Abstract
Genetic variation harbors signatures of natural selection driven by selective pressures that are often unknown. Estimating the ages of selection signals may allow reconstructing the history of environmental changes that shaped human phenotypes and diseases. We have developed an approximate Bayesian computation (ABC) approach to estimate allele ages under a model of selection on new mutations and under demographic models appropriate for human populations. We have applied it to two resequencing data sets: An ultra-high depth data set from a relatively small sample of unrelated individuals and a lower depth data set in a larger sample with transmission information. In addition to evaluating the accuracy of our method based on simulations, for each SNP, we assessed the consistency between the posterior probabilities estimated by the ABC approach and the ancient DNA record, finding good agreement between the two types of data and methods. Applying this ABC approach to data for eight single nucleotide polymorphisms (SNPs), we were able to rule out an onset of selection prior to the dispersal out-of-Africa for three of them and more recent than the spread of agriculture for an additional three SNPs.
Keywords: approximate Bayesian computation, onset of selection, selective pressures, autoimmune disease
Introduction
Over the past decade, much effort has been devoted to the discovery of selection signals using genetic variation data in humans (Sabeti et al. 2007; Coop et al. 2009; Pickrell et al. 2009; Chen et al. 2010; Grossman et al. 2010, 2013; Yi et al. 2010; Hancock et al. 2011). Selection scans have employed a variety of population genetic approaches that exploit different aspects of observed genetic variation. Most approaches for inferring positive selection are powered to detect events from a limited period of time (usually recent) (Sabeti et al. 2006), but they have seldom been used in a way that explicitly accounts for differential power and that connects selected alleles to specific hypotheses. As a result, we know little about how selective pressures changed over time as a consequence of cultural and environmental transitions. Estimating the ages of selection signals may facilitate inferences about the history of human functional and phenotypic evolution, especially in the case of advantageous alleles associated with specific traits or diseases.
Among the many environmental shifts that occurred during human evolution, the dispersal out-of-Africa and the spread of agriculture and animal farming are recognized as the transitions associated with the most dramatic environmental changes and the onset of strong new selective pressures. With the dispersal out-of-Africa sometime earlier than 40,000 years ago (Benazzi et al. 2011; Higham et al. 2011; Mellars 2011), humans encountered—among other environmental differences—colder climates, reduced levels of ultraviolet radiation, and lower pathogen diversity. In contrast, the spread of agriculture and animal farming around 12,800 years ago (Gamble et al. 2004) led to dramatic increases in the amounts and types of pathogens and, due to the higher population densities, in their transmission rates (Mira et al. 2006; Harper and Armelagos 2010; Mummert et al. 2011). In addition, major dietary changes occurred, including the use of milk in adult diets and an increase in the consumption of complex carbohydrates from cultivated and processed grains (Luca et al. 2010). These two environmental transitions offer a framework for testing hypotheses about the impact of natural selection on human phenotypes by asking whether the estimated age of an advantageous allele associated with a phenotype is compatible with an onset of selection during the dispersal out-of-Africa, the spread of agriculture, or both.
Here, we developed an approximate Bayesian computation (ABC) method for estimating the ages of the selected alleles under complex demographic models. This approach, which generates a posterior probability distribution for parameters of interest based on simulations, has been widely applied to inference problems in population genetics (Beaumont et al. 2002; Slatkin 2008; Peter et al. 2012), and it allows one to consider the effects of selection in a complex, and more realistic, demographic setting. We initially applied this approach to two resequencing data sets: A newly generated one obtained by sequence capture followed by sequencing (CapSeq) to obtain ultra-high depth data, and a lower depth shotgun sequencing data set generated by complete genomics (CG) for a larger sample of individuals. Because both data sets provided similar estimates, we applied our ABC approach to the CG data for five additional single nucleotide polymorphisms (SNPs) previously reported to be associated with selective sweep signals. Finally, by comparing our results with the rapidly growing ancient DNA data sets, we determined that the ABC estimates are compatible with the ancient DNA record.
Results
Ultra-High Depth Sequencing Survey and Comparison with CG Sequence Variation Data
Several aspects of the empirical data may affect allele age estimation, including the accuracy of genotype calling, which in turn depends on the sequencing depth, the accuracy of phasing, and the sample size. In order to compare sequencing data sets with different features for the purpose of allele age estimation, we generated ultra-high depth sequence data obtained by sequence capture (CapSeq) for genomic segments spanning three SNPs (rs6822844, rs3184504, and rs12913832) associated with selection signals in Europeans. These SNPs are polymorphic almost exclusively in Western Eurasia, making it likely that selection acted on a new advantageous mutation rather than on standing variation (supplementary fig. S1, Supplementary Material online). Two of them are associated with several immune-mediated diseases; however, the advantageous allele at rs6822844 protects against disease (Todd et al. 2007; van Heel et al. 2007; Wellcome Trust Case Control Consortium 2007; Zhernakova et al. 2007; Adamovic et al. 2008; Hunt et al. 2008; Liu et al. 2008; Albers et al. 2009; Daha et al. 2009; Festen et al. 2009; Marquez et al. 2009; Teixeira et al. 2009; Hollis-Moffatt et al. 2010; Maiti et al. 2010; Petukhova et al. 2010; Warren et al. 2011), while at rs3184504 the advantageous allele associates with increased disease risk (Wellcome Trust Case Control Consortium 2007; Hunt et al. 2008; Barrett et al. 2009; Alcina et al. 2010; Dubois et al. 2010; Stahl et al. 2010; Zhernakova et al. 2011; de Boer et al. 2014). The third SNP (rs12913832) is known to influence eye and hair color in Europeans and to a lower extent also skin pigmentation (Sulem et al. 2007; Han et al. 2008; Kayser et al. 2008; Branicki et al. 2009; Cook et al. 2009; Eriksson et al. 2010; Liu et al. 2010; Zhang et al. 2013). For a full description of the SNPs, associated traits or diseases and selection signals, see supplementary text S1, Supplementary Material online.
The region spanning these SNPs were captured and sequenced in 14 unrelated CEU (Centre d'Etude du Polymorphisme Humain from Utah of Northern European descent) samples (supplementary table S1, Supplementary Material online). The average coverage depth per sample was very high (219×, 292×, and 307× for the rs6822844, rs3184504 and rs12913832 regions, respectively) (supplementary figs. S2 and S3 and tables S2 and S3, Supplementary Material online). We observed that on average ∼53% of the reads at heterozygous positions were identical to the reference suggesting that there is a small bias against the nonreference allele (supplementary fig. S4, Supplementary Material online), but no evidence of significant allelic dropout at heterozygous positions, consistent with previous reports for libraries prepared by array capture (Hedges et al. 2011; Kiialainen et al. 2011).
In order to assess the accuracy of the CapSeq genotype calls, we compared the CapSeq genotypes with the HapMap (HapMap_2010-08_phaseII+III) genotypes for all 23,488 overlapping SNPs and we found 162 discordant genotype calls (supplementary tables S4, Supplementary Material online). We then used the Phase 3 1000 Genome (1KG) data to investigate the sources of error. Of the 162 inconsistent genotype calls, 68 of them had been called also in the 1KG data and all of them were consistent with our calls (supplementary table S5, Supplementary Material online) supporting the high accuracy in the CapSeq genotype calls.
