Abstract
Microaspiration is a common phenomenon in healthy subjects, but its frequency is increased in chronic inflammatory airway diseases, and its role in inflammatory and immune phenotypes is unclear. We have previously demonstrated that acellular bronchoalveolar lavage samples from half of the healthy people examined are enriched with oral taxa (here called pneumotypeSPT) and this finding is associated with increased numbers of lymphocytes and neutrophils in bronchoalveolar lavage. Here, we have characterized the inflammatory phenotype using a multi-omic approach. By evaluating both upper airway and acellular bronchoalveolar lavage samples from 49 subjects from three cohorts without known pulmonary disease, we observed that pneumotypeSPT was associated with a distinct metabolic profile, enhanced expression of inflammatory cytokines, a pro-inflammatory phenotype characterized by elevated Th-17 lymphocytes and, conversely, a blunted alveolar macrophage TLR4 response. The cellular immune responses observed in the lower airways of humans with pneumotypeSPT indicate a role for the aspiration-derived microbiota in regulating the basal inflammatory status at the pulmonary mucosal surface.
Culture-independent techniques have challenged the preconception that the lower airways are normally sterile; the lungs of healthy individuals frequently harbour DNA of oral anaerobes such as members of the genera Prevotella and Veillonella, although at lower levels compared with the upper respiratory tract (URT)1–4. Because the oropharynx and the tracheobronchial tree are contiguous, continuing microaspiration probably seeds the lungs with oral bacteria5–7. Bacterial DNA of common oral taxa detected in the lower airways of a proportion of healthy people could represent the vestiges of aspirated oral bacteria, either nonviable or in the process of dynamic clearance, or could reflect a viable community of microorganisms, including aspirated oral bacteria, living in dynamic equilibrium with host defences.
Determination of the pulmonary microbial ecology and host-microbiome interaction has been limited to a description of the taxa present in the lower airways in health and disease. In an earlier report3, we observed two distinct lung microbiomes in acellular bronchoalveolar lavage (BAL) samples of healthy subjects that we termed ‘pneumotypes’: pneumotypeSPT, characterized by high bacterial load and supraglottic predominant taxa (SPT) such as the anaerobes Prevotella and Veillonella; and pneumotypeBPT, with low bacterial burden and background predominant taxa (BPT) found in the saline lavage and bronchoscope. We proposed that pneumotypeSPT reflects the presence of authentic URT-derived microbes, whereas pneumotypeBPT predominantly reflects an absence of lung-derived bacterial DNA, enabling a strong representation of environmental sequences present in instruments and reagents. Here, we have determined the prevalence of these two contrasting lung microbiome types, in a multi-centre study. Using a multi-omic approach, we tested the hypothesis that pneumotypeSPT is associated with a characteristic microbial metabolism profile and an inflammatory lower airway immune phenotype.
Results
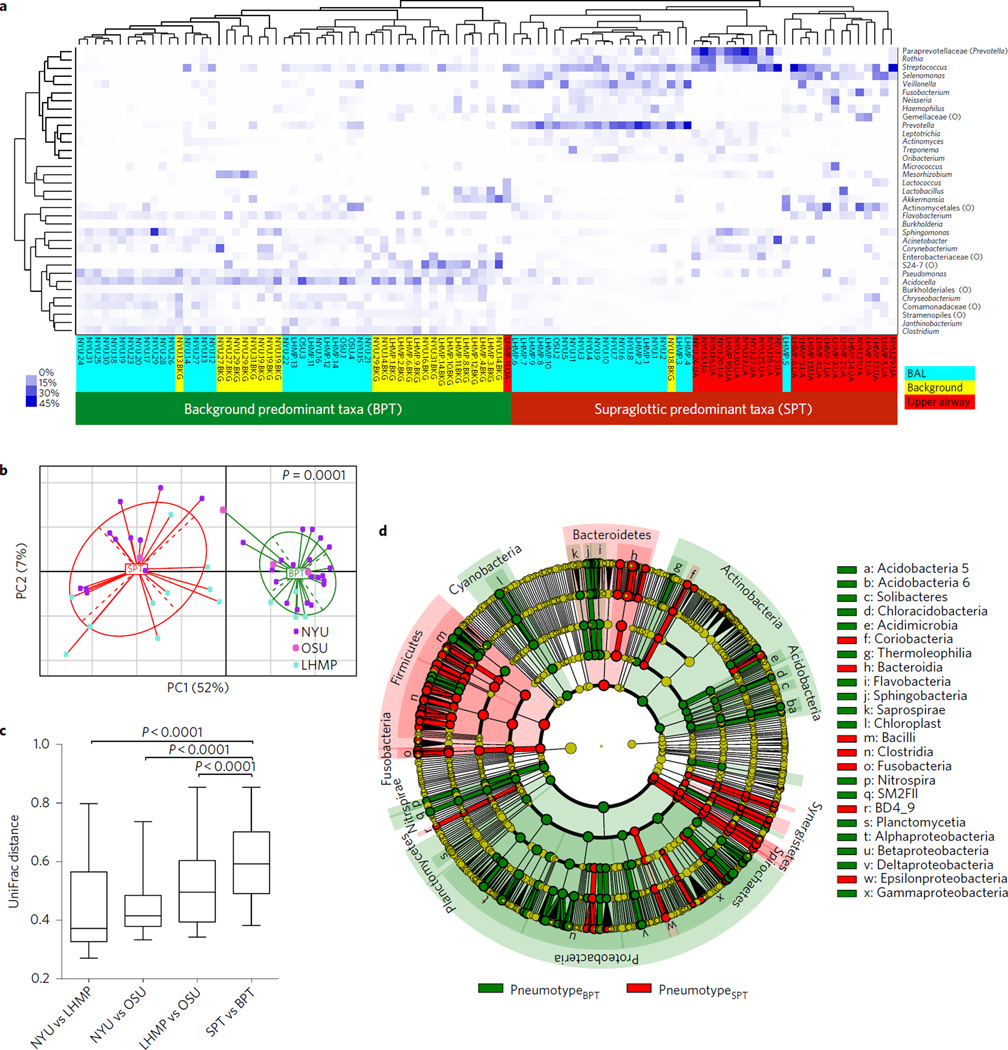
To test the generalizability of our earlier observation that two alternative lower airway pneumotypes exist among the studied subjects, we used acellular BAL samples obtained from 49 subjects recruited from three separate cohorts (Table 1). Analysis of the microbiome for 28 out of 49 acellular BAL samples using a 454 platform targeting the V1–V2 variable region has been reported previously (see Supplementary Information)3. For this report, all acellular BAL samples for 49 samples were processed at the same time at New York University (NYU) for DNA isolation, amplicon library preparation targeting the V4 variable region and sequencing using a MiSeq platform. When acellular BAL samples were compared with upper airways and background samples, unsupervised hierarchical clustering based on relative abundance of most prevalent taxa (>3% in any given sample) clustered the BAL samples into two groups (Fig. 1a). In one cluster, 27 out of 49 BAL samples clustered tightly with most of the background samples and one upper airway sample. We therefore denominated this group background predominant taxa. The other cluster of BAL samples constituted the remaining 22 out of 49 BAL samples and one background sample and was characterized by enrichment with taxa commonly seen in the upper airways. Two major distinct lung microbiomes (pneumotypes)3 were thus defined. PneumotypeSPT is characterized by enrichment with SPT including Prevotella and Veillonella, and pneumotypeBPT with BPT including Acidocella and Pseudomonas, consistent with the taxa identified as the most discriminant and abundant for upper airways and background, respectively (Supplementary Fig. 1). This dichotomy was present in acellular BAL samples obtained from subjects drawn from all three cohorts (although a small number of samples were obtained from Ohio State University). No demographic, clinical, smoking status or pulmonary function data distinguished subjects with pneumotypeSPT from those with pneumotypeBPT (Table 1). The results of the analysis of β-diversity, based on weighted UniFrac distances, showed a distinct clustering of samples identified as pneumotypeSPT and pneumotypeBPT (Fig. 1b). Importantly, BAL samples from each cohort were present in both pneumotypes, indicating that there was no cohort bias. Comparison of mean UniFrac pairwise distances showed greater divergence between samples of the two pneumotypes than between cohorts (Fig. 1c).
