF2 ChIP-Seq peaks was analyzed by HOMER 4.7. The association of USF1, USF2, BPTF and H3K4me3 with global gene expression (GEO accession number: GSE53983) was performed by ‘ChIPpeakAnno’ R package (32). The ChIP-seq data were visualized by UCSC genome browser (33) and IGV browser (34,35).
For MNase-seq, CD34+ and CD36+ cells were treated with MNase to generate approximately 80% mononucleosomes and 20% dinucleosomes. The DNA fragments of ∼150 bp were isolated from agarose gels, blunt-ended, ligated to Solexa adaptors and sequenced. Nucleosome tag-enriched regions were identified by the NPS Python software package (36). Nucleosome profiles were obtained by applying a simple scoring function to the sequenced reads. A sliding window of 10 bp was applied across all chromosomes and at each window all reads mapping to the sense strand 80 bp upstream of the window and reads mapping to the antisense strand 80 bp downstream of the window contributed equally to the score of the window. The program identified 1.7, 2.2 and 2.3 million nucleosome tag-enriched regions with a length varying from 80 to 250 bp for HSCs (P < 0.05) and CD36+ cells (P < 0.01), To separate TF sites based on whether or not the sites are occupied by positioned nucleosomes, we determined the most likely nucleosome position in a nucleosome-enriched region j as pj = sum of pji*nji/sum of nji, where nji is the number of tags at position pji in region j. The binding sites were occupied by nucleosomes if the distance to the nearest nucleosome is <75 bp. The normalized nucleosome positioning around USF1 ChIP-seq proximal peaks was analyzed by HOMER 4.7 (31).
Formaldehyde-assisted isolation of regulatory elements (FAIRE)
Formaldehyde-assisted isolation of regulatory elements (FAIRE) assays were performed as described (37,38). Briefly, 1 × 107 cells were cross-linked with 1% formaldehyde for 10 min, and then cell lysate was sonicated to achieve an average DNA fragment size of ∼200–400 bp. FAIRE DNA was extracted from the upper aqueous phase by phenol/chloroform/isoamyl alcohol followed by chloroform and purified. Quantitative PCR was used to detect FAIRE enrichment.
Chromosome conformation capture (3C) assay
The chromosome conformation capture (3C) assay was performed as described previously with minor modifications (39). In brief, 2 × 107 cells were cross-linked with 2% formaldehyde for 10 min and stopped by the addition of glycine at a final concentration of 0.125M. Cells were pelleted and washed twice with cold PBS. Cells were collected and washed with appropriate 1× restriction buffer and then resuspended in 1× restriction enzyme buffer containing 0.3% SDS at 37° with overnight shaking. Triton X-100 was then added to a final concentration of 2% to sequester SDS at 37°C for 1.5 h with shaking. Chromatin was than digested with 800U of BglII at 37°C overnight with shaking. Next day the reaction was stopped by adding SDS to a final concentration of 1.6% at 65°C for 20 min. The digested chromatin was then diluted in 1 ml of T4 DNA ligation buffer (NEB) containing 1% Triton X-100 and incubated at 37°C for 1.5 h with shaking. A total of 400 U of T4 DNA ligase (NEB) was added and the ligation was carried out at 16°C for 3 days followed by 1 h at room temperature. The ligated chromatin was then reverse crosslinked overnight by adding 200 μg of Proteinase K (Invitrogen) at 65°C followed by phenol chloroform extraction to purify 3C DNAs. Purified 3C ligated DNA was amplified using PCR and the products were cloned into pCR-TOPOII vector (Invitrogen) for sequencing. Relative crosslinking frequencies were calculated and plotted after normalization to the loading control (40).
Conditional knockout mice and peripheral blood profile analysis
Setd1afl/fl mice (20) were intercrossed with ErGFPcre transgenic mice (41) to generate erythroid-specific ErGFPcre:Setd1afl/fl mice. Aliquots of 106 spleen cells were stained with a combination of labeled antibodies on ice in PBS plus 4% FBS for 30 min and analyzed using a LSR-II flow cytometer (BD Biosciences). Blood samples were collected in EDTA-containing Microtainer tubes (BD Biosciences) and analyzed on a HemaTrue analyzer (HESKA, Inc). Animal protocols were approved by the University of Florida Institutional Animal Care and Use Committee.
RESULTS
Loss of Setd1a impaired erythropoiesis in a Setd1a knockout mouse model
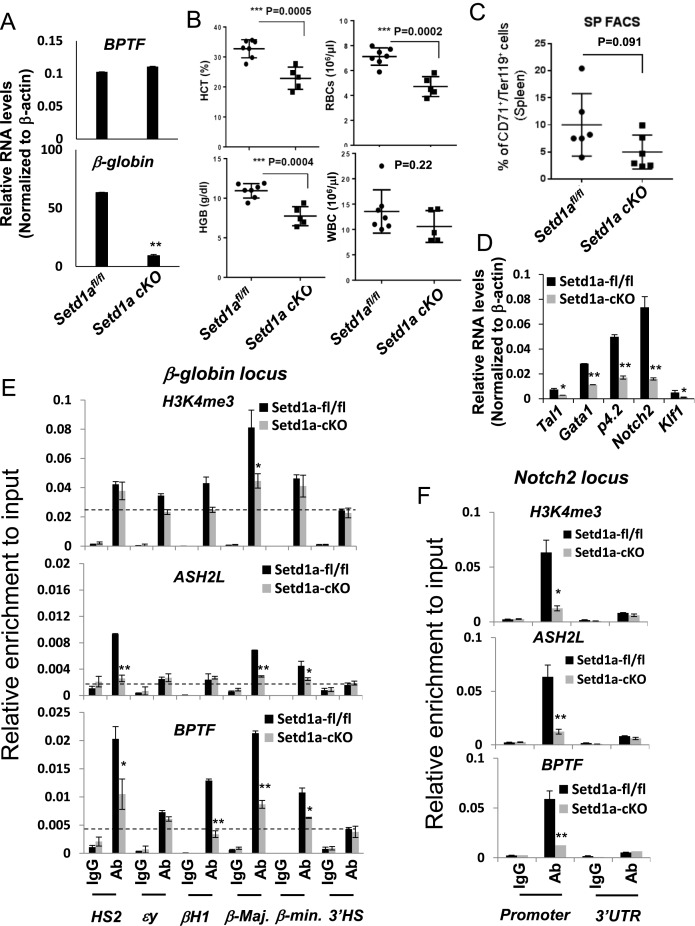
We recently reported that knockout of Setd1a in murine BM resulted in splenomegaly with blocked lymphopoiesis and perturbed myelopoiesis and erythropoiesis (20). To assess the role of Setd1a in erythropoiesis, we crossed the Setd1afl/fl mice (20) with the ErGFPcre knock-in mice in which green fluorescent protein (GFP)/cre fusion protein is controlled by the EPO receptor promoter and cre-mediated recombination is limited to erythroid progenitors in the BM (41). We generated nine ErGFPcre:Setd1afl/fl (CKO) mice that showed a significant decrease in expression of β-globin, but not the bromodomain PHD finger TF (bptf), in BM cells (Figure 1A). The mice were viable and appeared normal. Biochemical analysis of peripheral blood of these mice revealed that the hematocrit (HCT) value was significantly decreased in Setd1a-CKO mice compared to the Setd1afl/fl mice in which mice were obtained from the same generation of the three months old pups with matched gender and age (Figure 1B). HGB levels and RBC counts were also significantly reduced in the Setd1a-CKO mice (Figure 1B). In contrast, there was no difference in white blood cell counts comparing Setd1a-CKO and Setd1afl/fl control mice (Figure 1B).
