Abstract
DNA damage can significantly modulate expression of the affected genes either by direct structural interference with transcription components or as a collateral outcome of cellular repair attempts. Thus, DNA glycosylases of the base excision repair (BER) pathway have been implicated in negative transcriptional response to several spontaneously generated DNA base modifications, including a common oxidative DNA base modification 8-oxoguanine (8-oxoG). Here, we report that single 8-oxoG situated in the non-transcribed DNA strand of a reporter gene has a pronounced negative effect on transcription, driven by promoters of various strength and with different structural properties, including viral, human, and artificial promoters. We further show that the magnitude of the negative effect on the gene expression correlates with excision of the modified base by OGG1 in all promoter constructs tested. Moreover, by using expression vectors with nuclease resistant backbone modifications, we demonstrate that OGG1 does not catalyse DNA strand cleavage in vivo. Rather, cleavage of the phosphate bond 5′ to 8-oxodG (catalysed by APE1) is essential and universally required for the onset of transcriptional silencing, regardless of the promoter structure. Hence, induction of transcriptional silencing emerges as a ubiquitous mode of biological response to 8-oxoG in DNA.
INTRODUCTION
Consequences of oxidative damage of nucleobases for physiological functions of DNA are of major toxicological concern, because generation of reactive oxygen species (ROS) and the resulting damage to genomic DNA are inevitably associated with endogenous physiological processes and can be manifold amplified in response to numerous chemicals and various forms of radiation (1,2). One of the most prevalent reactions of DNA with ROS is oxidation of guanine to 8-oxo-7,8-dihydroguanine (8-oxoG), a pre-mutagenic lesion capable to mispair with adenine during replication (3,4), eventually leading to G to T transversion mutations and cancer (5,6). This is counteracted by very efficient repair, which ensures a reasonably low steady state level of 8-oxoG in the chromosomal DNA, estimated in the order of a few modifications per million guanine bases (7). Base excision repair (BER) initiated by the specific 8-oxoguanine DNA glycosylase (OGG1) effectively protects from accumulation of the characteristic mutations in growing organs of young mice and regenerating liver (8–10), being the only comprehensively documented pathway for the removal of 8-oxoG from chromosomal DNA in mammals. Human OGG1 is a bifunctional DNA glycosylase, which first generates an apurinic (AP) site by catalysing hydrolysis of the N-glycosidic bond. This can be followed by either beta-elimination of the 3′ phosphate by an intrinsic beta lyase activity of the enzyme (11,12) or endonucleolytic hydrolysis of the 5′ phosphodiester bond by the AP endonuclease APE1 (13,14). In cell-free cleavage reactions, APE1 strongly stimulates the DNA glycosylase activity and improves OGG1 dissociation from the AP site, thus favouring the second scenario (15,16); however, it is not known which of the two mechanisms actually prevails in cells. Subsequent repair steps follow the unified BER scenario, reviewed elsewhere (17,18).
In the absence of replication of DNA, 8-oxoG apparently does not induce mutations, however it can interfere with the process of transcription, which makes this DNA lesion potentially dangerous to non-proliferating cells as well (19). At least three modes of interference of 8-oxoG with gene transcription have obtained experimental support. The first is direct modulation of the strength of binding of several transcription factors to their target DNA sequences (20–22). The second is an increased rate of erroneous incorporation of ribo-ATP opposite 8-oxoG during RNA polymerase II transcription, sometimes called ‘RNA mutagenesis’ or ‘transcriptional mutagenesis’ (23,24). The third is suppression of transcriptional activity of affected genes, which can be induced by 8-oxoG situated at apparently any location in the gene sequence (25–28). Remarkably, already a single 8-oxoG suffices to significantly reduce the gene's transcriptional output (26,29), which makes this mode conceivably the most relevant one at physiologically low 8-oxoG densities, albeit the precise mechanism is not yet clarified.
Thorough expression analyses of reporter vectors containing 8-oxoG revealed that the negative effect of the lesion on transcription is robustly manifested in host cells harbouring a functional OGG1 gene, but never in the absence of OGG1 (27,29). This is true even for constructs in which synthetic 8-oxoG was deliberately positioned in the transcribed DNA strand and even in host cells deficient in transcription-coupled repair (29), clearly testifying that the lesion is efficiently bypassed by RNA polymerase II in vivo. These results further suggested that excision of 8-oxoG plays a decisive role in the mechanism of transcriptional gene inactivation, plausibly via interaction of transcription components with BER intermediates. In support of this hypothesis, it was shown that the magnitude of the inhibition of transcription in genetically manipulated host cells extremely well correlates with the OGG1 protein amounts and the actual 8-oxoG excision efficiencies (30). Moreover, suppression of transcription appeared to be a common feature of several BER substrates besides 8-oxoG; and in all cases investigated so far the respective specific DNA glycosylases (either mono- or bifunctional) have been clearly implicated ((30,31,32) and unpublished results). All these observations strongly indicate that some common post-excision BER intermediate plays a critical role for the inhibition of transcription in cells. This intermediate does not necessarily have to block the elongating RNA polymerase, since the lesions in the transcribed and the non-transcribed DNA strands are equally harmful (30,32). Rather, it should elicit a signal leading to transcriptional repression or silencing of the gene's promoter (28).
To understand the mechanism of transcriptional inactivation by BER substrates, it is necessary to figure out which events in the chain of enzymatic BER steps (generation of AP site, strand incision by bifunctional DNA glycosylases or APE1, processing of DNA ends, repair patch synthesis, ligation) are critical for the manifestation of transcriptional inactivation of damaged genes. On the other hand, it is important to investigate whether intrinsic gene-specific peculiarities of transcriptional regulation have an impact on transcriptional responses to DNA lesions. An idea that promoter strength and structure might determine the magnitude of transcriptional inhibition by 8-oxoG was previously proposed in attempt to reconcile the heterogeneous observations in different experimental systems (26,33), however no systematic research has been performed in this direction. The aims of present work were to measure the impact of single 8-oxoG on transcription, driven by promoters of various strength and with different structures, and to identify the harmful repair intermediate.
MATERIALS AND METHODS
Cell lines
T-REx™-HeLa cells were purchased from Life Technologies GmbH (Darmstadt, Germany). HeLa/OGG1sh cells (clone 12), in which the OGG1 protein expression was 70% reduced by stable expression of the specific shRNA, and the corresponding isogenic cell line HeLa/pEpS with normal OGG1 expression were generated and characterised previously (30). HeLa cells stably overexpressing human OGG1 protein fused to GFP (34) were kindly provided by Pablo Radicella (CEA, Fontenaix au Roses).
Synthetic oligonucleotides
Synthetic 18-mer 5′-TGAGCACCCAGTCC[8oG]CCC deoxyribo-oligonucleotides, containing 8-oxodG, as well as 5′-CATTGCTTC[AP]CTAGCACG, containing a stable AP site analogue tetrahydrofuran (AP), were from BioSpring GmbH (Frankfurt am Main, Germany). When specified, 5′ and/or 3′ phosphodiester bonds flanking the modified nucleotide were substituted for the phosphorothioate linkages (a random combination of the R- and S-stereoisomers). The respective control deoxyribo-oligonucleotides without base or backbone modifications (5′-TGAGCACCCAGTCCGCCC and 5′-CATTGCTTCGCTAGCACG) were from Eurofins MWG Operon (Ebersberg, Germany). All oligonucleotides used for incorporation into vector DNA and subsequent gene expression analyses were HPLC-purified and validated by electrospray ionisation mass spectrometry. High purity salt free grade oligonucleotides used for cloning purposes and as PCR primers were from Eurofins MWG Operon (Ebersberg, Germany).
Expression vectors
Expression vectors pZAJ, pZAJ-5W and pZAJ-5C suited for incorporation of synthetic oligonucleotides with desired base modifications into the coding region of the enhanced green fluorescent protein (EGFP) gene (p-ZAJ vector) or into the 5′-untranslated gene region as an additional option (pZAJ-5W and pZAJ-5C) were described previously (30). Vectors carrying the (EGFP) reporter gene under the control of various promoters were generated from pZAJ or pZAJ-5C vectors, in which the cytomegalovirus immediate early promoter (CMV-IE) was mutated, deleted or replaced, as described in Supplementary Methods. All vectors retained the entire EGFP coding region, including the tandem Bpu10I sites suited for the incorporation of synthetic oligonucleotides (29).