We also compared the CapSeq data with the CG data set, which includes a total of 32 CEU trios sequenced at ∼30× depth. To examine the concordance between CapSeq and CG genotype calls, we focused on the six individuals who were sequenced in both data sets (i.e., NA10855, NA11830, NA12489, NA10864, NA10839, and NA12762) (supplementary figs. S5 and S6, Supplementary Material online). Even though the concordance rate of genotypes in the overlapping SNPs is ∼100% (1 discordant genotype call out of 1,189 calls across the 3 genomic regions), the results in supplementary figure S5, Supplementary Material online, show a significantly lower number of variable sites in the CG data than in the CapSeq data in the six overlapping samples (32%, 41%, and 36% for rs6822844, rs3184504, and rs12913832, respectively); the lower number of variable sites in the CG data could be due to the lower sequence coverage (∼30× for GC vs. >200× for CapSeq) and/or to more aggressive data filtering in CG. In addition to read depth and variant filtering approach, the CapSeq and the CG data differ also with regard to sample size (64 for CG vs. 14 for CapSeq) and to phasing, with the CG data probably having fewer switching errors (although CG haplotypes were used as a scaffold to increase the accuracy of phasing the CapSeq data; see Materials and Methods for “Sequence capture and sequencing”).
An Approximate Bayesian Computation Approach to Estimate Age of Beneficial Alleles
To estimate allele ages, we developed an ABC approach (supplementary texts S2–S4 and figs. S7–S12, Supplementary Material online) and, to explore whether the different features of the CapSeq and CG data affect the inference of allele ages, we applied it to both data sets. To this end, we ran 1.5 M simulations of natural selection acting on a new mutation matching the features of each of the three regions for the CapSeq as well as the CG data sets (supplementary fig. S2 and table S6, Supplementary Material online). The selection signals based on extended haplotype homozygosity (EHH) could result from weakly negative rather than positive selection acting on these SNPs (Sabeti et al. 2006; Nielsen et al. 2007), because, conditional on the current allele frequency, positively and negatively selected alleles are expected to have the same genomic signature (Maruyama 1974). For this reason, we simulated selection coefficients (s) that included positive as well as slightly negative values (−0.01 ≤ s ≤ 0.1). We defined the selected region as the segment over which the EHH is greater than 0.05, and we calculated three partially nonredundant summary statistics (SSs) for each of the selected regions: The inverse of the genetic length of the selected region (1/LH), the average number of mutations accumulated in the haplotypes carrying the selected alleles divided by the physical length of the selected region (MH), and the number of singleton variants divided by the total number of segregating sites in these haplotypes (RH) (see Methods; supplementary texts S2–S3, Supplementary Material online). The same SSs were calculated for the CapSeq and CG data sets (shown in table 1; see supplementary table S7, Supplementary Material online, for correlation with the log(t) and the correlation between the SSs, where t is the allele age). Next, for each region, the 1,500 simulations with SS values closest to those observed in the CaqSeq or the CG data were used to estimate the posterior joint probability distribution for the selection coefficient and the age of each of the three alleles (supplementary fig. S8, Supplementary Material online).
Table 1.
Summary Statistics of Sequence Variation in the CG Data and, in Parentheses, in the CapSeq Data.
| rs6822844 | rs3184504 | rs12913832 | |
|---|---|---|---|
| Length (kb) | 680 | 1600 | 350 |
| No. of selected allelesa | 17 (17) | 53 (19) | 99 (25) |
| No. of nonselected allelesb | 111 (11) | 75 (9) | 29 (3) |
| πc | 195.029 (248.847) | 189.429 (227.008) | 63.188 (86.675) |
| Sd | 1,394 (1,015) | 2,561 (1,394) | 625 (492) |
| RH | 0.583 (0.295) | 0.594 (0.479) | 0.481 (0.448) |
| MH | 0.00012 (0.00020) | 0.00011 (0.00024) | 0.00015 (0.00016) |
| 1/LH | 13.311 (20.356) | 8.446 (15.589) | 5.307 (11.158) |
aNumber of sequences carrying the selected allele.
bNumber of sequences carrying the nonselected allele.
cNucleotide diversity.
dNumber of segregating sites.
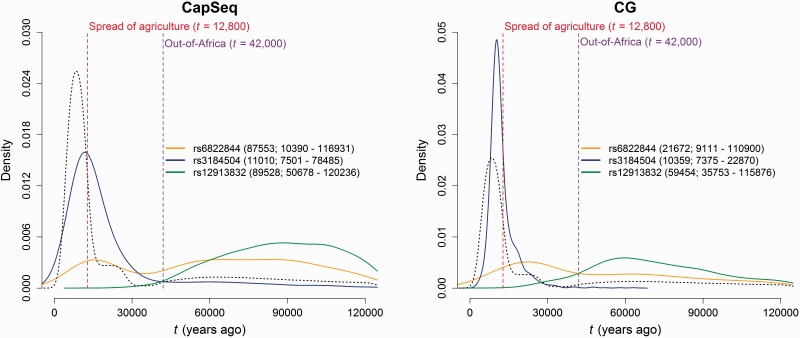
Although these alleles have been considered advantageous, the posterior distributions of selection coefficients included negative values; a larger proportion of negative s values was obtained in the CapSeq versus the CG data sets (table 2). Still, the vast majority of the retained simulations had a positive s value, with the highest proportion for the pigmentation SNP and the lowest for the rs6822844 immunity SNP. Therefore, even though we cannot rule out that weak purifying selection underlies the observed population genetic signals, positive selection is by far the most likely explanation. As expected, the plots of the joint posterior probability distributions show that s is inversely related to t in both data sets (supplementary fig. S8, Supplementary Material online). The joint posterior distributions occupy a different space compared with the priors, which confirms that the sequence variation data contain information for estimating t (figs. 1 and 2). Because our main goal is to estimate t and compare it with historical information, in the results that follow we integrate over s to get the marginal posterior distribution of t (Slatkin 2008). Based on the posterior marginal distributions of t (fig. 1), we estimated the age of the three alleles from the mode of the distributions for both CapSeq and the CG data sets. The CapSeq and CG data yield a similar point estimate for rs3184504*T, while those for rs6822844*G and rs12913832*G are markedly different. This is probably due to the flat shape of the posterior distribution for the latter two variants, suggesting that there is limited information in the data to estimate the allele age with confidence. Significant overlap between the 95% credible intervals (CIs) for the CapSeq and the CG is observed for all SNPs (fig. 1). We note that the upper boundary for the CI of the oldest variant, that is, 120,236 years, coincides with the upper end of the range of the prior on t in our simulations; therefore, the true upper boundary may be older than what we estimated.
Table 2.
Posterior Probability that the Age of an Allele Falls within Three Major Time Periods Based on the CG Data and, in Parenthesis, on the CapSeq Data.
|
t |
s > 0 | Phenotypes associated with Selected Alleles | |||
|---|---|---|---|---|---|
| <12.8 kya | 12.8–42 kya | >42 kya | |||
| rs4988235 | 99.73% | 0.27% | 0.00% | 100.00% | Lactase persistence |
| rs12913832 | 0.08% (0.00%) | 9.89% (1.81%) | 90.03% (98.19%) | 99.83% (99.98%) | Blue eyes and blond hair color |
| rs1426654 | 0.04% | 76.74% | 23.22% | 100.00% | Light skin pigmentation |
| rs16891982 | 0.02% | 1.37% | 98.61% | 100.00% | Light skin pigmentation |
| rs6822844 | 12.43% (9.62%) | 37.22% (20.71%) | 50.35% (69.67%) | 85.75% (74.79%) | Autoimmune disease protection |
| rs17810546 | 95.44% | 4.56% | 0.00% | 99.95% | Autoimmune disease risk |
| rs3184504 | 68.37% (42.23%) | 31.19% (45.86%) | 0.44% (11.90%) | 99.49% (94.93%) | Autoimmune disease risk |
| rs2188962 | 28.55% | 58.30% | 13.15% | 98.22% | Autoimmune disease risk |
Fig. 1.
Posterior probability distribution for the age (t) of the selected alleles at rs6822844, rs3184504, and rs12913832 in the CapSeq data and CG data. The posterior mode and 95% credible interval are shown in the caption with parenthesis. The red dashed line marks the spread of agriculture (12,800), the purple dashed line marks the out-of-Africa (42,000), and the black dashed line represents the prior probability distribution for t that is proportional to a demographic model estimated by a pairwise sequentially Markovian coalescent (PSMC) approach (Li and Durbin 2011).