Figure 1.
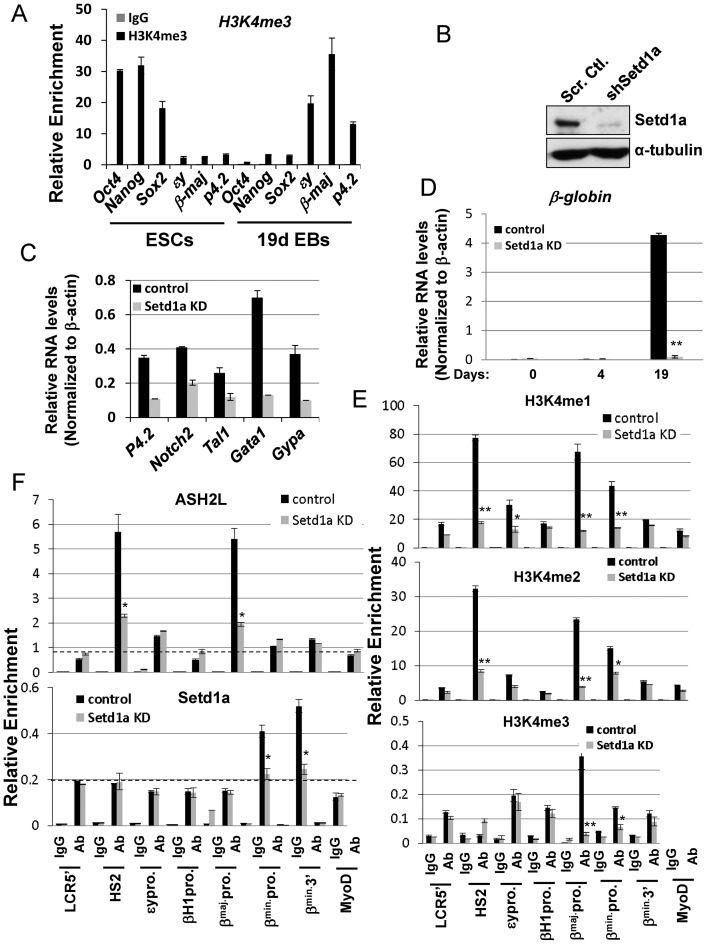
Loss of Setd1a in the erythroid compartment impaired erythropoiesis (A) qRT-PCR analysis of bptf and β-globin expression comparing two represented pairs of the Setd1afl/fl and Setd1a CKO mice. qRT-PCR analysis for each pair has been repeated three times. Data are shown as mean ± SD. *P < 0.05; **P < 0.01 by student t-test. (B) Peripheral blood analysis of hematocrit (HCT), red blood cells (RBCs), white blood cells (WBCs) and hemoglobin (HGB) levels in the seven controls and the five Setd1a KO mice. The unpaired t-test was performed to determine the statistical significance. *P < 0.05; **P < 0.01; ***P < 0.001. (C) FACS analysis of CD71 and Ter119 positive erythroblast in spleen comparing six pairs of the Setd1afl/fl and Setd1a KO mice in which two male mice and four female mice at three months of age were used. The unpaired t-test was performed to determine the statistical significance. *P < 0.05; **P < 0.01; ***P < 0.001. (D) qRT-PCR analysis of genes important for erythroid differentiation comparing two represented pairs of the Setd1afl/fl and Setd1a CKO mice. qRT-PCR analysis for each pair has been repeated three times. Data are shown as mean ± SD. *P < 0.05; **P < 0.01 by student t-test. (E and F) ChIP analysis of H3K4me3, ASH2L occupancy and BPTF recruitment at β-globin (E) and Notch2 (F) loci in one represented pair of the Setd1afl/fl and Setd1a CKO BM cells with three technical repeats. Data are shown as mean ± SD. *P < 0.05; **P < 0.01 by student t-test.
Next, we examined if splenic erythroid progenitors are impaired by the Setd1a-CKO. Splenic cells were analyzed for CD71high/Ter119high expression using FACS assays comparing the Setd1afl/fl and Setd1a-CKO mice. In both control and mutant groups, we used matched age and gender mice in which two male mice and four female mice at three months of age were used. CD71 is a transferrin receptor and Ter119 encodes a protein associated with glycophorin A (GYPA) and specifically marks the late stages of murine erythroid lineage. Both are surface markers of erythroid precursors. Activation of both markers indicates differentiation of proerythroblasts into more matured erythroblasts just prior to enucleation (42). Setd1a-CKO led to a moderate, but consistent decrease in numbers of CD71high/Ter119high erythroblasts (Figure 1C). Consistent with a defective erythroid phenotype, genes critical for the erythroid transcription program such as Tal1 (T-cell acute lymphoblastic leukemia 1), Gata1, Notch2, p4.2 (erythrocyte membrane protein band 4.2) and Klf1 were affected by the Setd1a KO in the erythroid compartment (Figure 1D). Setd1a is a major HMT required for promoter H3K4 trimethylation (43,44). We next examined whether promoter-associated Setd1a KO impairs recruitment of the Setd1a components and H3K4 trimethylation at promoters in total BM of the Setd1a KO mice. Setd1a KO disrupted recruitment of ASH2L and perturbed H3K4me3 levels at promoters of erythroid-specific genes such as β-globin, p4.2 and Notch2 (Figure 1E and F; Supplementary Figure S1A and B). Interestingly, promoter recruitment of BPTF, a core component of the NURF complex, was also impaired by the Setd1a-CKO, but its expression was not (Figure 1A, E and F; Supplementary Figure S1C). Thus, Setd1a may play an important role in regulating erythropoiesis, perhaps through its H3K4 methyltransferase activity. It is possible that the decrease in the recruitment of chromatin modifying complexes and levels of H3K4me3 at erythroid promoters was in part the result of a decreased number of erythroblasts in the Setd1a KO BM. Nevertheless, the data reveal an important role of Setd1a in erythroid cell differentiation.