Incorporation of synthetic oligonucleotides containing 8-oxodG and adjacent backbone modifications into vector DNA
Double incisions were generated in the non-transcribed DNA strand of the EGFP gene with the Nt.Bpu10I nicking endonuclease (Thermo Fisher Scientific, Bonn, Germany), after which synthetic oligonucleotides with or without modifications were incorporated into the vector DNA by a straightforward strand-exchange procedure (31). Completeness of the strand exchange reactions was monitored by the inhibition of formation of covalently closed DNA in the absence of polynucleotide kinase, as described previously (31). Care was taken that all vector preparations carrying the specified modifications contain the same proportion of nicked DNA as the respective control constructs. The presence of 8-oxoG was further verified by detection of DNA strand scission by the Fpg DNA glycosylase. To this end, 100 ng covalently closed plasmid DNA in a 15 μl reaction volume were incubated with 2 units Fpg (NEB GmbH, Frankfurt am Main, Germany) in buffer containing 10 mM HEPES (pH 7.5), 200 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), and 0.1 mg/ml nuclease-free bovine serum albumin (NEB) 1 h at 37°C. Analogously, the presence of synthetic AP site and the impact of adjacent phosphorothioates on the APE1 strand cleavage activity were verified by incubation with 0.65 units human APE1 (NEB) in the 1× NEB4 buffer for 1 h at 37°C. Reactions were followed by heat-inactivation for 20 min at 65°C and electrophoresis in agarose gels in the presence of 0.5 mg/l ethidium bromide.
Quantification of in vitro incision activities at 8-oxodG
OGG1 incision activity was measured in 10 mM HEPES (pH 7.5), 200 mM NaCl, 1 mM EDTA, and 0.1 g/l nuclease-free bovine serum albumin (NEB). Vector DNA constructs (100 ng in 15 μl) containing single 8-oxodG flanked by the specified phosphate backbone modifications were incubated with the specified amounts of recombinant human OGG1 (NEB) for 1 h at 37°C, followed by heat-inactivation for 20 min at 65°C. Protein extracts of OGG1-GFP overexpressing HeLa cells for the in vitro cleavage assay were prepared by the protocol described previously (30). Reaction conditions were the same as for recombinant OGG1. Prior to gel electrophoresis, samples were adjusted to 0.1% SDS and heated to 50°C for 3 min. The extent of DNA strand cleavage was determined from the relative intensities of DNA bands by the GelDoc™ XR+ molecular imager and Image Lab™ software (Bio-Rad Laboratories, GmbH, Munich, Germany), as described previously (30). Since the incised substrate yields stronger ethidium bromide fluorescence than the same amount of covalently closed DNA (30) we have further applied the experimentally determined correction factor of 2.4 (specific for pZAJ vector) for calculation of relative abundance of the analysed DNA forms.
Transient transfections for the quantitative EGFP expression analyses
HeLa cells were co-transfected with equal copy numbers of the EGFP reporter constructs and the pDsRed-Monomer-N1 vector (Clontech, Saint-Germain-en-Laye, France). Since EGFP and DsRed are detected in separate fluorescence channels of the flow cytometer, DsRed was used as a marker for selective gating of the transfected cells, which allows precise quantitative measurement of the EGFP expression (28). This principle was applied to measure strengths of the newly constructed promoters and to measure the impact of 8-oxoG (or AP site) on the EGFP expression, as explained in the following section.
Cells were transfected in six-well plates (Nunc, Wiesbaden, Germany) with the help of Effectene (Qiagen, Hilden, Germany), as described previously (29). Transfections were always done in parallel with equal amounts of vector constructs containing the specified modifications (8-oxoG, AP site, phosphorothioates) and the reference construct harbouring unmodified synthetic oligonucleotide (always mixed with the same amount of the unmodified reference vector, encoding for DsRed). Inducible promoters were activated by addition of 1 mg/l tetracycline (pCMV_TetO2) or 100 nM dexamethasone (GR_SynthRE). Both reagents were from Sigma-Aldrich GmbH (Seelze, Germany). PARP inhibitors olaparib and ABT-888 (both from Absource Diagnostics GmbH, Munich, Germany) were dissolved at 10 mM dimethylsulfoxide (DMSO) and applied at 30 μM. Trichostatin A (Sigma-Aldrich) was dissolved at 3.3 mM in ethanol. Inhibitors or inducers were added at the time of transfection, unless otherwise specified. For time course analyses, cells were gently detached at either 6 or 8 h post-transfection (as indicated) and a fraction of them was fixed with 1% formaldehyde, as described previously (29). The remainder was split into several parts for further cultivation. Further samples were harvested at the specified time intervals and formaldehyde-fixed for flow cytometry (see below) or lysed for DNA isolation (Supplementary Methods).
EGFP expression analyses by flow cytometry and data presentation
The method for quantitative determination of EGFP expression in transiently transfected cells by flow cytometry was described and validated previously (28). Formaldehyde-fixed cells were equilibrated in phosphate-buffered saline (PBS) and analysed using FACSCalibur™ and the CellQuest™ Pro software (Beckton Dickinson GmbH, Heidelberg, Germany). dDsRed was used as gating marker to trace effectively transfected cells, as described and illustrated previously (28,35). After the exclusion of untransfected cells, EGFP fluorescence (FL1-H) distribution plots were generated and average EGFP expression per cell determined as the median of the distribution. To assess variability between independent experiments, relative expression level of each construct with a modification was calculated in each experiment relative to the expression of the control construct as a ratio of the respective FL1-H values. Thereby, relative expression of DNA without modification was arbitrarily set to 1. Mean relative expression, errors and P-values were calculated based on data of n independent experiments (as indicated in the figure legends). Further, for better visual appreciation of the impacts of analysed DNA modifications on the EGFP gene expression, we always present exemplary raw data as overlays of two individual fluorescence distribution curves—one for transfection with a construct carrying the specified DNA modifications and another for parallel transfection with corresponding control construct with incorporated unmodified synthetic oligonucleotide (as indicated by labelling).
RESULTS
Transcriptional inhibition by 8-oxodG requires a 5′ nucleolytic cleavage
To understand which proteins and structural transitions of DNA are critical for the initiation of gene repression following the recognition of 8-oxoG by OGG1, we sought for possibilities to modulate the efficiencies of single BER reactions. We have designed oligonucleotides containing phosphorothioate bonds next to 8-oxodG and incorporated the modified oligonucleotides into the pZAJ expression vector (Figure 1A–C) by the technique described previously (31). Substitution of phosphodiester bonds with phosphorothioate linkages renders nucleic acids resistant to hydrolysis by various types of nucleases, therefore we hoped that a phosphorothioate ester immediately 3′ to the lesion would inhibit the beta-lyase activity of OGG1. On the other hand, the 5′ phosphorothioate was meant to inhibit the strand cleavage by APE1, based on the reported potent inhibition of strand scission by a phosphorothioate situated 5′ to an AP site analog (36). Because surrounding DNA sequence can have a strong influence on the excision efficiency by OGG1 (29,30), we chose to place 8-oxoG in the sequence context where the excision efficiency and the capacity to inhibit the gene expression were maximal, based on previous results. To prevent that transcribing RNA polymerase complexes run into a BER intermediate, which might cause elongation block or RNA mutagenesis and could be potentially mistaken for transcriptional repression, 8-oxoG was placed in the non-transcribed DNA strand of the gene.
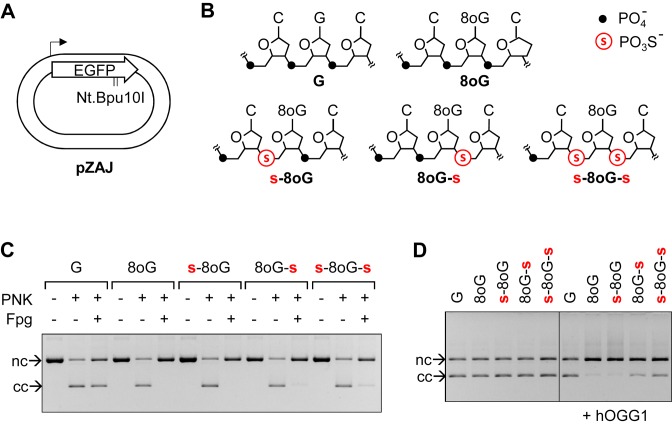
Figure 1.
Construction of plasmid vectors containing the indicated DNA base (8-oxoG) and backbone (phosphorothioate) modifications in the non-transcribed strand of the reporter EGFP gene. (A) Out of scale scheme of the pZA expression vector showing the EGFP coding sequence (signed open arrow), transcription start (broken arrow) and tandem Nt.Bpu10I nicking sites used for site-specific insertion of synthetic oligonucleotides. (B) Overview of synthetic oligonucleotides and the contained modifications. (C) Verification of the incorporation of the indicated synthetic DNA strands into vector DNA. Inhibition of ligation reaction in the absence of polynucleotidekinase (PNK) is an indicator of the successful strand exchange reaction, as described previously (31). The presence of 8-oxoG is confirmed by DNA strand scission by bacterial Fpg. Positions of covalently closed (cc) and the nicked circular (nc) forms are shown. (D) Incision of plasmid vectors containing the specified DNA modifications with 0.5 units human OGG1.
