Abstract
Deinococcus bacteria are extremely resistant to radiation, oxidation, and desiccation. Resilience to these factors has been suggested to be due to enhanced damage prevention and repair mechanisms, as well as highly efficient antioxidant protection systems. Here, using mutation-accumulation experiments, we find that the GC-rich Deinococcus radiodurans has an overall background genomic mutation rate similar to that of E. coli, but differs in mutation spectrum, with the A/T to G/C mutation rate (based on a total count of 88 A:T→G:C transitions and 82 A:T→C:G transversions) per site per generation higher than that in the other direction (based on a total count of 157 G:C→A:T transitions and 33 G:C→T:A transversions). We propose that this unique spectrum is shaped mainly by the abundant uracil DNA glycosylases reducing G:C→A:T transitions, adenine methylation elevating A:T→C:G transversions, and absence of cytosine methylation decreasing G:C→A:T transitions. As opposed to the greater than 100× elevation of the mutation rate in MMR− (DNA Mismatch Repair deficient) strains of most other organisms, MMR− D. radiodurans only exhibits a 4-fold elevation, raising the possibility that other DNA repair mechanisms compensate for a relatively low-efficiency DNA MMR pathway. As D. radiodurans has plentiful insertion sequence (IS) elements in the genome and the activities of IS elements are rarely directly explored, we also estimated the insertion (transposition) rate of the IS elements to be 2.50 × 10−3 per genome per generation in the wild-type strain; knocking out MMR did not elevate the IS element insertion rate in this organism.
Keywords: Deinococcus radiodurans, mutation accumulation, mutation rate, mutation spectrum, DNA mismatch repair
Introduction
Because mutations are the ultimate source of genetic variation, information on their rate and molecular spectra is essential for understanding most evolutionary processes. G/C and A/T nucleotides vary in mutation rate, with a majority of surveyed organisms exhibiting a G/C→A/T mutation bias (Lynch 2007; Hershberg and Petrov 2010). Because the A/T mutation bias often exceeds the standing genome-wide nucleotide content of these organisms, it has been suggested that GC-content variation among species is strongly influenced by natural selection and/or biased gene conversion (Petrov and Hartl 1999; Keightley et al. 2009; Balbi et al. 2009; Hershberg and Petrov 2010; Hildebrand et al. 2010; Lynch 2010b). However, the mutation spectrum (i.e., the frequency distribution of mutation types) has been directly observed in only a few model organisms (Ossowski et al. 2010; Denver et al. 2009, 2012; Lee et al. 2012; Sung, Ackerman, et al. 2012; Schrider et al. 2013; Zhu et al. 2014). To fully understand the variation in genome-wide nucleotide content that exists across the Tree of Life, it is necessary to expand experimental assays to more diverse organisms.
Two biochemical properties of DNA can contribute to a G/C→A/T mutation bias. First, in vivo evidence shows that spontaneous hydrolytic deamination of cytosine or 5-methyl cytosine, which, respectively, produce C:G→U:G or C:G→T:G mismatches (Duncan and Miller 1980; Ehrlich et al. 1986; Lind and Andersson 2008; Hershberg and Petrov 2010; Lee et al. 2012), and if uncorrected lead to G:C→A:T transitions in the next round of DNA replication. In most organisms, erroneous uracils are removed by uracil-DNA glycosylases (UDG) (McCullough et al. 1999; Kavli et al. 2002). Second, formation of G:C→T:A transversions can be driven by the oxidation of guanine to 8-oxoguanine by reactive oxygen species (ROS), a ubiquitous mechanism that exists in all organisms (see review in Grollman and Moriya 1993). 8-oxoguanine derived from a resident guanine can pair with adenine, generating G:C→T:A transversions (Cheng et al. 1992; Michaels et al. 1992; Grollman and Moriya 1993; Osterod et al. 2001; Lind and Andersson 2008; Lee et al. 2012). Given that these biochemical processes are mutagenic, bacteria have evolved multiple repair mechanisms to remove 8-oxoguanines from unincorporated nucleotides and DNA strands (Grollman and Moriya 1993).
Deinococcus radiodurans is a particularly interesting bacterium for studying mutation and repair processes. Its extreme resistance to radiation, desiccation, and oxidation has lead researchers to propose that this organism harbors highly refined damage prevention and repair mechanisms (Anderson et al. 1956; White et al. 1999; Cox and Battista 2005; Krisko and Radman 2013). Such mechanisms are the ROS-scavenging/detoxifying manganese complex and proteins with Fe-S clusters that can be used to highly efficiently remove ROS (Mennecier et al. 2004; Cox and Battista 2005; Zahradka et al. 2006; Slade et al. 2009; Slade and Radman 2011; Krisko and Radman 2013). Surprisingly, a critical DNA repair system, DNA mismatch repair (MMR), appears to be less efficient in D. radiodurans than in other organisms. For example, a knockout experiment removing the critical MMR enzyme mutL resulted in only a 5-fold elevation in the mutation rate of D. radiodurans (Mennecier et al. 2004), whereas the same knockout in Escherichia coli yields a 138-fold elevation of the mutation rate (Lee et al. 2012).
To further evaluate whether the unique biology of D. radiodurans gives rise to unusual aspects of replication fidelity, we performed mutation-accumulation (MA) experiments using D. radiodurans MMR+ and MMR− strains. In an MA experiment, replicate cell lines are repeatedly taken through single-cell bottlenecks, allowing for the accumulation of all but the most deleterious mutations. For the wild-type D. radiodurans strain BAA-816, we find that mutations are biased toward G/C instead of A/T, and we propose that this unusual feature is likely a consequence of the evolution of multiple UDGs, DNA cytosine methyltransferase (Dcm) absence, DNA adenine methylase (Dam) presence, and additional antioxidation mechanisms. Despite this change in mutation spectrum relative to other studied bacteria, D. radiodurans has an overall mutation rate comparable to other microbes. This suggests that although differences in the fidelity of different mechanisms can exist, the limit to overall DNA replication fidelity may be restricted by other forces, for example, those proposed by the drift barrier hypothesis (Lynch 2012; Sung, Ackerman, et al. 2012).
