Abstract
The source of genetic novelty is an area of wide interest and intense investigation. Although gene duplication is conventionally thought to dominate the production of new genes, this view was recently challenged by a proposal of widespread de novo gene origination in eukaryotic evolution. Specifically, distributions of various gene properties such as coding sequence length, expression level, codon usage, and probability of being subject to purifying selection among groups of genes with different estimated ages were reported to support a model in which new protein-coding proto-genes arise from noncoding DNA and gradually integrate into cellular networks. Here we show that the genomic patterns asserted to support widespread de novo gene origination are largely attributable to biases in gene age estimation by phylostratigraphy, because such patterns are also observed in phylostratigraphic analysis of simulated genes bearing identical ages. Furthermore, there is no evidence of purifying selection on very young de novo genes previously claimed to show such signals. Together, these findings are consistent with the prevailing view that de novo gene birth is a relatively minor contributor to new genes in genome evolution. They also illustrate the danger of using phylostratigraphy in the study of new gene origination without considering its inherent bias.
Keywords: BLAST, gene age, new genes, phylostratigraphy, proto-gene, yeast.
Introduction
Different species tend to have different numbers of genes. The human genome, for instance, has somewhere between 19,000 and 25,000 protein-coding genes (Hattori 2005; Ezkurdia et al. 2014). By contrast, there are approximately 13,000 protein-coding genes in the genome of the fruit fly Drosophila melanogaster (Misra et al. 2002). There is some amount of overlap between these two gene sets, but there are also genes unique to each of the two organisms. The question of how these differences in gene number and content arise has been an area of interest and investigation for decades (Nei 1969; Ohno 1970; Wolfe 2001; Long et al. 2003; Zhang 2003, 2013; Kaessmann et al. 2009). In general, these differences are attributable to differential gene gains and losses in different evolutionary lineages. In terms of gene gains, three distinct mechanisms are known: Horizontal gene transfer, gene (and genome) duplication, and de novo gene birth. Although the first two mechanisms and their contributions to organismal adaptation have been abundantly documented (Koonin et al. 2001; Pál et al. 2005; Zhang 2013; Qian and Zhang 2014), the arising of genes from nongenic material through de novo gene birth (Tautz and Domazet-Lošo 2011) was thought nigh-impossible for a long time (Jacob 1977). Although the last decade has seen the discovery of de novo gene birth in several species (Levine et al. 2006; Begun et al. 2007; Cai et al. 2008; Heinen et al. 2009; Knowles and McLysaght 2009; Xiao et al. 2009; Li, Zhang, et al. 2010; Wu et al. 2011; Yang and Huang 2011), the number of reported cases remains small. Because horizontal gene transfer merely transfers genes between species, gene duplication is commonly regarded as the dominant source of new genes whereas de novo gene birth is thought to have a minimal contribution.
The above view was recently challenged by Carvunis et al. (2012), who claimed that de novo gene birth is common in evolution and is a larger source of new genes than gene duplication. Specifically, they proposed that nongenic sequences are spuriously transcribed and translated, and the protein products may by chance possess biological functions, which could be selected for, resulting in a gradual enhancement of the protein function in evolution. They named the open-reading frames (ORFs) that are transcribed and translated but have not fully established their functions as proto-genes. They asserted that their model predicts a number of trends as proto-genes gradually age, including, for example, increases in ORF length, expression level, codon usage bias, and probability of being under purifying selection. The ideal test of their hypothesis would be to conduct laboratory evolution experiments and watch in real time how a nongenic sequence turns into a functional protein-coding gene. But because such evolutionary events are expected to be rare and the evolutionary processes slow, the authors took an indirect approach by comparing various properties among different age groups of proto-genes and genes from the genome of the budding yeast Saccharomyces cerevisiae, where gene ages were estimated using phylostratigraphy (Domazet-Lošo et al. 2007). In phylostratigraphy, the age of a gene from a focal species is defined by the time since the divergence between the focal species and its most distantly related taxon in which a homolog of the gene is found by a commonly used homology detection tool such as BLAST. Carvunis et al. reported that multiple trends predicted by their model were observed. The same claim was made in a similar study of vertebrates (Neme and Tautz 2013). Carvunis et al. further noted that 143 proto-genes originated in S. cerevisiae since its divergence from its sister species S. paradoxus and 19 of them are under purifying selection in S. cerevisiae. By contrast, they noted that no more than five genes were estimated to have been generated by gene duplication in the same period of time. These results led Carvunis et al. to conclude that de novo gene birth is widespread and is a bigger source of new genes than is gene duplication. A subsequent study based on a similar analysis of age distributions of gene properties suggested that proto-genes are gradually integrated into cellular networks by for instance gradual gains of protein interactions and genetic interactions (Abrusán 2013).
Although nothing is wrong with the theoretical model of de novo gene birth, whether the reported genomic patterns signify de novo gene birth and subsequent evolution is questionable for two reasons. First, some of the asserted predictions from the de novo gene birth model do not seem to be definitive. For example, it is unclear why the ORF of a gene should continually increase in length with time. Although it is easy to imagine scenarios where length increases are beneficial, one can also come up with situations where length reductions are advantageous. Because of the frequency of stop codons in random sequences, it is likely that a de novo gene is short and will increase in length in its early lifespan as a proto-gene. But it is not clear that this trend would be monotonic or prolonged for hundreds of millions of years. Once a function is established, why would increasing rather than decreasing its length tend to enhance or refine its function? Even if increasing the ORF length is beneficial to the functional refinement of a proto-gene, why should the length continue to rise even long after the proto-gene has become a well-established gene (e.g., when the gene is over 500 My old), as was observed by Carvunis and colleagues? Second, phylostratigraphy tends to underestimate gene age and the probability and amount of underestimation differ among genes (Moyers and Zhang 2015). For example, the probability of age underestimation decreases with the increase of ORF length, which could in principle explain Carvunis et al.’s observation of a gradual increase in ORF length with the estimated gene age. In this work, we show that the age distributions of various gene properties supporting widespread de novo gene birth are in fact largely attributable to age estimation errors created by phylostratigraphy. As such, there is no valid evidence to date for a larger contribution of de novo gene birth than gene duplication to new gene origination.