USF1 positions SETD1A and NURF complexes at TSSs of active erythroid genes
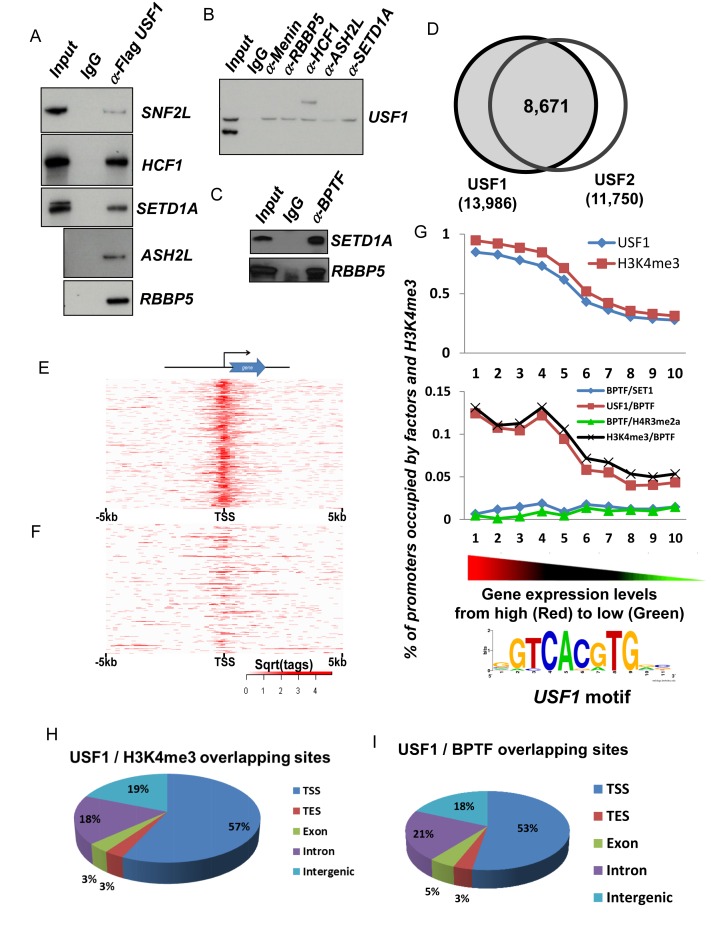
SETD1A is the enzymatic component of the trithorax (TrxG) H3K4 HMT complex and it is ubiquitously required for promoter activity (45). In K562 cells, USF1 associates with enzymatically active SETD1A and NURF complexes (6) and both complexes are critical for chromatin structure alterations at proximal promoters of active genes (16). To test if the SETD1A HMT complex physically associates with the NURF chromatin remodeling complex, we purified USF1 associated protein complexes in Flag-tagged USF1 expressing HeLa cells. USF1 associated with SETD1A complex and with SNF2L, an enzymatic subunit of the NURF complex (Figure 2A). Furthermore, all components of the SETD1A complex co-immunopricipitated with USF1 in nuclear extract (Figure 2B). Importantly, a BPTF antibody specifically precipitated the SETD1A complex (Figure 2C) indicating that USF1, SETD1A and NURF complexes physically interact with each other.
Figure 2.
Correlation of USF1, H3K4me3 and NURF complex genome wide occupancy and gene expression patterns of erythroid genes (A and B) western blot analysis of reciprocal co-immunoprecipitation performed in Hela NEs with antibodies against flag-tagged USF1 (A) or a component of the Setd1a complex (B). (C) Western blot analysis of Setd1a and RBBP5 protein immunoprecipitated by BPTF antibody, a specific component of the NURF complex. (D) Global colocalization of USF1 and USF2 at proximal promoters of ChIP-seq identified genes in erythroblast cells. (E and F) Heat map showing USF1 localization at top 1000 expressed erythroid genes (E) and bottom 1000 silenced genes (F). (G) Colocalizion of USF1, H3K4me3 and NURF (BPTF) complexes at gene promoters correlates positively with gene expression levels in erythroid cells. The USF1 motif was predicted by MEME analysis suite using the top 1000 USF1 binding peaks (according to the intensity of binding profiles). (H) Global colocalization of USF1 and H3K4me3 in erythroid cells. TSS, transcription start site; TES, transcription termination site. (I) Global colocalization of USF1 and NURF complexes in erythroid cells. For ChIP-seq assays, average reads per sample for BPTF, H3K4me3, SETD1A, USF1 and USF2 were 28.04, 28.38, 33.75, 10.38 and 8.33 million reads. The ChIP-seq peaks were identified using MACs with a (default) cutoff of 1.00e-05.
Next, we thought to determine how SETD1A regulates erythroid gene expression during erythropoiesis. The presence of SETD1A and NURF in the USF1-associated complexes suggests that their action in erythroid cells could be mediated by TF USF1, which plays an important role in erythropoiesis (6,46). Ablation of SETD1A blocked differentiation of hematopoietic stem and progenitor cells (HSPCs) into CFU-E (colony forming unit-erythroid lineage) and BFU-E (erythroid burst-forming unite) but not into CFU-GEMM (CFU-granulocyte, erythrocyte, monocyte, and megakaryocyte) and CFU-GM (CFU-granulocyte and monocyte) colonies (47). To examine the relationship of USF, SETD1A and NURF, human CD34+ HSCs were differentiated into primary erythroid cells and subjected to ChIP-Seq using USF1/2, SETD1A, BPTF and H3K4me3 specific antibodies. Two replicates of the USF1 and USF2 ChIP-Seq revealed reproducible genome-wide interactions, including associations with the β-globin locus (Supplementary Figure S2). We found 13,986 USF1 and 11,750 USF2 binding peaks located near TSSs. The majority (73.8%) of the USF1 and USF2 peaks overlapped at proximal promoters suggesting that USF1 and USF2 act as a heterodimer at promoters (Figure 2D), binding to consensus E-box motifs. Analyzing the binding of USF1 in the top 1000 highly expressed erythroid genes and the bottom 1000 silenced genes revealed USF1/2 bound to the consensus E-box motif at proximal TSSs in highly transcribed genes (Figure 2E–G).
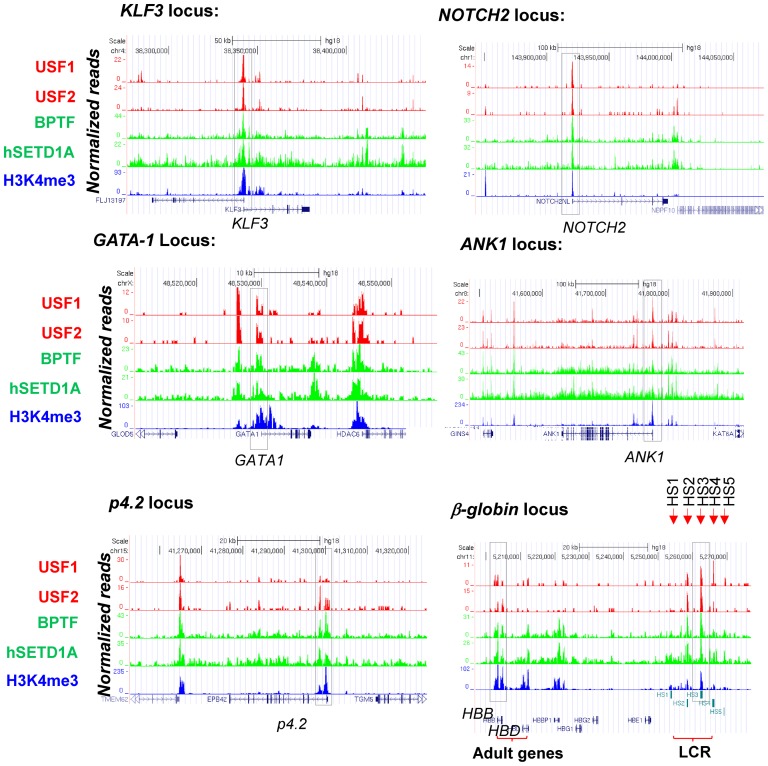
ChIP-seq data were correlated with global transcriptome profiles (5) and composite profiles of H3K4me3 modifications as well as USF and NURF occupancy at the TSSs were created for highly expressed erythroid genes, medium expressed genes and silent genes (Supplementary Figure S3A and B). Notably, H3K4me3 and BPTF significantly colocalized with USF1/2 at predicted E-box motifs in proximal promoters (Figure 2G–I) and co-occupancy positively correlated with levels of erythroid gene expression (Figure 2, Supplementary Figure S3). More than 50% of sites co-occupied by USF1 and H3K4me3 or BPTF were located near TSSs of active erythroid genes (Figure 2H and I) suggesting that NURF and H3K4me3 may be important for erythroid promoter activation, likely through interactions with USF1/2. When we examined their occupancy and histone modification at erythroid-specific genes, we found that USF1/2, SETD1A, BPTF and H3K4me3 colocalized with many promoters of highly expressed erythroid genes including β-globin, NOTCH2, P4.2, GATA-1, Ankyrin 1 (ANK1) and KLF3 in erythroblasts (Figure 3). The ChIP-seq data have either been repeated or confirmed by ChIP-qPCR assays. Thus, the global binding and histone modification data revealed that USF1 preferentially co-occupied promoter-chromatin sites together with the SETD1A and NURF complexes.