Results
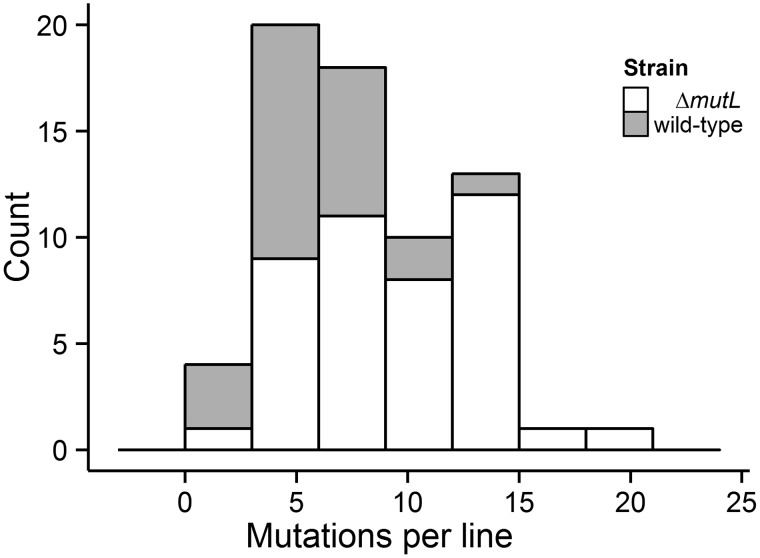
All MA lines in each strain were initiated from a single cell, and repeatedly bottlenecked by streaking single colonies, every 2 days onto fresh nutrient agar plates supplemented with 1% glucose. For the wild-type BAA-816, 43 MA lines were initially established. Cells from a single colony near the end of the streaking were streaked onto a new plate and incubated at 30 °C. The cell lines went through 250 transfers over 636 days, each undergoing approximately 23.8 generations per transfer, yielding 5,961 generations per line. For the mutL− strain, 24 MA lines were initiated from a single mutL− progenitor cell, and maintained with the same culturing and transferring techniques that were applied to the wild-type. Each mutL− growth cycle represented about 23.7 generations, with 42 transfers performed in 84 days (993 total generations per line).
Chromosomal Mutation Rates
Deinococcus radiodurans has two chromosomes, one plasmid (CP1) and one megaplasmid (MP1). In total, 388 base-pair substitutions (BPSs; figs. 1 and 2, open bars; supplementary table S1 and fig. S1, Supplementary Material online), partitioned into 245 transitions and 143 transversions (transition/transversion ratio 1.71), accumulated on the two chromosomes across 43 wild-type MA lines, yielding a genome-wide BPS mutation rate of 4.99 × 10−10 per site per generation, with a 95% Poisson CI of (4.50 × 10−10, 5.51 × 10−10) (table 1) or 0.0015 BPS mutations per genome per generation. Chromosome 1 and chromosome 2, respectively, accumulated 331 (transition/transversion ratio 1.65) and 57 (transition/transversion ratio 2.17) BPSs, leading to BPS mutation rates of 4.91 × 10−10 and 5.47 × 10−10 per site per generation, which are not significantly different from each other (χ2 = 0.46, df = 1, P = 0.50). In total, 17 small indels (<10 bp; supplementary table S1, Supplementary Material online) were also detected on the two chromosomes, 4.20% of all detected chromosomal mutations, implying a small-indel mutation rate of 2.17 × 10−11 (1.47 × 10−11, 3.06 × 10−11) per site per generation (table 1). 52.94% of the small indels occurred in simple sequence repeats (SSRs), for example, homopolymer runs (supplementary table S2, Supplementary Material online). We also performed a fluctuation test on the wild-type strain using rifampicin reporter construct, finding a mutation rate of 3.28 × 10−9 per locus per generation, which is one order of magnitude lower than previously reported (Mennecier et al. 2004). The per site mutation rate of 0.99 × 10−10 is calculated by dividing 3.28 × 10−9 by 33, the number of sites that lead to rifampicin resistance in D. radiodurans (Kim et al. 2004); and this value is one-fifth of the above 4.99 × 10−10 per site per generation mutation rate from whole-genome sequencing of MA lines, consistent with the 1/6 ratio found in E. coli (Lee et al. 2012).
Fig. 1.
Frequency distribution of mutations in the wild-type and mutL− MA lines.
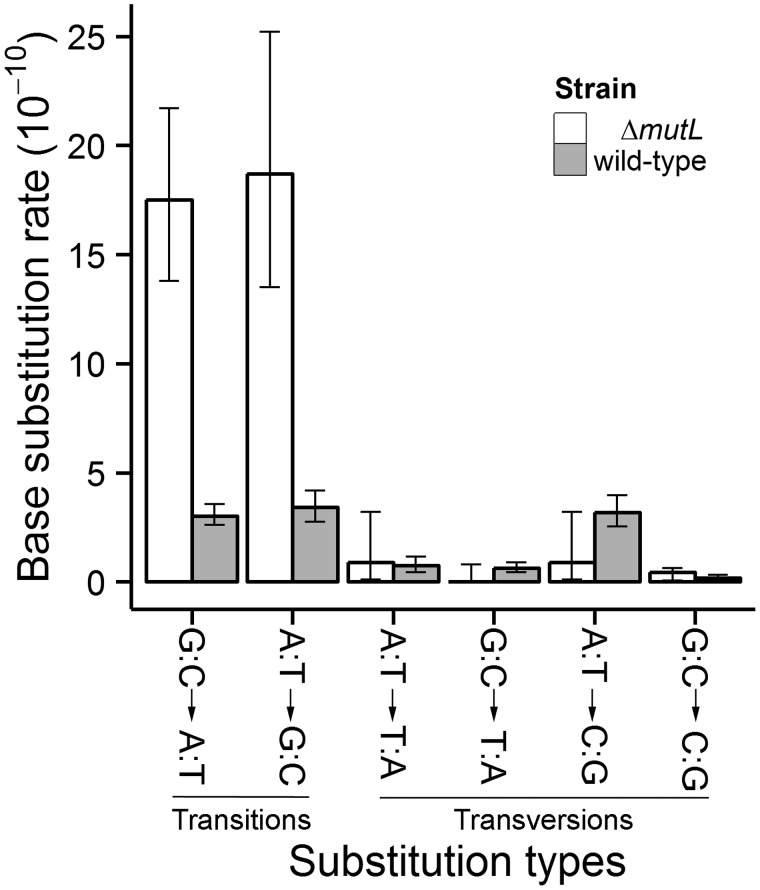
Fig. 2.
Conditional mutation rates of different types of base substitutions in wild-type and mutL− Deinococcus radiodurans. Error bars are 95% Poisson confidence intervals.
Table 1.