Results
Phylostratigraphy of Simulated Genes
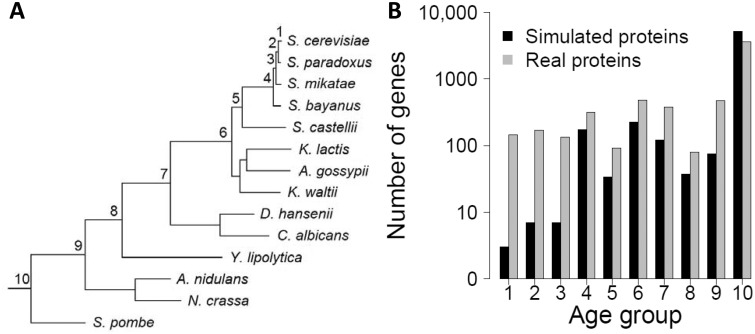
To examine whether gene age estimation error caused by phylostratigraphy could create spurious age distributions of gene properties resembling Carvunis et al.’s observations, we conducted a computer simulation of the evolution of all S. cerevisiae protein sequences along the tree shown in figure 1A using protein-specific parameters for site-specific rates and overall evolutionary rate. All S. cerevisiae protein sequences were simulated to have orthologs in all of the species shown in the tree (fig. 1A). That is, they all have the same age of 10, and there is no de novo gene origination in our simulation. We then applied phylostratigraphy to estimate the ages of the S. cerevisiae proteins by BLASTing them against the simulated sequences in all other species. These ages are referred to as estimated ages of simulated proteins (fig. 1B). We subsequently computed age distributions of various properties of S. cerevisiae proteins using the above estimated ages (figs. 2 and 3). Note that we used the properties provided by Carvunis et al. for each S. cerevisiae protein in these distributions; the only difference is the estimated gene age. In other words, we ask what would be the observed age distributions of gene properties if all S. cerevisiae genes have the same true age with no de novo gene birth. If the age distributions we observed resemble what Carvunis et al. observed, their observations cannot be used to support the de novo gene birth hypothesis because these observations are expected even in the absence of de novo gene birth.
Fig. 1.
Computer simulation for examining phylostratigraphic errors. (A) Tree used in the simulation of protein sequence evolution. The tree, including relative branch lengths, follows Wapinski et al. (2007). Node label refers to the age group corresponding to that node. (B) Numbers of genes estimated to belong to each age bin for real and simulated protein data. Numbers of genes in bins 1–10 for simulated protein data are 2, 6, 6, 171, 33, 222, 119, 36, 74, and 5,209, respectively. Numbers of genes in bins 1–10 for real data, as provided by Carvunis et al., are 143, 169, 133, 314, 90, 476, 381, 78, 469, and 3,625, respectively. Carvunis et al. arbitrarily assigned 107,425 smORFs to bin 0, which is not shown here.
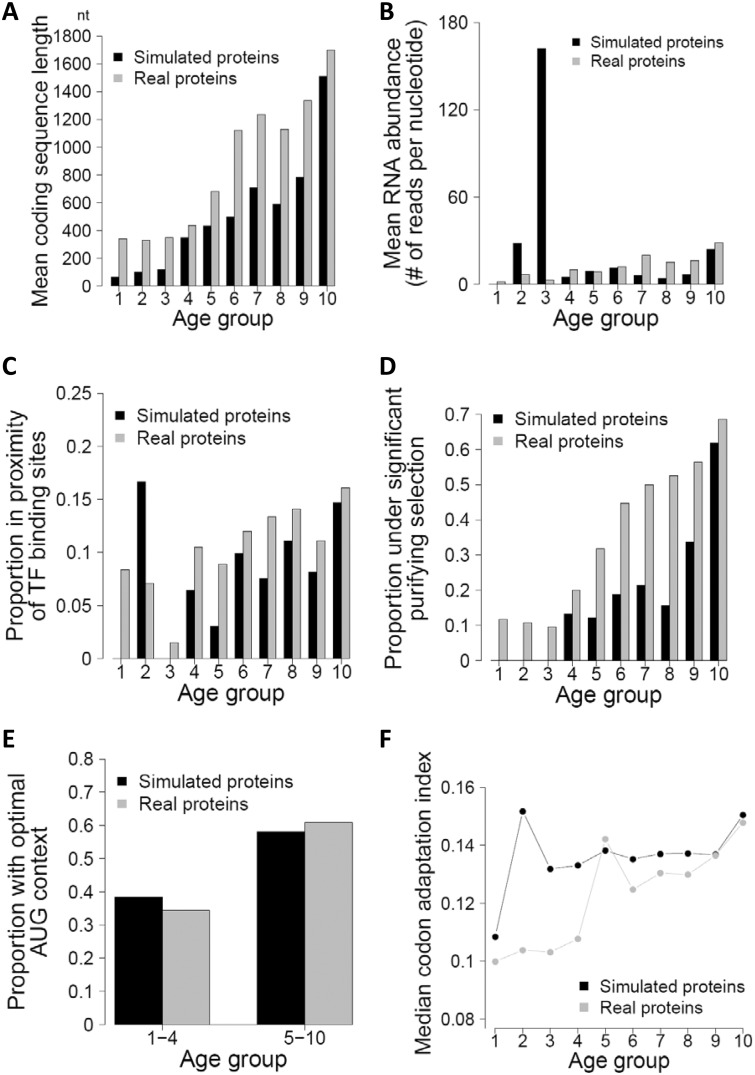
Fig. 2.
Age distributions of six gene properties in real and simulated proteins. (A) Average coding sequence length of genes in each age bin. Interestingly, although the same lengths are used for the real and simulated proteins, mean length is lower for simulated than real proteins in each bin. This is an example of Simpson’s paradox in statistics and is not due to mistakes in our analysis. (B) Mean expression level of genes in each age bin. (C) Proportion of genes having a TF-binding site within 200 bp of the translation start site for each age bin. (D) Proportion of genes under purifying selection for each age bin. (E) Proportion of genes with optimal AUG context for each age bin. (F) Median CAI for each age bin.
To derive protein-specific parameters for simulation, we acquired 5,261 published orthologous protein sequence alignments from five sensu stricto yeast species (S. cerevisiae, S. paradox, S. mikatae, S. kudriavzevii, and S. bayanus) (Scannell et al. 2011). For each of these proteins, we estimated the mean substitution rate per amino acid site and the substitution rate at each site relative to the mean rate of the protein (see Materials and Methods). These parameters were used in the simulation of the evolution of the protein (see Materials and Methods). For 619 S. cerevisiae proteins that do not have homologs in all five sensu stricto yeast species, we simulated their evolution in a conservative manner by sampling rate heterogeneity patterns and mean evolutionary rates from sensu stricto restricted proteins (see Materials and Methods). In all, we simulated the evolution of all 5,878 proteins present in the Carvunis et al. data set. The genetic distance of simulated orthologous proteins matches well that of real proteins (supplementary fig. S1, Supplementary Material online).