The Nt.Bpu10I excised native strand fragment of the vector DNA (Figure 1A) was substituted for a matching unmodified synthetic deoxyribo-oligonucleotide or for one of the synthetic oligonucleotides containing 8-oxodG surrounded by different combinations of phosphate and phosphorothioate linkages (Figure 1B). Successful incorporation of each of the five synthetic strands into vector DNA was confirmed by inhibition of ligation reactions in parallel samples incubated without polynucleotide kinase (Figure 1C). The presence of 8-oxoG in the vector DNA was additionally verified by incision with saturating amounts of the Fpg DNA glycosylase, which resulted in conversion of covalently closed vector DNA into the open circular form (Figure 1C). The incision was slightly inhibited in both substrates containing phosphorothioate 3′ to 8-oxodG, as can be judged from the small residual amounts of covalently closed DNA. Also incision by human OGG1 was partly inhibited in both substrates containing phosphorothioate bonds in the 3′ position, but not in the substrate containing a combination of 5′ phosphorothioate with 3′ phosphate linkages (Figure 1C), indicating that the beta lyase activity of OGG1 is inhibited, but not completely abolished by the presence of a phosphorothioate ester in the 3′ position.
Next, we transfected human host cells with vectors in which none, one or both phosphodiester bonds flanking the 8-oxodG nucleotide were replaced with phosphorothioate esters. Quantitative EGFP expression analyses in fully repair-proficient HeLa cells by flow cytometry (Figure 2A, B) showed that all constructs were expressed at equal strengths at early time (6 h) after transfections. In line with previous reports, this clearly indicates that 8-oxoG does not directly inhibit gene expression (29,30). The negative effect of 8-oxoG on the EGFP expression was progressively building up with the course of time in the construct that contained no phosphorothioate, as it was expected to take place in consequence to the 8-oxoG excision by OGG1 (29). A very similar expression pattern was observed for the construct in which 8-oxodG was flanked by unmodified phosphodiester bond on the 5′ and phosphorothioate on the 3′. In contrast, in cells transfected with constructs containing a 5′ phosphorothioate (combined with either type of the bond on the 3′), the inhibition of the gene expression by 8-oxodG was abolished or, at least, very significantly retarded (Figure 2A and B). These results indicate that incision of phosphodiester linkage on the 5′ side of 8-oxodG is required for the manifestation of the inhibitory effect on transcription of the damaged gene. Based on the knowledge of BER mechanism, this incision has to be done by APE1.
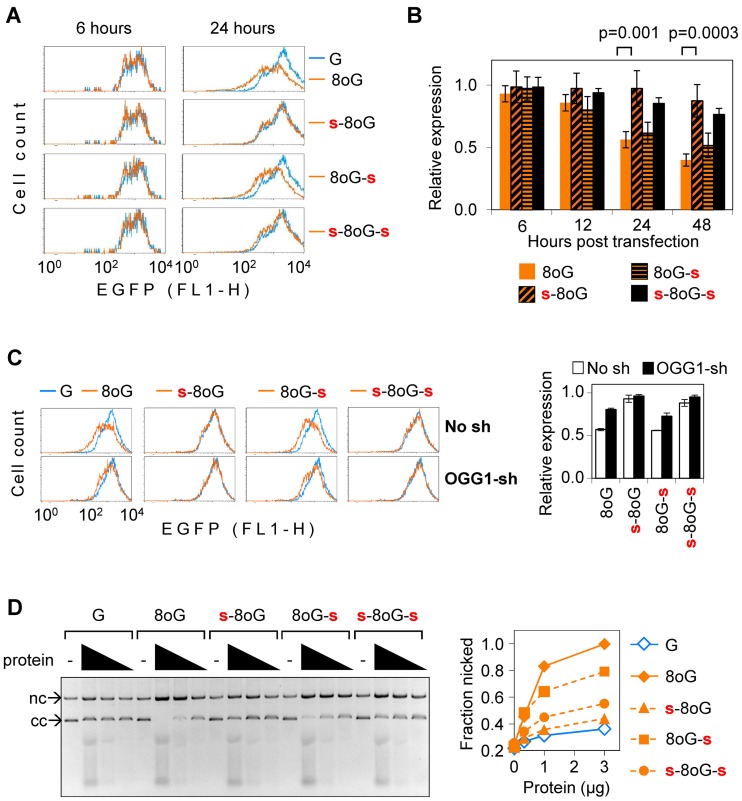
Figure 2.
Effects of 5′ and 3′ phosphorothioate bonds on the magnitude of inhibition of the EGFP gene expression by single synthetic 8-oxodG in HeLa cells. (A) Representative EGFP fluorescence distribution plots of cells transfected with vectors containing 8-oxodG in combination with the specified DNA backbone modifications (amber lines). Cells in the reference sample (overlaid blue line) were transfected with a control vector containing the corresponding synthetic oligonucleotide without 8-oxoG. (B) EGFP expression in cells transfected with the specified constructs, each normalised relative to the reference construct without 8-oxoG, transfected in parallel. Summary of 6 independent experiments (mean ± SD). P-values: Student's two-tailed t-test. (C) EGFP expression in the OGG1 knockdown (OGG1-sh) HeLa cells and the isogenic control cell line (No sh) analysed at 24 h post-transfection, as described in (A). Error bars show data range (n = 2). (D) DNA strand cleavage activities in the OGG1-overexpressing cell extracts towards plasmid substrates containing the specified modifications. Agarose gel and quantification of the nicked fraction.
Since phosphorothioate 3′ to 8-oxodG inhibits the DNA lyase activity of OGG1 (Figure 1D) we questioned whether OGG1 is still required for the inhibition of transcription by this combination of DNA modifications. We therefore investigated the effect of OGG1 knockdown in HeLa cells on the expression constructs containing 8-oxodG surrounded by all four combinations of phosphodiester and phosporothioate bonds (Figure 2C). We found that OGG1 knockdown clearly moderates the strength of transcriptional inhibition inflicted by 8-oxodG with a phosphodiester bond on the 5′ side, regardless of the nature of the linkage 3′ to the modified nucleotide. This suggests that the 5′ incision is dependent on the specific OGG1-8-oxoG interaction, and this interaction takes places also in the presence of the 3′ phosphorothioate. We have further verified the integrity of the expression constructs in both cell types by re-isolation of DNA from transfected cells and real-time quantitative PCR with primers enclosing the modification site (Supplementary Methods). Previously, we have shown that single 8-oxoG does not result in degradation of analogous expression constructs in HeLa cells at least over 24 h post-transfection (29). Now we have performed the same type of analyses in isogenic cell lines with different OGG1 expression statuses containing 8-oxodG and various combinations of adjacent phosphodiester and phosporothioate linkages over the extended time of 48 h (Supplementary Figure S1). We observed very minor (within the error range of the method) stochastic variation in the amounts of recovered DNA. The EGFP gene copy numbers recovered from cells were similar, regardless of the type of modification present, and there was no difference between the control and OGG1-knockdown cell lines. Nevertheless, the protein expression was strongly affected in cells transfected with both constructs containing 8-oxodG with unprotected 5′-linkages, and these effects were partly attenuated by the knockdown of OGG1.
The observation that 5′ phosphorothioate protects from the negative effect of 8-oxodG on the gene transcription even if a normal phosphodiester bond is retained on the 3′ side (Figure 2A and C) made us suggest that the 3′ bond might not be cleaved at all under physiological BER conditions. We measured nicking activities in extracts of OGG1 overexpressing HeLa cells towards the same 8-oxodG-containing circular DNA constructs, which were used for the aforementioned gene expression analyses. At equivalent cell extract concentrations, the substrate containing phosphorothioate 3′ to 8-oxodG was nicked with 70–85% efficiencies, compared to the substrate with phosphodiester linkages on both sides of 8-oxodG. Under the same conditions, the specific cleavage was almost completely inhibited in both substrates containing phosphorothioate 5′ to 8-oxodG. Even if the 3′ phosphodiester bond was retained, the yield of nicked DNA was almost as low as in the absence of 8-oxoG (Figure 2D). It is striking that the 3′ bond cleavage was not favoured over the 5′ incision, in spite of the elevated >10-fold OGG1 expression and activity in these cell extracts (30,34). Such mode of strand cleavage was also observed when vector constructs containing 8-oxodG and the specified phosphorothioates were incubated with limited amounts of OGG1 in the presence of recombinant human APE1 (Supplementary Figure S2). In summary, these results demonstrate that DNA strand cleavage under cell-free conditions takes place preferably at the linkage 5′ to 8-oxodG, which strengthens the earlier conclusion about the requirement of APE1 for strand cleavage in vivo. Such a mechanism is in agreement with the crucial role of the APE1-generated repair intermediate for the inhibition of transcription in cell transfection assays, described above (Figure 2A–C).