Fig. 2.
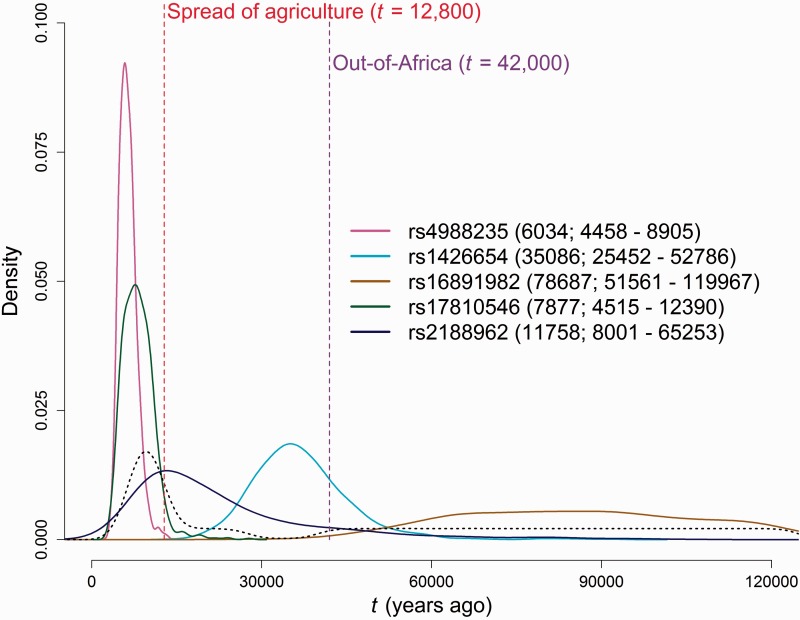
Posterior probability distribution for t of the selected allele at five SNPs (rs4988235, rs1426654, rs16891952, rs17810546, and rs2188962) for which only CG data were available. The posterior mode and 95% credible interval are shown in the caption with parenthesis. The red dashed line marks the spread of agriculture (12,800), the purple dashed line marks the out-of-Africa (42,000), and the black dashed line represents the prior probability distribution for t that is proportional to a demographic model estimated by a PSMC approach (Li and Durbin 2011).
To infer the onset of selection acting on these alleles in relation to the main transitions during human evolution, we used the posterior probability distribution to calculate the probability that the age of each allele is more recent than the spread of agriculture (<12,800 years ago) (Pinhasi et al. 2005) or older than the dispersal out-of-Africa and into Europe (>42,000 years ago) (Benazzi et al. 2011; Higham et al. 2011; Mellars 2011). As shown in table 2, most of the probability mass for the light pigmentation and the autoimmunity protective alleles is observed for values of t older than the dispersal out-of-Africa. In contrast, the autoimmunity risk allele at rs3184504 is substantially younger and incompatible with an onset of selection prior to the out-of-Africa migration (probability that t>42 kya is 0.44%).
The posterior probability distributions are more peaked for the CG than the CapSeq data for all 3 SNPs and in particular for rs3184504. This suggests a greater amount of information in the CG than the CapSeq data, probably due to the larger sample size and the greater phasing accuracy. In general, at a qualitative level, we conclude that there is good agreement between the results obtained using these two data sets (supplementary fig. S8, Supplementary Material online). For this reason, we used the CG data to estimate the age of five additional alleles associated with selection signals in European populations.
Estimating the Age of Five Additional Alleles using CG Data
As for the three SNPs above, the five additional SNPs are polymorphic almost exclusively in European populations (supplementary fig. S1, Supplementary Material online). Three of them are associated with well-established adaptive phenotypes: Lactase persistence (rs4988235) (Enattah et al. 2002) and skin pigmentation (rs1426654 at SLC24A5 and rs16891982 at SLC45A2) (Newton et al. 2001; Nakayama et al. 2002; Graf et al. 2005; Lamason et al. 2005). The remaining two SNPs, rs17810546 and rs2188962, are strongly associated with immune-mediated diseases (Barrett et al. 2008; Hunt et al. 2008; Dubois et al. 2010; McGovern et al. 2010; Jostins et al. 2012); in both these cases, the allele associated with the selection signal increases disease risk. For a full description of the selection and association signals for these five SNPs, see supplementary text S1, Supplementary Material online.
When we applied our ABC approach to these SNPs, the highest observed posterior probability for negative values of s was 1.88% (compared with 9% in the prior) (table 2), supporting the notion that these five alleles are advantageous. With a mode at 6,034 years ago, the posterior distribution of t for rs4988235 at the LCT gene points to a recent origin and spread for lactase persistence (fig. 2). Although rs1426654 and rs16891982 are both associated with skin pigmentation, the two selected alleles are estimated to have different ages (fig. 2), with the allele at SLC24A5 showing a modal value between 30,000 and 40,000 years ago and the posterior distribution for the SLC45A2 allele showing a relatively flat distribution with a weak mode at around 80,000 years ago. Both autoimmunity SNPs, rs17810546 at IL12A and rs2188962 at IRF1, showed modal values of t between 7,000 and 12,000 years ago (fig. 2).
We further calculated the posterior probability that t falls in each of the three periods defined by the two major environmental transitions, as described above (table 2). The lactase persistence allele and autoimmunity risk allele at rs17810546 significantly support a time of onset more recent than the spread of agriculture with a cumulative posterior probability (p. p.) of >95%. For the other autoimmunity risk SNP (rs2188962), the distribution is shifted toward slightly older ages compared with the other two, but the mode coincides with the spread of agriculture. In contrast, both skin pigmentation alleles are estimated to be older than the spread of agriculture (cumulative p. p. ≥ 99.06%), where rs1426654 is likely to be due to a selective event between the two transitions and rs16891982 is estimated to be older than the dispersal out-of-Africa (cumulative p. p. ≥ 98.61%).
Comparison of Allele Age Estimates with the Ancient DNA Record
The rapidly growing ancient DNA data sets, especially for European populations, offer an opportunity to compare allele age estimates with inferences based on the archaeological record. To this end, we downloaded aligned sequence data from four recent ancient DNA studies (Lazaridis et al. 2014; Skoglund et al. 2014; Allentoft et al. 2015; Haak et al. 2015). The samples were grouped into five periods based on the upper bound of the specimen age estimate: 8-7, 6-5, 5-4, 4-3, and 3-1 thousand years ago (kya). We then estimated the allele frequency in each period using a maximum-likelihood method described in Mathiesson et al. (2015) (fig. 3).
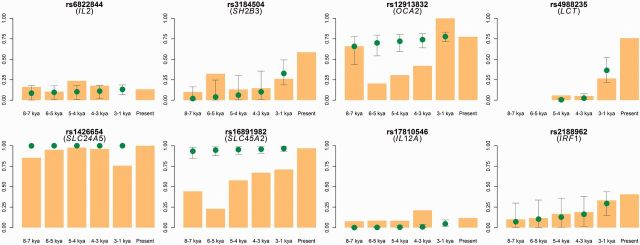
Fig. 3.
Comparison between the ABC simulated allele frequency (dots) and the allele frequency observed in the ancient DNA data (bar graphs) for all eight SNPs. The line plots show the mean allele frequencies at the five time points with 95% confidence intervals in 10,000 simulated trajectories. The bar graphs show maximum-likelihood estimates of allele frequency in the ancient DNA data. The frequency at the present is calculated from the CG data. Because the lactase allele is absent in the 8-7 and 6-5 kya periods, the allele frequency is only shown for three time points: 4, 3, and 1 kya. The 95% confidence intervals for the SLC24A5 SNPs are very narrow because the advantageous allele was fixed in most of the simulations.