Because the true ages are 10 for all genes in the simulation (fig. 1A), any observed age distribution in which not all genes are in age group 10 is spurious. We found that, for 11.4% of simulated proteins, a homolog could not be found in the most distant species considered (Schizosaccharomyces pombe) (fig. 1A), which was estimated to diverge from S. cerevisiae approximately 788 Ma (Heckman et al. 2001; Hedges et al. 2006). The error rate of 11.4% is likely an underestimate, because a portion of our genes were evolved in a conservative manner (see Materials and Methods) and because we assumed that each site has a fixed substitution rate throughout its evolution, which is known to result in an underestimation of the error rate (Moyers and Zhang 2015). Of the 669 simulated proteins whose ages were underestimated by phylostratigraphy, 185 had estimated ages of 1–4 (fig. 1B). These genes would therefore be considered “candidate proto-genes” under Carvunis et al.’s definition, although they originated hundreds of millions of years ago in our simulation. Most strikingly, phylostratigraphy determined that two of these genes are S. cerevisiae-specific, despite that they originated in the common ancestor of S. cerevisiae and Sc. pombe. Nevertheless, the number of genes with estimated age 1–9 is greater in the actual data than in the simulated data (fig. 1B). Although this disparity may indicate the presence of some de novo genes, it may also be due to the fact that our simulation is conservative. That is, evolutionary processes that are not simulated here, such as gene duplication followed by rapid divergence and changes in the evolutionary rate of a site during evolution, could be responsible for this disparity.
Age Distributions of Six Gene Properties with Statistical Support
We next compared the age distributions between the real genes and simulated genes for each gene property used by Carvunis et al. as evidence for their model of widespread de novo gene birth. If the age distributions for a gene property are similar between the real genes and simulated genes, the age distribution observed by Carvunis et al. for the real genes can be explained by phylostratigraphy errors and hence cannot be used to support their model.
We first examined the six trends for which statistical support was previously provided (Carvunis et al. 2012). These trends are significant increases in ORF length (fig. 2A), mRNA abundance (fig. 2B), proportion of genes in proximity of transcription factor (TF)-binding sites (fig. 2C), proportion of genes under significant purifying selection (fig. 2D), proportion of genes with optimal AUG context (fig. 2E), and codon adaptation index (CAI) (fig. 2F) with gene age estimated through phylostratigraphy. Here, proportion of genes under significant purifying selection was determined by testing the action of purifying selection on each gene based on sequence polymorphisms among eight S. cerevisiae strains. All gene properties are defined as in Carvunis et al. (2012) and the property data were acquired from the authors. We found that, although qualitative appearances differed between the real and simulated data in these age distributions (fig. 2), statistical trends, quantified by Kendall’s τ as in (Carvunis et al. 2012), were almost identical between the two (table 1). Kendall’s τ was used following Carvunis et al. Using Spearman’s ρ did not alter our results. Both effect size (i.e., correlation coefficient) and significance level were reasonably well matched. This implies that the observed statistical trends of various gene properties with regard to gene age can be largely explained by gene age estimation errors.
Table 1.
Correlations (Kendall’s τ) between Estimated Gene Age and Various Gene Properties for Real and Simulated Proteins.
| Comparison | ORF Length | RNA Abundance | Proximity of TF-Binding Sites or Not | CAI | Purifying Selection or Not | Optimal AUG Context |
|---|---|---|---|---|---|---|
| Age groups 0–10a | ||||||
| Real proteins | 0.31** | 0.27** | 0.11** | 0.12** | 0.45** | 0.14** |
| Simulated proteins | 0.31** | 0.27** | 0.11** | 0.12** | 0.45** | 0.14** |
| Simulated proteins (assuming oldest paralog agesb) | 0.31** | 0.27** | 0.11** | 0.12** | 0.45** | 0.14** |
| Age groups 1–10c | ||||||
| Real proteins | 0.39** | 0.26** | 0.08* | 0.31** | 0.32** | 0.13** |
| Simulated proteins | 0.33** | 0.26** | 0.06* | 0.21** | 0.27** | 0.12** |
| Simulated proteins (assuming oldest paralog agesb) | 0.31** | 0.21** | 0.04* | 0.22** | 0.26** | 0.13** |
*P < 0.05;
**P < 1E-16.
aAnalysis includes all smORFs.
bThe age of a gene is assumed to equal that of the oldest gene in the same gene family. See main text for details.
cAnalysis excludes all smORFs.
Carvunis et al. included in their analysis approximately 108,000 so-called small ORFs (smORFs) that were arbitrarily assigned the age of 0. These S. cerevisiae smORFs are not annotated genes, are at least 30 nt long, and are free from overlap with annotated features on the same strand. The similarity in the above six trends between real and simulated data holds whether or not these smORFs were included in our analysis (table 1).
Some of the S. cerevisiae genes analyzed are paralogous to one another, but our simulation and subsequent phylostratigraphy treated them as unrelated genes, rendering our result from the simulated data not directly comparable with that from the real data. To solve this problem, we performed an all-against-all BLASTP search of the original S. cerevisiae proteins and recorded paralogous relationships. From this information, we used the oldest age among each gene family as the age of all genes in that family. This modification of phylostratigraphically estimated gene age on our simulated data did not change our results on the genomic trends studied above (table 1).
Age Distributions of Four Gene Properties without Statistical Support
Carvunis et al. (2012) also reported four additional trends without providing statistical support, including changes in amino acid usage, hydropathicity, proportion of transmembrane regions, and proportion of disordered regions with estimated gene age. For the majority of these, the simulated data do not qualitatively match the real data (fig. 3A–C). A notable exception is the patterns found in amino acid usage, where simulated data match real data quite closely (fig. 3D). Note, however, no explicit explanation was provided by Carvunis et al. why these observed trends are expected from the de novo gene birth model (see Discussion). As such, we do not see these trends as evidence for or against the de novo gene birth model.
Fig. 3.
Age distributions of four additional gene properties in real and simulated proteins. (A) Mean hydropathicity value for each age bin. (B) Mean proportion of transmembrane regions for each age bin. (C) Mean proportion of disordered regions for each age bin. (D) Amino acid frequency ratios between age groups.
Age Distributions of Gene Properties Reflecting Genetic Integrations
Subsequent to Carvunis et al.’s study, Abrusán used Carvunis et al.’s data to examine the phylostratigraphy-based age distributions of a number of additional gene properties that he proposed to reflect gradual genetic integrations of de novo genes into cellular networks or maturation of protein structures (Abrusán 2013). These properties included genetic coregulation, number of protein–protein interactions, number of genetic interactions, number of feed-forward loops regulating a gene, number of TFs regulating a gene, epistatic effects, percent of a protein made up of alpha-helices or beta-sheets, and the propensity of a protein to aggregate. Interestingly, all significant trends he found in real genes are also significant in simulated genes, except for the case of alpha helices (table 2). We note that, in several but not all cases, effect sizes are comparable as well (table 2). Even in those cases where the effect size appears quite different between real data and simulated data, the differences do not necessarily support the de novo gene birth model, because the differences may be attributable to new genes created through gene duplication in the real data (He and Zhang 2005). Furthermore, it is unclear whether several of the trends observed (e.g., decrease in percent in beta sheets) indicate structure maturation of de novo genes. These appear to be post hoc explanations rather than a priori predictions of the de novo gene birth model (see Discussion).