8-OxoG inhibits expression of genes transcribed at different rates
Sarasin et al. have previously suggested that 8-oxoG might only affect expression of genes containing sufficiently strong promoters, as were two viral promoters used by their group (26). The new knowledge that the inhibition of transcription requires a BER-generated single strand break (ssb) could rationalise this hypothesis, assuming that the probability for transcribing RNA polymerase to encounter the repair intermediate is proportional to the frequency of transcription cycles. Since EGFP transcription was driven by a potent CMV-IE promoter in the pZAJ vector described above, we wondered whether decreasing the promoter strength would obliterate or modulate the indirect inhibition of transcription by 8-oxoG. To slow down the transcription rate without changing the promoter structure, we introduced two TetO2 sequence elements into the gene's 5′-untranslated region and performed further expression analyses in T-REx™-HeLa cells (Figure 3). These cells stably express the TetR protein, whose binding to TetO2 restrains the rate of transcription to a very low level. Addition of tetracycline to the culture medium resulted in a >20-fold induction of the EGFP expression in cells transiently transfected with the TetO2-regulated reporter (calculated from Figure 3C), which corresponded to 40–60% strength of the unmodified CMV-IE promoter. When 8-oxoG was introduced into the EGFP gene, the expression levels where negatively affected at both inductive and repressive conditions (Figure 3C). Correction of the values obtained under each conditions with respect to the expression levels of the reference construct (without 8-oxoG) revealed that relative strength of the negative effect of 8-oxoG was the same in cells incubated with or without tetracycline. Measured 24 h post-transfection, the reduction of the EGFP expression was in the range of 40–45% (Figure 3D), which is also very close to values previously obtained for the unrestrained CMV-IE promoter (Figure 2D). Eight hours post-transfection (the earliest time point when the expression could be reproducibly measured in T-REx™-HeLa cells), the harmful effect was not yet detectable, again resembling the situation in HeLa and thus suggesting that processing of 8-oxoG is not perturbed by the TetR expression in cells. In summary, the results show that 8-oxoG is harmful for gene expression within the broad (at least 20-fold) range of transcription rates.
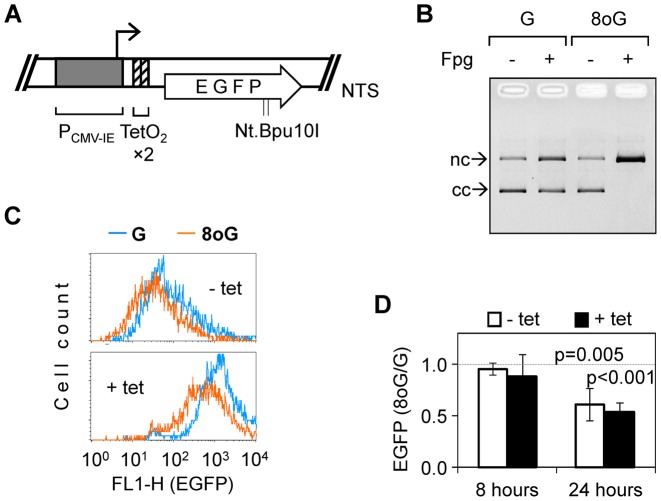
Figure 3.
Inhibitory effect of single 8-oxoG on the expression of the inducible EGFP gene. (A) Scheme of the tetracycline-regulated (tet-on) EGFP expression vector. Two TetR binding motifs (TetO2 ×2) were introduced into the 5′-untranslated region without affecting the protein-coding sequence. Tandem Nt.Bpu10I nicking sites were retained and used for incorporation of synthetic oligonucleotides, as above. (B) Verification of the incorporation of 8-oxoG into the tet-on vector. DNA strand scission analysis of constructs produced with unmodified synthetic oligonucleotide (G) or the oligonucleotide containing single 8-oxoG (8oG). (C) Representative fluorescence distribution plots of T-REx™-Hela cells 24 h post-transfection with the expression constructs containing synthetic oligonucleotides with or without 8-oxoG. Cells were incubated in the absence (- tet) or in the presence of tetracycline (+ tet). (D) Relative EGFP expression calculated at the uninduced (- tet) and induced (+ tet) conditions (n = 6, ± SD).
Deletion mutants of the CMV-IE promoter are susceptible to the inhibitory effect of 8-oxoG
The results described above strongly suggest that the observed negative effect of 8-oxoG on the gene transcription is mediated not at the level of elongation, but rather initiation. This would require a signalling mechanism, which would lead to gene repression—either by promoting an inactive chromatin structure incapable of binding transcriptional activators or by modulating interactions of an activator or repressor with a specific site in the promoter. To inspect whether such regulatory DNA region is present in the CMV-IE promoter, we investigated the effect of single 8-oxoG on the expression driven by several truncated versions of the promoter. Of the multiple transcription factor-binding motifs present in the DNA sequence, cAMP response elements (CRE) are of primary importance for high transcriptional activity of CMV-IE promoter in HeLa cells (37). We generated promoter deletions in which 1, 2, or 3 of 4 available full-length 5′-TGACGTCA-3′ CRE sites were eliminated (Figure 4A). As expected, promoter strength progressively declined with the increasing deletion scale. Judged on the measured EGFP expression, CMV_1111 promoter (in which all four canonical CRE sites were preserved) was the most active, followed by CMV_1011 (3 CRE sites), CMV_0011 (two CRE sites) and finally CRE-Uno (consisting of a single CRE site), which produced the lowest, but still measurable, EGFP expression level (Figure 4B).
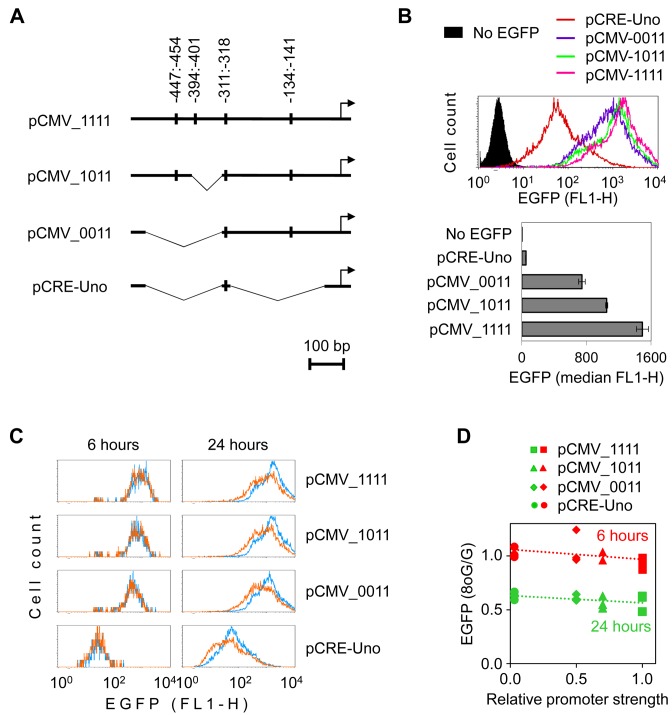
Figure 4.
Deletion mutants of the CMV-IE promoter are susceptible to the inhibition of the reporter gene expression by single 8-oxoG. (A) List of vectors containing the modified CMV_1111 promoter or its truncated versions together with the map of the introduced deletions. Batons show the canonical CRE sequences. (B) Fluorescence distribution plots and quantification of EGFP expression in HeLa cells 24 h post-transfection with the specified promoter constructs without artificially introduced base modifications. (C) Overlaid EGFP fluorescence distribution plots of cells transfected with vectors containing synthetic DNA strand with one 8-oxoG (amber) and those containing the respective unmodified oligonucleotide (blue). (D) Impact of single 8-oxoG on the gene expression as a function of promoter strength. Data of three independent experiments and the best-fit linear regression lines. EGFP expression was measured at 6 and 24 h post-transfection and calculated relative to expression of the matched construct without 8-oxoG.