To evaluate the agreement between our estimates and the ancient DNA data, we simulated trajectories, conditional on the current frequency, by sampling t and s from the joint posterior distribution for each SNP (supplementary figs. S8 and S12, Supplementary Material online) and calculated the mean frequency of the derived alleles in each of the five periods (fig. 3). These mean frequencies over time show a remarkably good fit to the pattern observed in the ancient DNA data for most SNPs (fig. 3), even though the simulations use a demographic model that does not account for the history of migrations in Europe (Allentoft et al. 2015; Haak et al. 2015). This oversimplification may explain the discrepancy between ancient DNA and our data for rs16891982 or rs12913832 in the transition from the 8-7 to 6-5 kya periods. Overall, the match between the allele frequency change over the past 8,000 years estimated by the ABC approach and that inferred by ancient DNA suggests that our allele age estimates are reasonably accurate.
Discussion
Although extensive catalogs of selection signals have been generated by genome scan studies (Vitti et al. 2013), limited effort has been devoted to estimating the age of selective events in a systematic manner and to developing a time frame for the history of the selective pressures that acted during human evolution (Peter et al. 2012; Beleza et al. 2013). Here, we developed an ABC approach for estimating the ages of new advantageous alleles. In addition to assessing its accuracy based on simulations, we have tested it on different types of empirical data. First, we have applied it to two resequencing data sets, allowing the comparison between ultra-high depth data from a relatively small sample of unrelated individuals and lower depth data in a larger sample with transmission information. The two data sets yielded qualitatively similar results, implying that the publicly available CG data are sufficiently accurate for allele age estimation. For each SNP, we also assessed the consistency between the posterior probabilities estimated by our ABC approach and the ancient DNA data, which showed good agreement between these two types of data and between inferential methods. Finally, to reconstruct the history of selective pressures in European populations, we used the posterior probability distributions to test if the estimated ages are compatible with two major environmental transitions during human evolution. Based on this analysis, we were able to rule out an onset of selection older than the dispersal out-of-Africa for three of the eight SNPs tested (rs3184504, rs4988235, rs17810546), while we ruled out an onset of selection more recent than the spread of agriculture for three other SNPs (rs12913832, rs1426654, and rs16891982).
Previously, a similar ABC approach was used in Peter et al. (2012) to distinguish between neutrality, selection on standing variation and selection on a de novo mutation. Even though allele ages and selection strengths were also estimated in that paper, the focus was on the distinction between different selection scenarios. Although we have a qualitative understanding of how selection on standing variation versus de novo mutations are expected to affect genetic diversity (Barrett and Schluter 2008), the expected patterns differ only slightly. As a consequence, the strategy of Peter et al. (2012) was to focus on very strongly selected alleles, where distinction is easiest, and to use a wide variety of SSs with the aim of capturing as much of the “signal” as possible for the ABC analysis. In contrast, here our main motivation is the estimation of the age of alleles for which we have a priori reasons to assume that selection acted on a new mutation. This problem has been extensively studied (Kimura and Ohta 1973; Slatkin and Rannala 1997, 2000; Slatkin 2000), which allowed us to design a relatively straightforward approach where the choice of SSs can be motivated directly by theory, and which allowed us to investigate also selection signals that are less strong.
Naively, it might seem desirable to include as many statistics as possible; however, there are well-established drawbacks to increasing the number of statistics used. In addition to increasing the run-time of the algorithm, having a large number of statistics implies that we aim at matching simulations and observations in a very high-dimensional space. This is problematic because more simulations are required to cover the space adequately, or a larger approximation error has to be introduced. In Peter et al. (2012), this was handled by using partial least squares to reduce the dimensionality; however, this introduces an additional layer to the algorithm, increasing complexity (Wegmann et al. 2009). A second issue of adding SSs with little marginal information is that each statistic adds statistical noise, thus requiring more simulations to obtain the same accuracy.
A further difference between our work and that of Peter et al. (2012) lies in the simulation scheme for the ABC estimation. Although Peter et al. (2012) reported allele age estimates, the allele age was not an independent parameter in their simulation framework, namely simulations were performed conditional on the allele frequency at present backward in time with a fixed selection coefficient. The time when the trajectory hit zero was recorded as the age of the allele. In contrast, here we simulate forward-in-time trajectories with the time of mutation drawn from an explicit prior distribution, and simulations are rejected if not consistent with the present allele frequency. For the purpose of estimating allele ages, the latter approach is preferable because it gives us direct control over the parameter and prior distributions. In particular, forward-in-time trajectories allow us to draw allele ages from a distribution that takes into account the fact that the influx of new mutations is proportional to the effective population size at a given time, and thus fewer mutations enter a population when the population size is low. For backwards-in-time trajectories, the proper way to do this is to use importance sampling (Slatkin 2001), which was not done in Peter et al. (2012), leading to inflated prior probabilities of allele ages in times when the effective population size is low. We note that even though our estimate of t for the LCT allele (fig. 2; 6,034 years ago; 95% CI 4,458–8,905) is compatible with that of Peter et al. (2012) (11,200 years ago; 95% CI 1,500–64,900), our estimate is closer to that obtained based on ancient DNA data (fig. 3).
An important feature of our study is that we combined information from genome-wide scans for selection and genome-wide association studies (GWAS) of common phenotypes to choose advantageous alleles with known phenotypic effects. Given the rapid growth of genome-scale sequence data and GWAS, this integrated approach promises to shed new light on the selective pressures that shaped different biological processes and phenotypes with the ultimate goal of reconstructing a broad narrative for the history of human functional evolution (Carroll 2003).
It has long been hypothesized that the spread of agriculture and the adoption of animal farming led to major increases in pathogen exposures and, as a consequence, in the frequency of alleles associated with stronger immune response (Armelagos and Harper 2005). Consistent with this proposal, most alleles associated with selection signals and with autoimmune diseases increase risk and only a minority of such advantageous alleles is protective (Barreiro and Quintana-Murci 2010; Casto and Feldman 2011). Our findings support this scenario in that the three advantageous alleles that increase disease risk are incompatible with an onset of selection at the dispersal out-of-Africa and are markedly more likely to have arisen during the spread of agriculture. Interestingly, we infer that an advantageous allele that protects against autoimmune diseases is older than the spread of agriculture, leading us to speculate that a less reactive immune system was beneficial in a different, earlier phase when human populations first expanded out-of-Africa into northern latitudes. This transition was likely associated with a reduction in pathogen levels, which in turn resulted in selection to change the set point for the immune response (Pennington et al. 2009). Interestingly, the correlation between pathogen diversity and latitude was shown to be due to the effects of climate factors, for example, temperature and precipitation rate (Guernier et al. 2004). Consistent with a role for climate factors as major determinants of pathogen-related selective pressures, many alleles associated with autoimmune diseases are also strongly correlated with climate variables (Hancock et al. 2011). In line with this proposal, several inflammation phenotypes are more common or more severe in individuals of African ancestry. These include benign neutropenia (Haddy et al. 1999; Hsieh et al. 2007), which was shown to be associated with an advantageous allele at near-fixation frequency in sub-Saharan Africa (Reich et al. 2009), a weaker response to the anti-inflammatory effects of glucocorticoids (Chan et al. 1998; Federico et al. 2005; Maranville et al. 2011), and higher levels of C-reactive protein, which is a biomarker of inflammation (Albert et al. 2004; Kelley-Hedgepeth et al. 2008).