Table 2.
Correlations (Kendall’s τ) between Estimated Gene Age and Gene Properties Purported in Abrusán (2013) to Reflect Genetic Integration or Protein Structure Maturation.
| Real Proteins | Simulated Proteins | |
|---|---|---|
| Genetic coregulation | 0.05* | 0.06* |
| % in alpha helices | 0.04* | −0.01 |
| % in beta sheets | −0.08* | −0.11** |
| Aggregation propensity | −0.14** | −0.15** |
| Number of protein–protein interactions | 0.22** | 0.11** |
| Number of genetic interactions | 0.14** | 0.08* |
| Average magnitude of epistasis | 0.13** | 0.08* |
| Number of feed-forward loops regulating a gene | 0.02 | 0.03* |
| Number of TFs regulating a gene | 0.02* | 0.03* |
*P < 0.05;
**P < 1E-16.
Number of Young Genes under Purifying Selection
Carvunis et al. (2012) noted that they observed 19 genes that are both S. cerevisiae-specific and under within-species purifying selection. Based on their new analyses (Carvunis A-R, personal communication), this number now drops to 16. The abundance of these genes was suggested by Carvunis et al. to be evidence of high rates of de novo (functional) gene birth in comparison to gene duplication (Gao and Innan 2004; Carvunis et al. 2012).
However, we noticed that 15 of the 16 genes are each overlapped with another gene on the opposite strand and the overlapping regions constitute between 73% and 93% of each of these 15 genes (table 3). The remaining gene, YOL166C, has no overlap with any annotated gene in S. cerevisiae. When searching for homologs in other fungal species, Carvunis et al. removed sections of query genes which overlapped. We searched for homologs using the full sequences of these query genes and discovered that many of them are present in other species (table 3). All hits occurred in true ORFs in the target sequence, which were at least 80 amino acids long and were frequently annotated and known to be transcribed. If these 15 genes are S. cerevisiae-specific, they are not expected to have long ORFs (≥80 codons) in other species even when the opposite strand has an overlapping gene. Thus, we conclude that these 15 genes are not S. cerevisae-specific and that Carvunis et al.’s results were erroneous because of their use of short query sequences that rendered BLAST powerless.
Table 3.
Reexamining Purported Saccharomyces cerevisiae-Specific Selected Genes.
| Gene | Age Based on Full Sequence | Nonoverlapped Length in Nucleotides (full length) | No. of Synonymous Polymorphisms in Nonoverlapped Region | No. of Nonsynonymous Polymorphisms in Nonoverlapped Region | P valuea |
|---|---|---|---|---|---|
| YBR232C | 6 | 55 (360) | 1 | 0 | 0.29 |
| YCL046W | 2 | 58 (324) | 0 | 0 | 1.00 |
| YDR537C | 7 | 47 (606) | 0 | 2 | 0.57 |
| YER087C-A | 7 | 62 (552) | 0 | 0 | 1.00 |
| YFL013W-A | 5 | 53 (804) | 1 | 1 | 1.00 |
| YGL152C | 6 | 71 (678) | 2 | 2 | 0.58 |
| YHL030W-A | 9 | 49 (462) | 0 | 2 | 0.57 |
| YIL071W-A | 6 | 111 (477) | 0 | 0 | 1.00 |
| YLR232W | 9 | 58 (348) | 1 | 2 | 1.00 |
| YLR358C | 6 | 50 (564) | 0 | 1 | 1.00 |
| YNL105W | 10 | 88 (429) | 0 | 0 | 1.00 |
| YNL109W | 8 | 50 (546) | 0 | 0 | 1.00 |
| YOL150C | 8 | 62 (312) | 0 | 0 | 1.00 |
| YOL166C | 1 | 339 (339) | 3 | 3 | 0.37 |
| YOR055W | 6 | 55 (435) | 0 | 0 | 1.00 |
| YOR135C | 10 | 91 (342) | 1 | 0 | 0.30 |
aBased on two-tailed Fisher’s exact test of the neutral hypothesis.
The gene of most interest is YOL166C, because it is not overlapped by any other gene and has no hit in any other sequenced species. There are two major questions to be addressed about this gene. First, is there a homologous sequence in S. paradoxus, the species known to be the closest to S. cerevisiae, such that one can identify the source of YOL166C? Second, is there direct evidence for translation of this gene? To approach the first question, we looked for the S. paradoxus genomic region aligned to S. cerevisiae chromosome 15, base pairs 1–2078, a region encompassing YOL166C. No such alignment exists in this region, according to the Saccharomyces Genome Resequencing Project (SGRP) Genome Browser. We further checked for the homologs of YOL166C’s neighboring genes TEL15L and YOL165C. TEL15L found a significant hit in the S. paradoxus retrotransposons Ty5-10p and Ty5-5p, but YOL165C had no hit in S. paradoxus. YOL165C and YOL166C are in the subtelomeric region of chromosome 15 in S. cerevisiae. These regions are generally quite unstable (Brown et al. 2010), so it is not surprising that an orthologous region could not be found. Additionally, when BLASTed against the S. cerevisiae genome, YOL166C only finds itself as a hit.
To approach the second question, we searched for direct evidence of translation of YOL166C. Carvunis et al. did not find evidence of the translation of this gene under either rich or starved conditions based on yeast ribosome profiling data (Ingolia et al. 2009). Several papers report changes in the transcript concentration of YOL166C under different conditions (Fisk et al. 2006), but there is no evidence that YOL166C is expressed at the protein level. Based on these analyses, YOL166C does not meet the strict definition of a de novo gene (see Discussion). However, it also does not appear to be an instance of gene duplication. This leaves open the possibility that this is an example of a de novo gene birth.