Figure 3.
Colocalization of USF1 and associated histone modifying and remodeling complexes at promoters of erythroid genes. ChIP-seq analyses of USF1/2 binding, hSETD1A recruitment, BPTF occupancy and H3K4me3 enrichment at erythroid specific gene loci including KLF3, NOTCH2, GATA-1, ANK1, p4.2 and β-globin. The data has either been repeated or confirmed by ChIP-qPCR assays.
Recruitment of NURF correlates with promoter/enhancer nucleosome repositioning
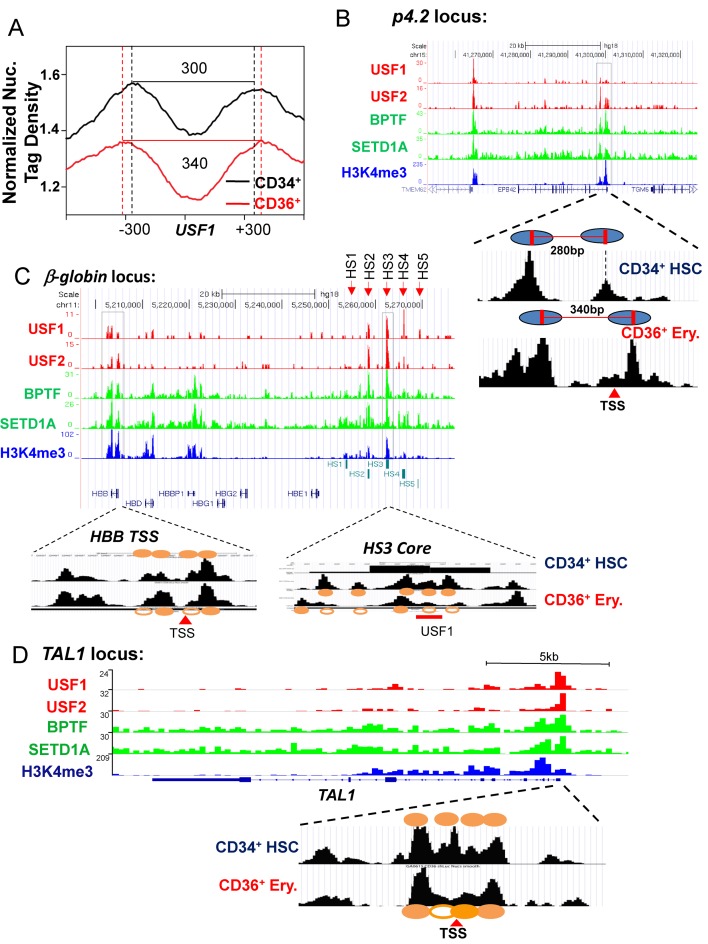
It has been shown by MNase-seq analysis that global position of nucleosomes surrounding enhancer-associated GATA motifs dramatically shift during erythroid differentiation (24). However, it remains unknown whether and how promoter proximal nucleosome positioning and accessibility are also altered to coordinate nucleosome position changes at the erythroid-specific TF bound enhancers during differentiation. Detailed comparisons between ChIP-seq analyses of H3K4me3, SETD1A and NURF occupancy in erythroblasts with MNase-seq data from CD34+ HSCs and CD36+ erythroid precursors revealed a positive correlation among the co-occupancy of USFs/SETD1A/NURF complexes, promoter H3K4me3 enrichments and repositioning of promoter/enhancer nucleosomes (Figure 4). The combined analyses of USF ChIP-seq data and nucleosome profiles surrounding the USF1 binding sites at promoters revealed a substantial increase of 40 bp nucleosome repeat length upon erythroid differentiation (Figure 4A). For example, the co-recruitment of USFs, SETD1A, H3K4me3 and BPTF at HS3 (hypersensitive site 3) of the LCR (locus control region), HBB promoter, TAL1 promoter 1 and the p4.2 promoter correlated with a lost/shift of nucleosome occupancy at the HS3 core region and the core promoters of HBB, TAL1 and p4.2 genes in erythroid cells compared to CD34+ HSCs (Figure 4B–D). The repositioning of nucleosomes created nucleosome-free accessible regions over these regulatory DNA elements in erythroid cells that may lead to gene activation (Figure 4). The data suggest that SETD1A and NURF complexes collaborate to establish local accessibility at USF binding sites at erythroid-specific promoters/enhancers.
Figure 4.
The recruitment of USF1 associated NURF complex in erythroid promoters positively correlates with promoter-associated nucleosome reorganization. (A) Normalized nucleosome tag densities obtained from MNase-Seq were plotted for the USF1 motif near proximal promoters of erythroid genes in HSCs (black line) and CD36 (red line) cells. The results show that in comparison to the CD34 positive HSCs, average nucleosome space surrounding the promoter associated USF1 motif of erythroid genes increases 40 bp in CD36 positive erythroid precursors upon erythroid differentiation. (B) Correlation of the binding of the USF1 associated complexes and nucleosome positioning at the P4.2 promoters during erythroid cell differentiation. The ovals indicated the positioned nucleosomes surrounding the P4.2 promoter. (C) Correlation of the binding of the USF1 associated complexes and altered nucleosome positioning at the β-major globin promoter and LCR element HS3 during erythroid cell differentiation. The orange closed ovals are positioned nucleosomes which are correlated with peaks of nucleosome tag density by deep sequencing and orange open ovals are nucleosome lost or shifted upon erythroid differentiation. (D) Correlation of the binding of the USF1 associated complexes and nucleosome positioning at the TAL1 promoter during erythroid cell differentiation. The orange solid circles are positioned nucleosomes which are correlated with peaks of nucleosome tag density by deep sequencing and orange open circles are nucleosome lost or shifted upon erythroid differentiation.
Setd1a mediated differential H3K4me3 is required for erythroid lineage differentiation
To further examine promoter associated H3K4me3 dynamics during lineage differentiation, we determined the enrichment of H3K4me3 at promoters of stemness genes, OCT4, Nanog, Sox2, as well as erythroid specific genes before and after EB erythroid differentiation by ChIP. We found that H3K4me3 is highly enriched at promoters of stemness genes, but not at erythroid-specific promoters at the ESC stage (Figure 5A). The enrichment of H3K4me3 is completely lost at the stemness genes upon EB erythroid differentiation and switched to promoters of erythroid-specific genes (Figure 5A). To test whether Setd1a is responsible for promoter associated H3K4me3 during erythroid differentiation, we silenced Setd1a in murine ESCs using a Tet-on doxycycline (Dox) inducible shRNA vector. Compared to vector control, Setd1a expression was substantially reduced in the Dox-induced Setd1a knockdown (KD) ESCs (Figure 5B). ESCs carrying the vector control or Setd1a-specific shRNAs were differentiated into erythroblast-like cells by adding cytokines and EPO following the withdrawal of LIF (Supplementary Figure S4A). Setd1a was silenced at day 10 of differentiation and erythroid gene expression patterns were examined in cells collected at different time points. Expression levels of definitive erythroid markers, β-globin, p4.2 and GYPA, gradually increased and reached peak levels at day19 while c-kit, a marker highly expressed at the HSPCs, was gradually silenced (Supplementary Figure S4B). Over the period of erythroid culture conditions, the expression of embryonic βH1 and ϵy gradually switched to adult β-major/minor globin (Supplementary Figure S4C) and the culture morphology gradually resembled definitive erythroblasts (Supplementary Figure S4D), indicating that ESCs were differentiating into cells with a definitive erythroid phenotype. Consistent with in vivo KO data (Figure 1D), Setd1a KD significantly inhibited expression of erythroid genes critical for the erythroid transcription program, Tal1, Gata1, Notch2, β-globin, p4.2 and Gypa (Figure 5C and D; Supplementary Figures S4C and 5A). The experiments have been carried out in two murine ES cell lines, R1/E and D3, with consistent results. Thus, the data suggest that erythroid gene transcription is regulated by Setd1a HMT.
