Abstract
Clostridium difficile is the most common hospital acquired pathogen in the United States, and infection is in many cases fatal. Toxins A and B are its major virulence factors, but increasingly a third toxin may be present, known as C. difficile transferase (CDT). An ADP-ribosyltransferase that causes actin cytoskeletal disruption, CDT is typically produced by the major, hypervirulent strains and has been associated with more severe disease. Here we show that CDT enhances the virulence of two PCR-ribotype 027 strains in mice. The toxin induces pathogenic host inflammation via a Toll-like Receptor 2 (TLR2) dependent pathway, resulting in the suppression of a protective host eosinophilic response. Finally, we show that restoration of TLR2 deficient eosinophils is sufficient for protection from a strain producing CDT. These findings offer an explanation for the enhanced virulence of CDT-expressing C. difficile and demonstrate a mechanism by which this binary toxin subverts the host immune response.
The Gram positive anaerobe C. difficile causes mild to severe antibiotic associated diarrhea, pseudomembranous colitis, toxic megacolon and death 1–2. The Rho-glucosylating Toxins A and B (TcdA and TcdB) cause host cell death and profound inflammation and are required for symptomatic infection 3–7. Production of the binary toxin CDT in addition to Toxins A and B by C. difficile is associated with higher mortality, increased peripheral white blood cell count, and elevated risk of recurrence in clinical studies 8–10. CDT expressing strains have also become increasingly common over the last ten years, paralleling the overall increase in incidence and severity of CDI, and now account for up to 20% of isolates in the hospital setting 11–14
CDT consists of two components which act cooperatively to intoxicate cells 14,15. CDTb, the binding component of CDT, is produced as a precursor protein and requires proteolytic cleavage prior to intoxication. Following cleavage, CDTb forms a heptamer and associates with the lipolysis stimulated lipoprotein receptor, or LSR 16,17. This receptor is highly expressed within the liver, small intestine, colon and various other tissues, and is thought to be involved in the uptake and removal of lipoproteins and in the formation of tricellular tight junctions 18,19. Following formation of the CDTb heptamer and LSR binding, CDTa, the enzymatic component of CDT, binds the CDTb heptamer. This complex is endocytosed, and endosomal acidification triggers insertion of the CDTb heptamer into the endosomal membrane, through which CDTa is released into the cytoplasm 20. CDTa then transfers an ADP-ribose moiety to globular actin, which subsequently acts as a capping protein to prevent actin filament elongation. This results in collapse of the actin cytoskeleton, allowing the formation of microtubule protrusions on the surface of host cells which are thought to increase C. difficile adherence 21,22.
Although CDT production is associated with more severe disease, the role of CDT during infection is not well understood. In a hamster model of CDI, CDT was shown to enhance virulence in the presence of Toxin A, but not Toxin B. In humans, the intensity of the host inflammatory response is critical in determining disease outcome, and in murine models, innate IL-23 production is detrimental during infection 23–26. Toxins A and B shift the immune response towards this pathogenic inflammatory state by inducing IL-1β secretion via activation of the inflammasome 26,27. Therefore, we hypothesized that CDT may play an additional role during infection by influencing host inflammatory signaling.
In order to investigate the role of CDT during disease, here we have utilized isogenic CDT mutants of two distinct PCR-ribotype 027 strains in a mouse model of C. difficile colitis (R20291 and M7404). While both strains are human isolates that express Toxins A and B as well as CDT, R20291 was originally isolated from an outbreak in the United Kingdom while M7404 originated in Canada 28,29. In this system, we have shown that CDT is a true virulence factor capable of enhancing disease severity in conjunction with Toxins A and B. We report that CDT increased pathogenic host inflammation via a novel Toll-like Receptor 2 dependent pathway, which was required for suppression of a protective host eosinophilic response during infection.
CDT production enhances the virulence of PCR-ribotype 027 C. difficile
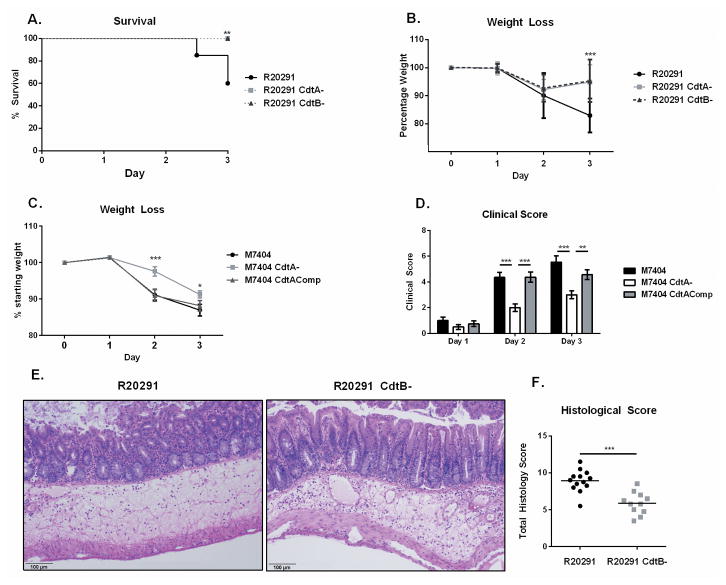
In order to test the role of CDT during disease, we infected mice with the PCR-ribotype 027 strain R20291 or one of the isogenic mutants R20291 CdtB- (lacking the binding component of CDT) or R20291 CdtA- (lacking the enzymatic component of CDT) 28,30. Infection with R20291 led to significantly greater mortality and weight loss than infection with an equivalent dose of R20291 CdtA- or CdtB- (60% survival vs 100% survival, n = 14, p = 0.001, Figure 1A–B, Supplementary Figure 1A–C). By day 3 of infection, mice infected with the CDT mutant strains began to recover weight and showed decreased clinical signs. At the same time point, groups infected with R20291 displayed increased weight loss and a high mortality rate. Thus, we concluded that the mutants lacking CDT were significantly attenuated at this dose.
Figure 1. CDT expression enhances the virulence of PCR-ribotype 027 C. difficile in a murine model of infection.
(A–B) 8 week old C57BL/6J mice underwent an antibiotic regimen prior to infection with 107 CFU of vegetative C. difficile strain R20291 or the isogenic mutants R20291 CdtA- or R20291 CdtB- (data shown combined from two independent experiments, n=15). (A) Animals were monitored for survival and (B) weight loss. (C–D) Mice were treated with the same antibiotic regimen before infection with 2×105 CFU of M7404, M7404 CdtA- or M7404 CdtAComp (data shown combined from two independent experiments (n=16). (C) Animals were monitored for weight loss and (D) clinical score. (E–F) Mice were sacrificed on day 2 of infection and cecal sections were isolated and fixed in Bouin’s solution for 18 hours before undergoing paraffin embedding, sectioning and haematoxylin & eosin staining. (F) Samples were scored blinded by 3 independent observers. (E) data shown are representative or (F) combined from two independent experiments (n=13). * = p value < 0.05, ** = p value < 0.01, *** = p value < 0.001 by Kaplan-Meier curve (A), two-tailed t-test (B–C) and Mann-Whitney test (D,F). NS = not significant. Error bars shown represent S.D. (B,C) or S.E.M. (D,F).