A major question remains about whether or not these 16 genes are under selective constraint. Carvunis et al. estimated the nonsynonymous to synonymous substitution rate ratio on a phylogeny of eight S. cerevisiae strains and found this ratio to be significantly lower than 1, an indication of the action of purifying selection. However, their method is commonly used for testing selection in gene sequences collected from different species and is inappropriate for testing selection in sequences from the same species, because, for intraspecific data, different regions of the genome can have different phylogenies due to recombination. Additionally, because the majority of the sequence was overlapped by another gene, inferring selective constraint can be confounded (Wei and Zhang 2015). So, in the cases of these genes, only their nonoverlapped portions should be used to infer selection. To increase the accuracy and power of selection detection, we used 38 S. cerevisiae strains in the SGRP (Cherry et al. 2012) and counted the number of synonymous and nonsynonymous polymorphisms in the region of a gene that is nonoverlapping with other genes (table 3). Using Fisher’s exact test, we then examined whether the ratio between the observed number of nonsynonymous polymorphisms to that of synonymous polymorphisms is significantly different from the corresponding ratio under neutrality, which was calculated from the potential numbers of nonsynonymous and synonymous sites in the same region (Zhang et al. 1998). In none of the 16 genes could the null hypothesis of neutrality be rejected in favor of the action of purifying selection or positive selection. This is probably unsurprising, because no evidence was found for their translation by Carvunis et al. and these genes probably bear no protein function. As a comparison, the same selection test was conducted for 100 randomly picked genes classified to age group 10 by Carvunis et al., and 86 of them were found to be under significant purifying selection. However, these genes are among the longest in the set. For the nonoverlapped region of an average gene in table 3, the probability of detecting significant purifying selection is about 2% even when all nonsynonymous mutations are strongly deleterious. In other words, there is virtually no power to detect purifying selection acting on such short sequences.
Discussion
The origin of new protein-coding genes from noncoding sequences is a fascinating hypothesis that has been supported by the discoveries of dozens of cases of de novo gene birth in human, Drosophila, yeast, and other species (Levine et al. 2006; Clark et al. 2007; Cai et al. 2008; Heinen et al. 2009; Knowles and McLysaght 2009; Xiao et al. 2009; Li, Zhang, et al. 2010; Wu et al. 2011; Yang and Huang 2011). Previous studies established a set of criteria for identifying de novo gene birth: 1) The candidate de novo protein-coding gene is transcribed and translated, 2) its homologous sequence can be found in the syntenic region in related species but the sequence has no protein-coding capacity, and 3) the sequence is ancestrally noncoding (Knowles and McLysaght 2009). One should add the fourth criterion of action of natural selection for a de novo gene to be considered functional. Satisfying all these criteria would prove de novo gene birth beyond reasonable doubt.
However, not all of the above criteria were used and satisfied in Carvunis et al.’s study. Instead, Carvunis et al. relied on estimating gene age by phylostratigraphy and using age distributions of various gene properties to test widespread de novo gene birth. For their approach to work, gene age estimation must be reliable and de novo gene birth must be widespread. Unfortunately, phylostratigraphy is known to be biased (Elhaik et al. 2006; Moyers and Zhang 2015). Thus, only those trends that are predicted by the de novo gene birth model but cannot be produced by phylostratigraphic bias may be used to support the model. But, we found that essentially every trend reported by Carvunis et al. (2012) and Abrusán (2013) is explainable at least to some extent by phylostratigraphic bias. One might argue that the age distributions observed from the actual data are not exactly the same as those observed from the simulated data, providing evidence for the de novo gene birth hypothesis. This argument is flawed for two reasons. First, a realistic simulation requires many parameters. Because not all parameters are known, we conducted conservative simulations. For example, the substitution rate of a site is unlikely to be constant in evolution (Fitch 1971; Penny et al. 2001; Zou and Zhang 2015) and this inconstancy increases phylostratigraphic error (Moyers and Zhang 2015). But because of the lack of information on the extent of this rate variation over time, we assumed no such variation in our simulation, rendering the phylostratigraphic error underestimated and our results conservative. Furthermore, the parameters chosen in simulating genes that are not found in all five sensu stricto yeast species also made the results conservative. Thus, the fact that the observed trends in real data are not exactly the same as in the simulated data does not necessarily indicate the existence of biological signals. Second, even if a biological signal truly exists, it does not necessarily support the de novo gene birth hypothesis. For instance, in figure 2B, one can see a gray peak at age 7, indicating that genes of age 7 have unusually high expressions. This feature in the real data is not present in the simulated data, so might mean a true biological signal. Nevertheless, this signal is not predicted by the de novo gene birth model and thus cannot be used to support the model.
A common pitfall of phylostratigraphy-based studies is to report whatever nonrandom trends observed and then provide post hoc explanations, as if all nonrandom trends have biological meanings. The problem of these kinds of explanations has been pointed out in other contexts (Pavlidis et al. 2012). Carvunis et al.’s and Abrusán’s studies also fall into this trap. Many of the trends they reported are not predicted a priori from the de novo gene birth model. These trends include ORF length in figure 2, all four properties in figure 3, genetic coregulation, % alpha helices, and % beta sheets in table 2. As mentioned, there is no particular reason why the refinement of the biological function of an ORF has to occur by increasing the ORF length rather than decreasing the length. Similarly, there is no prediction that as proto-genes age and mature, the mean hydropathicity should decrease, trans-membrane fraction of the protein should decrease, disordered fraction should increase, and certain amino acid frequencies should increase or decrease. In fact, the authors offer no explanation of why these trends are expected under the de novo gene birth model. Even for the trends that may be predicted by the de novo gene birth model, one cannot explain why some of them continue even for genes with age 10 (e.g., expression level and CAI), as if the maturation of de novo genes takes more than 500 My. Phylostratigraphic error remains the simplest and best explanation of the observed trends, whether or not they are predicted from the de novo gene birth model.
One might ask why phylostratigraphic error could result in seemingly nonrandom age distributions of so many gene properties. Based on the property of BLAST search, we previously predicted and demonstrated that gene age underestimation in phylostratigraphy is more severe when the protein under investigation is shorter or evolves faster (Moyers and Zhang 2015). Thus, the increase in ORF length with age observed in the simulated data (fig. 2A) is a known bias of phylostratigraphy. Lower protein evolutionary rates are caused by stronger purifying selection, so it is unsurprising that phylostratigraphic error causes a positive correlation between gene age and proportion of genes under purifying selection (fig. 2D). Because protein evolutionary rate is strongly negatively correlated with its mRNA expression level (Zhang and Yang 2015), mRNA expression level must also impact phylostratigraphic error, as seen in our simulated data (table 1). Hence, a positive correlation between gene age and expression level (fig. 2B) reflects an expected bias of phylostratigraphy. Phylostratigraphic error is also expected to create a positive correlation between gene age and CAI (fig. 2F), because CAI is positively correlated with gene expression level (Sharp and Li 1987). Because the expression level of a gene is positively correlated with the probability that the gene is in proximity of TF-binding sites (Wong et al. 2015) (τ = 0.094 in our data, P < 1E-300), phylostratigraphic error also causes a positive correlation between gene age and proportion in proximity of TF-binding sites (fig. 2C). It was reported (Miyasaka et al. 2002) and confirmed here that the expression level of a gene is positively correlated with the probability that the gene has an optimal AUG context (τ = 0.057, P < 1E-300), potentially explaining why a positive correlation between gene age and proportion in optimal AUG context is created by phylostratigraphic error (fig. 2E). Amino acid usage is known to be correlated with gene expression level (Akashi and Gojobori 2002), potentially explaining the observed trends in figure 3D. In fact, we found that all gene properties examined by Carvunis et al. are significantly correlated with one or more of the three factors that impact phylostratigraphic bias: ORF length, evolutionary rate, and expression level (table 4 and supplementary table S1, Supplementary Material online).