Table 1.
Demographics and pulmonary function.
| Cohort | PneumotypeBPT | PneumotypeSPT | P-value | |
|---|---|---|---|---|
| n | 49 | 27 | 22 | |
| Cohort (%) | NS | |||
| NYU | 63 | 64.5 | 35.5 | |
| OSU | 8 | 75 | 25 | |
| LHMP | 29 | 26.8 | 71.4 | |
| Age (years) | 53.0 (39.8–63.4) | 52.3 (36.8–62.7) | 55.0 (43.4–65.0) | NS |
| Male (%) | 64 | 63 | 77 | NS |
| Caucasian (%) | 77 | 81 | 91 | NS |
| BMI | 27 (23–31) | 26 (23–31) | 27 (24–31) | NS |
| Smoking status (%) | ||||
| Current | 26 | 33 | 18 | NS |
| Former | 37 | 33 | 41 | NS |
| Never | 37 | 33 | 41 | NS |
| Pack-years (smokers) | 25 (5–43) | 25 (5–43) | 39 (19–49) | NS |
| PFT* | ||||
| Spirometry | ||||
| FVC† | 98.0 (89.5–115.0) | 100.0 (91.7–116.0) | 98.9 (86.8–108.5) | NS |
| FEV1† | 90.0 (81.0–108.0) | 92.9 (85.5–100.0) | 90.0 (78.9–104.7) | NS |
| FEV1 /VC | 75.0 (67.0–79.7) | 75.5 (69.0–80.0) | 74.0 (68.9–79.6) | NS |
| Lung volumes | NS | |||
| TLC† | 100 (85–107) | 99 (85–107) | 100 (80–107) | NS |
| FRC† | 87 (75–107) | 86 (75–108) | 90 (75–106) | NS |
| RV/TLC | 0.32 (0.26–0.37) | 0.32 (0.23–0.36) | 0.34 (0.30–0.40) | NS |
| DLCO† | 84 (75–94) | 90 (72–96) | 82 (77–92) | NS |
Data presented as percentages or median (IQR). P-value for comparison between pneumotypeBPT vs pneumotypeSPT. Abbreviations: NYU, New York University; OSU, Ohio State University; LHMP, Lung HIV Microbiome Project; BMI, body mass index; FVC, forced vital capacity; FEV1, forced expiratory volume in 1 s; TLC, total lung capacity; FRC, functional residual capacity; RV, residual volume; DLCO, diffusing capacity of the lungs for carbon monoxide; NS, not significant.
PFT, pulmonary function testing according to standard ATS/ERS guidelines20
% predicted.
Figure 1. Major differences in microbial composition of the lower airways are driven by enrichment with either supraglottic taxa or background taxa.
a, Unsupervised hierarchical clustering of most abundant taxa (relative abundance ≥3% in any sample) for BAL samples, upper airway and background samples. Upper airway samples were obtained by oral wash or by separate bronchoscopy. BAL samples were obtained after passing the upper airways without suctioning and wedging in a subsegment of the lower airways. The dendrogram indicates two well-separated clusters, one dominated by background samples and 27 of the 49 BAL samples, and a second dominated by upper airway samples plus 22 of the 49 BAL samples. The heat map shows that the first cluster is enriched with Acidocella, Pseudomonas and Sphingomonas and the second cluster is enriched with taxa most commonly found in the upper airways such as Prevotella, Rothia and Veillonella. b, Principal coordinates analysis (PCoA) based on weighted UniFrac distances demonstrate that pneumotypeSPT BAL samples clustered separately from pneumotypeBPT BAL samples. Samples from all three cohorts can be found in both pneumotype clusters. c, Comparison of UniFrac distance between paired acellular BAL samples from New York University (NYU) (n = 31), the Lung HIV Microbiome Project (LHMP) (n = 14) and Ohio State University (OSU) (n = 4) show that there is greater UniFrac distance between pneumotypeSPT and pneumotypeBPT than between different institutions (represented as median (IQR), statistical significance of sample groupings based on Adonis). d, Cladogram representing results from calculated LDA LEfSe comparing taxonomic composition of BAL samples from pneumotypeSPT and pneumotypeBPT. Multiple significant taxonomic differences were observed at different phylogenetic levels, with labels in the cladogram written for differences at the phylum level and indicated by letters for differences at the class level.
Using linear discriminant analysis (LDA) effect size (LEfSe) analysis to compare the two pneumotypes, multiple taxonomic differences were found: pneumotypeSPT was enriched with operational taxonomic units (OTUs) from the phyla Firmicutes, Fusobacterium and Bacteroidetes, whereas pneumotypeBPT was enriched with several OTUs belonging to the phyla Proteobacteria, Actinobacteria, Acidobacteria and others (Fig. 1d).
Bacteria and bacteriophages exist, in general, in very specific relationships, with most bacteriophages exhibiting fairly narrow host ranges8. To further characterize the differences between the two pneumotypes, we analysed the DNA phageome. Consistent with the 16S rRNA gene data, pneumotypeSPT was enriched with sequences most closely related to phages infecting Firmicutes and Bacteroidetes (for example, five different Streptococcus phages, Lactococcus phage KSY1 and Cellulophaga phage φ17 2, Supplementary Fig. 5). The overlap between some of the phages found enriched in BAL samples with pneumotypeSPT (for example, the Streptococcus phages and Streptococcus genus) is consistent with infection of specific taxa with the appropriate phage.