We confirmed this phenotype in a second PCR-ribotype 027 strain, M7404, using an isogenic mutant lacking CdtA (M7404 CdtA-) as well as a third, CdtA complemented strain (M7404 CdtAComp) 29. Although we initially utilized a wide range of infectious doses (1×105 – 1×107 CFU/mouse) in an attempt to determine an LD50 inoculum, infection with M7404 did not result in significant mortality in our model. However, we did observe differences in weight loss and clinical scores between the groups. We found that M7404 CdtA- caused significantly less weight loss and lower disease severity scores on days 2 and 3 of infection than either the wild type or CDT complemented strains (Figure 1C–1D). These data suggest that although M7404 and R20291 may inherently differ in virulence in this model, CDT was able to enhance disease severity in both strains.
To further characterize the differences in disease manifestation in the presence or absence of CDT, we focused on strain R20291 as a model of severe C. difficile infection due to its significant mortality. The R20291 CdtB- mutant was chosen as a comparison to eliminate any potential effects of the known pore forming ability of CdtB alone 18. We examined haematoxylin and eosin stained histopathological sections of the ceca from both groups (Figure 1E–F). Sections taken from mice infected with R20291 displayed significantly more overall tissue damage in the form of epithelial barrier disruption (characterized by disorganization and sloughing of epithelial cells), submucosal edema, and lumenal exudate than those infected with R20291 CdtB- (Supplementary Figure 1D). We next measured the production of Toxins A and B in these mice on day 3 of infection, as differential expression of these major virulence factors could explain the differences observed in tissue pathology. However, we did not observe differences in production of Toxins A and B (Supplementary Figure 1E), suggesting that the difference in mortality rates was driven by other factors. We also assessed C. difficile burden in cecal contents on day 3 of infection, but did not observe a difference at this time point (Supplementary Figure 1F). We next assessed translocation of commensals from the gut to other organs as a possible consequence of epithelial damage. This process has been suggested to drive mortality in some models of CDI 25. However, we did not observe differences in translocation of commensals to the liver or spleen (Supplementary Figure 1G). We conclude that tissue pathology due to CDT may influence disease outcome independent of Toxin A and B production, C. difficile burden, or liver commensal burden.
CDT causes increased inflammation
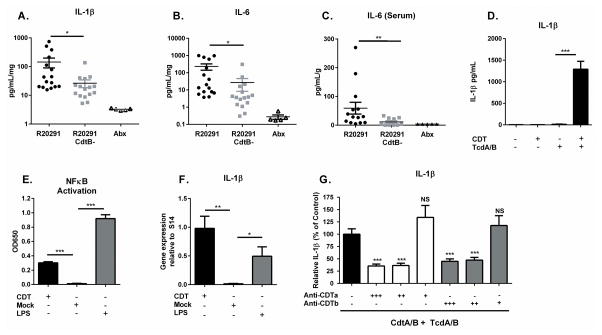
Based on clinical data demonstrating a correlation between the host inflammatory response and disease severity, we hypothesized that a systemic inflammatory response could result from increased tissue pathology and contribute to mortality during infection with R20291. To investigate this possibility, we quantified the inflammatory cytokines IL-1β and IL-6 within cecal tissue of C57BL6 mice infected with R20291 or R20291 CdtB- on day 3 post infection. Both IL-1β and IL-6 are known to be highly upregulated during CDI and to influence the type of immune response which develops. We found that R20291 induced significantly more cecal IL-1β and IL-6 than R20291 CdtB- (Figure 2A–B). Similarly, we observed significantly elevated serum IL-6 in R20291 infected mice (Figure 2C). These results suggest that CDT production by R20291 results in a stronger local and systemic inflammatory response within the murine host.
Figure 2. CDT promotes host inflammatory signaling.
(A–B) Mice infected with the indicated strain or uninfected and treated with antibiotics only (Abx) were sacrificed on day 3 of infection and cecal cytokines assessed by lysing whole cecal sections and quantifying protein via ELISA (data combined from two independent experiments, shown normalized to total protein concentration, n=16). (C) Serum IL-6 was measured via ELISA at the same time point (n=14). (D) Bone marrow derived dendritic cells were treated with 200 ng/mL purified CDTa and CDTb (CDT) or 2 ng/mL Toxin A and 2 ng/mL Toxin B (TcdA/B) for 24 hours. Secreted IL-1β was measured by ELISA. (E) NFκB activation was detected in a Raw Blue NFκB reporter cells by measuring Secreted Embryonic Alkaline Phosphatase (SEAP) in the culture media. (F) BMDCs were treated with 200 ng/mL CDTa and 200 ng/mL CDTb or with 100 ng/mL LPS as a positive control for 8 hours. Pro-IL-1β gene expression was assessed by qRT-PCR and is shown normalized to S14 housekeeping gene. (G) BMDCs were treated with 200 ng/mL CDTa and 200 ng/mL CDTb plus 2 ng/mL Toxin A and 2 ng/mL Toxin B in combination with decreasing amounts of anti-CDTa nanobody or anti-CDTb nanobody as indicated. +++ = 200 ng/mL, ++ = 20 ng/mL, + = 2 ng/mL. Secreted IL-1β was measured by ELISA. Data shown combined from 3 independent experiments with 3 replicates each (D–G). * = p value < 0.05, ** = p value < 0.01, *** = p value < 0.001 by Welch’s unequal variance t-test (A–F) or Mann-Whitney test (G). NS = not significant. Error bars shown represent S.E.M.
We next sought to determine how CDT signaling directly influences inflammatory cytokine production. To investigate this, we exposed bone marrow derived dendritic cells (BMDCs) generated from C57BL6 mice to purified CDT in the presence or absence of 2 ng/mL Toxins A and B, and measured the amount of IL-1β secreted into the culture medium as a readout of inflammatory cytokine production (Figure 2D). As we and other groups have shown previously, Toxins A and B are sufficient to activate the inflammasome at this concentration, but a prior “priming” signal is required for robust IL-1β secretion 27. We found that CDT alone did not induce IL-1β secretion at this relatively long time point, even at a much higher concentration than Toxins A and B (200 ng/mL). However, CDT did enhance IL-1β release in the presence of Toxins A and B. These data suggest that CDT acts as a priming signal prior to inflammasome activation by Toxins A and B. To investigate this further, we examined the ability of CDT to activate NFκB in a RAW macrophage reporter cell line. CDT alone was able to significantly activate NFκB over mock treated cells (Figure 2E). Additionally, CDT alone induced significant pro-IL-1β gene expression in C57BL6 BMDCs as measured by qRT-PCR (Figure 2F). In order to determine whether this phenotype was specific to CDT activity, we utilized anti-CDT targeted nanobodies which have previously been demonstrated to block CDT intoxication 31. At concentrations of 200 ng/mL (12 nM) and 20 ng/mL (1.2 nM), the anti-CDTa and anti-CDTb nanobodies both significantly decreased the amount of IL-1β secreted by BMDCs (Figure 2G). Thus, we conclude that exposure to CDT is sufficient to activate NFκB and enhance inflammatory cytokine production.