Table 4.
Correlations (Kendall’s τ) between Various Gene Properties and Three Properties Known to Bias Phylostratigraphy, Using Genes in Age Groups 1–10.
| Evolutionary Rate | ORF Length | Expression Level | |
|---|---|---|---|
| TF-binding sites | −0.09* | 0.02* | 0.08* |
| CAI | −0.33** | 0.15** | 0.26** |
| Optimal AUG context | −0.14** | 0.05* | 0.14** |
| Purifying selection | −0.22** | 0.37** | 0.09** |
| Mean hydropathicity | 0.03* | −0.14** | −0.10** |
| Percent in disordered regions | 0.05* | 0.13** | 0.01 |
| Percent in transmembrane regions | 0.07* | −0.07* | −0.07* |
| Genetic coregulation | −0.10** | 0.03* | 0.07* |
| Number of TFs | −0.07* | 0.02* | 0.02* |
| Number of feed-forward loops | −0.07* | 0.02 | 0.03* |
| Percent alpha helices | −0.05* | −0.07* | 0.09** |
| Percent beta sheets | −0.01 | −0.22** | 0.03* |
| Aggregation propensity | 0.05* | −0.06* | −0.11** |
| Number of protein–protein interactions | −0.23** | 0.11** | 0.15** |
| Number of genetic interactions | −0.11** | 0.11** | 0.04* |
| Average magnitude of epistasis | −0.12** | 0.05* | 0.10** |
*P < 0.05;
**P < 1E-16.
The contribution of de novo gene birth compared with gene duplication to the origin of new (functional) genes is an important subject of evolutionary genomics. Carvunis et al. suggested that there have been 16 de novo births of functional genes in S. cerevisiae since its split from S. paradoxus. They compared this with a suggested five genes formed by duplication in the same time period (Gao and Innan 2004), though this duplicate gene number has since been challenged (Casola et al. 2012). If correct, Carvunis et al.’s comparison would contradict the paradigm that duplication is the primary source of new genes. We found that 15 of the 16 genes claimed by Carvunis et al. to be S. cerevisiae-specific and under selection have homologous ORFs in at least one other species and that none of the 16 bear significant signals of natural selection or have evidence for translation. To our knowledge, there are only two verified instances of functional de novo gene births in S. cerevisiae (Cai et al. 2008; Li, Dong, et al. 2010), whereas approximately 144 functional duplications occurred in that time based on the inference from gene family expansions since the common ancestor of sensu stricto yeasts (Hahn et al. 2005). Although these estimates may not be precise, gene duplication appears to surpass de novo gene birth by 2 orders of magnitude in terms of contribution to the number of new functional genes. Of course, apart from this rate difference, the two mechanisms of new gene origination may supply different kinds of genetic materials. Gene duplication confers a functional gene structure to the daughter gene, whereas de novo gene birth provides something closer to a blank slate, a near-random form and function that may or may not be useful. It is possible that de novo gene births offer a greater degree of novelty, even if they contribute less frequently to the genome.
The investigation of de novo gene birth mechanisms brings up the question of what is meant by a (functional) gene. There is no shortage of answers to this question (Demerec 1933; Gerstein et al. 2007). Clearly, in the de novo gene birth model discussed here, what is meant is a functional, protein-coding gene. It is thus important to prove the functionality of a gene by demonstrating that it is under purifying or positive selection. Given the widespread transcription of intergenic sequences in eukaryotes (Johnson et al. 2005) and widespread translation of noncoding RNAs (at least based on ribosome profiling data) (Ingolia et al. 2014), it is probably not rare for a random noncoding sequence to be spuriously transcribed and translated. For example, over 100 human pseudogenes were reported to be translated, but the vast majority of them are not under purifying selection at the protein level (Xu and Zhang 2016). If one starts to call all such sequences as de novo genes, de novo gene birth rate is expected to be high, even if only a tiny fraction of them are functional. The real question is the birth rate of de novo genes that have selected functions. It is thus imperative to require the fourth criterion (natural selection) in identifying de novo genes. Nonetheless, we recognize that statistical tests of natural selection may be powerless for species-specific genes because only intraspecific polymorphism data may be used and because newly created de novo genes may be short. Thus, it appears that a more productive approach to estimating the rate of de novo gene birth is to identify de novo genes that arose in the common ancestor of a few closely related species such as that of S. cerevisiae and S. paradoxus rather than in S. cerevisiae. Although Carvunis et al. and this study focused on protein-coding genes, noncoding RNAs may also play important biological functions. It is possible that the larger part of genetic novelty in evolution is in the aspect of noncoding RNA genes. When searching for de novo genes in the future, it may be beneficial to expand the scope of “gene” to include this group.
In conclusion, it is clear that de novo gene birth plays some role in the formation of new genes in yeast, given previously identified cases. However, compared with gene duplication, the relative contribution of de novo gene birth to new genes is minor. Moving forward, evidence for de novo gene birth will need to be evaluated gene by gene based on the criteria mentioned rather than in aggregate, because current genomic studies for these trends are insufficient and confounded by phylostratigraphic error.
Materials and Methods
Yeast Genes
For simulation of sequence evolution, we acquired 5,261 orthologous sequence alignments in protein format from the sensu stricto group of yeast species from http://www.saccharomycessensustricto.org/current//aligns/coding_allfiles.fasta.tgz last accessed January 25, 2016 (Scannell et al. 2011). Except for two alignments, all contain five orthologous sequences from five sensu stricto yeast species. The simulation of the 5,259 genes that have alignments of five sequences used parameters estimated from the alignments. The simulation of other genes in S. cerevisiae used parameters estimated from a set of sensu stricto restricted genes.