We further incorporated synthetic oligonucleotide containing single 8-oxoG into the same position in the EGFP gene as described previously (Figure 1A). After the presence of 8-oxoG was verified in all constructs (Supplementary Figure S3), we performed expression analyses in HeLa cells transfected in parallel with each promoter construct containing either single 8-oxoG or no base modification in the inserted DNA strand. The negative effect of 8-oxoG on the EGFP expression levels measured at 24 h post-transfection was clearly present in all analysed promoter constructs (Figure 4C). As observed earlier with the CMV-IE promoter, this effect did not yet manifest at 6 h post-transfection, which indicates that the same amounts of transcription-competent ‘G’ and ‘8-oxoG’ constructs have been delivered to cells by transfection. Again, as in case of the CMV-IE promoter, the strength of the negative effect of 8-oxoG was moderated by the OGG1 knockdown in all promoter variants, regardless of the deletion size (Supplementary Figure S4). These results show that excision of 8-oxoG is universally required for the inhibition of transcription and that the mechanism of transcriptional repression is not limited to a peculiar sequence element within the CMV promoter. Best-fit lines generated by plotting the effect of 8-oxoG against the promoter strength (Figure 4D) had slopes close to zero, further indicating that the rate of transcription does not have a major effect on the magnitude of the inhibitory effect of 8-oxoG—in agreement with results obtained with the tetracycline-inducible promoter, described previously (Figure 3).
Repression of transcription in response to 8-oxoG does not require the CRE motif
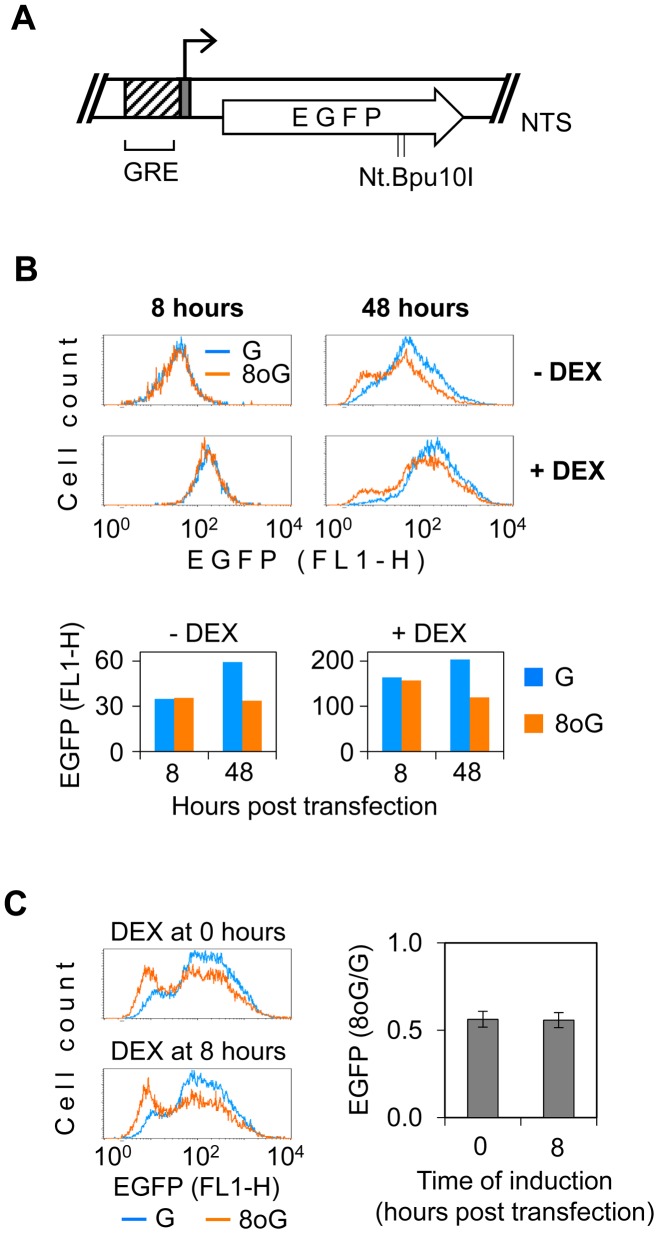
CRE is the response element for the CREB/CREM/ATF1 family of multifunctional transcription factors, which are involved in both positive and negative transcriptional regulatory mechanisms. Since at least one CRE site was present in all hitherto analysed promoters, we questioned whether transcriptional silencing by 8-oxoG is mediated by a specific repressor binding to CRE sequence. We therefore substituted the entire CMV-IE promoter with an artificial promoter, which consists of a glucocorticoid response element (GRE) fused to a minimal TK promoter and does not contain the consensus CRE sequence (Figure 5A). In the absence of dexamethasone, both TK and GRE-TK promoters yielded very low, but still robustly measurable, EGFP expression levels, similar to those produced by CRE-Uno promoter (data not shown). Dexamethasone induced a >4-fold increase of the EGFP expression in the GRE-TK construct. We inserted synthetic oligonucleotides with and without 8-oxoG into the pGRE-TK vector (Supplementary Figure S3) and measured the EGFP expression both in the absence and presence of dexamethasone. The negative effect of 8-oxoG on the gene expression was clearly manifested at both conditions and its magnitude did not notably differ between the induced and uninduced states (Figure 5B). As in the case of CMV promoter and its modified variants described hitherto, the inhibitory effect of 8-oxoG required some latency time to develop, which is consistent with the necessity of BER incision to take place. Under the induced conditions, transcriptional repression was not initiated by altered recruitment of glucocorticoid receptor to DNA containing 8-oxoG, since it did not matter, whether dexamethasone was added to the medium immediately or with a delay of 8 h (Figure 5C). The results thus indicate that impaired recruitment of activatory factors (glucocorticoid receptor or CREB-family TF) to their target sequences is not a primary mechanism of transcriptional repression by 8-oxoG located outside of the promoter region or by its ongoing BER. Based on the capacity of 8-oxoG to induce silencing of structurally heterologous promoters, we further conclude that the underlying mechanism is likely not mediated by binding of a sequence specific repressor in the proximity to the transcription start site.
Figure 5.
Effect of 8-oxoG on expression controlled by the artificial GR-TK promoter. (A) Elements of the glucocorticoid-regulated EGFP expression vector: minimal TK promoter with transcription start (grey box with broken arrow); EGFP coding sequence (open arrow); two Nt.Bpu10I nicking sites in the NTS, used for the insertion of synthetic oligonucleotides; and glucocorticoid receptor synthetic response element (GRE). (B) Expression of vector constructs containing ‘G’ and ‘8-oxoG’ (overlaid) at the uninduced (- DEX) and induced (+ DEX) conditions. Representative fluorescence distribution plots of cells transfected with the indicated constructs and the median fluorescence values (bar graphs below). (C) EGFP expression under the conditions of co-transfectional (0 h) and delayed (8 h) induction. Representative fluorescence distribution plots and mean EGFP expression, relative to the ‘G’ construct (n = 3, ±SD).