CDT suppresses protective colonic eosinophilia
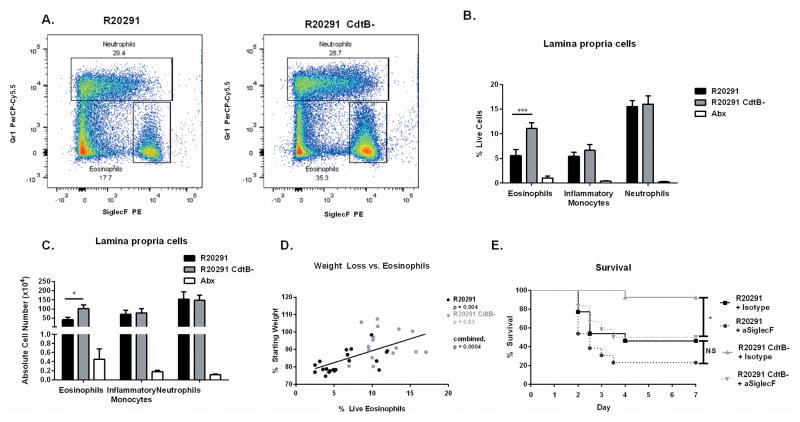
Because cytokine production shapes the immune response by influencing cell development, recruitment, proliferation and survival, we next utilized flow cytometry to examine the composition of effector cells in the colon during murine infection with R20291 and R20291 CdtB-. As expected, neutrophils (CD45+ CD11b+ Ly6G+ Ly6C+) dominated the innate compartment of both infected groups, representing roughly 20% of live cells in the colon (Figure 3A–C). Monocytes (CD45+ CD11b+ Ly6Chi Ly6G−) were also significantly elevated in the colon of both infected groups compared to uninfected, antibiotic treated controls. However, no difference was observed between neutrophils and monocytes in R20291 versus R20291 CdtB- infected groups, either as a percentage of live cells or by total cell number, suggesting that differences in neutrophil and monocyte recruitment were not responsible for infection outcome. In contrast, eosinophils were significantly elevated in R20291 CdtB- infected mice compared to R20291 infected animals (Figure 3A–C). As with neutrophils and monocytes, eosinophils were significantly increased in both groups compared to uninfected, antibiotic treated controls.
Figure 3. CDT production suppresses protective colonic eosinophilia.
(A) Mice infected with the indicated strain, or uninfected and treated with antibiotics only (Abx) were sacrificed on day 3. Colon tissue was isolated and processed to a single cell suspension and stained for flow cytometry. Representative flow plots depicting neutrophils and eosinophils are shown (two independent experiments, n=10). (B–C) Eosinophils (CD45+ CD11b+ SiglecF+), monocytes (CD45+ CD11b+ Ly6Chi) and neutrophils (CD45+ CD11b+ Ly6G+) were quantified. All three cell types are significantly elevated in both infected groups compared to Abx treated controls (data shown combined from two independent experiments, n=10). (D) Weight loss and percentage live colon eosinophils were compared using data combined from four independent experiments (n=17). (E) Colonic eosinophils were depleted using 40 ug of an anti-SiglecF targeted antibody or an isotype control antibody one day prior and one day following infection with C. difficile. Animals were monitored for clinical symptoms and humanely euthanized when required. Data shown combined from two independent experiments (n = 13). * = p value < 0.05, ** = p value < 0.01, *** = p value < 0.001 by Mann-Whitney test (B–C), linear regression (D) or Kaplan-Meier curve (E). NS = not significant. Error bars shown represent S.E.M.
Studies from our lab demonstrated a protective role for eosinophils after recombinant IL-25 administration 32. Because the protected, R20291 CdtB- infected group showed increased eosinophils compared to R20291, we hypothesized that high eosinophil numbers may correlate with protection from mortality. To examine this more closely, we used weight loss as a measure of disease severity and found a significant correlation between increased weight loss and lower eosinophil percentages, suggesting that eosinophils were associated with protection from disease (Figure 3D).
We next sought to determine whether eosinophils were causative in protection or merely associated with better outcome. To address this question, we depleted eosinophils in mice infected with R20291 or R20291 CdtB- using an anti-SiglecF targeted antibody which results in eosinophil apoptosis 33 (Figure 3E, Supplementary Figure 2). As expected, animals infected with R20291 CdtB- and treated with an isotype control antibody were relatively protected from mortality. In contrast, depletion of eosinophils with anti-SiglecF prior to and during infection with R20291 CdtB- significantly enhanced mortality. In R20291 infected mice, depletion of eosinophils did not significantly enhance mortality, perhaps because eosinophil counts are already relatively low in this group. Overall, these results demonstrate that an eosinophilic response is protective during CDI and that CDT production by a PCR-ribotype 027 strain has a suppressive effect on eosinophils in the colon, likely contributing to the enhanced virulence of this strain.
CDT induces apoptosis of blood eosinophils during infection
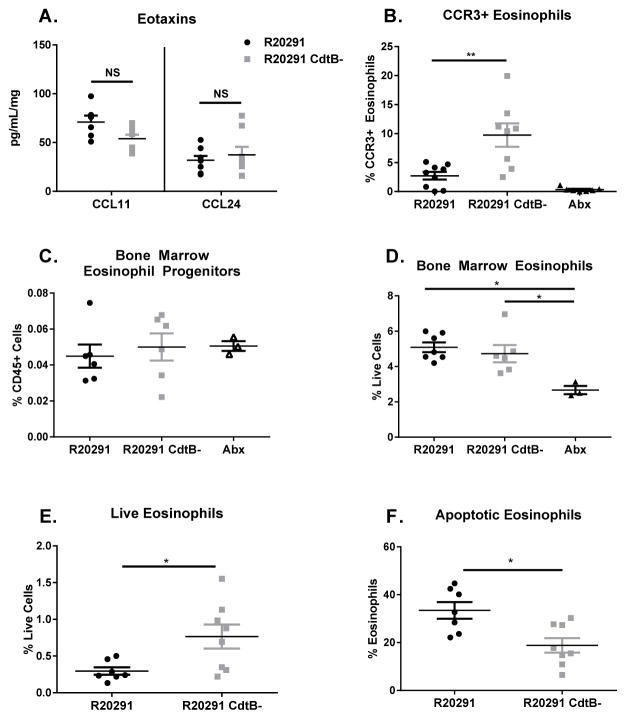
Next, we investigated the cause of the decrease in eosinophils observed in R20291 infected mice. We hypothesized that eosinophil recruitment signals or growth factors would be decreased in mice infected with the CDT producing strain. However, R20291 and R20291 CdtB- infected animals demonstrated equivalent levels of the eosinophil-specific chemokines eotaxin-1 and eotaxin-2 (Figure 4A). Eosinophil-promoting growth factors, such as IL-5, GM-CSF, IL-13, IL-33, and thymic stromal lymphopoietin (TSLP), were similar or elevated in R20291 infected mice, suggesting these factors were not responsible for the observed decrease in eosinophils (Supplementary Figure 3A–B) 34–36.
Figure 4. CDT production by C. difficile promotes eosinophil apoptosis.
(A) Mice infected with the indicated strain, or uninfected and treated with antibiotics (Abx) were sacrificed on day 3. Cecal eotaxin levels were assessed by lysing whole cecal sections and quantifying protein via ELISA (data combined from two independent experiments, shown normalized to total protein concentration, n=16). (B) Colon tissue was isolated and processed to a single cell suspension and stained for flow cytometry. CCR3 staining on eosinophils was assessed by first gating on total eosinophils (identified as CD45+ CD11b+ SiglecF+ SSChi cells) (data shown are representative of 2 independent experiments, n=8). (C–D) Mice were sacrificed on day 3 and bone marrow harvested for flow cytometry. Eosinophil progenitors were identified as Lin- CD34+ Sca-1− IL-5Rα+ cKitint cells by flow cytometry. Mature bone marrow eosinophils were quantified as CD45+ CD11b+ SiglecF+ SSChi cells (data shown combined from 2 independent experiments, n=6). (E–F) Blood eosinophils were assessed on day 3 of infection by flow cytometry following cardiac puncture, red blood cell lysis, and staining. Live eosinophils were identified at CD45+ CD11b+ SiglecF+ Live deadneg and apoptotic eosinophils identified as CD45+ CD11b+ SiglecF+ Annexin V+ Live deadneg cells. Data shown combined from two independent experiments. (n=7) * = p value < 0.05, ** = p value < 0.01, *** = p value < 0.001 by Mann-Whitney test (A, C–D, F) or Welch’s unequal variance t-test (B, E). NS = not significant. Error bars shown represent S.E.M.