Base Substitution and Small-Indel Mutation Rates for Deinococcus radiodurans Chromosomes and Plasmids.
| Mutations | Strains | Chromosomes | CP1 | MP1 |
|---|---|---|---|---|
| BPSs | mutL+ | 4.99 (4.50, 5.51) | 15.27 (8.89, 24.45) | 7.21 (4.93, 10.18) |
| mutL− | 18.59 (15.50, 22.12) | 10.66 (0.27, 59.37) | 12.48 (4.05, 29.12) | |
| Indels | mutL+ | 0.22 (0.15, 0.31) | 0.89 (0.02, 5.00) | 0.23 (0.006, 1.26) |
| mutL− | 4.68 (3.20, 6.61) | 0.00 (0.00, 3.31) | 0.68 (0.14, 1.98) |
Note.—All numbers are in units of 10−10 per site per generation; 95% Poisson confidence intervals are in parentheses.
For the mutL− lines, 127 chromosomal BPSs accumulated over 993 generations (fig. 1, gray bars; supplementary table S3 and fig. S1, Supplementary Material online), yielding a BPS mutation rate of 1.86 × 10−9 (95% CI: 1.55 × 10−9, 2.21 × 10−9) per site per generation, 3.75 × higher than the wild-type mutation rate (table 1). This elevation in mutation rate is consistent with the 5-fold inflation observed in the same mutL− strain using fluctuation tests with rifampicin (Mennecier et al. 2004). In these lines, chromosome 1 and chromosome 2 accumulated 102 (98 transitions, 4 transversions) and 25 (23 transitions and 2 transversions) BPSs, yielding different chromosome-specific BPS mutation rates of 1.73 × 10−9 and 2.70 × 10−9, respectively. In total, 32 small indels (<7 bp) were also detected across the two chromosomes (supplementary table S4, Supplementary Material online), leading to a small-indel mutation rate of 4.68 × 10−10 (3.20 × 10−10, 6.61 × 10−10) per site per generation (table 1), and equivalent to 20% of all detected mutations. As with the wild-type strain, a large fraction (90.6%) of the small indels involved SSRs. The small-indel mutation rate in this MMR knockout strain is 21.57 × higher than that of the wild-type rate.
Mutation Spectrum
Among the BPS mutations in the wild-type MA lines, there are 157 G:C→A:T transitions and 33 G:C→T:A transversions at G:C sites, yielding a mutation rate in the A/T direction of = 3.62 × 10−10 per site per generation. In total, 88 A:T→G:C transitions and 82 A:T→C:G transversions yield a mutation rate in the G/C direction of = 6.56 × 10−10 per site per generation (fig. 2, gray bars; table 2), which is significantly higher than (χ2 = 32.14, df = 1, P = 1.44 × 10−8). Given these differences in the conditional A/T↔G/C mutation rates, the expected GC content at mutation equilibrium is 64% with a standard error of 8% (see notes on calculation in Statistics and Calculations in Materials and Methods), not significantly different from the actual chromosomal GC content of 66.97%.
Table 2.
Chromosomal Base-Substitution Mutation Spectrum in Deinococcus radiodurans.
| Substitutions | Counts | Fraction | Counts | Fraction |
|---|---|---|---|---|
| (mutL+) | (mutL+) | (mutL−) | (mutL−) | |
| Intergenic regions | 57 | 0.147 | 16 | 0.125 |
| Coding regions | 331 | 0.853 | 111 | 0.874 |
| Synonymous | 93 | 0.240 | 39 | 0.305 |
| Nonsynonymous | 236 | 0.608 | 71 | 0.559 |
| Overlapping CDS | 2 | 0.005 | 1 | 0.008 |
| Transitions | 245 | 0.631 | 121 | 0.953 |
| A:T→G:C | 88 | 0.227 | 42 | 0.328 |
| G:C→A:T | 157 | 0.405 | 79 | 0.622 |
| Transversions | 143 | 0.369 | 6 | 0.047 |
| A:T→T:A | 19 | 0.049 | 2 | 0.016 |
| A:T→C:G | 82 | 0.211 | 2 | 0.016 |
| G:C→C:G | 9 | 0.023 | 2 | 0.016 |
| G:C→T:A | 33 | 0.085 | 0 | 0.000 |
Note.—Mutations in overlapping CDS refer to base substitutions falling on nucleotides in overlapping reading frames, which are distinguished from nonsynonymous/synonymous mutations. Fraction is the fraction of all chromosomal base substitution mutations (388 for wild-type, 127 for mutL−).
On the mutL− MA-line chromosomes, 121 transitions and 6 transversions accumulated over the experiment. In these lines, the conditional mutation rates of = 1.72 × 10−9 and = 1.96 × 10−9 per site per generation are not significantly different from each other (χ2 = 0.33, df = 1, P = 0.56), and the expected GC content at mutation equilibrium is 53.16%. Relative to the wild-type rates, the G:C→A:T transition rate is elevated to 1.72 × 10−9 (5.75 × greater than wild-type) and the A:T→G:C transition rate to 1.88 × 10−9 (5.53 × greater than wild-type) per site per generation (fig. 2, blank bars; table 2). In contrast, the mutation rates of global transversions (wild-type Poisson 95% confidence interval 1.55 × 10−10, 2.17 × 10−10 overlaps with that of mutL−: 0.32 × 10−10, 1.91 × 10−10) and individual types of transversions do not differ significantly between MMR+ and MMR− MA lines (all Poisson 95% confidence intervals overlap, supplementary tables S1 and S3, Supplementary Material online; fig. 2).
Minimal Selection during the MA Experiments
The motivation for the MA design, now a standard procedure in experiments of this sort (Kibota and Lynch 1996; Halligan and Keightley 2009), is that intense population bottlenecking results in the fates of spontaneous mutations being primarily determined by random genetic drift. To confirm that the influence of selection was indeed minimal during the MA experiment, we identified the functional context of each mutation.
For the mutL+ lines, 236 nonsynonymous and 93 synonymous mutations were observed (supplementary tables S1 and S5, Supplementary Material online). Given the codon usage in D. radiodurans and the observed transition to transversion ratio (1.71, supplementary table S1, Supplementary Material online), the expected nonsynonymous:synonymous mutation ratio is 2.61 in the absence of selection (supplementary table S6, Supplementary Material online), which is not significantly different from the observed ratio of 2.54 (χ2 = 0.006, df = 1, P = 0.94). Thus, selection had a minimal effect on the accumulation of mutations, as has been found in all prior studies of this sort (see reviews in Halligan and Keightley 2009; Lynch 2010a).
The mutL− MA experiment yielded an elevated transition/transversion ratio (20.17 vs. 1.71 in the wild-type MA lines), and because transitions are more prone to be synonymous, this causes the neutral expectation for the nonsynonymous:synonymous mutation ratio to be 1.97 (supplementary table S7, Supplementary Material online). In these lines, 71 nonsynonymous and 39 synonymous mutations accumulated (supplementary table S8, Supplementary Material online), and the ratio (1.82) is again not significantly different from the random expectation (χ2 = 0.02, df = 1, P = 0.89).