Effect of 8-oxoG on transcription driven by human promoters
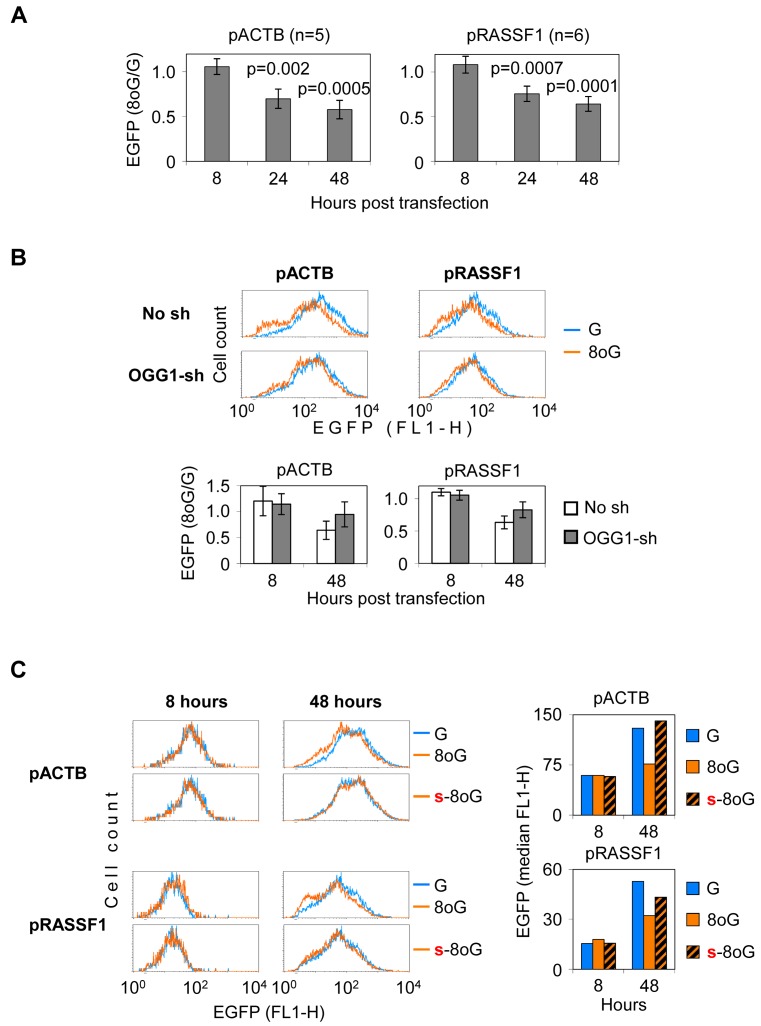
Previous data show that promoters of varying structure and strength are susceptible to silencing, induced by excision repair of 8-oxoG situated in the gene body. Since 8-oxoG is ubiquitously generated in chromosomal DNA during life, there is a risk that transcription of human genes could be affected in a similar way. We therefore questioned whether intracellular processing of 8-oxoG could induce silencing of human promoters. We chose several human promoters which do not contain Bpu10I sites and cloned them upstream of the reporter EGFP gene. These included promoters of tumour suppressor genes often silenced in cancers (APC, RASSF1, RB1) and of the housekeeping gene ACTB. Of these promoters, only ACTB and RASSF1 produced measurable EGFP expression levels (data not shown). Synthetic oligonucleotides with and without 8-oxoG were efficiently incorporated into the respective vectors (Supplementary Figure S3). In both ACTB and RASSF1 constructs transfected into HeLa cells, single 8-oxoG caused a pronounced, progressive decline in the EGFP expression (Figure 6A), indicating that both promoters can be efficiently silenced, just alike the viral and artificial promoters examined previously. As judged from unaltered EGFP expression 8 h post-transfection, unprocessed 8-oxoG did not perturbe the vector delivery to cells or interfere with transcription directly. As in the case of viral CMV promoter, the negative effect of 8-oxoG on the gene expression was attenuated by OGG1 knockdown (Figure 6B), thus indicating that BER is universally required for the manifestation of the reporter gene silencing. Moreover, substitution of the phosphate linkage 5′ to 8-oxoG with a nuclease-resistant phosphorothioate prevented the 8-oxoG-dependent decrease of transcription (Figure 6C), demonstrating that ssb generation is critical for the onset of transcriptional silencing in constructs controlled by human promoters as well.
Figure 6.
Effect of 8-oxoG on expression controlled by human promoters. (A) Mean EGFP expression in HeLa cells transfected with the specified constructs (±SD, Student's two-tailed t-test). (B) EGFP expression in the OGG1 knockdown (OGG1-sh) HeLa cells and the isogenic control cell line (No sh) measured at 48 h post-transfections. Representative fluorescent distribution plots and mean relative expression (bar charts below, n = 4, ±SD). (C) Effect of 5′ phosphorothioate on the expression of the pACTB and pRASSF1 constructs containing single 8-oxodG (8oG, s-8oG). Blue colour shows the control ‘G’ construct. Result representative of three independent experiments.
Release from the 8-oxoG-induced transcriptional repression by trichostatin A
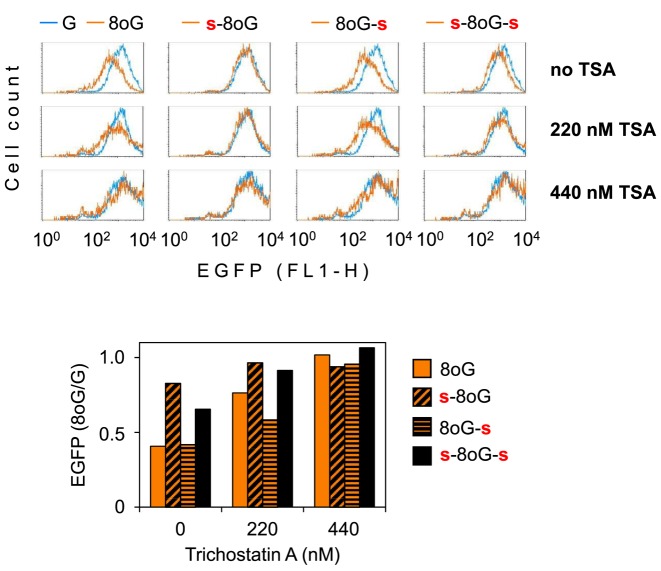
Previously, we have reported that the presence of photooxidative DNA lesions favours histone H4 deacetylation in promoters of expression vectors transfected to cells and concomitantly induces transcriptional silencing, which could be mitigated by the histone deacetylase inhibitor Trichostatin A (28). Therefore, we wanted to know whether transcriptional silencing of the EGFP gene induced by the base excision intermediate of single 8-oxoG (described in the present study) could be as well reversed by this inhibitor. Strikingly, addition of Trichostatin A to cells at the time of transfection dose dependently improved the EGFP expression, up to a complete relief from the negative impact of 8-oxoG (Figure 7). Concentration-dependent attenuation of the negative effect of 8-oxoG by TSA was reproducibly observed in multiple experiments, with some quantitative variation of the magnitude, allegedly because different batches of the inhibitor were used. Full abrogation of the negative effect of 8-oxoG on the gene expression was typically reached in the concentration range between 220 and 440 nM. The results thus confirm that 8-oxoG (or its processing) does not result in a permanent loss of function of the EGFP gene (e.g. by degradation of vector DNA or gene rearrangement) and further support the involvement of a gene silencing mechanism, which could be mediated by histone deacetylases.
Figure 7.
Reversal of the 8-oxoG-induced transcriptional repression by Trichostatin A (TSA). Fluorescence distribution plots of HeLa cells incubated 24 h in the presence of the specified concentrations of TSA. Amber lines show EGFP expression in cells transfected with the specified pZAJ expression constructs containing 8-oxodG flanked by phosphorothioate linkages, as explained in Figure 1. Overlaid blue line shows the result of parallel transfection with the control construct devoid of 8-oxoG. Bar graph below shows quantification of the median EGFP fluorescence. For each TSA concentration, values are normalised relative to the reference construct without 8-oxoG (‘G’). Representative result of at least three (constructs with phosphorothioates) or more independent experiments.
Impacts of AP sites on the gene transcription and the role of APE1
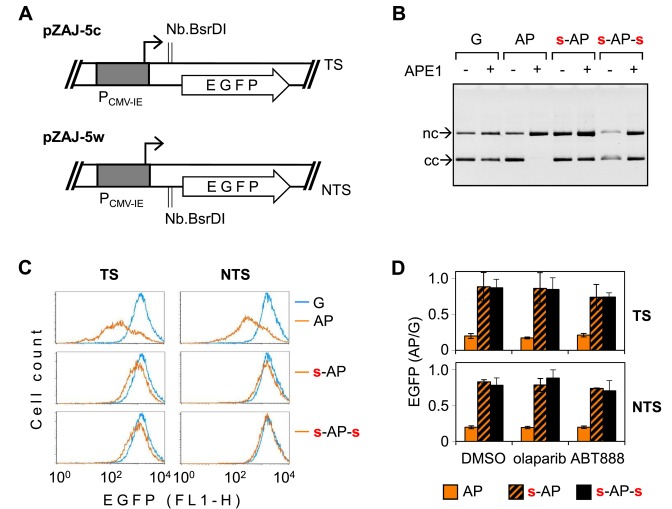
Repeated under all conditions tested above, the observation that 8-oxoG affects the gene expression only in the presence of unprotected bond 5′ to the modified nucleotide supports our previous conclusion that the negative impact on transcription requires APE1 activity. Therefore, it was important to directly verify this critical role of APE1. Because APE1 is an essential enzyme in mammals (both at the organismal and cellular levels) and since the efficient knockdown could not be achieved without inflicting cell toxicity (unpublished results and (38)), we have generated new expression constructs dedicated to this purpose. We incorporated synthetic oligonucleotides containing a synthetic AP site analogue tetrahydrofuran into either the transcribed or the non-transcribed strand of the EGFP gene (Figure 8A). Tetrahydrofuran is a very good substrate of APE1 (36) but, differently from the natural abasic site, resistant to beta elimination reactions (catalyzed by lyases) because of the absence of the aldehydic C-1′ carbonyl group—properties ideally suited for the investigation of the consequences of cleavage by APE1. In vitro cleavage analyses of the generated constructs incubated with human APE1 confirmed a very efficient incorporation of synthetic AP sites into vector DNA as well as a strong inhibition of the APE1 endonuclease activity by the 5′ phosphorothioate (Figure 8B). Expression analyses of constructs with one incorporated synthetic AP site in either the transcribed or the non-transcribed DNA strand in HeLa cells revealed a powerful negative effect of the modification on the EGFP expression (Figure 8C). Furthermore, in both strands this effect was almost entirely obliterated when the AP site was protected from the APE1 cleavage by a phosphorothioate linkage at the 5′ side. Incoporation of the second phosphorothioate 3′ to the AP site did not have any additional influence on the gene expression. In summary, the results indicate that uncleaved AP sites in the non-coding region barely have any significant impact on the gene expression and clearly attribute the decline of transcription to the AP endonuclease activity at the unprotected 5′ bond.