Because the expression of CCR3, the cognate receptor for the eotaxins, has been shown to influence eosinophil recruitment 37, we hypothesized that different levels of this receptor could influence responsiveness to equivalent signals. We evaluated eosinophil surface expression of CCR3 during infection with R20291 and R20291 CdtB- (Figure 4B) by gating on CD45+ CD11c− CD11b+ SiglecF+ cells and assessing CCR3 staining. We observed a significant decrease in the number of CCR3+ eosinophils in the colon of R20291 infected mice compared to R20291 CdtB- infected animals. These results could indicate that eosinophils in R20291 infected mice are less responsive to eotaxins, thereby impacting recruitment to the gut. However, CCR3 upregulation is also associated with eosinophil maturity and egress from the bone marrow 38. Additionally, both CCR3+ and CCR3− eosinophils have been identified within the colon, suggesting that this receptor not absolutely required for colonic eosinophil recruitment 33. Therefore, we hypothesized that a decrease in colonic CCR3+ eosinophils could also result from other causes, such as a decrease in the number of mature eosinophils in the colon due to cell death.
To further investigate this possibility, we sought to determine where the eosinophil defect began. Eosinophils develop within the bone marrow from eosinophil progenitors (EoPs), defined as Lineage−, CD34+, Sca-1−, IL-5Rα+, cKitint cells 33,39–41 (Supplementary Figure 3C). We did not observe a significant difference in the number of bone marrow EoPs in R20291 or R20291 CdtB- infected mice (Figure 4C), consistent with the similar levels of eosinophil-promoting growth factors in these groups. When we examined mature bone marrow eosinophils, we noted that both infected groups had significantly more eosinophils than antibiotic treated controls (Figure 4D). However, there was no significant difference in the number of mature eosinophils between the two infected groups. In contrast, when we examined eosinophil numbers in the peripheral blood of infected mice, we noted a significantly decreased percentage of live eosinophils in R20291 infected mice (Figure 4E). This mirrored the phenotype observed in the colon and indicated that the eosinophil defect was systemic, but did not originate in the bone marrow, suggesting that a recruitment defect was not responsible for the observed decrease in colonic eosinophils.
We also observed that a higher percentage of blood eosinophils in mice infected with R20291 stained positive for Annexin V, a marker of apoptosis, which could contribute to the observed defect in mature eosinophils (Figure 4F). Taken together, these data suggest that eosinopoiesis functions similarly within both groups of infected mice, but that mature eosinophils which exit the bone marrow in R20291 infected mice undergo increased rates of apoptosis, resulting in the observed decrease in live eosinophils in both the blood and colon of these animals.
CDT recognition by TLR2 mediates pathogenic eosinophil suppression
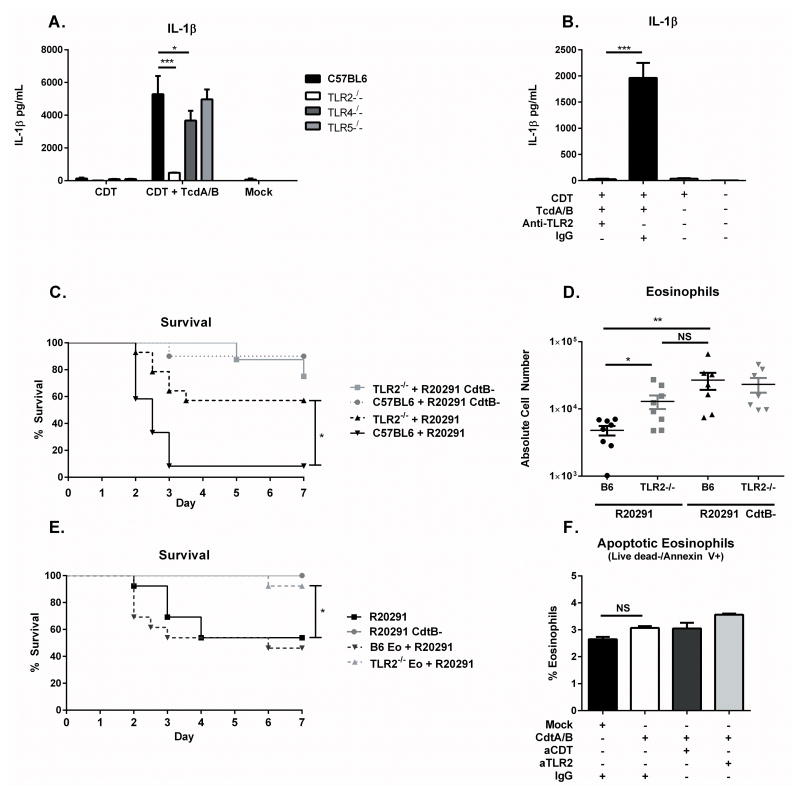
Because activation of NFκB is classically associated with Pattern Recognition Receptor (PRR) signaling, we next asked whether these signaling pathways were involved in recognition of CDT to mediate eosinophil apoptosis. We analyzed the ability of BMDCs generated from whole bone marrow of TLR2−/−, TLR4−/−, and TLR5−/− mice to secrete IL-1β following exposure to CDT and Toxins A and B. We found that TLR2−/− BMDCs were essentially unable to respond to CDT, suggesting that this pathway may be involved in CDT recognition (Figure 5A). To corroborate the role of TLR2, we treated wild type C57BL6 BMDCs with CDT and Toxins A and B following pre-treatment with an anti-TLR2 neutralizing antibody or an isotype control. The anti-TLR2 antibody significantly reduced the release of IL-1β (Figure 5B). To determine whether this interaction occurs in vivo, we next infected TLR2−/− mice alongside C57BL/6 wild-type controls with R20291 as well as R20291 CdtB-. Neither TLR2−/− nor C57BL/6 mice experienced significant mortality after infection with R20291 CdtB- (Figure 5C). However, TLR2−/− mice were significantly protected from mortality after infection with CDT-producing R20291, suggesting that TLR2 signaling enhances C. difficile virulence in the presence of CDT (Figure 5C).
Figure 5. TLR2 mediates CDT recognition and is required for eosinophil suppression.