Figure 5.
Depletion of Setd1a inhibits β-globin gene transcription as well as recruitment of the Setd1a complex and H3K4me3 levels at the β-globin promoter. (A) Comparison of H3K4me3 enrichment at stemness promoters and erythroid promoters in ES cells and differentiated EBs. (B) Western blot analysis of Setd1a protein levels in R1E ES cells harboring shRNA against Setd1a compared to control; α-tubulin served as a loading control. (C) Stable ESC clones harboring vector control or shSetd1a-expressing constructs were induced with EPO and other cytokines and total RNA was extracted at day 19. Expression of genes important for erythroid differentiation was analyzed by qRT-PCR. (D) Stable ESC clones harboring vector control or shSetd1a-expressing constructs were induced with EPO and other cytokines for erythroid differentiation and total RNA was extracted at day 0, 4 and 19. The βmaj-globin transcripts were analyzed by qRT-PCR and normalized with β-actin. (E) ChIP analysis of H3K4me1, me2 and me3 patterns over the β-globin locus at day 19 upon the Setd1a KD. (F) ChIP analysis of ASH2L (TOP) and Setd1a (Bottom) recruitment across the β-globin locus at days 19 upon Setd1a KD. Data are collected from at least three independent experiments and shown as mean ± SD. *P < 0.05; **P < 0.01 by student t-test.
To test if Setd1a KD could lead to a loss of H3K4me3 at target genes and thus perturb transcriptional activation, ChIP-qPCR was performed by comparing the vector control and Setd1a KD cells at day 19 of erythroid differentiation. Although levels of H3K4me1/me2 were decreased at the LCR and at promoters across the β-globin locus, the H3K4me3 levels were significantly reduced only at the β-major/β-minor promoters upon Setd1a depletion (Figure 5E), consistent with the decrease in Setd1a occupancy at the β-globin promoters (Figure 5F, bottom). However, ASH2L bound to both HS2 and the β-globin promoters and the binding was also decreased upon KD (Figure 5F, top) suggesting that other MLL complexes may also be involved in regulating LCR function (48). Similarly, reduced H3K4me3 levels were also observed at two other erythroid promoters, p4.2 and Notch2 (Supplementary Figure S5B). Setd1a KD had similar effects on H3K4me3 levels at the β-globin promoter and β-globin expression in the DMSO-induced MEL differentiation system (Supplementary Figure S6A–D). Thus, the combined in vivo KO (Figure 1) and in vitro KD (Figure 5, Supplementary Figure S6) data indicate that recruitment of Setd1a and its HMT activity plays a critical role in activating genes important for erythroid transcription and differentiation.
Setd1a-mediated H3K4me3 is required for BPTF recruitment at erythroid promoters and chromatin accessibility
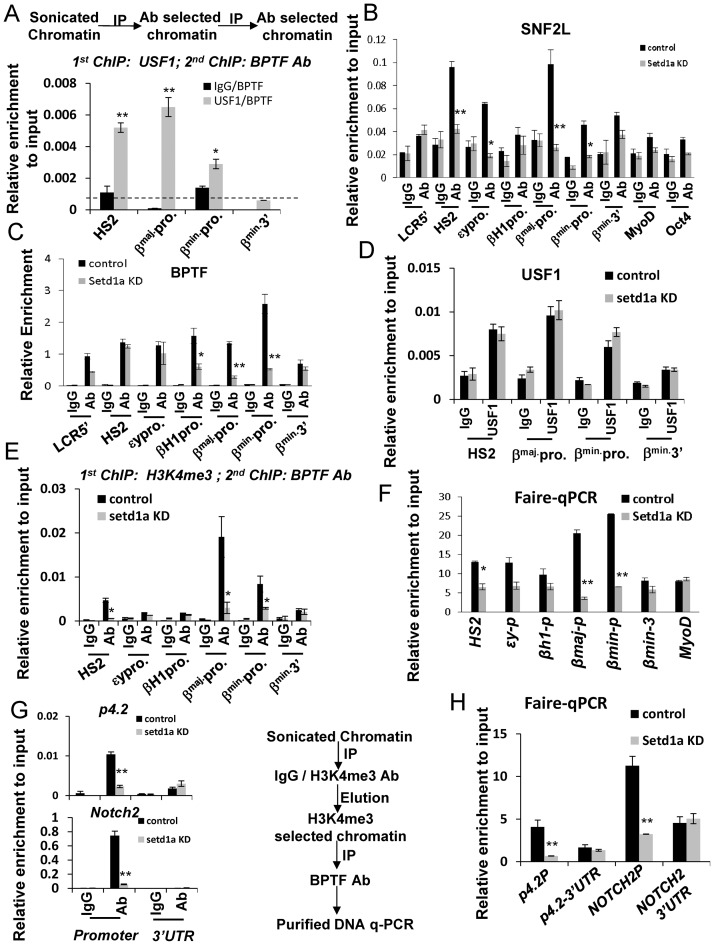
Given that USF1 binds specifically to erythoid specific promoters including β-globin, p4.2 and Notch in erythroid progenitor cells (Figures 2 and 3), we hypothesized that USF1 may act as the primary TF important for the recruitment of the Setd1a and NURF protein complexes at promoter chromatin sites to coordinate with the GATA-1 mediated accessible enhancer domains. We examined the binding relationship among USF1, Setd1a and NURF at erythroid promoters during EB erythroid differentiation using sequential ChIP analysis comparing control and Setd1a KD EBs. Sequential-ChIP indicated that USF1 and BPTF co-occupied HS2 of the LCR and adult promoters of the β-globin locus in wild-type cells (Figure 6A). Depletion of Setd1a led to a decrease in the occupancy of SNF2L at the LCR and the globin promoters, especially at the β-major/β-minor promoters. The occupancy of BPTF was specifically reduced at the globin promoters upon Setd1a KD (Figure 6B and C). In contrast, loss of Setd1a did not impair USF1 binding to HS2 and the β-globin promoters (Figure 6D), suggesting that although Setd1a and NURF can be targeted by USF1 to erythroid genes, Setd1a itself or its mediated H3K4me3 at promoters is essential for recruiting the NURF complex. To distinguish between these two possibilities, we carried out sequential-ChIP to examine whether H3K4me3 is required for BPTF recruitment, because the PHD domain of BPTF has been reported to recognize and couple H3K4me3 with chromatin remodeling (22). Both H3K4me3 and BPTF co-occupied β-globin promoters (Figure 6E), and other erythroid promoters, such as p4.2 and Notch2 (Figure 6G). Consistent with decreases in H3K4me3 at promoters of erythroid genes (Figure 5E, Supplementary Figure S5), loss of H3K4me3 at promoters by Setd1a KD impaired BPTF recruitment (Figure 6E and G). It should be noted that the H3K4me3 levels were very low at LCR HS2 (Figure 5E) suggesting that SNF2L is recruited to this site by other mechanisms. The NURF complex is an ATP-dependent chromatin remodeling complex that catalyzes promoter nucleosome sliding, thereby exposing DNA sequences for the recruitment of transcription complexes (21,49). Thus, the interaction of Setd1a-mediated H3K4me3 and BPTF at promoters may regulate promoter chromatin accessibility, perhaps through nucleosome repositioning during erythroid differentiation (Figure 4). We further tested if Setd1a is required for DNA sequence exposure at promoters of erythroid genes by FAIRE, which is based on differences in cross-linking efficiencies between free DNA and nucleosome-occupied DNA, therefore enriching DNA encompassing active promoters and TSSs (38). Consistent with the reduction of NURF complex occupancy at the β-globin locus (Figure 6B–C; Supplementary Figure S6E), KD of Setd1a significantly reduced the fraction of nucleosome-free accessible DNA regions at the β-major and β-minor promoters in both erythroid differentiated ESCs (Figure 6F) and DMSO-induced MEL cells (Supplementary Figure S6F). The accessible DNA regions were also lost at the promoters of the P4.2 and NOTCH2 genes (Figure 6H). Thus, these data revealed an important role of Setd1a in NURF recruitment and generation of promoter chromatin accessibility during erythroid gene activation.