To identify sensu stricto restricted genes, we acquired protein databases of four yeast species outside of the sensu stricto group. These species were S. castellii and S. kluyveri, downloaded from the Saccharomyces Genome Database (SGD) at http://www.yeastgenome.org/download-data/sequence last accessed January 25, 2016 (Cherry et al. 2012), as well as Kluyveromyces thermotolerans and Zygosaccharomyces rouxii, acquired from the Genolevures Consortium (Souciet et al. 2009). Using the alignments acquired from Scannell et al.(2011), we created five databases, one for each of the sensu stricto species. We then performed a BLASTP (E value = 0.01, in following with Carvunis et al.) search using each of these individually as a query, and the target being an aggregate of the S. castellii, S. kluyveri, K. thermotolerens, and Z. rouxii proteins. We identified proteins for which none of the five sensu stricto yeast homologs found a hit in the target database, amounting to 148 genes. These 148 genes exist in all five sensu stricto yeasts but are not found in the four outgroup species. Although homology detection error may explain the apparent restriction of these genes to the sensu stricto group, this is not a problem for our simulation, because it is exactly our goal to identify patterns of genes that appear to be sensu stricto restricted, whether or not they are in reality.
Main Simulation of Evolution
The evolutionary tree including the relative branch lengths used in simulation was from a previous study of yeast genes (Wapinski et al. 2007). For each of the 5,259 proteins with alignments of five sequences, we used TreePuzzle (Schmidt et al. 2002) to classify all sites into 16 equal-sized rate bins according to a discrete gamma model of among-site rate heterogeneity and estimated the relative rates of the 16 bins. We also inferred the mean evolutionary rate across all sites of the protein between S. cerevisiae and S. bayanus; all branch lengths for the protein concerned were then estimated using the relative tree branches aforementioned. Using all of these parameters, we simulated the evolution of these proteins using ROSE (Stoye et al. 1998), which allows the evolutionary rate for each site to be specified by the user, along the tree in figure 1A. ROSE evolves sequences through amino acid substitutions and insertions and deletions (indels). For each branch of the tree, ROSE first performs the amino acid substitution function, and then performs the indel function. If the branch is an internal branch in the tree, it then copies the resulting amino acid sequence to the base of each of the two branches after the split.
We used the JTT (Jones, Taylor, and Thorton)-f model in the ROSE simulation of protein sequence evolution, where “f” refers to the amino acid compositions of the protein concerned (Nei and Kumar 2000). Each site along the protein has a particular relative rate. The relative rate for a site is multiplied by the length of the branch to obtain the expected amount of evolution along the branch at the site. ROSE makes substitutions based on this expected amount of evolution and the substitution matrix supplied. This is repeated for all sites along the amino acid sequence.
For indels, there are two parameters that determine indel formation in ROSE, the indel threshold and the indel function. The indel threshold measures how frequently indels occur and was determined in the following manner. Taking the alignments of the yeast sensu stricto orthologs acquired from Scannell et al. (2011) and using a custom script, we determined the minimum number of indels necessary to produce the observed gapped alignments. From this information, we determined the number of indels per amino acid, averaged over all proteins. This indel threshold was then applied to all proteins in simulation. The indel function is a vector that sums to 1 and gives, at each vector site i, the probability of an indel of size i, given that an indel is occurring. For the indel function, we took the observed frequencies of indel sizes from 1 amino acid to 30 amino acids long (accounting for >99% of all observed indels), and adjusted these frequencies to sum to 1. Sequence simulation was performed once for each protein.
Simulation of Other Proteins
Sequences were acquired as described above, but we could not determine evolutionary rate or rate heterogeneity for proteins lacking an alignment or the two proteins from Scannell et al. (2011) that do not have alignments of all five orthologous sequences. We used parameters estimated from the group of sensu stricto limited genes to simulate these proteins. To do this, we took each protein in this group and multiplied the relative rates of all sites by the average evolutionary rate for the protein. This gave us an absolute evolutionary rate for each site. We then concatenated these numerical vectors into a single vector from which we could sample rates for each protein (supplementary fig. S2, Supplementary Material online). We specifically sampled the inferred absolute substitution rates of a contiguous set of sites. From there, we performed a simulation of evolution as described above. This simulation likely rendered our estimate of phylostratigraphic error rate conservative, because on average sensu stricto limited genes are expected to evolve more slowly than the 619 genes which do not have homologs in all sensu stricto species, as fast evolution is a reason for an apparently young gene age (Moyers and Zhang 2015). Note that smORF sequences were not simulated. Instead, they were universally assigned to age group 0, as in Carvunis et al. (2012).
Protein Phylostratigraphy
To perform protein phylostratigraphy, we used BLASTP with a permissive E value of 0.01, following the methods of Carvunis et al. (2012). We used the simulated sequences corresponding to S. cerevisiae as the query, and each other species as an independent database. We ran BLASTP searches for each simulated species independently rather than as a single aggregate database to increase sensitivity of homology detection.
Carvunis et al. conducted BLASTP, TBLASTX, and TBLASTN searches; the latter two searches require the use of DNA sequences. We chose not to simulate the evolution of protein-coding DNA sequences because realistic simulation of codon sequence evolution is difficult and because protein-based homology searches are generally much more sensitive than DNA-based homology searches.
NCBI Homology Searches
We acquired from SGD the DNA and protein sequences of Carvunis et al.’s 16 genes of age group 1 that were purported to be under purifying selection. We used the NCBI BLAST tool to perform BLASTN, TBLASTN, and TBLASTX searches against the full nonredundant database of all species. We restricted results to a permissive E value of 0.01, and only considered hits that had at least 40% query coverage.
Testing Purifying Selection in 16 Young Genes
We downloaded the reference sequence for each of the 16 young genes in question from the SGD, and noted exactly which nucleotides were not overlapped by another annotated ORF. We then acquired single nucleotide polymorphisms (SNPs) for all chromosomes in all strains, available at ftp://ftp.sanger.ac.uk/pub/users/dmc/yeast/latest/cere_matches.tgz last accessed January 25, 2016. We extracted the SNPs of 38 strains present in both the SGRP data and the phylogeny in Liti et al. (2009). We extracted only those SNPs for which quality score was 55 or greater, following Carvunis et al. (2012). We modified the reference sequence for each strain, producing FASTA files containing each strain’s sequence. We removed all sections of the sequence which were overlapped with another ORF. In order to retain full codons, we removed any codon which had even partial overlap with another ORF. We then aligned these sequences using MUSCLE (Edgar 2004). We performed Fisher’s exact test using the observed numbers of synonymous and nonsynonymous SNPs and the potential numbers of synonymous and nonsynonymous sites estimated assuming 70% of random mutations are nonsynonymous (Zhang et al. 1997). In no case was the result significantly different from the neutral expectation.
The 38 strains used are as follows: DBVPG6040, NCYC361, S288c, W303, 378604X, YJM789, YS2, YS4, YS9, 273614N, YIIc17_E5, RM11_1A, YJM975, YJM978, YJM981, DBVPG1853, 322134S, BC187, DBVPG6765, DBVPG1788, L-1374, L-1528, DBVPG1106, DBVPG137, SK1, DBVPG6044, NCYC110, Y55, UWOPS87_2421, UWOPS83_787_3, UWOPS03_461_4, UWOPS05_227_2, UWOPS05_217_3, K11, Y12, Y9, YPS606, and YPS128.