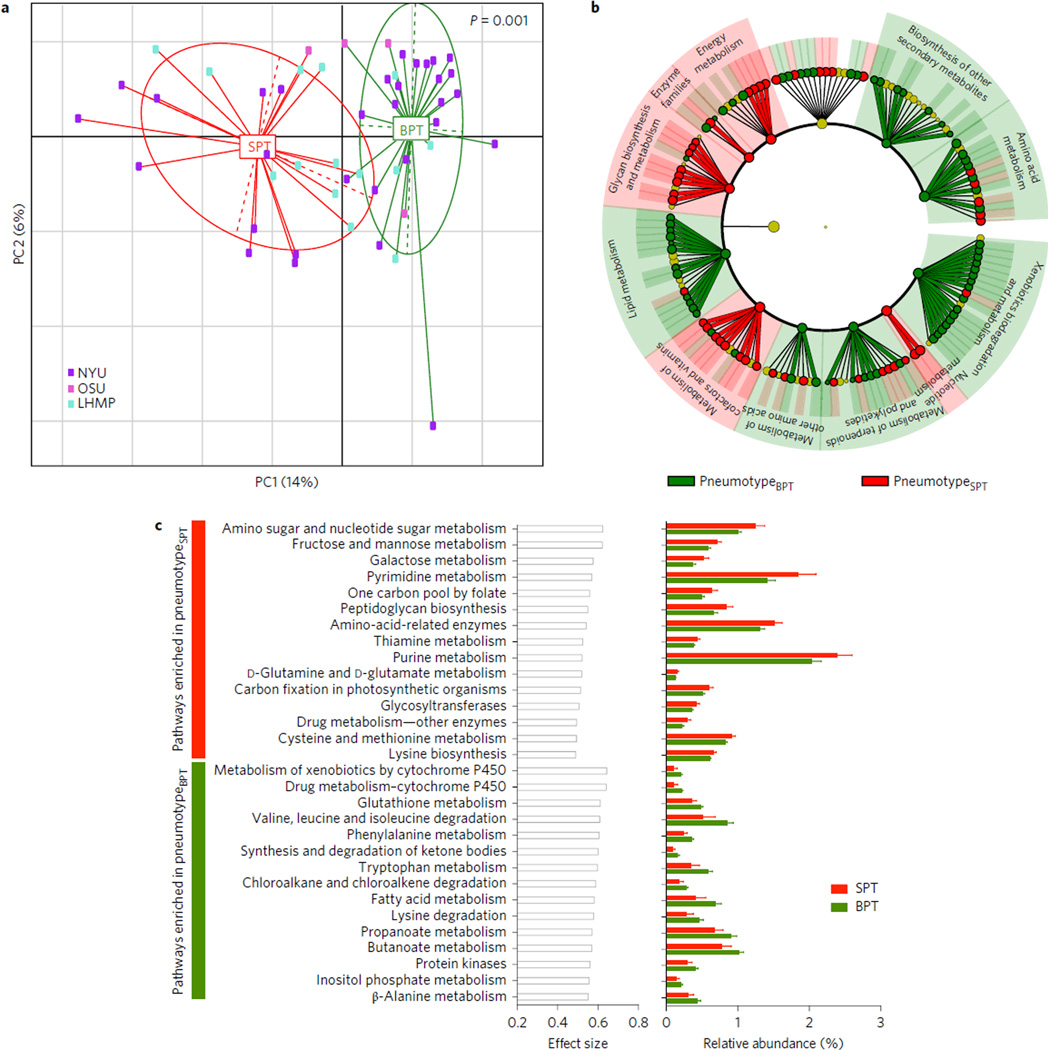
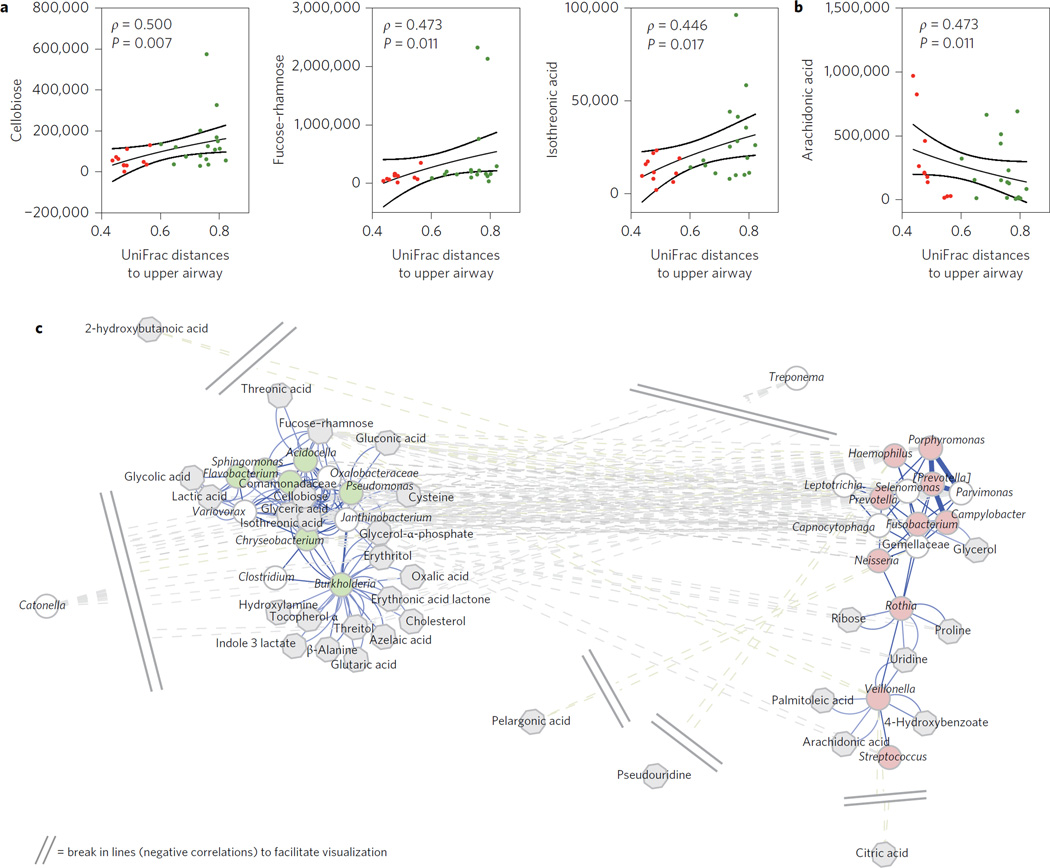
A limitation of taxonomic assessment of microbial communities is the lack of functional information. We therefore used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) to infer the relative abundance of protein-coding genes based on the 16S rRNA taxonomic assignment described above, allowing us to examine each pneumotype9. Multiple significant differences in coding potential between pneumotypeBPT and pneumotypeSPT were noted (Fig. 2 and Supplementary Table 1). To examine whether the differences in genomic composition of the metabolic pathways observed in the two pneumotypes were consistent with different metabolic environments in the lower airways, metabolites in BAL fluid were assayed for 29 subjects from the NYU cohort by gas chromatography– time of flight (GC-TOF) mass spectrometry, and then correlated with the 16S data. We proposed the hypothesis that metabolome/ microbiome correlations would be present, indicating active microbial metabolism. Among those metabolites, we were interested in those of bacterial origin (such as rhamnose, a substrate for lipo-polysaccharides) or those related to fatty acids found at high levels in the lower airway environment that may have important immunological functions. Because our pneumotype classification is mainly driven by the extent of similarity of the lower and upper airway microbiomes, we quantified the similarity between samples from BAL and the upper airway by calculating mean pairwise UniFrac distances. The BAL UniFrac distance to the upper airway correlated with six out of 83 metabolites (Supplementary Table 3), with a positive Spearman’s (ρ) with fucose-rhamnose, cellobiose, isothreonic acid (Fig. 3a), threonic acid and glyceric acid, and negative Spearman’s ρ with arachidonic acid (Fig. 3b).
Figure 2. Comparison of inferred metagenomes of pneumotypeSPT and pneumotypeBPT.
a, PCoA based on Jensen–Shannon divergence shows that the metagenome of pneumotypeSPT is significantly different from the metagenome of pneumotypeBPT. b, LEfSe analysis was performed using the summarized functional annotation for the KOs annotated to metabolism inferred for each BAL sample. This analysis showed multiple functional differences in the genomic composition of pneumotypeSPT as compared with pneumotypeBPT. c, STAMP was used to determine metabolic pathways differentially enriched (P < 0.05) and their effect size (η2). The 15 top metabolic pathways for each pneumotype are represented with effect size and relative abundance.
Figure 3. Correlation between the lower airway microbiome and metabolome.
a,b, Average UniFrac distances between pairs of BAL samples and upper airways was positively (a) or negatively (b) correlated with levels of metabolites in BAL fluid. Red symbols represent BAL samples identified as pneumotypeSPT and green symbols represent BAL samples identified as pneumotypeBPT (lines represent medians and standard error, SE, P values are based on Spearman’s ρ). c, A co-occurrence network for genus-level summarized taxa was built using SparCC as described in the Methods. Genera (circles) were then correlated with levels of metabolites, and significantly correlated metabolites (grey octagons) are represented in the network. Genera identified as markers for pneumotypeBPT are in light green and genera identified as markers for pneumotypeSPT are in light red. Cytoscape 3.2.158 was used to visualize the network with a prefuse force-directed layout, with the length of edges being 1 – ρ for positive correlations and absolute (ρ) for negative correlations. Nodes in close proximity are therefore highly positively correlated, and nodes further apart are highly negatively correlated.