Mutations in Plasmids
The small plasmid (CP1) has a size of 46 kb and the megaplasmid (MP1) 177 kb. In both the wild-type and mutL− MA lines, CP1 and MP1 have on average 1.26× and 1.68× the sequencing depth of the primary chromosomes, respectively. Thus, based on a previous finding that the two primary chromosomes are generally present in 4−10 copies per cell (Makarova et al. 2001), we estimate that CP1 has approximately 5−13 and MP1 approximately 7−17 copies per cell, which is consistent with a prior estimate of six CP1 copies per cell (Daly et al. 1994). In the wild-type MA lines, the BPS mutation rates of CP1 and MP1 are, respectively, 3.06× and 1.44× higher than that of the primary chromosomes (table 1 and supplementary tables S9−S12, Supplementary Material online).
The small-indel mutation rate of CP1 is 3.87 × higher than that of the chromosomes, whereas that of the MP1 is almost identical to the chromosome rate (table 1). After mutL was knocked out, the small-indel mutation rate of MP1 was elevated to 2.96 × that of the wild-type MP1, although the CP1 small-indel mutation rate in the mutL− MA lines is not significantly different from that in wild-type lines (table 1; 95% Poisson confidence intervals overlap). The small-indel mutation rate of both plasmids is lower than that of the chromosomes in mutL− lines.
Owing to the fact that the nontranscribed strand in coding regions remains unprotected and single-stranded during transcription, the mutation rate in coding regions might be higher than that of intergenic regions (Datta and Jinks-Robertson 1995; Beletskii and Bhagwat 1996). However, we did not observe any elevation of the mutation rate in coding relative to intergenic regions, in either the chromosomes or plasmids (table 3), suggesting that if transcription-induced mutagenesis occurs in this bacterium, it has only a small quantitative effect.
Table 3.
Mutation Rates in Different Genomic Regions of Wild-Type and mutL− Lines.
| Strains | P | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr1 | mutL+ | 52 | 7.05 | 279 | 4.61 | 287,564 | 2,362,343 | 0.65 (0.48, 0.87) | 0.002* | |
| mutL− | 13 | 18.97 | 89 | 15.82 | 287,564 | 2,362,343 | 0.83 (0.47, 1.49) | 0.54 | ||
| Chr2 | mutL+ | 5 | 5.69 | 52 | 5.37 | 34,289 | 379,268 | 0.94 (0.38, 2.35) | 0.89 | |
| mutL− | 3 | 4.03 | 22 | 2.27 | 34,289 | 379,268 | 0.76 (0.23, 2.54) | 0.66 | ||
| CP1 | mutL+ | 10 | 31.49 | 7 | 8.03 | 12,391 | 34,018 | 0.25 (0.10, 0.66) | 0.002* | |
| mutL− | 0 | — | 1 | 12.33 | 12,391 | 34,018 | — | — | ||
| MP1 | mutL+ | 9 | 9.52 | 23 | 6.33 | 36,870 | 141,790 | 0.66 (0.31, 1.43) | 0.29 | |
| mutL− | 1 | 11.38 | 4 | 11.83 | 36,870 | 141,790 | 1.04 (0.12, 9.31) | 0.97 |
Note.—All mutation rates are in units of 10−10 per site per generation. , number of mutations in intergenic regions; , intergenic region mutation rate; , number of mutations in coding regions; , coding region mutation rate; , number of intergenic sites; , number of coding sites excluding sites covered by more than one reading frames; numbers in parentheses are 95% confidence intervals from the odds ratio logit method.
*One-sided P values from the odds ratio logit method which shows a significantly lower mutation rate in coding versus intergenic regions, all other P values were from a two-sided odds ratio logit method.
Insertion Sequence Mutations
Insertion sequence (IS) elements, the simplest mobile genetic elements widely found in prokaryotic genomes, play important evolutionary roles; see review by Mahillon and Chandler (1998). Deinococcus radiodurans has a substantial number of IS elements in the genome—more copies per genome size than Escherichia coli K-12 MG1655 (White et al. 1999; Makarova et al. 2001). In D. radiodurans, our methods detected 640 novel IS insertions in 43 wild-type MA lines and 9 in 19 mutL− MA lines. The overall IS insertion rates (i.e., transposition) for wild-type and mutL− are thus 2.50 × 10−3 and 4.77 × 10−4 insertions per genome per generation (supplementary tables S13–S15, Supplementary Material online). No sole deletion (excision) was found other than fixed deletions compared with the reference genome. The IS insertion rate for wild-type is almost one order of magnitude higher than 2.77 × 10−4 insertions per genome per generation in E. coli wild-type MA lines (6,000 generations) (Lee et al. 2014). This difference in the IS insertion rates between E. coli and D. radiodurans may possibly be from the different IS elements composition in the genomes and the fact that different IS elements have greatly different activities (Lee et al. 2014). Among the 640 IS insertions recovered from wild-type lines, insertion sites of 428 were in protein-coding regions and 212 were in noncoding regions. Taking into account of the relative genome occupancy of each site category and assuming that IS elements insert equally likely into protein-coding and noncoding regions, there is a strong bias toward noncoding regions (binomial test, P = 1.39 × 10−52) (supplementary table S16, Supplementary Material online). Such bias is caused by certain conservative structural motifs, instead of purifying selection (Lee H, unpublished data). Among in-gene truncating insertions, insertion events near 5′ and 3′ ends of genes seem to be elevated compared with other regions of genes (supplementary fig. S2, Supplementary Material online).
Mutation-Rate Alterations at Dam Sites
Target sites of Dam (canonical site: 5′-GATC-3′) and Dcm (5′-CCWTT-3′) are known mutational hotspots in bacteria (Lindahl and Nyberg 1972; Farabaugh et al. 1978), with 6-methylated adenine depurination generating transitions or transversions after postreplicative repair and 5-methylated cytosine deamination generating C→T transitions. Though Dam activity has been reported in D. radiodurans (Loeb and Preston 1986; Prasad et al. 2005), unlike in most bacteria, DNA methylation at Dcm sites has not been found in D. radiodurans (Makarova et al. 2001). Thus, we expect that in comparison to most other microbes with functional Dcm, the C→T mutation rate would be decreased in this organism due to the lack of 5-methyl cytosines mutation hotspots.