An onset of selection more recent than the spread of agriculture can be ruled out for all the pigmentation SNPs, and for two of these three SNPs (i.e., rs12913832 and rs16891982), selection prior to the dispersal out-of-Africa is by far the most likely scenario, consistent with the idea that light pigmentation alleles were driven to high frequency when humans moved to higher latitudes. The ages of skin pigmentation alleles have been previously estimated based on microsatellite (Beleza et al. 2013) and sequence variation (Soejima et al. 2006) data. Our estimate for rs1426654 at SLC24A5 is consistent with those obtained by Beleza et al. (2013), but it is not for rs16891982 at SLC45A2, with our estimate being substantially older. Soejima et al. (2006) estimated the age of the AIM1 allele to be relatively young, that is, 10,965 years, but the confidence intervals for this estimate are compatible with selection acting as early as 39,000 years ago (Soejima et al. 2006), similar to our estimates for the pigmentation alleles we tested. More importantly, at least two of the pigmentation SNPs show marked discrepancies with the ancient DNA data. This is not surprising given that we used a simplified model of European history that does not take into account the complex ancestries of these populations (Allentoft et al. 2015; Haak et al. 2015) and the distribution of selective pressures across Europe. In particular, recent work has shown that substantial geographic structure was present with regard to pigmentation among European hunter-gatherers as well as between these and the farmer populations (Mathieson et al. 2015). Therefore, there is still substantial uncertainty about the history of selective pressures acting on human pigmentation traits. Additional contemporary and ancient DNA data may help clarify the tempo and mode of evolution of these traits.
An example of how to use ancient DNA data to learn about allele age estimates was provided by the study of Sams et al. (2015), which estimated the ages of neutral alleles and tested the accuracy of these estimates using data from a single ancient DNA sample. However, for positively selected alleles, t and s need to be considered jointly to model the allele frequency trajectory (Slatkin and Rannala 2000). The ABC approach we developed here is appropriate for this goal because it generates a joint posterior probability distribution of t and s for each SNP from which parameters can be sampled to obtain expected allele frequencies at any given time point in the past. The expected allele frequencies can then be compared with those estimated based on ancient DNA data. In the case of the SNPs tested here, this comparison increases the confidence in our age estimates.
In summary, our study is an example of how sequence variation data coupled with inferential approaches that account for the complexities of human population history may begin to illuminate hypotheses about the evolution of selective pressures acting on different phenotypes. The growing catalog of GWAS provides invaluable information regarding the genetic bases of many phenotypes that were likely targets of natural selection during human evolution, but little is known about the history of these selective pressures. The analysis of a larger group of variants associated with a specific phenotype (e.g., risk to autoimmune diseases, height, etc.) could reveal broadly consistent patterns of allele ages, thus providing strong evidence for a given evolutionary scenario. Investigation of other GWAS SNPs will also require extending the ABC approach to cases in which selection acted on standing variants that previously were not or were only weakly affected by selection, as already investigated by Peter et al. (2012).
Materials and Methods
Sequence Capture and Ultra-High Depth Sequencing of the Regions Spanning rs6822844, rs3184504, and rs12913832
DNA of 14 unrelated CEU individuals was purchased from the Coriell Cell Repository (http://ccr.coriell.org). In order to improve the accuracy of the allele age estimation, individuals were chosen to oversample the selected allele at each of the targeted SNPs, that is, rs6822844, rs3184504, and rs12913832 (supplementary table S1, Supplementary Material online). Despite the nonrandom sampling, the average levels of linkage disequilibrium (LD) between the two autoimmunity SNPs, as measured by the correlation coefficient r2, were similar to those within the complete sample of unrelated CEU individuals.
The genomic segments to be captured were defined based on the decay of LD surrounding each targeted SNP. More specifically, r2 values were calculated using the CEU HapMap data between each targeted SNP and all the SNPs within 2 Mb to obtain a map of LD decay. The genomic segment to be captured was chosen so that no SNP outside the segment had an r2 value greater than 0.2 with the focal SNP. The coordinates of the three segments were (hg_18) as follows: chr4:122729299–123782528 (rs6822844; 1.053 Mb); chr12:109769404–111681303 (rs3184504; 1.911 Mb), and chr15:25542017–26213429 (rs12913832; 0.671 Mb) (supplementary fig. S1 and table S2, Supplementary Material online). Unique probes were designed using the SSAHA algorithm (Ning et al. 2001) with an average length of ∼85 bp to capture the targeted regions avoiding repetitive elements. Of the targeted regions, 72.3% were directly covered by probes, and 86.5% of the regions were either directly covered or within 100 bp of a probe. The standard NimbleGen array capture protocol (Albert et al. 2007) was modified to allow sequencing on the Illumina Genome Analyzer II (Almomani et al. 2011). For each sample, an Illumina paired end (PE) library was created following the manufacturer’s instructions with minor modifications. Sequence capture was done according to Roche NimbleGen’s instructions. Each sequencing library was run on a lane of an Illumina Genome Analyzer II, which generated an average of 70 M 76 bp PE reads per lane (supplementary table S3, Supplementary Material online).
Sequence Data Processing
BWA 0.5.9rc1 (Li and Durbin 2009) was used to align the sequence reads to the human reference genome sequence (build National Center for Biotechnology Information 36; NCBI36) that was obtained from the Genome Analysis Tool Kit (GATK) website. Alignment and variant calling was performed following GATK’s “Best practice for variant detection v. 2” and using default parameters except that –a 600 was used for the alignment step (bwa sampe). The duplicate reads (i.e., read pairs with the same orientation and alignment position) were removed using Picard. Genotype calls that did not fulfill the following criteria were removed: (1) QUAL ≥ 50; (2) HRun ≤ 5; (3) MQ0 ≤ 4 or MQ0 ≤ 0.1 × DP; (4) PLAA/PLAB or BB ≥ 20, where AA is the most likely genotype; (5) “not in cluster” of 3 SNPs within 10 bases; and (6) not inside an indel. Genotype data were imputed and phased using IMPUTE2 (Howie et al. 2011) with the CG data as the reference panel; first, we used the CG haplotypes to help phase our sequence data at shared SNP sites between the two data sets. Second, we imputed our data based on the CG haplotypes and used the haplotypes phased in the first step as a scaffold for phasing private SNP sites to our data. Third, we removed the imputed SNP sites (i.e., private SNP sites to CG) from the phased haplotypes.
We downloaded the CG data for CEU including 32 trios from ftp.1000genomes.ebi.ac.uk:/vol1/ftp/release/20130502/supporting/cgi_variant_calls/filtered_calls/ (last accessed July 2015). First, we extracted a region included within 2.5 Mb upstream and downstream to each of eight focal SNPs. We then removed SNPs with missing genotypes or Mendelian errors. Third, we phased the data for the parents of each trio using the SHAPEIT2 with the pedigree information (Delaneau et al. 2012, 2013), thus providing a total of 64 unrelated individuals. This strategy increases phasing accuracy by combining both transmission and LD information, and it mirrors the approach used to generate the high-quality HapMap CEU haplotypes (Marchini et al. 2006). To compare sequence data for three regions including rs6822844, rs3184504, and rs12913832 between CG and CapSeq, we prepared the CG data that had the same coordinates as CapSeq. For the five SNPs analyzed using only the CG data, we used the sequence data for a region 500 kb upstream and downstream of the focal SNP site and processed the data as for the initial set of three SNPs.
Allele Age Estimation by ABC
We developed an ABC method to estimate the age of a new selected allele by incorporating coalescent-based simulations into the local linear regression (Beaumont et al. 2002). We provide details of the ABC method in supplementary text S2, Supplementary Material online. Briefly, simulations of natural selection acting on a new mutation were run using mssel (program available from R. Hudson upon request), a modified version of ms (Hudson 2002), which allows recombination and complex demography. First, we generated trajectories for a focal SNP by Wright–Fisher forward simulations. Every trajectory started at frequency 1/2Ne using a t value that was sampled from a prior proportional to the population size under the demographic model inferred by Li and Durbin (2011) for a CEU individual. The selection coefficient s was sampled from a uniform distribution between −0.01 and 0.1. We only accepted a trajectory if the frequency at present was compatible with observed one. Otherwise, we sampled a new value of s. This process generates a conditional prior on s that we use in the subsequent step of the ABC estimation of t. Second, we generated neutral variation around the SNP by coalescent simulations conditional on the trajectory. For each simulation, we chose 28 and 128 chromosomes, respectively, for the CapSeq or the CG data, with the frequency of the selected allele matching the frequency of the targeted alleles in the corresponding data set (supplementary table S6, Supplementary Material online).