We then investigated how different taxa and metabolites co-occurred, and whether those relationships were conserved across pneumotypes. A co-occurrence network of the 16S data at the genus level was performed using SparCC, which greatly reduces artefactual correlations in compositional data10. Consistent with the results of previous analysis (Fig. 1), marker taxa for pneumotypeSPT and pneumotypeBPT co-occurred with other marker taxa within, but not across, the pneumotypes. These taxa were then considered in relation to the 83 Kyoto Encyclopedia of Genes and Genomes (KEGG)-annotated metabolites. In the microbiome/metabolome correlation network that includes the most highly correlated metabolites (Fig. 3c), background-characteristic taxa such as Pseudomonas, Sphingomonas, Chryseobacterium, Burkholderia and Janthinobacterium were associated with glyceric acid, isothreonic acid, erythritol, threitol, cholesterol and fucose-rhamnose. In contrast, supraglottic-characteristic taxa, such as Prevotella, Rothia and Veillonella, were associated with palmitoleic acid, arachidonic acid, 4-hydroxybenzoate and glycerol. Furthermore, the genomic potential (metagenome) and end product (metabolome) levels were significantly correlated, especially in pneumotypeSPT (Supplementary Fig. 6), providing further evidence for active microbial metabolism in the pulmonary milieu.
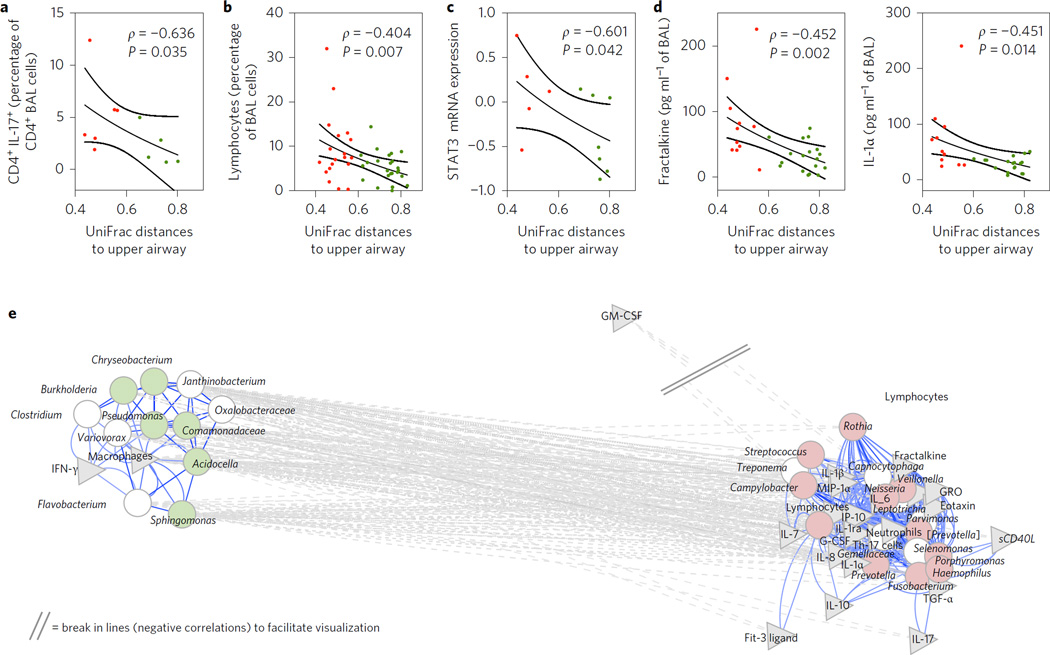
We then tested for associations between the lung microbiome and host immune phenotypes. The BAL UniFrac distance to the upper airway showed a significant inverse correlation with the percentage of IL-17+ CD4+ cells in BAL (Fig. 4a); thus, pneumotypeSPT was associated with a higher frequency of CD4+ IL-17+ cells than pneumotypeBPT (4.9% (1.9 to 5.7) vs 1.0% (0.9 to 2.4) respectively, P = 0.04). The BAL UniFrac distance to the upper airway significantly correlated with the percentage of lymphocytes in BAL (available for the NYU and LHMP cohorts, Fig. 4). Evaluation of the bronchial epithelial cell transcriptome in a subset of 12 subjects showed that 2,834 out of 54,675 mRNAs were statistically significantly different between the two pneumotypes (P < 0.05), including genes related to innate or adaptive immunity (see Supplementary Information). Importantly, the expression of STAT3, an important transcription factor for Th17 differentiation, tended to be higher in pneumotypeSPT than in pneumotypeBPT (0.12 (−0.30 to 0.51) vs −0.50 (−0.78 to 0.24), respectively, P = 0.14). Furthermore, the BAL UniFrac distance to the upper airway was significantly and inversely correlated with bronchial epithelial cell STAT3 expression (Fig. 4c). Many STAT3 downstream molecules (FST, LYZ, HP, SNAI2 and LEPR)11–16 were also present at significantly higher levels in pneumotypeSPT than in pneumotypeBPT (Supplementary Fig. 8). Taken together, these data indicate that microbes present in pneumotypeSPT or their products are linked to activation of these pathways and the lung mucosal Th17 response.
Figure 4. Similarity of the lower airway microbiome with the upper airway microbiome is associated with the percentage of lymphocytes in BAL.
a,b, Average UniFrac distances between pairs of BAL samples and upper airways were negatively correlated with the percentage of CD4+ IL17+ cells (a) and the percentage of lymphocytes (b) in BAL. c, Similarity of the lower airway microbiome with the upper airway microbiome is associated with increased expression of STAT3 mRNA in bronchial epithelial cells. d, Negative significant correlations were found between average UniFrac distances of BAL to the upper airway and fractalkine and IL-1α. Red symbols represent BAL samples identified as pneumotypeSPT and green symbols represent BAL samples identified as pneumotypeBPT (lines represent median and SE, P value based on Spearman’s ρ). e, Network analysis built around co-occurrent taxa as defined previously (see Fig. 3). Taxa (circles) remaining in the model were then correlated with levels of cells and cytokines in BAL (grey triangles). Genera identified as markers for pneumotypeBPT are shown in light green and genera identified as markers for pneumotypeSPT are shown in light red. Significant correlations are shown in the network. The network was visualized with Cytoscape with the same parameters as previously defined. GM-CSF, granulocyte-macrophage colony-stimulating factor; G-CSF, granulocyte colony-stimulating factor; GRO, growth-related oncogene-α.
To further characterize the mechanisms involved in the lower airway immune phenotypes associated with each pneumotype, BAL cytokine levels were measured for the 29 subjects from whom sufficient BAL fluid was available (Supplementary Table 4). The BAL UniFrac distance to the upper airway negatively correlated with levels of IL-1α and fractalkine, two cytokines involved in Th17 differentiation and recruitment (Fig. 4d) as well as with IL-1ra, IL-8, growth-related oncogene-α (GRO), Eotaxin, FGF-2, epidermal growth factor (EGF), MIP-1α, TGF-α and granulocyte colony-stimulating factor (G-CSF) (Supplementary Table 5).