(A) BMDCs were generated from TLR2−/−, TLR4−/−, and TLR5−/− mice before treatment with 200 ng/mL CDTa and 200 ng/mL CDTb (CDT) and 2 ng/mL Toxin A and 2 ng/mL Toxin B (TcdA/B) for 24 hours. IL-1β secretion was assessed by ELISA. (B) BMDCs were treated with 200 ng/mL CDTa and 200 ng/mL CDTb (CDT) and 2 ng/mL Toxin A and 2 ng/mL Toxin B (TcdA/B) for 24 hours in the presence of a TLR2 neutralizing antibody or an isotype control. IL-1β was assessed by ELISA. Data shown combined from three independent experiments of 3 replicates each (A–B). (C) 8 week old TLR2 knockout mice or C57BL/6J mice were infected with R20291 or R20291 CdtB- and monitored for survival (data combined from two independent experiments, n=14). (D) Mice were sacrificed on day 3 of infection and colonic eosinophils (CD45+ CD11b+ SiglecF+ SSChi) were measured by flow cytometry following tissue processing and staining (data combined from two independent experiments, n=8). (E) C57BL/6 mice received 4×105 TLR2−/− or B6 bone marrow-derived eosinophils (TLR2−/− Eo or B6 Eo) via IP injection one day prior and for 3 subsequent days following infection with R20291 or R20291 CdtB-. Mice were monitored daily for survival (data combined from two independent experiments, n=13). (F) BM Eos were incubated for 8 hours with 200 ng/mL CDTa and 200 ng/mL CDTb in the presence or absence of anti-TLR2 neutralizing antibody (aTLR2) or anti-CDT neutralizing nanobody (aCDT). Eosinophils were stained with Live dead or Annexin V and cell death was assessed by flow cytometry, data shown are representative of 3 independent experiments assayed in duplicate. * = p value < 0.05, ** = p value < 0.01, *** = p value < 0.001 by Mann-Whitney test (A), Welch’s unequal variance t-test (B, D, F) or Kaplan-Meier Analysis (C, E). NS = not significant. Error bars shown represent S.E.M.
We next asked whether TLR2 may be involved in the marked decrease in colonic eosinophils observed during infection with R20291. We assessed eosinophilia in the colonic lamina propria via flow cytometry, and found that TLR2−/− mice displayed significantly higher numbers of colonic eosinophils than wild-type mice after infection with R20291. In contrast, no significant difference was noted after infection with R20291 CdtB-, suggesting that the increase in colonic eosinophils in TLR2−/− mice specifically occurred in the presence of CDT (Figure 5D). These results suggest that recognition of CDT via TLR2 leads to suppression of eosinophilia in the colon and enhanced mortality.
Restoration of TLR2−/− eosinophils protects against CDT+ C. difficile
To determine whether preventing TLR2 signaling on a subset of eosinophils was sufficient to provide protection from R20291, we performed an adoptive transfer of bone marrow derived eosinophils (BM Eos) generated from either C57BL/6 or TLR2−/− mice. BM Eo cultures routinely consisted of greater than 75% CD45+ CD11b+ SiglecF+ SSChi mature eosinophils (Supplementary Figure 4A). Mice that received TLR2−/− BM Eos were significantly protected from R20291-mediated mortality compared to mice that received C57BL/6 BM Eos and to wild type controls (Figure 5E), suggesting that restoration of eosinophils is protective only when the eosinophils are unable to signal via TLR2. We next investigated the possibility that this could be due to direct killing of eosinophils by CDT. We incubated BM Eos with CDT in vitro and assessed cell death by Live/dead and Annexin V staining measured by flow cytometry. Treatment with CDT did not significantly enhance the number of apoptotic (Figure 5F) or dead eosinophils detected (Supplementary Figure 4B). CDT likewise did not enhance apoptosis or death of eosinophils when added in conjunction with Toxins A and B (Supplementary Figure 4C–D). Therefore, we conclude that direct killing of eosinophils by CDT is unlikely, suggesting that TLR2 signaling on eosinophils influences host survival by another mechanism, perhaps by modifying the protective function of these cells or their recruitment capability.
Discussion
Despite understanding the enzymatic function of CDT, the role of this toxin in enhancing C. difficile virulence remains incompletely characterized, and the influence of CDT on the host immune response has not been examined. We found that CDT was able to enhance the virulence of two PCR-ribotype 027 C. difficile strains, resulting in increased tissue pathology. CDT skewed the host inflammatory response by suppressing protective eosinophils within the colon and blood via the indirect induction of eosinophil apoptosis. Additionally, purified CDT was able to activate NFκB and to induce inflammatory IL-1β production in conjunction with Toxins A and B. This required TLR2 signaling, and TLR2-deficient mice were protected against the CDT-producing strain R20291, correlating with increased colonic eosinophils compared to wild type mice. Finally, adoptive transfer of TLR2 deficient eosinophils was sufficient to protect mice against CDT+ R20291, confirming an unexpected role for eosinophils in protection from CDI.
Although we demonstrated that CDT is able to activate NFκB and enhance inflammatory cytokine production, it is unclear where this signaling occurs in vivo. CDT may intoxicate epithelial cells to promote recruitment of immune cells, which may in turn enhance the inflammatory response. Alternatively, CDT may act directly on immune cells to enhance NFκB activation. The role of the lipolysis-stimulated lipoprotein receptor in mediating an immune response is also unknown. It is conceivable that LSR and TLR2 mediate separate pathways in response to CDT. Alternatively, these receptors may cooperate to permit CDT recognition and intoxication.
Another major question concerns the mechanism of eosinophil killing by CDT. Although our data suggest that CDT does not directly kill eosinophils, intoxication by CDT and the subsequent increase in inflammatory cytokine production by innate cells may shape the inflammatory environment to support eosinophil apoptosis. Indeed, innate inflammation has long been known to suppress eosinophilia by unknown mechanisms 42,43. TLR2-dependent eosinophil suppression is also not unprecedented, as multiple groups have reported in different murine models of allergic inflammation 44–46. Administration of a TLR2 agonist has been shown to reduce eosinophilia by inducing T cell apoptosis in a model of allergic conjunctivitis, as well as by enhancing T regulatory cells and inducing Th1 cytokines in murine asthma models. It remains to be investigated whether these mechanisms underlie eosinophil suppression during CDI. The protective role of eosinophils is likewise unknown, and clarifying how TLR2 signaling impacts the function of these cells may provide clues to their activity during infection.
Understanding the mechanism by which CDT enhances C. difficile virulence is essential to understanding the virulence of PCR-ribotype 027 strains, and of other hypervirulent strains which express CDT. These isolates are increasingly common and their spread likely contributes to the overall increase in CDI incidence and severity. Our data clarify the protective role eosinophils play during CDI and suggest that targeting CDT in the development of vaccines and therapeutic inhibitors is essential to successfully treating infection with these strains. Our knowledge of C. difficile virulence factors which result in pathogenic immune responses during CDI continues to evolve, presenting new potential drug targets for treating this common and life threatening infection.
Methods
Bacterial Strains and Culture
C. difficile strains R20291 CdtA- and R20291 CdtB- was generated using the ClosTron system and functional inactivation of the targeted genes confirmed by Western blot as previously described 28. Strains M7404 CdtA- and M7404 CdtComp were generated using the targetron system as previously described 29. To prepare the infection inoculum, strains were inoculated onto BHI agar supplemented with the appropriate selective antibiotic from frozen stocks and incubated at 37°C overnight in an anaerobic work station (Shel Labs). Single colonies were inoculated into BHI medium supplemented with cycloserine and cefoxitin (C. difficile supplement, Sigma) and grown anaerobically overnight at 37°C. The next day, cultures were spun for 1 minute at 6,000 × g and washed twice in anaerobic PBS. The optical density of the cultures were measured and culture density adjusted to 1×108 CFU/mL (R20291 strains) or 2×106 CFU/mL (M7404 strains) in sterile, anaerobic PBS. Syringes were loaded with the inoculum and sealed in airtight bags before and during transport to the infection facility. Mice received 100 μl of inoculum each by oral gavage. To enumerate C. difficile in cecal samples, cecal contents were resuspended by weight in pre-reduced PBS. Resuspended cecal contents were serially diluted in PBS and plated on BHI agar supplemented with 1% Sodium Taurocholate and 1 mg/mL D-cycloserine and 0.032 mg/mL cefoxitin (Sigma) before incubating overnight anaerobically at 37°C. Liver bacterial burden was determined by homogenizing whole liver samples in sterile, aerobic PBS. Liver homogenate was serially diluted and plated on non-selective BHI agar before incubating aerobically overnight at 37°C. Liver CFU are reported according to liver sample weight.