For the three regions analyzed in both CapSeq and CG data sets (rs6822844, rs3184504, and rs12913832), we took into account the features of the genomic regions and the data processing (i.e., gap exclusion, phasing) in the coalescent simulations. Specifically, we simulated a subset of the surveyed length (i.e., subregions), which was delimited either by the edge of the surveyed segment or by the presence of a strong recombination hotspot (i.e., a region with >30× the background recombination rate). The length of the simulated subregions was 680 kb, 1.6 Mb, and 350 Kb for rs6822844, rs3184504, and rs12913832, respectively (supplementary fig. S2, Supplementary Material online, and table 1). For the five SNPs analyzed using only the CG data (rs4988235, rs1426654, rs16891982, rs17810546, and rs2188962), we first retrieved 1 Mb region centered on each focal SNP site and then removed flanking regions if there are recombination hotspots (supplementary fig. S2, Supplementary Material online). To take into account the uncertainty in the mutation rate (μ), we sampled μ from a truncated normal distribution in which the variance was the square of the mean and the tails were truncated at half of the mean and at 1.5 times the mean. The mean of the distribution was set at the estimate based on the number of fixed derived alleles in the 1KG data identified using the alignment of human, chimpanzee, orangutan, and rhesus macaque, assuming a divergence time between human and chimpanzee of 5 Ma and an ancestral population size of 12,500 diploid individuals (Patterson et al. 2006; Hobolth et al. 2007, 2011; Scally et al. 2012). The estimates for the population recombination rate parameter for each region were obtained from Myers et al. (2005) inferred from HapMap data; because the genomic segments that we considered do not contain strong hot spots, these estimates were assumed to apply uniformly in our simulations. The parameter values used in the simulations are given in supplementary table S6, Supplementary Material online. We tested the sensitivity of the inference to various assumptions and aspects of the data (i.e., the effect of complex demography, sample size, phasing uncertainty, and missing data) in supplementary text S4, Supplementary Material online.
We retained the 1,500 simulations (from the 1.5M simulations) that were closest to our observed data based on three SSs: 1/LH, MH, and RH. These SSs were chosen based on an initial exploration of a broad range of SSs to identify a subset that contains information about allele ages (supplementary text S3, Supplementary Material online). These SSs were calculated only for the haplotypes carrying the derived allele at the selected site, which will be referred to as “selected haplotypes”, and for the genomic region defined by the positions away from the selected allele where the EHH declines to 0.05, which will be referred to as the “selected region.” 1/LH is defined as the inverse of the genetic length of the selected region (Sabeti et al. 2002). MH is defined as the average number of mutations within the selected region divided by the physical length of the region. MH is proportional to the estimator of age described by Thomson et al. (2000) and is proportional to the number of mutations that have occurred on the selected haplotypes since the most recent common ancestor of these haplotypes. RH is defined as the number of singletons divided by the total number of segregating sites in the selected haplotypes over the selected region. Therefore, 1/LH captures the idea of the “haplotype homozygosity decay” method (Sabeti et al. 2002), while MH is related to the information captured by the “counting” method (Thomson et al. 2000). RH was included because the allele frequency spectrum, and in particular the number of singletons, depends on both demography and selection; because all simulations assumed the same demographic model, RH reflects the effect of selection.
The accepted values of t and s for each region were adjusted by local linear regression, as implemented in the R package “abc” (Csilléry et al. 2012). Then, we applied the Gaussian kernel density estimation with a bandwidth chosen following Silverman’s “rule of thumb” (Silverman 1986) to generate the posterior distributions. To evaluate the fit of the model with the observation from each of the SNPs, we generated distributions of SSs by simulating data from the posterior distributions that we estimated by ABC and tested if the observed SSs were included within the variation of simulated SSs (supplementary text S2 and figs. S9–S10, Supplementary Material online). We further investigated if the estimated posterior distributions depend on the prior by comparing our results with those obtained using a uniform prior distribution (supplementary fig. S11, Supplementary Material online).
Comparison with the Ancient DNA Record
We estimated the past frequency of eight SNPs using ancient DNA data. First, we retrieved aligned sequence reads (BAM files) of ancient European samples from different ages (Lazaridis et al. 2014; Skoglund et al. 2014; Allentoft et al. 2015; Haak et al. 2015). Second, the age of each sample was defined as the one reported by the original publication by either using radiocarbon dating or from other archaeological evidence; samples were excluded from downstream analysis if age estimates were unavailable. Samples were then classified in five different periods based on the upper value of the age estimates. The total number of samples in each period was 31 in 8-7 kya, 25 in 6-5 kya, 72 in 5-4 kya, 36 in 4-3 kya, and 6 in 3-1 kya. Third, we estimated the reference allele frequency in each period using a likelihood function, described by Mathieson et al. (2015) (fig. 3). The CG allele frequency represents the frequency in the contemporary populations. All the allele counts in the ancient DNA data sets are available at the Ancient Human Genome Archive (http://genome-data.cri.uchicago.edu/ahga/; last accessed July 2015).
To evaluate if the frequency observed in the ancient DNA data is consistent with the estimates from our ABC, we simulated 10,000 trajectories for each SNP, conditional on their current frequency, by sampling t and s from the joint posterior distribution (supplementary figs. S8 and S12, Supplementary Material online). The trajectory started at t with the frequency 1/2Ne, and the frequency changed under the Wright–Fisher model with the corresponding value of s. We calculated the average allele frequencies at the lower bound of each period (7, 5, 4, 3, or 1 kya) as well as 95% confidence intervals to compare with the allele frequency estimated using ancient DNA data (fig. 3). If we estimated the allele frequencies at each time point by using frequency as a parameter in the ABC, we obtained virtually identical posterior distributions (data no shown).
Data Access
All raw sequencing reads can be retrieved from the NCBI Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRP017764.
Supplementary Material
Supplementary texts S1–S4, figures S1–S12, and tables S1–S7 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Joanna Kelley, John Novembre, Molly Przeworski, Daniel Wegmann, and Michael Zwick for helpful discussions and to David Witonsky for computational support. This work was supported by the National Institutes of Health (grant R01GM10168 to A.D.). This project was supported by the University of Chicago Comprehensive Cancer Center Support Grant (#P30 CA14599), with particular support from the Genomics Core Facility. S.N. was supported by a Grant-in-Aid for the Japan Society for the Promotion of Science Research Fellow (24-3234).