We find that the mutation rate at 5′-CCWTT-3′ Dcm motifs is not significantly elevated from the baseline mutation rate when the target nucleotide’s flanking nucleotides are taken into account (5′-CCW-3′ in this case) (log-likelihood ratio test, P > 0.05, supplementary table S17, Supplementary Material online; Long et al. 2015). In contrast, mutation rates at internal adenines of three noncanonical Dam target sites 5′-GACC-3′, 5′-GACG-3′, 5′-GACT-3′ (Schlagman and Hattman 1989) and three other unexplored motifs 5′-CACC-3′, 5′-CAGC-3′, 5′-GAGC-3′ are significantly higher than the baseline mutation rate with nucleotide contexts considered (log-likelihood ratio test, P < 0.05; efforts to identify extended motifs were unsuccessful; supplementary table S17, Supplementary Material online), whereas the internal adenine in the canonical Dam site 5′-GATC-3′ does not have an elevated mutation rate relative to the baseline context mutation rate (log-likelihood ratio test, P = 0.31; supplementary table S17, Supplementary Material online). Transversions at the six above-mentioned motifs account for 50% of all A:T→C:G transversions (41 of 82; table 2), suggesting that the high transversion rate may be due to methylation at these sites. This is consistent with a direct demonstration of Dam activity in D. radiodurans, with the canonical 5′-GATC-3′ site used in other bacteria being only poorly methylated, but 6-methylated adenines being present in other sites (Prasad et al. 2005). The mutL− MA lines showed a similar mutation pattern at potential Dcm and Dam target sites, with no mutations occurring at internal cytosines of potential Dcm sites and the only two A:T→C:G transversions both occurring at internal adenines of noncanonical potential Dam sites 5′-GACC-3′ and 5′-CATC-3′. This also indicates that more mutations are needed to test whether Dam sites are mutation hotspots in the mutL− MA lines. Taken together, these results suggest an absence of Dcm in D. radiodurans, but possible functioning of Dam at noncanonical sites.
Discussion
Despite D. radiodurans being well-known for its resistance to ionizing radiation (Anderson et al. 1956), it does not exhibit an unusually low background mutation rate, so replication fidelity does not appear to be exceptionally refined in this organism. Furthermore, the mere 4-fold elevation in genomic mutation rate after MMR is knocked out suggests that DNA MMR in this organism is not associated with the extreme radiation resistance. Thus, mechanisms other than those involved in DNA replication and MMR must be responsible for resistance to ionizing radiation in D. radiodurans.
Point mutations biased in the direction of G/C production have not been seen previously in any wild-type bacteria before this study, and there are several possible causes for this unique mutation spectrum. First, it is likely that the abundant UDGs in D. radiodurans effectively recognize and remove uracils originating from cytosine deamination. There are four functional groups within the UDG super family: The uracil-DNA N-glycosylases, the mismatch-specific uracil DNA glycosylases, the single-strand selective non-functional UDGs, and the uracil-DNA glycosylases containing a 4Fe-4S cluster (Aravind and Koonin 2000; Pearl 2000). Although most prokaryotic genomes contain just a single type of UDG, the number varies across bacterial phyla, with only a few species having all four UDGs (Aravind and Koonin 2000). The D. radiodurans genome contains homologs of all four major UDGs, three of which have been confirmed to have UDG activity (Aravind and Koonin 2000; Sandigursky et al. 2004; Leiros et al. 2005), the combination of which would enhance the ability for D. radiodurans to remove erroneous uracils, thus decreasing the rate of C→T transitions when compared with other bacteria. Second, the absence of Dcm (Makarova et al. 2001) could reduce the number of G:C→A:T transition hotspots, whereas methylation of potential Dam target sites could elevate the number of A:T→C:G transversions.
The mere 4-fold elevation of the BPS mutation rate upon MMR deficiency confirms the low efficiency of the D. radiodurans MMR system when compared with other bacteria (Mennecier et al. 2004). One possible explanation for this unexpected observation is related to the concept of evolutionary layering and the drift-barrier hypothesis (Lynch 2012). Selection operates to lower the total genome-wide mutation rate to the point that the selective advantage from any further drop in the mutation rate cannot overcome the power of genetic drift (Sung, Ackerman, et al. 2012). Once the drift barrier has been reached, alternative replication-fidelity pathways are free to covary in complementary manners so long as they are not pushed to their biophysical/biochemical limits. Because D. radiodurans has evolved multiple antimutation pathways like the abundant uracil-DNA glycosylases and the highly efficient antioxidation mechanisms (Pearl 2000; Slade and Radman 2011), the unusually low efficiency of the D. radiodurans mismatch-repair pathway might be simply compensated for by alternative mechanisms with higher levels of refinement (including the DNA polymerases, themselves). In addition to the relatively low elevation of the total BPS mutation rate, knocking out MMR only elevates transitions but not transversions, suggesting that D. radiodurans might use another mismatch-repair pathway independent of mutL for transversions, as reported in E. coli (Liu and Chang 1988).
Similar to E. coli (Lee et al. 2012), the lower elevation of in D. radiodurans mutL− than in wild-type MA lines indicates that MMR could potentially bias mutations toward G/C. However, as the D. radiodurans mutL− strain has only 4 × elevation in the mutation rate and 993 generations’ mutation accumulation time was allowed for the 24 MA lines, only 127 chromosomal mutations were accumulated. This limited number of mutations caused mutL− mutation types to be less representative of the mutational pattern than those sampled in the wild-type, for example, no G:C→T:A transversions were observed in the mutL− MA lines. One of our ongoing larger mutation accumulation experiments may provide a conclusion for this.
The higher mutation rate in plasmids relative to chromosomes might suggest that the efficiency of DNA replication differs between chromosomes and plasmids, that is, plasmid premutations may not be repaired efficiently by the MMR system. The different nucleotide composition between plasmids (CP1 GC content: 56.18%; MP1 63.19%) and chromosomes (chromosome 1: 67.01%; chromosome 2: 66.69%) may also partly explain this, as A/T has a higher mutation rate than G/C in this organism.
The relatively high mutation rate per site and low DNA mismatch-repair efficiency of D. radiodurans do not explain the extreme resistance to radiation and desiccation. This is in line with the recent hypothesis that extreme resistance to radiation in this species may be a consequence of highly efficient protection against damage to the proteome, but not genome (Krisko and Radman 2013). Our study also suggests that low variation in genomic mutation rate in bacteria is determined by other general forces like selection and genetic drift, rather than by the diverse DNA repair systems.