CDT Complementation
The recombinant plasmid used for complementation of the cdtA mutant was pJIR3107, described previously 47. C. difficile strains were grown in TY broth (3.0% tryptone, 2.0% yeast extract and 0.1% sodium thioglycollate) at 37°C in a Don Whitley A35 Anaerobic Workstation in an atmosphere of 10% (v/v) H2, 10% (v/v) CO2 and 80% (v/v) N2. Toxins were partially purified and concentrated eight-fold from 72 hour C. difficile culture supernatants by methanol-chloroform precipitation 48. Protein concentrations were determined using the BCA protein assay kit (Pierce) as per the manufacturer’s instructions. Concentrated supernatant proteins (10 μg) were separated by 10% (w/v) SDS-PAGE and transferred onto a nitrocellulose membrane (Whatman). Membranes were detected as previously described 29. CDTa and CDTb were detected using a CDTa-specific antibody and C. perfringens Ib-specific antibody that is cross reactive with CDTb 49, respectively. CDTa and CDTb antibodies were detected using horseradish peroxidase conjugated anti-rabbit goat antibodies (Millipore) The Western Lightning Chemiluminescence reagent kit (Perkin-Elmer) was used to detect the blots, following the manufacturer’s instructions and blots were recorded by exposure to X-ray film (Fujifilm).
Cell culture
Murine BMDC were generated as previously described with minor modifications 50. Briefly, femurs and tibia were removed and bone marrow flushed with PBS. Cells were counted and viability assessed by Trypan Blue staining and resuspended in RPMI 1640 media (Life Technologies) containing 10% Fetal Bovine Serum, 2 mM L-glutamine and 100 U/ml Penicillin and 100 U/ml Streptomycin. Media was supplemented with 10 ng/ml GM-CSF (Peprotech) and 55 μM β-mercaptoethanol (Gibco), and 3×106 cells were seeded into a T75 vent cap tissue culture flask. Cells were cultured for 7 days, and supplemented with fresh media on days 2 and 4. On day 7 cells were harvested for stimulation. For stimulation, BMDCs were detached with a cell scraper and resuspended to 1.1×106 cells/mL in fresh media. 180 ul of cell suspension was added per well of a 96 well plate. Cells were stimulated with the indicated concentrations of CDT or Toxins A and B in a total volume of 20 ul complete media. After the indicated incubation time, cells were spun down and supernatant removed and frozen at −80 °C for later analysis. For cell stimulation, TLR-ligand tested lipopolysaccharide (Sigma) was used as a positive control. Anti-TLR2 antibody and IgG isotype control were obtained from Invivogen and eBioscience, respectively. Anti-CDT nanobodies were generated as previously described 31.
Raw Blue NFκB reporter cell were obtained from Invivogen and grown in DMEM supplemented with 4.5 g/L glucose and 10% FBS. Cell responsiveness to TLR stimulation and mycoplasma status was assessed by Invivogen. For stimulation, cells were detached using a cell scraper and resuspended to a density of 5.5 × 105 cells/mL in fresh media. 180 ul of cell suspension was added per well of a 96 well plate, and cells were stimulated with CDT as indicated. Secreted Embryonic Alkaline Phosphatase (SEAP) secretion was quantified by spinning down cells, removing the culture supernatant, and incubating 50 ul of supernatant with 150 ul of Quanti-Blue detection media (Invivogen) for 30 minutes at 37°C prior to reading in a spectrophotometer. For treatment of BM Eos with CDT, cells were harvested from culture on day 10 and plated in 96 well plates in fresh media before addition of CDT and neutralizing antibodies. Cells were harvested at 8 hours and stained for flow cytometry to assess eosinophil viability.
Toxins
Purified CdtA and CdtB were expressed in Bacillus megaterium as previously reported 22. CdtB was previously activated by protease cleavage (0.2 μg of trypsin/μg of protein for 30 minutes at 37°C) prior to addition of trypsin inhibitor. Mock buffer used contained identical concentrations of trypsin and trypsin inhibitor to mimic the purified proteins. Purified Toxins A and B were purified from C. difficile strain VPI 10463 and were obtained as a kind gift from Techlab, Inc. (Blacksburg, VA). Toxins were detected within cecal contents using the C. difficile TOXA/B ELISA according to manufacturer instructions, also kindly donated by Techlab, Inc.
Cytokine detection
IL-1β and IL-6 were detected in protein supernatants from BMDCs, serum and tissue lysates using the Mouse IL-1β and IL-6 Ready-Set-Go! ELISA kit (eBioscience) according to manufacturer’s instructions. Eotaxin-1 and Eotaxin-2 were detected in cecal lysates using R&D Systems DuoSet ELISA kits according to manufacturer’s instructions. BMDC pro-IL-1β production was assessed by quantitative reverse transcription PCR. RNA was isolated using the RNeasy isolation kit (Qiagen). Contaminating genomic DNA was digested using the Turbo DNA-free kit (Ambion) and RNA reverse transcribed with the Tetro cDNA synthesis kit (Bioline) according to manufacturer instructions. The resulting cDNA was purified using Qiagen’s PCR purification kit. IL-1b gene expression was quantified by Quantitect Primer assay (Qiagen) using Sensifast SYBR & Fluorescein Mix (Bioline) using the Quantitect 2-step amplification protocol. Gene expression was normalized to the S14 housekeeping gene (forward primer - TGGTGTCTGCCACATCTTTGCATC, reverse primer – AGTCACTCGGCAGATGGTTTCCTT).
Mice and infection
Experiments were carried out using sex matched 8–12 week old C57BL6, TLR2−/−, TLR4−/−, and TLR5−/− mice from The Jackson Laboratory. Experiments contained no fewer than 4 mice, with a goal of 8 mice, per experimental group. Based on an expected protection from 50% to 0% mortality, 6 mice per group will have > 90% power to detect a statistically significant difference in mortality. All animals were housed under specific pathogen free conditions at the University of Virginia’s animal facility and procedures were approved by the Institutional Animal Care and Use Committee at the University of Virginia. No animals were excluded from the study, and sample harvesting was randomized between groups. Blinded scoring of animals was not possible due to separate equipment used to prevent contamination between C. difficile strains. Prior to infection, bedding was exchanged every two days between TLR2−/− and C57BL6 mice for a minimum of two weeks to equilibrate microbiota between strains. Mice were infected using a modified version of a previously published model for CDI 30. Briefly, mice were given an antibiotic cocktail consisting of 45 mg/L Vancomycin (Mylan), 35 mg/L Colistin (Sigma), 35 mg/L Gentamicin (Sigma), 215 mg/L Metronidazole (Hospira) ad libitum for 3 days (Days −6 to −4). Mice were then switched back to regular drinking water for two days (Days −3 onward), followed by an IP injection (0.032 mg/g) of Clindamycin (Hospira) on Day −1. On Day 0, mice were gavaged with C. difficile. Mice were monitored twice daily throughout the course of the infection and immediately euthanized if severe illness developed according to clinical scoring parameters. Scores were based on weight loss, coat condition, activity level, diarrhea, posture and eye condition for a cumulative clinical score between 1 and 20.