Figure 8.
Inhibition of the EGFP gene expression by single apurinic site (AP). (A) Vectors used for site-specific insertion of synthetic oligonucleotides containing a single AP lesion (tetrahydrofuran) on the place of dG in the transcribed (TS) or the non-transcribed (NTS) strand of the EGFP gene. (B) Activity of APE1 toward plasmid substrates containing single synthetic AP lesion and the effects of the specified phosphorothioate linkages. (C) Fluorescence distribution plots of HeLa cells 24 h post-transfection with the expression constructs containing the indicated modifications in the specified DNA strand (TS or NTS), compared to the ‘G’ construct (overlaid blue line). Analogous plots for cells incubated in parallel with PARP inhibitors are shown in Supplementary Figure S5. (D) Mean relative EGFP expression of the specified constructs in the absence (DMSO) and in the presence of the indicated PARP inhibitors (n = 3, ±SD).
BER of several types of DNA lesions can be hindered by the poly(ADP-ribose) polymerase (PARP) inhibitor olabarib, which stabilises binding of PARP1 and PARP2 to the ssb intermediate (39,40). To test whether such a mechanism is of relevance for the strand-cleaved intermediate generated by the AP endonuclease, we also measured the expression of constructs containing synthetic AP sites in cells treated with PARP inhibitors olaparib or ABT-888 (which differ in their capacities to induce PARP trapping on DNA (39)). We observed no modulation of the magnitude of the negative effect of synthetic AP sites on the gene expression, indicating that PARP catalytic activity is not required for either sustainment or repression of transcription in the presence of the AP endonuclease-induced ssb (Figure 8C and Supplementary Figure S5). Even though PARP activity was not important for the processing of the synthetic AP sites, its potential relevance for a naturally generated BER intermediate has not yet been excluded, because PARP1 also can form covalent complexes with DNA by the beta elimination mechanism of the deoxyribose phosphate (41), which would not work with the tetrahydrofurane AP site analogue. For this reason, we have further tested both PARP inhibitors on the expression constructs containing 8-oxoG. However, as in the case of synthetic AP sites, the inhibitors did not at all modulate the magnitude of the effect of 8-oxoG on the gene expression (Supplementary Figure S6). Likewise, the construct with phosphorothioate 3′ to 8-oxodG showed the same degree of the inhibition of the EGFP expression as the construct with unmodified backbone. The results thus indicate that PARP activity is not relevant for the processing of 8-oxoG in transcribed DNA.
DISCUSSION
Sustainment of transcription is an essential function of DNA in all cell types. There is massive experimental evidence in vitro and in vivo that damage to DNA affects this housekeeping function (19), however, transcriptional responses of genes to physiologically low levels of DNA lesions in unstressed cells remain largely unexplored. Expression vectors carrying structurally defined nucleobase modifications provide a valuable tool for closing this knowledge gap (23,24,29,35,42). We previously demonstrated that, in repair-proficient cells, OGG1-dependent suppression of transcription of gene constructs containing synthetic 8-oxoG strongly prevails over the potential phenotypic effects of erroneous transcriptional bypass of the lesion or direct blockage of elongating RNA polymerase complexes (29,30). The requirement of OGG1 suggested that the inhibition of transcription is initiated either by unproductive binding of OGG1 to 8-oxoG in DNA or by one of the post-excision products.
In order to identify the exact nature of the harmful BER intermediate, we artificially intervened with the efficiencies of individual endonucleolytic reactions, catalysed by different enzymes of the BER pathway, by the means of substitution of phosphodiester linkages on either side of 8-oxodG with nuclease-resistant phosphorothioates. This approach turned out to be very useful, since the results showed that, on the one hand, the presence of phosphorothioates was compatible with gene expression analyses in transfected cells, without influencing the gene expression to any significant extent (Figure 2A, early time point). On the other hand, phosphorothioates in respective positions substantially inhibited backbone incisions 3′ to 8-oxodG (by OGG1 in the beta-lyase mode) and 5′ to the lesion (by cell extracts or by purified APE1 in combination with OGG1), as desired (Figures 1 and 2, Supplementary Figure S2). Combined with the effects of 8-oxodG in different backbone configurations on the gene expression in cells, these results lead to the conclusion that DNA strand cleavage in vivo has a strong preference to the linkage 5′ to 8-oxodG and, consequently, has to be done by APE1. Moreover, we deduce from the gene expression data that strand cleavage never occurs 3′ to the lesion, because this would lead to generation of maleficent strand break with blocked 3′ end in the case of the substrate with 5′ phosphorothioate and 3′ phosphate linkages. Such futile repair intermediate would inevitably impede transcription (43), which is clearly not the case (Figure 2A). Consequently, OGG1 does not manifest a beta-lyase activity in vivo, in this resembling monofunctional DNA glycosylases. For the same reason, any significant contribution of the Nei-homologues NEIL1 and NEIL2 to cellular repair of 8-oxoG should also be excluded (because catalytic mechanism of these bifunctional DNA glycosylases comprises the beta-lyase reaction). In summary, the results provide evidence for a concerted action of OGG1 with APE1. Although the strand cleavage is catalysed by APE1, the overall efficiency of incision is limited by the availability of OGG1, as documented by the OGG1 knockdown.
In order to exert an inhibitory effect on the gene transcription, the 5′ cleaved repair intermediate of 8-oxodG does not have to be situated in the transcribed DNA strand (where it would be encountered by transcribing RNA polymerase II) or in the gene promoter (where it could directly impede or promote binding of transcription factors or co-activators/co-repressors). The observed mode of action would require signal transduction from the damage/repair site to the gene regulatory region, i.e. over the distance of at least 650 nucleotides. The nature of this signal (e.g., topology of DNA itself or molecular interactions of some ssb recognition proteins) remains to be determined. What is clear is that transcriptional repression following the strand incision at 8-oxodG is not mediated by altered binding of a sequence specific protein to DNA, because it affects various promoters with most unrelated structures and regulation principles. Previously, we have demonstrated that plasmid vectors transfected to cells undergo packaging into chromatin (28). It is therefore plausible that transcriptional silencing is mediated by histone modifiers or chromatin remodelling factors, especially considering the observation that the negative effect of 8-oxoG on the gene expression can be prevented by the histone deacetylase inhibitor (Figure 7). Notably, all promoters tested in this study were susceptible to transcriptional repression induced by the BER intermediate of 8-oxoG, which suggests its potential relevance for transcriptional regulation of human genes and is in agreement with a chromatin-mediated silencing mechanism. However, further identification of molecular components of the gene repression mechanism triggered by the specified BER intermediate would be required to judge about the relevance of present findings to regulation of expression of chromosomal DNA. It is important to note that monofunctional DNA glycosylases UNG1/2 and SMUG1 were also shown to elicit transcriptional inactivation of genes containing their respective substrates (32). As in the case of OGG1, BER initiated by these DNA glycosylases requires the APE1 strand cleavage step; therefore, various BER substrates might be capable to induce gene silencing in analogous fashion. In summary, we suggest that induction of transcriptional silencing is one of the most prominent phenotypic consequences of 8-oxoG in DNA and, possibly, of other endogenously generated DNA base modifications processed by BER.
With respect to the role of oxidative DNA base modifications in the regulation of gene transcription, it is important to mention that generation of 8-oxoG (44) and 5-hydroxymethyl uracil (45) in mammalian cells can be coupled with enzymatically catalysed oxidative demethylation of histones (by the LSD1 histone demethylase) and 5-methylated pyrimidines in DNA (by TET1/TET2 dioxygenases). Moreover, it was shown in the estrogen receptor- (ER-) and MYC-activated gene expression models that specific recruitment of LSD1 to the regulatory gene elements in response to the physiological stimuli provides a mechanism for targeted induction of 8-oxoG in the promoter regions (44,46). Strikingly, subsequent recruitment of OGG1 to the DNA damage sites was found essential for transcriptional activation of the target genes in these studies. OGG1 has been further implicated in the induction of proinflammatory genes by at least two different mechanisms. In trans, OGG1 in complex with 8oxoG was shown to stimulate Ras family GTPases (47). In cis, similarly to the ER- and MYC-driven transcription models described above, OGG1 recruitment to promoters enhanced the tumour necrosis factor alpha-induced transcriptional activation (48); however, in the latter case the induction of 8-oxoG in promoter DNA has been ascribed to the increased cellular ROS levels rather than a targeted recruitment of oxidases. Taken together, these findings have marked a new turn in our understanding of the role of oxidatively generated DNA damage and BER proteins in the mechanisms of transcriptional control. We believe that deciphering of these mechanisms and identification of molecular switches that determine the decision between transcriptional activation and repression in cells is an exciting and challenging task in the field.