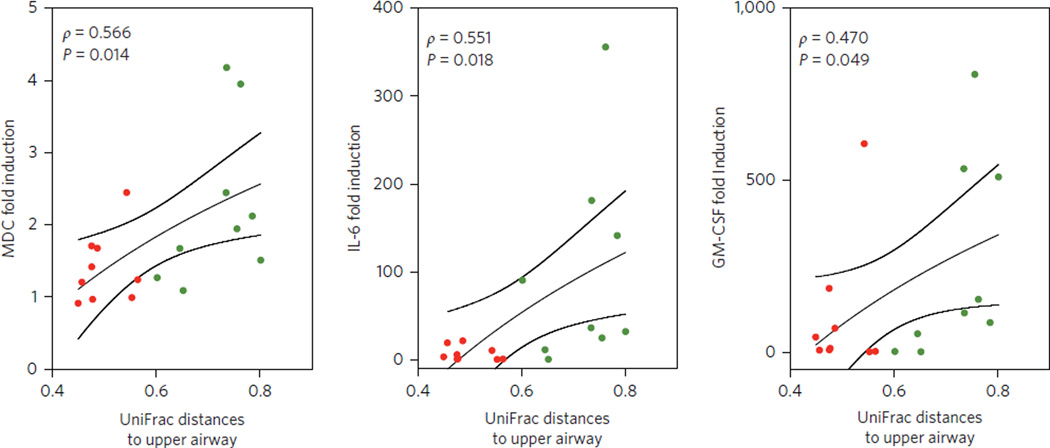
To address which component of the lower airway microbiome might be most relevant for the local host immune phenotypes, we performed a network analysis between co-occurring taxa (summarized at the genus level) and BAL cytokines/cells (Fig. 4e). Using the previously constructed co-occurrence network (as shown Fig. 3c), we now searched for those taxa most highly correlated with particular BAL cytokines/cells. Among the significant correlations, SPT-characteristic taxa, including Prevotella, Rothia and Veillonella, were positively correlated with levels of multiple cytokines including Th17 cytokines, such as IL-1α, IL-1β, IL-6, fractalkine and IL-17, and with both Th17 cells and neutrophils. In contrast, BPT-characteristic taxa, including members of the genera Pseudomonas, Sphingomonas, Chryseobacterium, Burkholderia and Janthinobacterium, only correlated with BAL macrophage percentage and levels of IFN-γ. To consider the innate responses in the lung to the two pneumotypes, we then evaluated the TLR4 responses of alveolar macrophages obtained from a subset of 18 subjects (eight from pneumotypeBPT and ten from pneumotypeSPT). Pairwise BAL UniFrac distance to the upper airway directly correlated with lipopolysaccharide (LPS)-induced increases in IL-6, macrophage-derived chemokine (MDC) and MIP-1α production (Fig. 5 and Supplementary Table 7). These data are indicative of the presence of a blunted TLR4 response in cells from subjects with pneumotypeSPT.
Figure 5. PneumotypeSPT is associated with a blunted TLR4 response of alveolar macrophages.
Alveolar macrophages of subjects with pneumotypeSPT or pneumotypeBPT were cultured for 24 h and exposed to 10 ng LPS or media alone. Cytokine production was compared for LPS/MA to calculate fold induction. Average UniFrac distances between pairs of BAL samples and upper airways correlated to fold induction of MDC, IL-6 and GM-CSF. Red symbols represent BAL samples identified as pneumotypeSPT and green symbols represent BAL samples identified as pneumotypeBPT (lines represent median and SE, P value based on Spearman’s ρ).
Discussion
In our previous work, we found that the lower airway microbiomes assessed in acellular BAL samples obtained from healthy individuals could be divided into those with higher levels of bacterial 16S rRNA in the lower airway, whose bacterial communities resembled those in the URT (pneumotypeSPT), and individuals with low bacterial copy number, whose BAL bacterial sequences resembled background environmental taxa (pneumotypeBPT). In this cross-sectional study using three independent cohorts, we confirmed that a lower airway microbiome enriched with upper airway microbes (pneumotypeSPT) was present in approximately 45% of the studied individuals. Interestingly, a similar proportion of healthy individuals have been shown to microaspirate based on detectable radiotracer5,17. The existence of this distinct microbial community is further supported by phage data, which constitute an indirect method of studying bacterial communities independent of the 16S analysis. Importantly, both the pneumotypeSPT, as a categorical classification, and overall similarity between BAL and URT communities, as measured by the UniFrac statistic, were associated with enhanced lung inflammation characterized by increased BAL lymphocytes and with increased Th17 cells (Fig. 4a and b), concentrations of Th17-chemoattractant cytokines (for example, IL-1α, IL-1β, fractalkine and IL-7, Fig. 4d,e and Supplementary Tables 4 and 5), expression of inflammatory pathway mRNA (data in Supplementary Information and Supplementary Figs 8 and 9), and free fatty acids with immunological properties (for example, arachidonic acid, data in Supplementary Information and Supplementary Tables 2 and 3) and with blunted TLR4 responses. These data indicate that, among the subjects studied, the presence in the lung of URT-derived bacteria and/or their metabolic products regulates the tonic level of airway inflammation and Th17 immune activation.
In this study, we have utilized both the pneumotypeSPT as a categorical classification, and the overall similarity between BAL and URT communities, as measured by the UniFrac distance between BAL upper airways. We consider that the pneumotype categorization is useful for the conceptualization of a distinct microbial community. Our co-occurrence network analysis indicates that upper airway taxa tend to coexist. However, it is important to note that this is likely to be in dynamic change and, as indicated by previous descriptions of the ‘enterotypes’ in the gut microbiome18, an individual’s clusters can be highly variable19. We therefore also used the measurement of the UniFrac distance between BAL and the upper airways as a continuous variable of mouth-lung similarity, which may be more representative of the continuum variability that occurs in the physiological scenario where the lower airways are in communication with the upper airways and subjected to frequent aspiration events.
Because little is known about functional aspects of the lower airway microbiome, we used data from metabolomic analysis and from the taxa-inferred metagenome to explore possible associations between taxa and microbial metabolism. An increased relative abundance of carbohydrate metabolism genes in pneumotypeSPT correlated with reduced levels of cellobiose and fucose-rhamnose, consistent with active bacterial metabolism in the lower airway. Taken together, these data indicate that indigenous lung microbiota enriched with oral taxa (pneumotypeSPT) form a metabolically active consortium that induces host cellular mucosal immunity of the Th17/neutrophilic phenotype, while suppressing innate immunity.
Taxa from the upper airways have been consistently observed in the lower airways in multiple lung microbiome studies1–4,20–23. However, across these studies, it has been difficult to establish whether the presence of upper airway taxa in BAL specimens is an artefact of carry-over of oropharyngeal microbes during sampling or is due to the authentic presence of such microbes in the lungs due to greater levels of microaspiration or inefficient microbial clearance. Microaspiration is common in healthy subjects5–7 and more frequent in those with lung disease, including chronic obstructive pulmonary disease (COPD), asthma, obstructive sleep apnoea, cystic fibrosis, pulmonary fibrosis, non-tuberculous mycobacteria and pneumonia6,24–28. Smoking or air pollution exposure further inhibit mucociliary clearance of bacteria29,30. Thus, commonly finding pneumotypeSPT in these asymptomatic subjects is not surprising. The topographical continuity of the upper and lower airways allows for microaspiration of upper airway anaerobes (for example, Prevotella or Veillonella, both markers for pneumotypeSPT). However, we also found differences between the upper airway microbiome and the microbiome observed in acellular BAL samples categorized as pneumotypeSPT (see Supplementary Information and Supplementary Fig. 4). Differences in the mucosal environment or immune response at the upper and lower airway may exert a selection pressure that is likely to affect the microbial community composition in each mucosal site. Furthermore, we have previously shown that pneumotypeSPT correlates with higher overall levels of bacteria based on 16S rRNA levels. Individuals with pneumotypeBPT are therefore likely to have less microaspiration and/or more effective microbial clearance. Consequently, BAL samples of pneumotypeBPT have greater phylogenetic differences from the microbiome present in the upper airways, as shown by higher BAL UniFrac distances to the upper airway. Using this continuous measurement complements earlier dichotomous pneumotype definitions3.
The strong correlation of the pneumotypeSPT microbial genomic potential and the lower airway metabolic environment indicate that microbial metabolism is active in the lungs of these subjects. In contrast, for subjects with pneumotypeBPT, the lower overall correlation between the metagenome and its metabolic environment indicates a lesser effect on the lower airway environment. However, the additive effects of the genomic potential of background taxa may dilute any microbial signal from resident microbes in this pneumotype. Background subtraction techniques optimized for samples with low signal-to-noise ratio (for example, samples from pneumotypeBPT) are needed to better understand the microbiome contributions to the host environment.