Materials and Methods
Strains and Media
The wild-type D. radiodurans strain BAA-816, for which a complete genome sequence exists (White et al. 1999), was ordered from the American Type Culture Collection (ATCC). The mutL− strain was constructed from the type strain R1 (ATCC13949) and kindly provided by Suzanne Sommer’s lab (Mennecier et al. 2004). The R1 and BAA-816 strains originate from a common culture and there are very few genetic differences between the two strains (White et al. 1999). The recommended growth conditions by ATCC, nutrient agar (Becton, Dickinson and Company, Sparks, MD) with 1% glucose, were used for MA line transfers.
Mutation Accumulation Transfers and High-Throughput Genome Sequencing
For the wild-type strain, 43 sequenced MA lines were established from a single progenitor wild-type cell. Individually for each MA line, cells from a single colony were streaked onto a new plate and incubated at 30 °C. Each month, for each of at least ten randomly selected MA lines of each strain, one single colony was cut out from the agar plate using a sterile scalpel, transferred to a sterile tube with PBS buffer (pH 7.5) and then vortexed and serially diluted. The colony forming units (CFU; denoted as N here) of the diluted cultures were counted and averaged. The number of generations (n) was then calculated by . For the wild-type strain, each growth cycle represents about 23.8 generations and the entire 250 transfers took 636 days (5,961 total generations for each line). For the mutL− strain, 24 MA lines were initiated from a single mutL− progenitor cell, using the same culturing and transferring techniques as for wild-type; each growth cycle represented about 23.7 generations, and on average 42 transfers were done in 84 days (993 total generations per line).
Genomic DNA was prepared using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI). DNA libraries for Illumina sequencing with an insert size of 350 bp were constructed by the Center for Genomics and Bioinformatics, Indiana University, Bloomington. Paired-end sequencing of wild-type MA lines was done by Beijing Genome Institute, Shenzhen, yielding a mean sequencing depth of 128.17 × across all lines (calculated after filtering out reads with quality score <20). On average, 99.12% of the chromosome positions in the reference genome were covered in each MA line. Sequencing of the mutL− lines was done at the Hubbard Center for Genome Studies, University of New Hampshire, yielding a depth of coverage of 83.12 × across all lines (93.62% of the chromosome positions were covered).
Fluctuation Test and Mutation Rate Estimation with Reporter Construct
The progenitor cell line (at generation 0) was thawed and revived in TGYX2 medium overnight at 30 °C. This culture was then diluted and plated onto TGY plates. Nineteen randomly selected single colonies were inoculated to 3 ml TGYX2 medium, and then grown to OD650 = 1 in about 24 h. Cell density was estimated from CFU on TGY plates. 1.4 ml cell culture from each replicate (on average 8.44 × 108 cells) was then centrifuged, resuspended into 150 µl PBS buffer (pH 7.5), plated onto TGY plates with 25 µg/ml rifampicin (Dot Scientific, Burton, MI), and incubated at 30 °C for 2 days. There were on average 9.63 rif-resistant colonies per plate (supplementary table S18, Supplementary Material online). The mutation rate and 95% confidence interval were then calculated using the Ma–Sandri–Sarkar Maximum-Likelihood Estimator method in FALCOR (Hall et al. 2009).
Mutations Analyses
A high-fidelity consensus approach for identifying fixed BPSs in the MA lines was modified from Sung, Tucker, et al. (2012). Paired-end reads from each MA line were mapped to the BAA-816 reference genome (GenBank accession numbers: NC_001263.1 for chromosome 1, NC_001264.1 for chromosome 2, NC_000959.1 for plasmid CP1, NC_000958.1 for plasmid MP1), and the resulting output was parsed with SAMTOOLS (Li et al. 2009). The following criteria were used for read mapping, designed specifically to reduce false positives that may arise from read mismapping or library amplification errors: 1) Two mapping programs, BWA (Li and Durbin 2009) and NOVOCRAFT, were used in two independent pipelines to reduce algorithm-specific read mapping errors; 2) a paired-end read was not allowed to map to multiple sites (BWA option: sampe –n 1; NOVOCRAFT option: novoalign –r None), with mapping/sequencing quality scores > 20; 3) greater than 80% of reads in a line are required to determine the line-specific consensus nucleotide; and 4) to reduce false positive calls due to errors in library construction or sequencing, three forward and three reverse reads are required to determine the line-specific consensus nucleotide.
Across all lines, the overall consensus nucleotide of a site was determined by greater than 50% read support from all MA lines. Although this criterion is not extremely stringent, it allows for the analysis of mutation hotspots, which reduces the read frequency of the major allele (Schrider et al. 2011). Mutations at each site were then called for the MA line(s) with a line consensus that is different from the overall consensus. The use of the consensus method to identify both BPS and small-indel mutations in high coverage MA lines has been validated in prior experiments, and has been shown to have a near zero false positive rate (Long et al. 2015).
Because D. radiodurans is polyploid (White et al. 1999), random assortment of alleles preexisting in the progenitor line could create false positives originating from ancestral heterozygosity. To filter out such false-positives, we also sequenced the genome of the progenitor lines and filtered out candidate mutation sites in the progenitor lines that were greater than 5% heterozygous, which greatly exceeds sequencing error rate (∼0.3%, see below). 97.03% of the final reported mutation sites are homozygous for the wild-type progenitor allele and 96.27% for the mutL− progenitor (>99% of all sites in the total genome are homozygous; supplementary table S1, Supplementary Material online). Sequencing error was estimated at mapped sites where the line-specific consensuses across all MA lines were identical to the overall consensus. For each individual nucleotide type, the pooled mean sequencing error rates were calculated by dividing the total number of discordant base reads by the total number of analyzed reads in all lines at all sites (wild-type lines 0.33%, 0.29% for mutL− lines).
Due to the polyploid nature of D. radiodurans, real mutations might be present at minor frequency in the chromosomes of one mutant line, and be incorrectly filtered by the above criteria. Therefore, for sites without mutation calls, we determined whether the site contained a minor allele that exceeded the average sequencing error rate by a considerable amount (∼15-fold). We then made sure that the minor allele was not also found in the progenitor line or any other MA line, ensuring that the minor allele arose in a single MA line during propagation. Using this analysis, we detected 12 additional mutations in wild-type MA lines and 3 additional mutations in mutL− MA lines. Overall, these mutations have little influence on overall genome-wide mutation rate (12 of 437, or 2.75% of wild-type mutations; 3 of 133, or 2.26% of mutL mutations). If we assume chromosomes are randomly transmitted to offspring cells and each generation starting from a single cell, in a finite “population” (ploidy) size of 4−10, the mean number of cell divisions for newly arisen neutral mutations to reach fixation is approximately two times the ploidy (Kimura and Ohta 1969), that is, 8−20 generations. Relative to the average 5,961 (wild-type) and 993 (mutL−) cell divisions that MA lines went through, heterozygous mutations could thus not be more than a tiny fraction of the total.