Tissue Protein and Histology
Mice were humanely euthanized and tissue was immediately removed for analysis. Total cecal lysate was generated by removing the ceca and rinsing gently with PBS. Tissue was bead beaten for 1 minute in 400 ul of Lysis Buffer I (1× HALT Protease Inhibitor (Pierce), 5 mM HEPES). 400 ul of Lysis Buffer II was added (1× HALT Protease Inhibitor (Pierce), 5 mM HEPES, 2% Triton X-100) and tubes inverted gently. Tissue samples were incubated on ice for 30 minutes, followed by a 5 minute spin at 13,000 × g at 4°C. Supernatant was removed to a fresh tube, and total protein concentration was assessed by BCA assay according to manufacturer’s instructions (Pierce). Cytokine concentration is shown relative to total protein concentration. To generate histological sections, cecal samples were placed in Bouin’s Solution (Sigma) for 18 hours. Tissue samples were moved to 70% Ethanol before paraffin embedding and sectioning. Sections were mounted on slides and stained with haemotoxylin and eosin prior to microscopic examination. Slides were scored blinded, with a score from 0 to 3 assigned based on 5 parameters: epithelial disruption, submucosal edema, inflammatory infiltrate, mucosal thickening and luminal exudate as previously reported 51. Scores were averaged between 3 independent, blinded observers. Image manipulation was limited to movement of scale bars to bottom left corner.
Eosinophil Depletion
Eosinophils were depleted using an anti-mouse SiglecF monoclonal antibody (R&D Systems) as previously shown 33. Control groups received Rag IgG2A isotype control antibody (R&D Systems). Both groups were given 40 ug of antibody per mouse on day −1 and day +1 of infection (80 ug total per mouse) via intraperitoneal injection. Eosinophil depletion was evaluated by flow cytometry, which showed an 82% average reduction in colonic eosinophils.
Eosinophil adoptive transfer
Bone marrow derived eosinophils were generated as previously described and examined on day 10 for eosinophil markers and viability via flow cytometry 52,53. Cultures routinely consisted of >75% live CD45+ CD11b+ SiglecF+ SSChi eosinophils. For adoptive transfer, cultures were synchronized such that each injection occurred on day 10 of culture. Cultures were collecting using a cell scraper and washed 2× in sterile PBS. 4×105 eosinophils were given intraperitoneally per mouse. Mice received 4 total injections of eosinophils, starting the day prior to infection and for 3 subsequent days.
Statistical Analysis
Statistical analysis was calculated and significance determined (p< 0.05) using Mann-Whitney test or other suitable analysis, including Welch’s t-test for unequal variance. All statistical analysis was performed using GraphPad Prism software.
Flow Cytometry
Colonic lamina propria was prepared for flow cytometry by thoroughly rinsing the tissue in Hank’s Balanced Salt Solution (HBSS) supplemented with 5% FBS and 25 mM HEPES. Epithelial cells were removed by gentle shaking for 40 minutes at 37°C in HBSS with 15 mM HEPES, 5 mM EDTA, 10% FBS and 1 mM DTT. Halfway through the incubation, colon samples were transferred to fresh buffer. Next, colon samples were thoroughly chopped using scissors and digested in RPMI 1640 containing 0.17 mg/mL Liberase TL (Roche) and 30 ug/mL DNase (Sigma). Samples were digested for 40 minutes at 37°C with gentle shaking. Samples were then spun down at 300 × g and resuspended in HBSS with 5% FBS and 25 mM HEPES before passage through a 100 uM cell strainer followed by a 40 uM cell strainer (both Fisher Scientific). Cells were counted and density adjusted to 1×107 cells/mL. 100 ul of cell suspension were aliquoted per well of a 96 well plate for antibody staining. For staining, cells were initially blocked with TruStain fcX (anti-mouse CD16/32 antibody, BioLegend) for ten minutes at room temperature. Cells were spun down and resuspended in LIVE/DEAD Fixable Aqua (Life Technologies) for 20 minutes at room temperature. Cells were washed twice and stained with fluorochrome conjugated antibodies. Flow cytometry was performed on an LSR Fortessa cytometer (BD Biosciences) and all data analysis performed via FlowJo (Tree Star Inc.). See Supplementary Table S1 for antibodies used.
Supplementary Material
Acknowledgments
We thank the UVA Research Histology and Flow Cytometry Cores for their assistance with sample preparation and analysis. We also thank TechLab, Inc. for providing purified Toxins A and B and TOX A/B ELISA kits. We thank A. Criss, J. Casanova, U. Lorenz and M. Kendall for helpful discussion. C.A.C. was supported by the Robert R. Wagner Fellowship from the University of Virginia School of Medicine and by NIH training grant 5T32AI07046-38. E.L.B. was supported by T32AI07496 and F31AI114203. M.M.S. was supported by 2T32GM008715-16. D.L. was supported by Future Fellowship FT120100779 (Australian Research Council). This work was supported by NIH grants R01AI124214, R01AI026649 and R21AI114734 to W.A.P.
Footnotes
Individual contributions: C.A.C. conceived and designed the experiments, performed the experiments, analyzed the data, and wrote the paper. E.L.B. performed the experiments, provided valuable advice and contributed materials. M.M.S., M.G.W., and S.L.B performed the experiments. S.A.K., C.S., A.M.E, F.K, D.L., K.A. and N.P.M. contributed materials and valuable advice on experimental design. W.A.P. assisted in experimental design and edited the paper.
References
- 1.Lessa FC, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ananthakrishnan AN. Clostridium difficile infection: epidemiology, risk factors and management. Nat Rev Gastroenterol Hepatol. 2010;8:17–26. doi: 10.1038/nrgastro.2010.190. [DOI] [PubMed] [Google Scholar]
- 3.Kuehne SA, et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 4.Carter GP, Rood JI, Lyras D. The role of toxin A and toxin B in Clostridium difficile-associated disease. Gut Microbes. 2010;1:58–64. doi: 10.4161/gmic.1.1.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47:349–352. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JY, et al. Clostridium difficile toxin A promotes dendritic cell maturation and chemokine CXCL2 expression through p38, IKK, and the NF-kappaB signaling pathway. J Mol Med Berl Ger. 2009;87:169–180. doi: 10.1007/s00109-008-0415-2. [DOI] [PubMed] [Google Scholar]
- 7.Bobo LD, et al. MK2 Kinase Contributes to Clostridium difficile-Associated Inflammation. Infect Immun. 2012 doi: 10.1128/IAI.00186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart DB, Berg A, Hegarty J. Predicting Recurrence of C. difficile Colitis Using Bacterial Virulence Factors: Binary Toxin Is the Key. J Gastrointest Surg. 2013;17:118–125. doi: 10.1007/s11605-012-2056-6. [DOI] [PubMed] [Google Scholar]
- 9.Bacci S, Mølbak K, Kjeldsen MK, Olsen KEP. Binary Toxin and Death after Clostridium difficile Infection. Emerg Infect Dis. 2011;17:976–982. doi: 10.3201/eid1706.101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbut F, et al. Clinical features of Clostridium difficile-associated infections and molecular characterization of strains: results of a retrospective study, 2000–2004. Infect Control Hosp Epidemiol. 2007;28:131–139. doi: 10.1086/511794. [DOI] [PubMed] [Google Scholar]
- 11.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 12.Bauer MP, et al. Clostridium difficile infection in Europe: a hospital-based survey. The Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 13.Spigaglia P, Mastrantonio P. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J Med Microbiol. 2004;53:1129–1136. doi: 10.1099/jmm.0.45682-0. [DOI] [PubMed] [Google Scholar]