The finding that inflammatory cytokines and lymphocytes inversely correlate with the β-diversity gradient indicates that an increasing presence of oral microbes (lower BAL UniFrac distance to the upper airway) induces more lower airway inflammation. Lipoprotein lipase (LPL) mRNA is elevated in pneumotypeSPT (Supplementary Fig. 8), and its metabolic product arachidonic acid (Fig. 3b) is associated with enrichment with supraglottic microbes in the lower airways, possibly contributing to the increased inflammation31. In the gut, the gastrointestinal tract microbiota is critical to the mucosal immune phenotype. Indeed, in mouse gut lamina propria, a single group—segmented filamentous bacteria (SFB)—is critical for Th17 cell differentiation32. Despite emerging data indicating that diverse microaspirated bacterial populations are normally present in association with airway mucosa in some people, little attention has been paid to the role of airway microbiota in Th17-mediated airway mucosal inflammation33. In the lung, the mechanisms through which commensal-derived signals regulate innate and adaptive immunity are not well defined. Compared with the blood, the lung had an increased Th17/Treg ratio (Supplementary Fig. 7), and pneumotypeSPT was associated with increased IL17+ CD4+ T cells (Fig. 4a). Furthermore, pneumotypeSPT and its marker taxa (for example, Veillonella and Prevotella) were associated with increased in vivo levels of several cytokines relevant for Th17 differentiation (IL-1β and IL-6) or chemotaxis (fractalkine). These findings are indicative of a role for the lung microbiome in regulating the pulmonary Th17 response.
A strength of this study is that our findings were relevant across three independent cohorts, two of them (NYU and LHMP) reasonably sized. Although metabolome and detailed host immune phenotyping was only available from one of the cohorts (NYU), all three cohorts showed the pneumotypeSPT/pneumotypeBPT dichotomy. Also, lymphocyte percentages (available for the NYU and LHMP cohorts) were higher in pneumotypeSPT (Supplementary Table 4) and inversely correlated with the BAL UniFrac distance to the upper airway (Supplementary Table 5 and Fig. 4b).
Our study has several limitations. The BAL samples utilized in this study were acellular BAL fluid. There is still controversy regarding whether acellular BAL, whole BAL or BAL cells should be used to evaluate the lower airway microbiome, because it has been noted that these different sample types yield different results34,35. Due to the low bacterial burden of these samples, especially in pneumotypeBPT samples, it is possible that changes in processing may affect our pneumotype interpretation. Thus, our distinct pneumotype designations will need to be validated using other lower airway samples (for example, lung tissue, airway brushes and whole BAL specimens). In this study, a cross-sectional design was used to examine samples obtained from one segment of the lung. It is likely that microbiome heterogeneity (and thus pneumotype categorization) occurs over time and across different segments of the lung36. Longitudinal studies and studies evaluating different lung segments will be needed to confirm this hypothesis. It is also important to note that because the metagenome data are predicted from 16S rRNA data (PICRUSt), it does not constitute an independent validation of the taxonomic clusters. Furthermore, we note that inferred metagenomic data should be interpreted with caution due to concerns pertaining to biased amplification of 16S rRNA sequences, and that predicted pathways of non-bacterial origin (for example, pathways related to human disease and drug development) need to be removed from the analysis37. PICRUSt predictions depend on the sequenced reference genomes, which might not be sufficient to distinguish genomic content between related bacterial strains. The associations observed between microbiome, metabolome and inflammation in the human lung are insufficient to determine the causal direction, which would require interventional studies utilizing antibiotics or other measures aimed at altering the microbiome33. The mRNA from airway brushings represents a mixture of cells that mediate the mucosal immune response; experiments with sorted cells will allow better determination of the contribution of epithelial, myeloid and lymphoid cells to the expression signatures observed in the pneumotypes. Ultimately, animal models that replicate observations in humans with pneumotypeSPT are needed to uncover the mechanisms of the lung microbiome, metabolome and immune response interactions. Despite the differences in inflammatory phenotypes between pneumotypeSPT and pneumotypeBPT, there were no differences in pulmonary function correlating with airway and/or parenchymal injury (recognizing that subjects were included only if they had no known clinical lung disease). We therefore interpret our findings as reflecting a sub-clinical inflammatory phenotype, which may be relevant to persistence. Our analysis was limited to the bacteriome (and its associated phages). Concurrent examination of the mycobiome and virome will be valuable38,39. Finally, a large percentage of subjects had significant smoking history (26% were current smokers and 37% were ex-smokers) and may not be representative of the general population. Although studies have not been able to show a direct effect of smoking on the lower airway microbiome3,4, smoking affects the host immune response. Although our multivariate analysis, considering smoking as a cofactor (see Supplementary Information and Supplementary Fig. 10), indicates that several of the associations observed between the microbiome and the host immune phenotype are independent of smoking, experimental studies are needed to discern the independent contribution of microbiome and smoking to the host immune phenotype.
In summary, our findings indicate that the basal level of lower airway mucosal Th17 immune activation is associated with compositional characteristics of local lung bacteria, where in some subjects this seems to be derived largely from the URT though microaspiration. Further studies will be needed to determine: the dynamics of entry; clearance and local replication by the microbial community; the stability of this phenotype over time within individuals; whether these levels of asymptomatic tonic Th17 inflammation contribute to long-term consequences for lung health; whether they interact with other environmental factors in the pathogenesis of inflammatory lung disease; and whether they regulate responses to recognized pulmonary pathogens.
Methods
Subjects
Forty-nine subjects were enrolled for research bronchoscopy as part of three cohorts: 31 from NYU, 14 from the healthy control group from the LHMP (four from the University of Pittsburgh, four from the University of California San Francisco and six from the University of California Los Angeles) and four from OSU. All subjects signed informed consent forms and the research protocol was approved by the human subjects review committees of the institutional review boards at each institution. Exclusion criteria included known underlying lung disease, treatment with antibiotics or steroids in the previous three months, cardiovascular, renal or liver disease, diabetes mellitus and heavy alcohol use (more than six beers daily). Before bronchoscopy, subjects underwent a screening visit where they were questioned about respiratory symptoms and were subjected to pulmonary function testing. None of the subjects complained of new respiratory symptoms (cough, wheezing or shortness of breath).
16S rRNA gene sequencing
Research bronchoscopy was performed by obtaining samples from background (sterile saline and saline through the bronchoscope before bronchoscopy), upper airways and lower airways using BAL as previously described3,4. Samples were stored at −80 °C until they were processed. DNA was extracted using an ion exchange column (Qiagen). High-throughput sequencing of bacterial 16S rRNA gene amplicons encoding the V4 region40 (150 bp read length, paired-end protocol) was performed using a MiSeq Illumina Sequencer. The 16S rRNA gene sequences were analysed using the Quantitative Insights into Microbial Ecology (QIIME) pipeline for analysis of microbiome data (see Supplementary Information)3,41–45. The proportion of reads at the OTU or genus levels was used as a measure of the relative abundance of each type of bacteria. Microbiome analysis of BAL samples was compared with 16S data from background and upper airways. Because lower airway samples have low biomass, we used SourceTracker to estimate the contribution of background microbiota to BAL samples46. In a subset of samples (n = 28) 16S rRNA gene sequences were previously obtained using a 454 platform and targeting the V1–V2 variable region and results have been reported elsewhere3. Procrustes analysis based on UniFrac distances was then performed to evaluate for the consistency of microbiome results. When samples were sequenced using two different approaches (454 and MiSeq) and targeting two different variable regions of the 16S rRNA gene (V1–V2 and V4), similar pneumotype allocation was found for each sample (see Supplementary Information and Supplementary Fig. 3 for more details). To determine the genomic potential of these two pneumotypes, we computationally predicted the metagenome using PICRUSt47. This software tool uses the obtained 16S rRNA gene sequence data to predict the functional profile of a bacterial community based on an existing reference genome database. Metagenomic pathway analysis was performed using STAMP with default parameters48.
Shotgun sequencing and phageome analysis
DNA was extracted from BAL samples as described for 16S sequencing above. Following fragmentation and library preparation, sequencing was performed on the Illumina MiSeq platform (2 × 250 bp paired-end reads). Sequences were analysed using VirusSeeker49. The DNA phageome was analysed using LEfSe to identify discriminant bacteriophage taxonomic markers.