A similar consensus approach was used for small-indel analysis: SAM-formatted paired-end reads from two different mappers were used to call indels with support by at least three forward and three reverse reads each, as well as supporting reads of greater than 30% (this criterion is lowered due to the read heterogeneity created from misalignment and indels usually occurring at hotspots, e.g., SSRs) of total reads at the site. In addition, BreakDancer 1.1.2 (Chen et al. 2009) and Pindel 0.2.4w (Ye et al. 2009)—programs to detect indel break points using a local sequence pattern growth approach and to reduce indel-induced artifacts—were also applied on the BAM- and SAM-formatted files from the BWA pipeline. The above methods have been validated to have high accuracy using Sanger sequencing in multiple MA projects (Denver et al. 2009; Ossowski et al. 2010; Sung, Tucker, et al. 2012; Long et al. 2015).
In order to call duplicative insertions that occurred during MA and fixed differences of IS elements between the progenitors and the reference genome, an Al-Bruijn graph (l = 50, error rate of 5%) was constructed based on the algorithms described in Lee et al. (2014) using the concatenated sequence of chromosomes and plasmids of BAA-816. For each MA line, the SAM-formatted paired-end mapping file generated using BWA was loaded onto the graph first to select discordant read pairs to infer novel insertions of IS elements during MA. Any clusters having at least ten read-pairs were called as discordant clusters. From the clustering results, deletions and duplicative insertions of IS elements were called. For the accurate calling of large-scale variation, higher depth of sequencing compared with what is typically required for smaller variants, we eliminated a total of five mutL− lines (M6, M7, M11, M20, and M24) from IS element analysis. In comparison to the reference genome, both wild-type progenitor (BAA-816) and mutL− progenitor (R1 ΔmutL) lines have identical fixed deletions and insertions of IS elements (supplementary tables S13 and S19, Supplementary Material online). ISDra2 and IS2621 had five and three copies deleted, respectively, in both, relative to the reference genome. ISDra3, IS1 (not annotated as an IS element in GenBank), IS2621, and ISDra6 each had one extra copy inserted in both wild-type and mutL− progenitors.
Context-Dependent Mutation Rate
The 5′ and 3′ flanking bases on the same strand as a focal base were defined as “context.” This analysis took leading strands of the left and the right replichores; the chromosomal origins and termini of replication were predicted using DoriC 5.0 (Gao et al. 2013). Different BPSs occurring at the focal base of all possible 64 contexts on each replichore were called. These BPS numbers were then normalized by dividing by the product of the number of MA lines, number of generations per line, and the total number of each context (trinucleotide) in one replichore, which was detected by a 3-bp sliding window with a 1-bp step.
Statistics and Calculations
The confidence intervals of mutation-rate estimates were calculated using the Poisson cumulative distribution function approximated by the χ2 distribution (Johnson et al. 1993): With k observed mutations following a Poisson distribution with mean µ, a 95% confidence interval for µ is then: , where χ2 is the quantile function, α is 0.05 for the 95% confidence interval, and the second argument in the first parentheses is the degrees of freedom, N is the number of analyzed sites and t is the total number of generations.
The odds ratio logit method (Morris and Gardner 1988) is briefly: The natural log odds ratio is approximately normally distributed and calculated by , where is the number of mutations in coding regions, NCDS is the number of coding sites excluding overlapping sites, is the number of intergenic mutations, NIG is the number of intergenic sites, the standard error of the log odds ratio is approximated by: , the 95% confidence interval is calculated by: , the two-sided P value is then , where P is the cumulative normal probability.
The log-likelihood ratio test was used to compare mutation rates at potential methylation target sites and in corresponding nucleotide contexts (e.g., 5′-CCA-3′ context corresponds to 5′-CCAGG-3′ methylation sites, underlined cytosines are the focal bases): The null model is that mutations at methylation target sites and those with corresponding contexts follow the same Poisson probability distribution with the grand average mutation rate as the mean; the alternative model is that they follow different Poisson probability distributions. The log-likelihood ratio statistic was calculated based on these two models, and the P value was approximated by the χ2 cumulative distribution function with one degree of freedom, using Wilks’s theorem (Wilks 1938).
The chromosomal GC content at mutation equilibrium is calculated as Lynch (2007): , where equals the number of A/T mutations at A:T sites resulting in an A/T→G/C change (including both A:T→G:C transitions and A:T→C:G transversions) divided by the product of number of A/T sites and the number of generations; and equals the number of G/C mutations at G:C sites resulting in G/C→A/T change (including both G:C→A:T transitions and G:C→T:A transversions) divided by the product of the number of G/C sites and the number of generations (supplementary table S1, Supplementary Material online). The standard error of the GC content at equilibrium is calculated using formula A1.19b, page 818 of Lynch and Walsh (1998).
Supplementary Material
Supplementary tables S1–S19 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by a Multidisciplinary University Research Initiative award (W911NF-09-1-0444) from the US Army Research Office. The authors thank Suzanne Sommer for kindly providing the mutL− strain. They also thank Chi-Chun Chen, Sam Miller, Weiyi Li, and Jean-Francois Gout for helpful discussions and technical support.