- 14.Popoff MR, Rubin EJ, Gill DM, Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun. 1988;56:2299–2306. doi: 10.1128/iai.56.9.2299-2306.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stiles BG, et al. Clostridium and Bacillus Binary Enterotoxins: Bad for the Bowels, and Eukaryotic Being. Toxins. 2014;6:2626–2656. doi: 10.3390/toxins6092626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemmasi S, et al. Interaction of the Clostridium difficile Binary Toxin CDT and its Host Cell Receptor LSR. J Biol Chem. 2015 doi: 10.1074/jbc.M115.650523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papatheodorou P, et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT) Proc Natl Acad Sci U S A. 2011;108:16422–16427. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT. Gut Microbes. 2014;5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higashi T, et al. Analysis of the ‘angulin’ proteins LSR, ILDR1 and ILDR2 – tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J Cell Sci. 2013;126:966–977. doi: 10.1242/jcs.116442. [DOI] [PubMed] [Google Scholar]
- 20.Barth H, Aktories K, Popoff MR, Stiles BG. Binary Bacterial Toxins: Biochemistry, Biology, and Applications of Common Clostridium and Bacillus Proteins. Microbiol Mol Biol Rev. 2004;68:373–402. doi: 10.1128/MMBR.68.3.373-402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwan C, et al. Clostridium difficile toxin CDT hijacks microtubule organization and reroutes vesicle traffic to increase pathogen adherence. Proc Natl Acad Sci U S A. 2014;111:2313–2318. doi: 10.1073/pnas.1311589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwan C, et al. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 2009;5:e1000626. doi: 10.1371/journal.ppat.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Feghaly RE, Stauber JL, Tarr PI, Haslam DB. Intestinal Inflammatory Biomarkers and Outcome in Pediatric Clostridium difficile Infections. J Pediatr. 2013 doi: 10.1016/j.jpeds.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abt MC, et al. Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium difficile Infection. Cell Host Microbe. 2015;18:27–37. doi: 10.1016/j.chom.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa M, et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity. 2014;41:620–632. doi: 10.1016/j.immuni.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buonomo EL, et al. Role of IL-23 signaling in Clostridium difficile Colitis. J Infect Dis. 2013:jit277. doi: 10.1093/infdis/jit277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowardin CA, et al. Inflammasome activation contributes to interleukin-23 production in response to Clostridium difficile. mBio. 2015;6 doi: 10.1128/mBio.02386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehne SA, et al. The importance of toxin A, toxin B and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2013 doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter GP, et al. Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. mBio. 2015;6:e00551–15. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Unger M, et al. Selection of Nanobodies that Block the Enzymatic and Cytotoxic Activities of the Binary Clostridium Difficile Toxin CDT. Sci Rep. 2015;5 doi: 10.1038/srep07850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buonomo EL, et al. Microbiota-regulated IL-25 increases eosinophil number to provide protection during Clostridium difficile infection. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griseri T, et al. Granulocyte Macrophage Colony-Stimulating Factor-Activated Eosinophils Promote Interleukin-23 Driven Chronic Colitis. Immunity. 2015;43:187–199. doi: 10.1016/j.immuni.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13:9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung Y, Rothenberg ME. Roles and Regulation of Gastrointestinal Eosinophils in Immunity and Disease. J Immunol. 2014;193:999–1005. doi: 10.4049/jimmunol.1400413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rådinger M, Lötvall J. Eosinophil progenitors in allergy and asthma - do they matter? Pharmacol Ther. 2009;121:174–184. doi: 10.1016/j.pharmthera.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Sehmi R, et al. Allergen-induced fluctuation in CC chemokine receptor 3 expression on bone marrow CD34+ cells from asthmatic subjects: significance for mobilization of haemopoietic progenitor cells in allergic inflammation. Immunology. 2003;109:536–546. doi: 10.1046/j.1365-2567.2003.01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palframan RT, et al. Mechanisms of Acute Eosinophil Mobilization from the Bone Marrow Stimulated by Interleukin 5: The Role of Specific Adhesion Molecules and Phosphatidylinositol 3-Kinase. J Exp Med. 1998;188:1621–1632. doi: 10.1084/jem.188.9.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denburg JA, Keith PK. Eosinophil progenitors in airway diseases: clinical implications. Chest. 2008;134:1037–1043. doi: 10.1378/chest.08-0485. [DOI] [PubMed] [Google Scholar]
- 40.Gauvreau GM, Ellis AK, Denburg JA. Haemopoietic processes in allergic disease: eosinophil/basophil development. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2009;39:1297–1306. doi: 10.1111/j.1365-2222.2009.03325.x. [DOI] [PubMed] [Google Scholar]
- 41.Smith SG, et al. Thymic stromal lymphopoietin and IL-33 modulate migration of hematopoietic progenitor cells in patients with allergic asthma. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2014.12.1918. [DOI] [PubMed] [Google Scholar]
- 42.Bass DA. Behavior of eosinophil leukocytes in acute inflammation. II Eosinophil dynamics during acute inflammation. J Clin Invest. 1975;56:870–879. doi: 10.1172/JCI108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan JE, Beeson PB. Experimental Observations on the Eosinopenia Induced by Acute Infection. Br J Exp Pathol. 1971;52:214–220. [PMC free article] [PubMed] [Google Scholar]
- 44.Fukushima A, Yamaguchi T, Ishida W, Fukata K, Ueno H. TLR2 agonist ameliorates murine experimental allergic conjunctivitis by inducing CD4 positive T-cell apoptosis rather than by affecting the Th1/Th2 balance. Biochem Biophys Res Commun. 2006;339:1048–1055. doi: 10.1016/j.bbrc.2005.11.114. [DOI] [PubMed] [Google Scholar]
- 45.Nawijn MC, et al. TLR-2 Activation Induces Regulatory T Cells and Long-Term Suppression of Asthma Manifestations in Mice. PLoS ONE. 2013;8:e55307. doi: 10.1371/journal.pone.0055307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel M, et al. TLR2 Agonist Ameliorates Established Allergic Airway Inflammation by Promoting Th1 Response and Not via Regulatory T Cells. J Immunol. 2005;174:7558–7563. doi: 10.4049/jimmunol.174.12.7558. [DOI] [PubMed] [Google Scholar]
- 47.Carter GP, et al. Binary Toxin Production in Clostridium difficile Is Regulated by CdtR, a LytTR Family Response Regulator. J Bacteriol. 2007;189:7290–7301. doi: 10.1128/JB.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PloS One. 2013;8:e79666. doi: 10.1371/journal.pone.0079666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff MR. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun. 1997;65:1402–1407. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross O. Measuring the inflammasome. Methods Mol Biol Clifton NJ. 2012;844:199–222. doi: 10.1007/978-1-61779-527-5_15. [DOI] [PubMed] [Google Scholar]
- 51.Pawlowski SW, et al. Murine Model of Clostridium difficile Infection with Aged Gnotobiotic C57BL/6 Mice and a BI/NAP1 Strain. J Infect Dis. 2010;202:1708–1712. doi: 10.1086/657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dyer KD, et al. Functionally competent eosinophils differentiated ex vivo in high purity from normal mouse bone marrow. J Immunol Baltim Md 1950. 2008;181:4004–4009. doi: 10.4049/jimmunol.181.6.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen T, Besse JA, Mingler MK, Fulkerson PC, Rothenberg ME. Eosinophil adoptive transfer system to directly evaluate pulmonary eosinophil trafficking in vivo. Proc Natl Acad Sci U S A. 2013;110:6067–6072. doi: 10.1073/pnas.1220572110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.