Measurement of metabolites in BAL fluid
For metabolomics analysis, we utilized BAL samples from 29 NYU subjects in which sufficient BAL fluid was available. Samples (4 ml) of BAL fluid were processed for GC–TOF metabolomics (see Supplementary Information for further details)50,51. Mass spectra were acquired for m/z 85–500 at 20 spectra s−1 and 1,750 V detector voltage. The resulting files were processed by the metabolomics BinBase database (University of California Davis). All database entries in BinBase were matched against the Fiehn mass spectral library of 1,200 authentic metabolite spectra using retention index and mass spectrum information or the NIST05 commercial library. Identified metabolites were reported if present in at least 50% of the samples per study design group (as defined in the software). Intensity data were mean-centred and divided by the standard deviation of each variable using MetaboAnalyst52.
Measurement of BAL cells
BAL cells were centrifuged (500g for 5 min) and washed twice. Cell viability was assessed by trypan blue exclusion, and cytospins were prepared. Total cell count and BAL cell differentials (500 cells counted) were performed to assess counts of macrophages, lymphocytes and neutrophils. To obtain measurement of T cell subsets, fluorescence-activated cell sorting (FACS) was performed as described with Treg defined as CD3+, CD4+ and FoxP3+ and Th17 defined as CD3+, CD4+ and IL-17+ (ref. 53).
Evaluation of transcriptome of bronchial epithelial cells
In a subset of subjects (n = 12), paired peripheral bronchial epithelial cells were obtained by airway brushing. The presence of club cells was used to confirm sampling of the small peripheral airways. RNA was then extracted using a Qiagen miRNeasy Mini Kit. To identify target genes, global gene expression profiling was performed with the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (HG-U 133 Plus 2.0). Array data were analysed by GeneSpring GX version 12.6.1 (Agilent Technologies) using Linear Models for Microarray Data (LIMMA). Hierarchical heat maps were generated by Java TreeView 3.0 using the complete linkage clustering method and squared Euclidean distance measure. Ingenuity pathway analysis (IPA) was used to identify top biological functions and disease/disorders, top regulator effect networks, and to generate network pathways. Networks generated by IPA grouped the differentially regulated genes according to previously known associations between genes or proteins54.
Measurement of in vivo cytokines in BAL fluid
For cytokines, we utilized BAL samples from 29 subjects from NYU in which a sufficient amount of BAL fluid was available. Thirty-nine cytokines were measured in the concentrated BAL with Luminex using Human Cytokine Panel I (Millipore) according to the manufacturer’s instructions in a Luminex 200IS system (Luminex). Because analytes in the epithelial lining fluid (ELF) are diluted with sterile saline during BAL, a concentration step was performed via dialysis and lyophilization, using albumin as an internal control. After approximately 75-fold concentration, 31 out of 39 cytokines were present at measurable levels (defined as levels above the lowest standard in >70% of the samples). These included interleukin(IL)-1 receptor antagonist (IL-1ra), IL-1α, IL-1β IL-5, IL-6, IL-7, IL-8 (CXCL8), IL-12 (p40), IL-12 (p70), IL-15, IL-17, interferon -γ-induced protein 10 (IP-10 or CXCL10), interferon-γ (IFN-γ), epidermal growth factor (EGF), eotaxin (CCL11), fibroblast growth factor-2 (FGF-2), Flt-3 ligand, fractalkine (CX3CL1), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), growth-related oncogene-α (GRO or CXCL1), monocyte chemotactic protein 1 (MCP-1 or CCL2), monocyte chemotactic protein 3 (MCP-3 or CCL7), macrophage-derived chemokine (MDC or CCL22), macrophage inflammatory protein 1α (MIP-1α or CCL3), macrophage inflammatory protein 1β (MIP-1β), transforming growth factor alpha (TGF-α), tumour necrosis factor α (TNF-α), vascular endothelial growth factor (VEGF), soluble CD40 ligand (sCD40 L), and soluble IL-2 receptor (sIL-2Rα). Average results from technical duplicates were utilized. The levels of the following cytokines were below the detection limit of the assay and are not included in the analyses: IFN-α, TNF-β IL-2, IL-3, IL-4, IL-9, IL-10, and IL-13. Data were analysed with MasterPlex TM QT software (version 1–2, MiraiBio).
Measurement of TLR4 response of alveolar macrophages
In a subset of samples from the NYU cohort (n = 18), alveolar macrophages were isolated for ex vivo TLR4 stimulation. BAL macrophages were isolated by plastic adhesion and cells (2 × 106) were incubated in 12-well plates with RPMI (Gibco), plus 10% fetal calf serum (HyClone) and 2% penicillin-streptomycin (Gibco) with or without 40 ng LPS ml−1 (Escherichia coli 0.55:B4 and B5, Sigma-Aldrich). After 24 h, supernatants were collected and assayed using Human Cytokine Panel I (Millipore) according to the manufacturer’s instructions in a Luminex 200IS system (Luminex). Data were analysed with MasterPlex TM QT software (version 1–2, MiraiBio). Average results from technical duplicates were utilized.
Statistical and multi-omic analysis
Because the distributions of microbiome data are non-normal, and no distribution-specific tests are available, we used non-parametric tests of association. For association with discrete factors, we used either the Mann-Whitney test (in the case of two categories) or the Kruskal–Wallis analysis of variance (in the case of more than two categories). For tests of association with continuous variables, we used non-parametric Spearman correlation tests. False discovery rate (FDR) was used to control for multiple testing55. To evaluate differences between groups of 16S data or inferred metagenomes, we used LDA LEfSe56. Features significantly discriminating among groups with LDA score >2.0 were represented as a cladogram, as produced by LEfSe with default parameters.
For multi-omic analysis, analysis was restricted to the samples obtained at NYU where all omic platforms where applied. Co-occurrence networks using 16S OTUs were calculated as previously described57. Briefly, 16S data were summarized at the genus level and co-occurrence was calculated using SparCC10, with results validated over ten rarefactions of the genus-level input table. Genera co-occurring significantly (P < 0.05) with ρ > 0.7 or ρ < −0.7 were conserved for Spearman correlation analysis with metabolome, cytokine and BAL cell data. To evaluate whether smoking status was a confounder for the observed differences in biomarkers (for example, metabolites, BAL cells and cytokines) noted to be associated with a distinct pneumotype, we used a multivariate logistic regression model, where biomarkers were considered as outcome (dichotomized as below or above the median), and pneumotype and smoking status (never smoker vs smoker) were predictors (covariates). This analysis was carried out using SPSS (IBM, version 20), and age and gender were forced in the model. Results were reported as odds ratio (95% confidence interval) for both pneumotypeSPT and smoking status.
Accession codes
All multi-omic data have been submitted to the Gene Expression Omnibus (GEO) under accession numbers GSE74395 (for microbiome and host immune response data) and GSE73585 (for transcriptome data).
Supplementary Material
Acknowledgments
Research support funding was provided by the National Institute of Allergy and Infectious Diseases (NIAID) K23 AI102970 (to L.N.S.); the National Heart, Lung and Blood Institute (NHLBI) R01 HL125816 (to S.B.K.); NIAID K24 AI080298 (to M.D.W.); the Clinical and Translational Science Institute (CTSI) grant no. UL1 TR000038; the Early Detection Research Network (EDRN) 5U01CA086137-13; the Diane Belfer Program for Human Microbial Ecology; the Michael Saperstein Scholarship Fund; the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01DK090989; the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) UH2 AR57506; NIAID U01AI111598; NHLBI R01 HL090339; NHLBI K24 HL123342 (to A.M.); NHLBI U01 HL098962 (to A.M. and E.G.); NHLBI K24HL123342; NHLBI K24 HL087713 (to L.H.); NIAID and the National Cancer Institute (NCI) UO1-AI-35042; 5-MO1-RR-00722 from the General Clinical Research Center (GCRC); UL1TR000124 from the University of California Los Angeles Clinical and Translational Research Center (UCLA CTRC); NIAID UO1-AI-35043; NIAID UO1-AI-37984; NIAID UO1-AI-35039; NIAID UO1-AI-35040; NIAID UO1-AI-37613; NIAID UO1-AI-35041 (Multicenter AIDS Cohort); NIAID and the National Institute of Child Health and Human Development (NICHHD) UO1-AI-35004; NIAID UO1-AI-31834; NIAID UO1-AI-34994; NIAID UO1-AI-34989; NIAID UO1-AI-34993; NIAID UO1-AI-42590; NICHHD UO1-HD-32632; the Women’s Interagency HIV Study (WIHS); NHLBI U01-HL098957 and NHLBI R01-HL113252 (to R.G.C).