Other Data Sets
We were provided with various gene properties from Carvunis et al. through email communication. We downloaded data sets used by Abrusan (2013) from the supplementary data of that paper. The definitions and measurements of all of these properties were detailed in the respective publications (Carvunis et al. 2012; Abrusan 2013).
Supplementary Material
Supplementary figures S1 and S2 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Anne-Ruxandra Carvunis and Marc Vidal for supplying their data reported in Carvunis et al. (2012) and Anne-Ruxandra Carvunis for sharing unpublished results, members of the Zhang lab for discussion, and Jian-Rong Yang and Zhengting Zou for valuable comments on the manuscript. This work was supported in part by the US National Institute of Health (NIH) grant R01GM103232 to J.Z. and by the NIH training grant in genome sciences (T32HG000040) to B.A.M.
References
- Abrusán G. 2013. Integration of new genes into cellular networks, and their structural maturation. Genetics 195:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A. 99:3695–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA, Kern AD, Jones CD. 2007. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176:1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CA, Murray AW, Verstrepen KJ. 2010. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 20:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W. 2008. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179:487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A-R, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Conant GC, Hahn MW. 2012. Very low rate of gene conversion in the yeast genome. Mol Biol Evol. 29:3817–3826. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. 2012. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 40:700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. [DOI] [PubMed] [Google Scholar]
- Demerec M. 1933. What is a gene. J Hered. 24:368–378. [Google Scholar]
- Domazet-Lošo T, Brajkovic J, Tautz D. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23:531–533. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaik E, Sabath N, Graur D. 2006. The “inverse relationship between evolutionary rate and age of mammalian genes” is an artifact of increased genetic distance with rate of evolution and time of divergence. Mol Biol Evol. 23:1–3. [DOI] [PubMed] [Google Scholar]
- Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML. 2014. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 23:5866–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry M, et al. 2006. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23:857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. 1971. Rate of change of concomitantly variable codons. J Mol Evol. 1:84–96. [DOI] [PubMed] [Google Scholar]
- Gao L-Z, Innan H. 2004. Very low gene duplication rate in the yeast genome. Science 306:1367–1370. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. 2007. What is a gene, post-ENCODE? History and updated definition. Genome Res. 17:669–681. [DOI] [PubMed] [Google Scholar]
- Hahn MW, De Bie T, Stajich JE, Nguyen C, Cristianini N. 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 15:1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M. 2005. Finishing the euchromatic sequence of the human genome. Nature 50:162–168. [PubMed] [Google Scholar]
- He X, Zhang J. 2005. Gene complexity and gene duplicability. Curr Biol. 15:1016–1021. [DOI] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129–1133. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22:2971–2972. [DOI] [PubMed] [Google Scholar]
- Heinen TJ, Staubach F, Häming D, Tautz D. 2009. Emergence of a new gene from an intergenic region. Curr Biol. 19:1527–1531. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJS, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 196:1161–1166. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE. 2005. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21:93–102. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 10:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A. 2009. Recent de novo origin of human protein-coding genes. Genome Res. 19:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 55:709–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A. 103:9935–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-Y, Zhang Y, Wang Z, Zhang Y, Cao C, Zhang P-W, Lu S-J, Li X-M, Yu Q, Zheng X, et al. 2010. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput Biol. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Dong Y, Jiang Y, Jiang H, Cai J, Wang W. 2010. A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res. 20:408–420. [DOI] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 4:865–875. [DOI] [PubMed] [Google Scholar]
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka H, Kanai S, Tanaka S, Akiyama H, Hirano M. 2002. Statistical analysis of the relationship between translation initiation AUG context and gene expression level in humans. Biosci Biotechnol Biochem. 66:667–669. [DOI] [PubMed] [Google Scholar]
- Moyers BA, Zhang J. 2015. Phylostratigraphic bias creates spurious patterns of genome evolution. Mol Biol Evol. 32:258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1969. Gene duplication and nucleotide substitution in evolution. Nature 224:177–178. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press. [Google Scholar]
- Neme R, Tautz D. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin (Germany: ): Springer-Verlag. [Google Scholar]
- Pál C, Papp B, Lercher MJ. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 37:1372–1375. [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Jensen JD, Stephan W, Stamatakis A. 2012. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol Biol Evol. 29:3237–3248. [DOI] [PubMed] [Google Scholar]
- Penny D, McComish BJ, Charleston MA, Hendy MD. 2001. Mathematical elegance with biochemical realism: the covarion model of molecular evolution. J Mol Evol. 53:711–723. [DOI] [PubMed] [Google Scholar]
- Qian W, Zhang J. 2014. Genomic evidence for adaptation by gene duplication. Genome Res. 24:1356–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the saccharomyces sensu stricto genus. G3 (Bethesda) 1:11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li W-H. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souciet J-L, Dujon B, Gaillardin C. 2009. Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19:1696–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J, Evers D, Meyer F. 1998. Rose: generating sequence families. Bioinformatics 14:157–163. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12:692–702. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. 2007. Natural history and evolutionary principles of gene duplication in fungi. Nature 449:54–61. [DOI] [PubMed] [Google Scholar]
- Wei X, Zhang J. 2015. A simple method for estimating the strength of natural selection on overlapping genes. Genome Biol Evol. 7:381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH. 2001. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2:333–341. [DOI] [PubMed] [Google Scholar]
- Wong ES, Thybert D, Schmitt BM, Stefflova K, Odom T, Flicek P. 2015. Decoupling of evolutionary changes in transcription factor binding and gene expression in mammals. Genome Res. 25:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP. 2011. De novo origin of human protein-coding genes. PLoS Genet. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S. 2009. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One 4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang J. 2016. Are human translated pseudogenes functional? Mol Biol Evol. 33:755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Huang J. 2011. De novo origin of new genes with introns in Plasmodium vivax. FEBS Lett. 585:641–644. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18:292–298. [Google Scholar]
- Zhang J. 2013. Gene duplication In: Losos J, editor. The Princeton guide to evolution. Princeton (NJ): Princeton University Press; p. 397–405. [Google Scholar]
- Zhang J, Kumar S, Nei M. 1997. Small-sample tests of episodic adaptive evolution: a case study of primate lysozymes. Mol Biol Evol. 14:1335–1338. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 95:3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang J-R. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16:409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Zhang J. 2015. Are convergent and parallel amino acid substitutions in protein evolution more prevalent than neutral expectations? Mol Biol Evol. 32:2085–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.