References
- Adamovic S, Amundsen SS, Lie BA, Gudjonsdottir AH, Ascher H, Ek J, van Heel DA, Nilsson S, Sollid LM, Torinsson Naluai A. 2008. Association study of IL2/IL21 and FcgRIIa: significant association with the IL2/IL21 region in Scandinavian coeliac disease families. Genes Immun. 9:364–367. [DOI] [PubMed] [Google Scholar]
- Albers HM, Kurreeman FA, Stoeken-Rijsbergen G, Brinkman DM, Kamphuis SS, van Rossum VA, Girschick HJ, Wouters C, Saurenmann RK, Hoppenreijs E, et al. 2009. Association of the autoimmunity locus 4q27 with juvenile idiopathic arthritis. Arthritis Rheum. 60:901–904. [DOI] [PubMed] [Google Scholar]
- Albert MA, Glynn RJ, Buring J, Ridker PM. 2004. C-reactive protein levels among women of various ethnic groups living in the United States (from the Women’s Health Study). Am J Cardiol. 93:1238–1242. [DOI] [PubMed] [Google Scholar]
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 4:903–905. [DOI] [PubMed] [Google Scholar]
- Alcina A, Vandenbroeck K, Otaegui D, Saiz A, Gonzalez JR, Fernandez O, Cavanillas, Cénit MC, Arroyo R, Alloza I, et al. 2010. The autoimmune disease-associated KIF5A, CD226 and SH2B3 gene variants confer susceptibility for multiple sclerosis. Genes Immun. 11:439–445. [DOI] [PubMed] [Google Scholar]
- Allentoft ME, Sikora M, Sjogren KG, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder J, Ahlström T, Vinner L, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522:167–172. [DOI] [PubMed] [Google Scholar]
- Almomani R, van der Heijden J, Ariyurek Y, Lai Y, Bakker E, van Galen M, Breuning MH, den Dunnen JT. 2011. Experiences with array-based sequence capture: toward clinical applications. Eur J Hum Genet. 19:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armelagos GJ, Harper KN. 2005. Genomics at the origins of agriculture, part two. Evol Anthropol. 14:109–121. [Google Scholar]
- Barreiro LB, Quintana-Murci L. 2010. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 11:17–30. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. 2009. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 41:703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 40:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Schluter D. 2008. Adaptation from standing genetic variation. Trends Ecol Evol. 23:38–44. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162:2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleza S, Santos AM, McEvoy B, Alves I, Martinho C, Cameron E, Shriver MD, Parra EJ, Rocha J. 2013. The timing of pigmentation lightening in Europeans. Mol Biol Evol. 30:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benazzi S, Douka K, Fornai C, Bauer CC, Kullmer O, Svoboda J, Pap I, Mallegni F, Bayle P, Coquerelle M, et al. 2011. Early dispersal of modern humans in Europe and implications for Neanderthal behaviour. Nature 479:525–528. [DOI] [PubMed] [Google Scholar]
- Branicki W, Brudnik U, Wojas-Pelc A. 2009. Interactions between HERC2, OCA2 and MC1R may influence human pigmentation phenotype. Ann Hum Genet. 73:160–170. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2003. Genetics and the making of Homo sapiens. Nature 422:849–857. [DOI] [PubMed] [Google Scholar]
- Casto AM, Feldman MW. 2011. Genome-wide association study SNPs in the human genome diversity project populations: does selection affect unlinked SNPs with shared trait associations? PLoS Genet. 7:e1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MT, Leung DY, Szefler SJ, Spahn JD. 1998. Difficult-to-control asthma: clinical characteristics of steroid-insensitive asthma. J Allergy Clin Immunol. 101:594–601. [DOI] [PubMed] [Google Scholar]
- Chen H, Patterson N, Reich D. 2010. Population differentiation as a test for selective sweeps. Genome Res. 20:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, Brown DL, Duffy DL, Patorino L, Bianchi-Scarra G, et al. 2009. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol. 129:392–405. [DOI] [PubMed] [Google Scholar]
- Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, Myers RM, Cavalli-Sforza LL, Feldman MW, Pritchard JK. 2009. The role of geography in human adaptation. PLoS Genet. 5:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csilléry K, François O, Blum MGB. 2012. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol Evol. 3:475–479. [Google Scholar]
- Daha NA, Kurreeman FA, Marques RB, Stoeken-Rijsbergen G, Verduijn W, Huizinga TW, Toes RE. 2009. Confirmation of STAT4, IL2/IL21, and CTLA4 polymorphisms in rheumatoid arthritis. Arthritis Rheum. 60:1255–1260. [DOI] [PubMed] [Google Scholar]
- de Boer YS, van Gerven NM, Zwiers A, Verwer BJ, van Hoek B, van Erpecum KJ, Beuers U, van Buuren HR, Drenth JP, den Ouden JW, et al. 2014. Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology 147:443–452.e445. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. 2012. A linear complexity phasing method for thousands of genomes. Nat Methods. 9:179–181. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. 2013. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 10:5–6. [DOI] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, et al. 2010. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 42:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. 2002. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 30:233–237. [DOI] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, Avey L, Wojcicki A, Pe'er I, Mountain J. 2010. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 6:e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federico MJ, Covar RA, Brown EE, Leung DY, Spahn JD. 2005. Racial differences in T-lymphocyte response to glucocorticoids. Chest 127:571–578. [DOI] [PubMed] [Google Scholar]
- Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, Lefèbvre C, Brant SR, Cho JH, Silverberg MS, et al. 2009. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut 58:799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble C, Davies W, Pettitt P, Richards M. 2004. Climate change and evolving human diversity in Europe during the last glacial. Philos Trans R Soc Lond B Biol Sci. 359:243–253; discussion 253–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf J, Hodgson R, van Daal A. 2005. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 25:278–284. [DOI] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Shlyakhter I, Karlsson EK, Bryne EH, Morales S, Frieden G, Hostetter E, Angelino E, Garber M, Zuk O, et al. 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327:883–886. [DOI] [PubMed] [Google Scholar]
- Guernier V, Hochberg ME, Guegan JF. 2004. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddy TB, Rana SR, Castro O. 1999. Benign ethnic neutropenia: what is a normal absolute neutrophil count?. J Lab Clin Med. 133:15–22. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, Hankinson SE, Hu Fb, Duffy DL, Zhao ZZ, et al. 2008. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 4:e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Alkorta-Aranburu G, Beall CM, Gebremedhin A, Sukernik R, UtermannG, Pritchard JK, Coop G, Di Rienzo A. 2011. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 7:e1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper K, Armelagos G. 2010. The changing disease-scape in the third epidemiological transition. Int J Environ Res Public Health. 7:675–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Guettouche T, Yang S, Bademci G, Diaz A, Andersen A, Hulme WF, Linker S, Mehta A, Edwards YJ, et al. 2011. Comparison of three targeted enrichment strategies on the SOLiD sequencing platform. PLoS One 6:e18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higham T, Compton T, Stringer C, Jacobi R, Shapiro B, Trinkaus E, Chandler B, Gröning F, Collins C, Hillson S, et al. 2011. The earliest evidence for anatomically modern humans in northwestern Europe. Nature 479:52–-524. [DOI] [PubMed] [Google Scholar]
- Hobolth A, Christensen OF, Mailund T, Schierup MH. 2007. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth A, Dutheil JY, Hawks J, Schierup MH, Mailund T. 2011. Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection. Genome Res. 21:349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis-Moffatt JE, Chen-Xu M, Topless R, Dalbeth N, Gow PJ, Harrison AA, Highton J, Jones PB, Nissen M, Smith MD, et al. 2010. Only one independent genetic association with rheumatoid arthritis within the KIAA1109-TENR-IL2-IL21 locus in Caucasian sample sets: confirmation of association of rs6822844 with rheumatoid arthritis at a genome-wide level of significance. Arthritis Res Ther. 12:R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. 2011. Genotype imputation with thousands of genomes. G3 (Bethesda) 1:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MM, Everhart JE, Byrd-Holt DD, Tisdale JF, Rodgers GP. 2007. Prevalence of neutropenia in the U.S. population: age, sex, smoking status, and ethnic differences. Ann Intern Med. 146:486–492. [DOI] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18:337–338. [DOI] [PubMed] [Google Scholar]
- Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, et al. 2008. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 40:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Liu F, Janssens AC, Rivadeneira F, Lao O, van Duijn K, Vermeulen M, Arp P, Jhamai MM, van Ijcken WF, et al. 2008. Three genome-wide association studies and a linkage analysis identify HERC2 as a human iris color gene. Am J Hum Genet. 82:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley-Hedgepeth A, Lloyd-Jones DM, Colvin A, Matthews KA, Johnston J, Sowers MR, Sternfeld B, Pasternak RC, Chae CU. 2008. Ethnic differences in C-reactive protein concentrations. Clin Chem. 54:1027–1037. [DOI] [PubMed] [Google Scholar]