Figure 6.
Setd1a is required for NURF complex recruitment at erythroid gene loci and establishment of chromatin accessibility. (A) Crosslinked chromatin from days 19 differentiated EB cells was first subjected to ChIP with IgG or USF1 antibodies. The antibody selected chromatin was sequentially immunoprecipitated with BPTF antibodies. Co-occupancy of USF1 and BPTF at the β-globin LCR HS2 and promoters was analyzed by qPCR. (B and C) ChIP analysis of SNF2L (B) and BPTF (C) recruitment over the β-globin locus at days 19 erythroid differentiation upon Setd1a KD. (D) ChIP analysis of USF1 binding over the β-globin locus at days 19 erythroid differentiation upon Setd1a KD. (E) Cross-linked chromatin from control and induced Setd1a KD cells was first ChIP with H3K4me3 antibodies. The H3K4me3 selected chromatin was sequentially immunoprecipitated with BPTF antibodies. Co-occupancy of H3K4me3 and BPTF at the β-globin promoters was impaired by the Setd1a KD. (F) FAIRE assay of β-globin locus at days 19 of erythroid differentiation upon Setd1a KD. (G) Cross-linked chromatin from control and induced Setd1a KD cells was first subjected to ChIP with H3K4me3 antibodies. The H3K4me3 selected chromatin was sequentially immunoprecipitated with BPTF antibodies. Co-occupancy of H3K4me3 and BPTF at the p4.2 and NOTCH2 promoters was impaired by the Setd1a KD. (H) Results from FAIRE assays of the p4.2 and NOTCH2 promoters at days 19 of erythroid differentiation upon Setd1a KD. Data are collected from at least three independent experiments shown as mean ± SD. *P < 0.05; **P < 0.01 by student t-test.
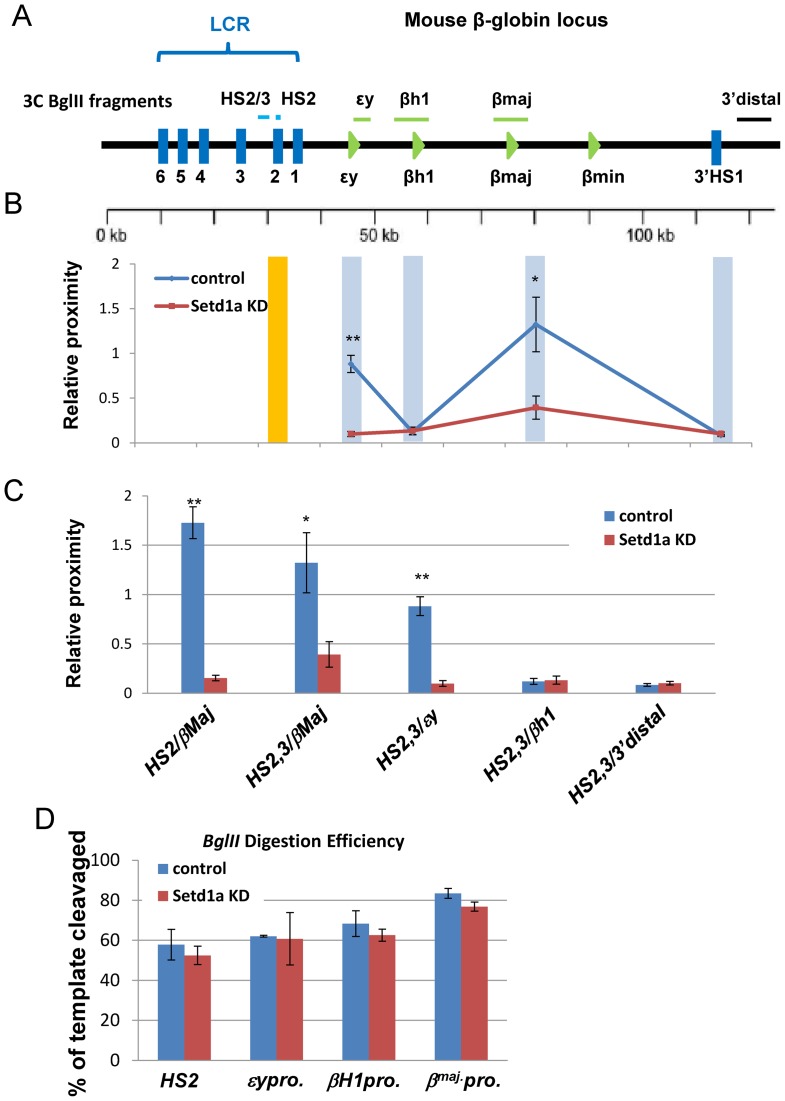
To further examine the consequence of reduced accessible chromatin regions at enhancers/promoters, we performed chromatin conformation capture (3C) assay using HS2 of the LCR as a bait for the interaction between the LCR and the individual globin genes across the entire β-globin locus comparing the vector control and Setd1a KD cells in erythroid cells 19 days after induction of differentiation (Figure 7A). Both HS2 core and HS2/3 of LCR interacted with the β-major promoter in differentiated cells as previously reported (39). The ϵy promoter was also shown to interact with HS2 core and HS2/3 because of its proximity to the LCR (Figure 7B and C). Setd1a KD resulted in a specific inhibition of looped interactions between HS2/3 or the HS2 core and the β-major promoter or ϵy promoter (Figure 7B and C), consistent with inhibition of β-globin gene expression. The reduction of ligation frequency between the LCR and the εγ promoter is likely due, at least in part, to reduced chromatin accessibility at the LCR core enhancers as revealed by the FAIRE assay. As a control, BglII efficiently cut across the β-globin locus in both vector control and Setd1a KD cells (Figure 7D). The effect of Setd1a loss on the long-range interactions between HS2 of the LCR and the β-major promoter was confirmed in DMSO-induced MEL cells (Supplementary Figure S7A and B). In addition to the β-globin locus, loss of Setd1a and its mediated H3K4me3 also impaired a long-range interaction between the +51-enhancer and promoter at the TAL1 locus in K562 cells (47). Together, these data indicate that disruption of Setd1a HMT activity affects erythropoiesis by perturbing histone modifications, promoter accessibility and enhancer/promoter interactions.