References
- Anderson AW, Nordan HC, Cain RF, Parrish G, Duggan D. 1956. Studies on a radio-resistant Micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 10:575–577. [Google Scholar]
- Aravind L, Koonin EV. 2000. The α/β fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 1:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi KJ, Rocha EP, Feil EJ. 2009. The temporal dynamics of slightly deleterious mutations in Escherichia coli and Shigella spp. Mol Biol Evol. 25:345–355. [DOI] [PubMed] [Google Scholar]
- Beletskii A, Bhagwat AS. 1996. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci U S A. 93:13919–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 6:677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 1992. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J Biol Chem. 267:166–172. [PubMed] [Google Scholar]
- Cox MM, Battista JR. 2005. Deinococcus radiodurans-the consummate survivor. Nat Rev Microbiol. 3:882–892. [DOI] [PubMed] [Google Scholar]
- Daly MJ, Ling O, Minton KW. 1994. Interplasmidic recombination following irradiation of the radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 176:7506–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. 1995. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268:1616–1619. [DOI] [PubMed] [Google Scholar]
- Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledó JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M, et al. 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci U S A. 106:16310–16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Wilhelm LJ, Howe DK, Gafner K, Dolan PC, Baer CF. 2012. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol Evol. 4:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan BK, Miller JH. 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287:560–561. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Norris KF, Wang RYH, Kuo KC, Gehrke CW. 1986. DNA cytosine methylation and heat-induced deamination. Biosci Rep. 6:387–393. [DOI] [PubMed] [Google Scholar]
- Farabaugh PJ, Schmeissner U, Hofer M, Miller JH. 1978. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 126:847–857. [DOI] [PubMed] [Google Scholar]
- Gao F, Luo H, Zhang CT. 2013. DoriC 5.0: an updated database of oriC regions in both bacterial and archael genomes. Nucleic Acids Res. 41:D90–D93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP, Moriya M. 1993. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9:246–249. [DOI] [PubMed] [Google Scholar]
- Hall BM, Ma C, Liang P, Singh KK. 2009. Fluctuation AnaLysis CalculatOR (FALCOR): a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25:1564–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD. 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu Rev Ecol Evol Syst. 40:151–172. [Google Scholar]
- Hershberg R, Petrov DA. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6:e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A. 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 6:e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NL, Kotz S, Kemp AW. 1993. Univariate discrete distributions. 2nd ed. New York: Wiley-Interscience. p. 171. [Google Scholar]
- Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. 2002. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem. 277:39926–39936. [DOI] [PubMed] [Google Scholar]
- Keightley P, Trivedi U, Thomson M, Oliver F, Kumar S, Blaxter M. 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19:1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibota T, Lynch M. 1996. Estimate of the genomic mutation rate deleterious to overall fitness in E. coli. Nature 381:694–696. [DOI] [PubMed] [Google Scholar]
- Kim M, Wolff E, Huang T, Garibyan L, Earl AM, Battista JR, Miller JH. 2004. Developing a genetic system in Deinococcus radiodurans for analyzing mutations. Genetics 166:661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ohta T. 1969. The average number of generations until fixation of a mutant gene in a finite population. Genetics 61:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko A, Radman M. 2013. Biology of extreme radiation resistance: the way of Deinococcus radiodurans. Cold Spring Harb Perspect Biol. 5:a012765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Popodi EM, Foster PL, Tang H. 2014. Detection of structural variants involving repetitive regions in the reference genome. J Comput Biol. 21:219–233. [DOI] [PubMed] [Google Scholar]
- Lee H, Popodi EM, Tang H, Foster PL. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 109:E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros I, Moe E, Smalas AO, McSweeney S. 2005. Structure of the uracil-DNA N-glycosylase (UNG) from Deinococcus radiodurans. Acta Crystallogr D Biol Crystallogr. D61:1049–1056. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000-Genome-Project-Data-Processing-Subgroup 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Andersson DI. 2008. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci U S A. 105:17878–17883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11:3610–3618. [DOI] [PubMed] [Google Scholar]
- Liu AL, Chang DY. 1988. Repair of single base-pair transversion mismatches of Escherichia coli in vitro: correction of certain A/G mismatches is independent of dam methylation and host mutHLS gene functions. Genetics 118:593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Preston BD. 1986. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 20:201–230. [DOI] [PubMed] [Google Scholar]
- Long H, Sung W, Miller SF, Ackerman M, Doak TG, Lynch M. 2015. Mutation rate, spectrum, topology, and context-dependency in the DNA mismatch repair (MMR) deficient Pseudomonas fluorescens ATCC948. Genome Biol Evol. 7:262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sunderland (MA): Sinauer Associates, Inc. [Google Scholar]
- Lynch M. 2010a. Evolution of the mutation rate. Trends Genet. 26:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2010b. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 107:961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2012. Evolutionary layering and the limits to cellular protection. Proc Natl Acad Sci U S A. 109:18851–18856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland (MA): Sinauer. [Google Scholar]
- Mahillon J, Chandler M. 1998. Insertion sequences. Microb Mol Biol Rev. 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 65:44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough AK, Dodson ML, Lloyd RS. 1999. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 68:255–285. [DOI] [PubMed] [Google Scholar]
- Mennecier S, Coste G, Servant P, Bailone A, Sommer S. 2004. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol Genet Genomics. 272:460–469. [DOI] [PubMed] [Google Scholar]
- Michaels ML, Cruz C, Grollman AP, Miller JH. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 89:7022–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Gardner MJ. 1988. Calculating confidence intervals for relative risks (odds ratios) and standardised ratios and rates. Br Med J. 296:1313–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana . Science 327:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterod M, Hollenbach S, Hengstler JG, Barnes DE, Lindahl T, Epe B. 2001. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis 22:1459–1463. [DOI] [PubMed] [Google Scholar]
- Pearl LH. 2000. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res. 460:165–181. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Hartl DL. 1999. Patterns of nucleotide substitution in Drosophila and mammalian genomes. Proc Natl Acad Sci U S A. 96:1475–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BJ, Sabnis K, Deobagkar DD, Deobagkar DN. 2005. Deinococcus radiodurans strain R1 contains N6-methyladenine in its genome. Biochem Biophys Res Commun. 335:412–416. [DOI] [PubMed] [Google Scholar]
- Sandigursky M, Sandigursky S, Sonati P, Daly MJ, Franklin WA. 2004. Multiple uracil-DNA glycosylase activities in Deinococcus radiodurans. DNA Rep. 3:163–169. [DOI] [PubMed] [Google Scholar]
- Schlagman SL, Hattman S. 1989. The bacteriophage T2 and T4 DNA-[N6-adnine] methyltransferase (Dam) sequence specificities are not identical. Nucleic Acids Res. 17:9101–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Houle D, Lynch M, Hahn MW. 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster . Genetics 194:937–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Hourmozdi JN, Hahn MW. 2011. Pervasive multinuclotide mutational events in eukaryotes. Curr Biol. 21:1051–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D, Lindner AB, Paul G, Radman M. 2009. Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136:998–1000. [DOI] [PubMed] [Google Scholar]
- Slade D, Radman M. 2011. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 75:133–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M. 2012. Drift-barrier hypothesis and mutation rate evolution. Proc Natl Acad Sci U S A. 109:18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W, Tucker AE, Doak TG, Choi E, Thomas WK, Lynch M. 2012. Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc Natl Acad Sci U S A. 109:19339–19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White O, Eisen JA, Heidelberg JF, Hickey EK, et al. 1999 Genome sequence of the radioresistant bacterium. Deinococcus radiodurans R1. Science 286:1571–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks SS. 1938. The large-sample distribution of the likelihood ratio for testing composite hypotheses. Ann Math Stat. 9:60–62. [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25:2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569–573. [DOI] [PubMed] [Google Scholar]
- Zhu YO, Siegal ML, Hall DW, Petrov DA. 2014. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci U S A. 111:E2310–E2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.