- Kiialainen A, Karlberg O, Ahlford A, Sigurdsson S, Lindblad-Toh K, Syvanen AC. 2011. Performance of microarray and liquid based capture methods for target enrichment for massively parallel sequencing and SNP discovery. PLoS One 6:e16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ohta T. 1973. The age of a neutral mutant persisting in a finite population. Genetics 75:199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310:1782–1786. [DOI] [PubMed] [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wollstein A, Hysi PG, Ankra-Badu GA, Spector TD, Park D, Zhu G, Larsson M, Duffy DL, Montgomery GW, et al. 2010. Digital quantification of human eye color highlights genetic association of three new loci. PLoS Genet. 6:e1000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J, Wise C, Miner A, Malloy MJ, Pullinger CR, et al. 2008. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 4:e1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F, Perry GH, Di Rienzo A. 2010. Evolutionary adaptations to dietary changes. Annu Rev Nutr. 30:291–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti AK, Kim-Howard X, Viswanathan P, Guillén L, Rojas-Villarraga A, Deshmukh H, Direskeneli H, Saruhan-Direskeneli G, Cañas C, Tobön GJ, et al. 2010. Confirmation of an association between rs6822844 at the Il2-Il21 region and multiple autoimmune diseases: evidence of a general susceptibility locus. Arthritis Rheum. 62:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranville JC, Baxter SS, Torres JM, Di Rienzo A. 2011. Inter-ethnic differences in lymphocyte sensitivity to glucocorticoids reflect variation in transcriptional response. Pharmacogenomics J. 13:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Cutler D, Patterson N, Stephens M, Eskin E, Halperin E, Lin S, Qin ZS, Munro HM, Abecasis GR, et al. 2006. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 78:437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez A, Orozco G, Martinez A, Palomino-Morales R, Fernández-Arquero M, Mendoza JL, Taxonera C, Díaz-Rubio M, Gómez-García M, Nieto A, et al. 2009. Novel association of the interleukin 2-interleukin 21 region with inflammatory bowel disease. Am J Gastroenterol. 104:1968–1975. [DOI] [PubMed] [Google Scholar]
- Maruyama T. 1974. The age of an allele in a finite population. Genet Res. 23:137–143. [DOI] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. doi:10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Jones MR, Taylor KD, Marciante K, Yan X, Dubinsky M, Ippoliti A, Vasiliauskas E, Berel D, Derkowski C, et al. 2010. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet. 19:3468–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellars P. 2011. Palaeoanthropology: the earliest modern humans in Europe. Nature 479:483–485. [DOI] [PubMed] [Google Scholar]
- Mira A, Pushker R, Rodriguez-Valera F. 2006. The Neolithic revolution of bacterial genomes. Trends Microbiol. 14:200–206. [DOI] [PubMed] [Google Scholar]
- Mummert A, Esche E, Robinson J, Armelagos GJ. 2011. Stature and robusticity during the agricultural transition: evidence from the bioarchaeological record. Econ Hum Biol. 9:284–301. [DOI] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310:321–324. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Fukamachi S, Kimura H, Koda Y, Soemantri A, Ishida T. 2002. Distinctive distribution of AIM1 polymorphism among major human populations with different skin color. J Hum Genet. 47:92–94. [DOI] [PubMed] [Google Scholar]
- Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH. 2001. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet. 69:981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. 2007. Recent and ongoing selection in the human genome. Nat Rev Genet. 8:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. 2001. SSAHA: a fast search method for large DNA databases. Genome Res. 11:1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. 2006. Genetic evidence for complex speciation of humans and chimpanzees. Nature 441:1103–1108. [DOI] [PubMed] [Google Scholar]
- Pennington R, Gatenbee C, Kennedy B, Harpending H, Cochran G. 2009. Group differences in proneness to inflammation. Infect Genet Evol. 9:1371–1380. [DOI] [PubMed] [Google Scholar]
- Peter BM, Huerta-Sanchez E, Nielsen R. 2012. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 8:e1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, Kim H, Singh P, Lee A, Chen WV, et al. 2010. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, Kudaravalli S, Li JZ, Absher D, Srinivasan BS, Barsh GS, Myers RM, Feldman MW, et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Fort J, Ammerman AJ. 2005. Tracing the origin and spread of agriculture in Europe. PLoS Biol. 3:e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, Mullikin J, Hsueh WC, Cheng CY, Coresh J, et al. 2009. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 5:e1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, LohmuellerJ, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander E. 2006. Positive natural selection in the human lineage. Science 312:1614–1620. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sams AJ, Hawks J, Keinan A. 2015. The utility of ancient human DNA for improving allele age estimates, with implications for demographic models and tests of natural selection. J Hum Evol. 79:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, et al. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 483:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman B. 1986. Density estimation. London: Chapman and Hall. [Google Scholar]
- Skoglund P, Malmstrom H, Omrak A, Maanasa R, Valdiosera C, Günther T, Hall P, Tambets K, Parik J, Sjögren KG, et al. 2014. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344:747–750. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 2000. Allele age and a test for selection on rare alleles. Philos Trans R Soc Lond B Biol Sci. 355:1663–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 2001. Simulating genealogies of selected alleles in a population of variable size. Genet Res. 78:49–57. [DOI] [PubMed] [Google Scholar]
- Slatkin M. 2008. A Bayesian method for jointly estimating allele age and selection intensity. Genet Res (Camb). 90:129–137. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Rannala B. 1997. Estimating the age of alleles by use of intraallelic variability. Am J Hum Genet. 60:447–458. [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Rannala B. 2000. Estimating allele age. Annu Rev Genomics Hum Genet. 1:225–249. [DOI] [PubMed] [Google Scholar]
- Soejima M, Tachida H, Ishida T, Sano A, Koda Y. 2006. Evidence for recent positive selection at the human AIM1 locus in a European population. Mol Biol Evol. 23:179–188. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, et al. 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 42:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, et al. 2007. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 39:1443–1452. [DOI] [PubMed] [Google Scholar]
- Teixeira VH, Pierlot C, Migliorini P, Balsa A, Westhovens R, Barrera P, Alves H, Vaz C, Fernandes M, Pascual-Salcedo D, et al. 2009. Testing for the association of the KIAA1109/Tenr/IL2/IL21 gene region with rheumatoid arthritis in a European family-based study. Arthritis Res Ther. 11:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson R, Pritchard JK, Shen P, Oefner PJ, Feldman MW. 2000. Recent common ancestry of human Y chromosomes: evidence from DNA sequence data. Proc Natl Acad Sci U S A. 97:7360–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. 2007. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 39:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnado MC, Bethel G, Holmes GK, et al. 2007. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 39:827–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitti JJ, Grossman SR, Sabeti PC. 2013. Detecting natural selection in genomic data. Annu Rev Genet. 47:97–120. [DOI] [PubMed] [Google Scholar]
- Warren RB, Smith RL, Flynn E, Bowes J, Eyre S, Worthington J, Barton A, Griffiths CE. 2011. A systematic investigation of confirmed autoimmune loci in early-onset psoriasis reveals an association with IL2/IL21. Br J Dermatol. 164:660–664. [DOI] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Excoffier L. 2009. Efficient approximate Bayesian computation coupled with Markov chain Monte Carlo without likelihood. Genetics 182:1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. 2010. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329:75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Song F, Liang L, Nan H, Zhang J, Liu H, Wang LE, Wei Q, Lee JE, Amos CI, et al. 2013. Genome-wide association studies identify several new loci associated with pigmentation traits and skin cancer risk in European Americans. Hum Mol Genet. 22:2948–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, Franke L, Posthumus MD, van Heel DA, van der Steege G, et al. 2007. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 81:1284–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Stahl EA, Trynka G, Raychaudhuri S, Festen EA, Franke L, Westra HJ, Fehrmann RS, Kurreeman FA, Thomson B, et al. 2011. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 7:e1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.