Figure 7.
Loss of Setd1a disrupts enhancer/promoter interactions in the β-globin gene locus. (A) Schematic representation of the 3C fragments after BglII digestion over the mouse β-globin locus. (B and C) 3C analysis of the interaction between the HS2/3 or HS2 enhancer and β-globin promoters in the β-globin locus at day 19 EBs upon Setd1a KD. Data are shown as PCR quantitation of the 3C products. (D) Digestion efficiency of ϵy, βH1, HS2 and βmaj BglII digested sites in the control and Setd1a KD EBs at day 19 of erythroid differentiation, HS4 as a loading control. Data are collected from at least three independent experiments shown as mean ± SEM. *P < 0.05; **P < 0.01 by student t-test.
DISCUSSION
We recently purified USF1-associated SETD1A/NURF complexes that exhibit H3K4 methyltransferase and ATP-dependent nucleosome sliding activities and act as chromatin barriers to prevent heterochromatin encroachment during terminal erythroid differentiation (6) suggesting that these complexes play an important role in erythroid transcription programs. Evidence from genome-wide studies showed that in erythroid cells USF1 predominantly binds to promoter proximal regions close to TSSs (23). We demonstrated that USF1 recruits SETD1A and NURF complexes to many promoter-associated USF sites of erythroid lineage-specific genes and that their co-occupancy positively correlates with transcription levels of erythroid genes upon lineage differentiation. Depletion of Setd1a reduced H3K4me3 at promoters of these erythroid genes, thereby decreasing NURF recruitment, promoter accessibility and gene transcription. Finally, KO of Setd1a in erythroid progenitors perturbed erythropoiesis in vivo. Thus, our data reveal a novel epigenetic mechanism by which USF1 cross-talks with Setd1a and NURF to coordinate erythroid gene expression and differentiation.
Setd1a predominantly trimethylates H3K4 at active promoter-associated nucleosomes near TSSs (16,44,50,51). BPTF contains two PHD domains and the second PHD domain recognizes the combined patterns of H3K4me3 and H4K16ac (52). Loss of Setd1a led to a decrease in H3K4me3 levels at active promoters, and subsequently resulted in reduced BPTF occupancy and nucleosome-free promoter DNA sequences further illustrating that Setd1a-mediated H3K4me3 plays a role in targeting the NURF complex to H3K4me3-enriched promoters leading to increased promoter accessibility in a cell context dependent manner. During differentiation, these chromatin modifying and remodeling complexes are recruited and targeted to erythroid specific promoters by USF TFs. This is illustrated by the fact that nucleosome spacing is substantially increased around the USF binding sites at promoters upon erythroid differentiation (Figure 4A). This notion is supported by the observation that loss of USF activity resulted in defective erythropoiesis in transgenic animals (46). Many lineage specific factors such as GATA1 and TAL1 are involved in regulating commitment and differentiation of erythroid cells by controlling enhancer accessibility (24). It is possible that USF1 mediated promoter chromatin structure opening coordinates and acts within the erythroid specific enhancer chromatin domain to confer erythroid-specific transcription program. In addition, it was recently reported that long-non-coding RNAs are also involved in targeting Setd1a to specify early lineage-specific chromatin structure alterations and differentiation (53). It remains to be determined whether the remodeling activity of NURF also stabilizes lineage specific factors, such as USF1, and Setd1a complexes at promoters. Nevertheless, our results support earlier observations showing that the NURF complex is involved in transcriptional activation by creating a nucleosome-free region at promoters (21,49).
Recent genome-wide studies indicate a correlation between different levels of histone modifications and gene enhancer/promoter activity (15,54) with specific histone modifications, such as H3K4 trimethylation involved in regulating enhancer/promoter communication (24,25,47). The chromatin remodeling complex Brg1 regulates nucleosome accessibility and subsequent binding of TFs at enhancers during erythroid differentiation (24). In the β-globin locus, long-range regulatory elements like the LCR interact with activate genes via formation of an active chromatin hub (55,56). In both α- and β-globin loci, Brg1 regulates nucleosome structure and long-range interactions to facilitate globin gene transcription (57,58). Long-range interactions between enhancers and promoters are dependent on active histone modifications (39,47) and we show that inhibition of Setd1a H3K4 trimethylase activity, which blocks recruitment of the NURF complex, prevents formation of nucleosome-free regions at promoters (Figure 6) and reduce long-range enhancer/promoter interactions in the β-globin and TAL1 loci (Figure 7) (47). These data reveal an important role of Setd1a and NURF in communications between distal enhancers and proximal promoters.
The trithorax HMT complexes play critical roles during early embryonic development and hematopoiesis. A recent large-scale study in zebrafish identified SET1 as one of the major regulators of hematopoiesis (59). Setd1a KO mice suffer from early embryonic lethality at the epiblast stage, while Setd1b KO mice appear normal and viable, suggesting a non-redundant role of Setd1a in embryonic development (19). Furthermore, hematopoietic-specific Setd1a-CKO blocks progenitor-B-to-precursor-B cell development by inhibiting H3K4me3 levels and IgH rearrangement (20). MLL1 KO mice exhibit a block in definitive hematopoiesis (10). Loss of Mll5 leads to pleiotropic hematopoietic defects and reduced neutrophil function (60,61). KO of Setd1a in the erythroid compartment impaired erythropoiesis (Figure 1) with numerous genes critical for erythroid differentiation affected (Figure 1D). It is possible that Setd1a may be more critical for the later phases of erythroid differentiation and maturation. Indeed, Setd1a-mediated chromatin barrier activity is required for protecting erythroid gene expression during late-stage erythropoiesis (6). It is worth noting that genetic alterations of MLLs have been observed in leukemia and lymphoma with frequent disruption of MLL1 by chromosomal translocations in acute leukemia (16,62). Combined with our finding that erythroid-specific deletion of Setd1a leads to a defect in erythropoiesis (Figure 1), these observations indicate that TrxG-related complexes play an essential role in hematopoiesis. However, it remains to be determined whether distinct branches of TrxG HMT complexes collaboratively or differentially target enhancers or promoters of hematopoietic genes, and if they play distinct or overlapping role in regulating different developmental hematopoietic pathways.
ACCESSION NUMBERS
The sequence data from this study have been deposited to the NCBI Gene Expression Omnibus (GEO) under accession numbers: GSE26501; GSE69347 and GSE43625.