Supplementary Material
Acknowledgments
The corresponding author thanks Thomas Lingg, Bork Lühnsdorf, Anna Campalans, Pablo Radicella, and Leon Mullenders for stimulating discussions at various stages of the work summarised in this manuscript.
Footnotes
Present address: Andriy Khobta, Institute of Toxicology, University Medical Center Mainz, Mainz 55131, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [KH 263/1 and KH 263/2]; Heisenberg Fellowship [KH 263/3 to A.K.]. Funding for open access charge: DFG (German Research Foundation).
Conflict of interest statement. None declared.
REFERENCES
- 1.Berquist B.R., Wilson D.M., 3rd Pathways for repairing and tolerating the spectrum of oxidative DNA lesions. Cancer Lett. 2012;327:61–72. doi: 10.1016/j.canlet.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dizdaroglu M. Oxidatively induced DNA damage and its repair in cancer. Mutat. Res. Reviews Mutat. Res. 2015;763:212–245. doi: 10.1016/j.mrrev.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Shibutani S., Takeshita M., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 4.Hsu G.W., Ober M., Carell T., Beese L.S. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 5.Sakumi K., Tominaga Y., Furuichi M., Xu P., Tsuzuki T., Sekiguchi M., Nakabeppu Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 2003;63:902–905. [PubMed] [Google Scholar]
- 6.Xie Y., Yang H., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D.E., Lindahl T., McIlhatton M., Fishel R., et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 7.Gedik C.M., Collins A., ESCODD Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 8.Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D.E. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arai T., Kelly V.P., Komoro K., Minowa O., Noda T., Nishimura S. Cell proliferation in liver of Mmh/Ogg1-deficient mice enhances mutation frequency because of the presence of 8-hydroxyguanine in DNA. Cancer Res. 2003;63:4287–4292. [PubMed] [Google Scholar]
- 10.Isogawa A. Functional cooperation of Ogg1 and Mutyh in preventing G: C–>T: a transversions in mice. Fukuoka igaku zasshi. 2004;95:17–30. [PubMed] [Google Scholar]
- 11.Nash H.M., Lu R., Lane W.S., Verdine G.L. The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: direct identification, ablation and chemical reconstitution. Chem. Biol. 1997;4:693–702. doi: 10.1016/s1074-5521(97)90225-8. [DOI] [PubMed] [Google Scholar]
- 12.Fromme J.C., Bruner S.D., Yang W., Karplus M., Verdine G.L. Product-assisted catalysis in base-excision DNA repair. Nat. Struct. Biol. 2003;10:204–211. doi: 10.1038/nsb902. [DOI] [PubMed] [Google Scholar]
- 13.Allinson S.L., Dianova II, Dianov G.L. DNA polymerase beta is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO J. 2001;20:6919–6926. doi: 10.1093/emboj/20.23.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morland I., Luna L., Gustad E., Seeberg E., Bjoras M. Product inhibition and magnesium modulate the dual reaction mode of hOgg1. DNA Repair. 2005;4:381–387. doi: 10.1016/j.dnarep.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Hill J.W., Hazra T.K., Izumi T., Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal A.E., Hickson I.D., Boiteux S., Radicella J.P. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegde M.L., Hazra T.K., Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y.J., Wilson D.M., 3rd Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khobta A., Epe B. Interactions between DNA damage, repair, and transcription. Mutat. Res. 2012;736:5–14. doi: 10.1016/j.mrfmmm.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh R., Mitchell D.L. Effect of oxidative DNA damage in promoter elements on transcription factor binding. Nucleic Acids Res. 1999;27:3213–3218. doi: 10.1093/nar/27.15.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramon O., Sauvaigo S., Gasparutto D., Faure P., Favier A., Cadet J. Effects of 8-oxo-7,8-dihydro-2′-deoxyguanosine on the binding of the transcription factor Sp1 to its cognate target DNA sequence (GC box) Free Radic. Res. 1999;31:217–229. doi: 10.1080/10715769900300781. [DOI] [PubMed] [Google Scholar]
- 22.Hailer-Morrison M.K., Kotler J.M., Martin B.D., Sugden K.D. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2′-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry. 2003;42:9761–9770. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 23.Saxowsky T.T., Meadows K.L., Klungland A., Doetsch P.W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bregeon D., Peignon P.A., Sarasin A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5:e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spivak G., Hanawalt P.C. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair. 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Pastoriza-Gallego M., Armier J., Sarasin A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb(-)/Ogg1(-) DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis. 2007;22:343–351. doi: 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- 27.Khobta A., Kitsera N., Speckmann B., Epe B. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA Repair. 2009;8:309–317. doi: 10.1016/j.dnarep.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Khobta A., Anderhub S., Kitsera N., Epe B. Gene silencing induced by oxidative DNA base damage: association with local decrease of histone H4 acetylation in the promoter region. Nucleic Acids Res. 2010;38:4285–4295. doi: 10.1093/nar/gkq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitsera N., Stathis D., Luhnsdorf B., Muller H., Carell T., Epe B., Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011;39:5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allgayer J., Kitsera N., von der Lippen C., Epe B., Khobta A. Modulation of base excision repair of 8-oxoguanine by the nucleotide sequence. Nucleic Acids Res. 2013;41:8559–8571. doi: 10.1093/nar/gkt620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luhnsdorf B., Kitsera N., Warken D., Lingg T., Epe B., Khobta A. Generation of reporter plasmids containing defined base modifications in the DNA strand of choice. Anal. Biochem. 2012;425:47–53. doi: 10.1016/j.ab.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Luhnsdorf B., Epe B., Khobta A. Excision of uracil from transcribed DNA negatively affects gene expression. J. Biol. Chem. 2014;289:22008–22018. doi: 10.1074/jbc.M113.521807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen E., Kwon K., Coin F., Egly J.M., Klungland A. Transcription activities at 8-oxoG lesions in DNA. DNA Repair. 2004;3:1457–1468. doi: 10.1016/j.dnarep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Amouroux R., Campalans A., Epe B., Radicella J.P. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitsera N., Gasteiger K., Luhnsdorf B., Allgayer J., Epe B., Carell T., Khobta A. Cockayne syndrome: varied requirement of transcription-coupled nucleotide excision repair for the removal of three structurally different adducts from transcribed DNA. PLoS One. 2014;9:e94405. doi: 10.1371/journal.pone.0094405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson D.M., 3rd, Takeshita M., Grollman A.P., Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 37.Schlabach M.R., Hu J.K., Li M., Elledge S.J. Synthetic design of strong promoters. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2538–2543. doi: 10.1073/pnas.0914803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vascotto C., Cesaratto L., Zeef L.A., Deganuto M., D'Ambrosio C., Scaloni A., Romanello M., Damante G., Taglialatela G., Delneri D., et al. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics. 2009;9:1058–1074. doi: 10.1002/pmic.200800638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murai J., Huang S.Y., Das B.B., Renaud A., Zhang Y., Doroshow J.H., Ji J., Takeda S., Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strom C.E., Johansson F., Uhlen M., Szigyarto C.A., Erixon K., Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad R., Horton J.K., Chastain P.D. 2nd, Gassman N.R., Freudenthal B.D., Hou E.W., Wilson S.H. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res. 2014;42:6337–6351. doi: 10.1093/nar/gku288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker D.J., Wuenschell G., Xia L., Termini J., Bates S.E., Riggs A.D., O'Connor T.R. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 43.Khobta A., Lingg T., Schulz I., Warken D., Kitsera N., Epe B. Mouse CSB protein is important for gene expression in the presence of a single-strand break in the non-transcribed DNA strand. DNA Repair. 2010;9:985–993. doi: 10.1016/j.dnarep.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 44.Perillo B., Ombra M.N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., Avvedimento E.V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 45.Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S.K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O., et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 46.Amente S., Bertoni A., Morano A., Lania L., Avvedimento E.V., Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 47.Boldogh I., Hajas G., Aguilera-Aguirre L., Hegde M.L., Radak Z., Bacsi A., Sur S., Hazra T.K., Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. J. Biol. Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ba X., Bacsi A., Luo J., Aguilera-Aguirre L., Zeng X., Radak Z., Brasier A.R., Boldogh I. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J. Immunol. 2014;192:2384–2394. doi: 10.4049/jimmunol.1302472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.