Supplementary Material
Acknowledgments
The authors thank their colleagues in the Huang, Gallagher, Bungert and Qiu laboratories for helpful discussions and support. We also thank Ms. Haoli Li for bioinformatics help and revising the figures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [R56DK101994 to S.H; R01HL106184 to P.G.G.; R01HL095674 to Y.Q.; R01DK052356 to J.B.; R01GM106174 to L.Z.]; Intramural research programs (to K.Z.); National Heart Lung and Blood Institute; National Institute of Health. Funding for open access charge: National Institute of Health [R56DK101994, R01HL106184].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hattangadi S.M., Wong P., Zhang L., Flygare J., Lodish H.F. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palis J., Segel G.B. Developmental biology of erythropoiesis. Blood Rev. 1998;12:106–114. doi: 10.1016/s0268-960x(98)90022-4. [DOI] [PubMed] [Google Scholar]
- 3.Kerenyi M.A., Orkin S.H. Networking erythropoiesis. J. Exp. Med. 2010;207:2537–2541. doi: 10.1084/jem.20102260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong P., Hattangadi S.M., Cheng A.W., Frampton G.M., Young R.A., Lodish H.F. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood. 2011;118:e128–e138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An X., Schulz V.P., Li J., Wu K., Liu J., Xue F., Hu J., Mohandas N., Gallagher P.G. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Wang S., Li Y., Deng C., Steiner L.A., Xiao H., Wu C., Bungert J., Gallagher P.G., Felsenfeld G., et al. Chromatin boundaries require functional collaboration between the hSET1 and NURF complexes. Blood. 2011;118:1386–1394. doi: 10.1182/blood-2010-11-319111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 8.Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 9.Steward M.M., Lee J.S., O'Donovan A., Wyatt M., Bernstein B.E., Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 10.Ernst P., Fisher J.K., Avery W., Wade S., Foy D., Korsmeyer S.J. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev. Cell. 2004;6:437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang P., Lin C., Smith E.R., Guo H., Sanderson B.W., Wu M., Gogol M., Alexander T., Seidel C., Wiedemann L.M., et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J., Saha P.K., Yang Q.H., Lee S., Park J.Y., Suh Y., Lee S.K., Chan L., Roeder R.G., Lee J.W. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19229–19234. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan M.A., Shilatifard A. Drosophila sets its sights on cancer: Trr/MLL3/4 COMPASS-like complexes in development and disease. Mol. Cell. Biol. 2013;33:1698–1701. doi: 10.1128/MCB.00203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng C., Li Y., Liang S., Cui K., Salz T., Yang H., Tang Z., Gallagher P.G., Qiu Y., Roeder R., et al. USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet. 2013;9:e1003524. doi: 10.1371/journal.pgen.1003524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 16.Mohan M., Lin C., Guest E., Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 17.Herz H.M., Mohan M., Garruss A.S., Liang K., Takahashi Y.H., Mickey K., Voets O., Verrijzer C.P., Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Bledau A.S., Schmidt K., Neumann K., Hill U., Ciotta G., Gupta A., Torres D.C., Fu J., Kranz A., Stewart A.F., et al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014;141:1022–1035. doi: 10.1242/dev.098152. [DOI] [PubMed] [Google Scholar]
- 20.Tusi B.K., Deng C., Salz T., Zeumer L., Li Y., So C.W., Morel L.M., Qiu Y., Huang S. Setd1a regulates progenitor B-cell-to-precursor B-cell development through histone H3 lysine 4 trimethylation and Ig heavy-chain rearrangement. FASEB J. 2014;28:1505–1515. doi: 10.1096/fj.14-263061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamiche A., Sandaltzopoulos R., Gdula D.A., Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 22.Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 23.Neph S., Vierstra J., Stergachis A.B., Reynolds A.P., Haugen E., Vernot B., Thurman R.E., John S., Sandstrom R., Johnson A.K., et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu G., Schones D.E., Cui K., Ybarra R., Northrup D., Tang Q., Gattinoni L., Restifo N.P., Huang S., Zhao K. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–1658. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui K., Zang C., Roh T.Y., Schones D.E., Childs R.W., Peng W., Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carotta S., Pilat S., Mairhofer A., Schmidt U., Dolznig H., Steinlein P., Beug H. Directed differentiation and mass cultivation of pure erythroid progenitors from mouse embryonic stem cells. Blood. 2004;104:1873–1880. doi: 10.1182/blood-2004-02-0570. [DOI] [PubMed] [Google Scholar]
- 27.Hu X., Li X., Valverde K., Fu X., Noguchi C., Qiu Y., Huang S. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10141–10146. doi: 10.1073/pnas.0900437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 31.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Shin H., Song J.S., Lei Y., Liu X.S. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon J.M., Giresi P.G., Davis I.J., Lieb J.D. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giresi P.G., Kim J., McDaniell R.M., Iyer V.R., Lieb J.D. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X., Hu X., Patel B., Zhou Z., Liang S., Ybarra R., Qiu Y., Felsenfeld G., Bungert J., Huang S. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood. 115:2028–2037. doi: 10.1182/blood-2009-07-236059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagege H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 41.Heinrich A.C., Pelanda R., Klingmuller U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood. 2004;104:659–666. doi: 10.1182/blood-2003-05-1442. [DOI] [PubMed] [Google Scholar]
- 42.Socolovsky M., Nam H., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 43.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salz T., Li G., Kaye F., Zhou L., Qiu Y., Huang S. hSETD1A regulates wnt target genes and controls tumor growth of colorectal cancer cells. Cancer Res. 2014;74:775–786. doi: 10.1158/0008-5472.CAN-13-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider J., Wood A., Lee J.S., Schuster R., Dueker J., Maguire C., Swanson S.K., Florens L., Washburn M.P., Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 46.Liang S.Y., Moghimi B., Crusselle-Davis V.J., Lin I.J., Rosenberg M.H., Li X., Strouboulis J., Huang S., Bungert J. Defective erythropoiesis in transgenic mice expressing dominant-negative upstream stimulatory factor. Mol. Cell. Biol. 2009;29:5900–5910. doi: 10.1128/MCB.00419-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel B., Kang Y., Cui K., Litt M., Riberio M.S., Deng C., Salz T., Casada S., Fu X., Qiu Y., et al. Aberrant TAL1 activation is mediated by an interchromosomal interaction in human T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:349–361. doi: 10.1038/leu.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demers C., Chaturvedi C.P., Ranish J.A., Juban G., Lai P., Morle F., Aebersold R., Dilworth F.J., Groudine M., Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol. Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao H., Sandaltzopoulos R., Wang H.M., Hamiche A., Ranallo R., Lee K.M., Fu D., Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 50.Schneider J., Wood A., Lee J.S., Schuster R., Dueker J., Maguire C., Swanson S.K., Florens L., Washburn M.P., Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 52.Ruthenburg A.J., Li H., Milne T.A., Dewell S., McGinty R.K., Yuen M., Ueberheide B., Dou Y., Muir T.W., Patel D.J., et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng C., Li Y., Zhou L., Cho J., Patel B., Terada N., Bungert J., Qiu Y., Huang S. HoxBlinc RNA recruits set1/MLL complexes to activate Hox gene expression patterns and mesoderm lineage development. Cell Rep. 2016;14:103–114. doi: 10.1016/j.celrep.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolhuis B., Palstra R.J., Splinter E., Grosveld F., de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 56.Ragoczy T., Bender M.A., Telling A., Byron R., Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim S.I., Bresnick E.H., Bultman S.J. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 2009;37:6019–6027. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S.I., Bultman S.J., Kiefer C.M., Dean A., Bresnick E.H. BRG1 requirement for long-range interaction of a locus control region with a downstream promoter. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2259–2264. doi: 10.1073/pnas.0806420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang H.T., Kathrein K.L., Barton A., Gitlin Z., Huang Y.H., Ward T.P., Hofmann O., Dibiase A., Song A., Tyekucheva S., et al. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nat. Cell Biol. 2013;15:1516–1525. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heuser M., Yap D.B., Leung M., de Algara T.R., Tafech A., McKinney S., Dixon J., Thresher R., Colledge B., Carlton M., et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113:1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 61.Madan V., Madan B., Brykczynska U., Zilbermann F., Hogeveen K., Dohner K., Dohner H., Weber O., Blum C., Rodewald H.R., et al. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 2009;113:1444–1454. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]
- 62.Muntean A.G., Hess J.L. The pathogenesis of mixed-lineage leukemia. Annu. Rev. Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.