The authors also thank HW. Virgin (Washington University School of Medicine), S. Stone, S. Fong, A. Malki and S. Tokman (University of California San Francisco (UCSF)), C. Kessinger, N. Leo, D. Camp, M.P. George, L. Lucht, M. Gingo, R. Hoffman, M. Fitzpatrick, J. Ries, A. Clarke (Pittsburgh) and J. Dermand and E. Kleerup (UCLA).
Computing was partially supported by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Some of the Pittsburgh LHMP data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS) with centres (Principal Investigators) at UCLA (R. Detels, U01-AI35040); University of Pittsburgh (C. Rinaldo, U01-AI35041); the Center for Analysis and Management of MACS, Johns Hopkins University Bloomberg School of Public Health (L. Jacobson, UM1-AI35043). MACS is funded primarily by NIAID, with additional cofunding from the NCI. Targeted supplemental funding for specific projects was also provided by the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute on Deafness and Communication Disorders (NIDCD). MACS data collection was also supported by UL1-TR000424 Johns Hopkins University Clinical and Translational Science Awards (JHU CTSA, https://statepi.jhsph.edu/macs/macs.html). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH).
Some of the Pittsburgh LHMP data in this manuscript were collected by the WIHS. WIHS (principal investigators): U01-AI-103408; Connie Wofsy Women’s HIV Study, Northern California (R. Greenblatt, B. Aouizerat and P. Tien). The WIHS is funded primarily by NIAID, with additional cofunding from the Eunice Kennedy Shriver NICHD, the NCI, the National Institute on Drug Abuse (NIDA) and the National Institute on Mental Health (NIMH). WIHS data collection was also supported by UL1-TR000004 (UCSF CTSA).
Footnotes
Author contributions
L.N.S., J.C.C., M.J.B. and M.D.W. conceived and designed the study. L.N.S., J.J.T., A.M., L.H., P.D. and W.R.W. acquired the data. L.N.S., J.C.C, J.J.T., S.B.K., B.G.W., Y.L., N.S., W.R.W., C.U., A.A., B.C.K., R.G.C., M.J.B. and M.D.W. analysed and interpreted the data. L.N.S., J.C.C., J.J.T., S.B.K., E.G., A.M., P.D., L.H., W.R.W., B.C.K., W.N.R., D.H.S., R.G.C., M.J.B. and M.D.W. drafted or revised the article. L.N.S., J.C.C., J.J.T., S.B.K., B.G.W., Y.L., N.S., E.G., A.M., P.D., L.H., W.R.W., C.U., A.A., B.C.K., W.N.R., D.H.S., R.G.C., M.J.B. and M.D.W. approved the final manuscript.
Additional information
Supplementary information is available online. Reprints and permissions information is available online at www.nature.com/reprints.
Competing interests
The authors declare no competing financial interests.
References
- 1.Charlson ES, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit.Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erb-Downward JR, et al. Analysis of the lung microbiome in the ‘healthy’ smoker and in COPD. PLoS ONE. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal LN, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris A, et al. Comparison of the respiratory microbiome in healthy non-smokers and smokers. Am. J. Respir. Crit. Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest. 1997;111:1266–1272. doi: 10.1378/chest.111.5.1266. [DOI] [PubMed] [Google Scholar]
- 6.Cvejic L, et al. Laryngeal penetration and aspiration in individuals with stable COPD. Respirology. 2011;16:269–275. doi: 10.1111/j.1440-1843.2010.01875.x. [DOI] [PubMed] [Google Scholar]
- 7.Rascon-Aguilar IE, et al. Role of gastroesophageal reflux symptoms in exacerbations of COPD. Chest. 2006;130:1096–1101. doi: 10.1378/chest.130.4.1096. [DOI] [PubMed] [Google Scholar]
- 8.Koskella B, Meaden S. Understanding bacteriophage specificity in natural microbial communities. Viruses. 2013;5:806–823. doi: 10.3390/v5030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang YJ, et al. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014;52:2813–2823. doi: 10.1128/JCM.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JH, et al. Signal transducer and activator of transcription-3 (Stat3) plays a critical role in implantation via progesterone receptor in uterus. FASEB J. 2013;27:2553–2563. doi: 10.1096/fj.12-225664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Baumann H. The carboxyl-terminal region of STAT3 controls gene induction by the mouse haptoglobin promoter. J. Biol. Chem. 1997;272:14571–14579. doi: 10.1074/jbc.272.23.14571. [DOI] [PubMed] [Google Scholar]
- 13.Zauberman A, Lapter S, Zipori D. Smad proteins suppress CCAAT/ enhancer-binding protein (C/EBP) β-, STAT3-mediated transcriptional activation of the haptoglobin promoter. J. Biol. Chem. 2001;276:24719–24725. doi: 10.1074/jbc.M005813200. [DOI] [PubMed] [Google Scholar]
- 14.Heinrich PC, et al. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang P, Li Z, Fu R, Wu H, Li Z. Pyruvate kinase M2 facilitates colon cancer cell migration via the modulation of STAT3 signalling. Cell. Signal. 2014;26:1853–1862. doi: 10.1016/j.cellsig.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Pathak RR, et al. Loss of phosphatase and tensin homolog (PTEN) induces leptin-mediated leptin gene expression: feed-forward loop operating in the lung. J. Biol. Chem. 2013;288:29821–29835. doi: 10.1074/jbc.M113.481523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huxley EJ, Viroslav J, Gray WR, Pierce AK. Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am. J. Med. 1978;64:564–568. doi: 10.1016/0002-9343(78)90574-0. [DOI] [PubMed] [Google Scholar]
- 18.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knights D, et al. Rethinking’enterotypes’. Cell Host Microbe. 2014;16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lozupone C, et al. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am. J. Respir. Crit. Care Med. 2013;187:1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS ONE. 2012;7:e47305. doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sze MA, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012;185:1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tunney MM, et al. Lung microbiota and bacterial abundance in patients with bronchiectasis when clinically stable and during exacerbation. Am. J. Respir. Crit. Care Med. 2013;187:1118–1126. doi: 10.1164/rccm.201210-1937OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morse CA, et al. Is there a relationship between obstructive sleep apnea and gastroesophageal reflux disease? Clin. Gastroenterol. Hepatol. 2004;2:761–768. doi: 10.1016/s1542-3565(04)00347-7. [DOI] [PubMed] [Google Scholar]
- 25.Teramoto S, et al. Obstructive sleep apnea syndrome may be a significant cause of gastroesophageal reflux disease in older people. J. Am. Geriatr. Soc. 1999;47:1273–1274. doi: 10.1111/j.1532-5415.1999.tb05216.x. [DOI] [PubMed] [Google Scholar]
- 26.Field SK, Underwood M, Brant R, Cowie RL. Prevalence of gastroesophageal reflux symptoms in asthma. Chest. 1996;109:316–322. doi: 10.1378/chest.109.2.316. [DOI] [PubMed] [Google Scholar]
- 27.Scott RB, O’Loughlin EV, Gall DG. Gastroesophageal reflux in patients with cystic fibrosis. J. Pediatr. 1985;106:223–227. doi: 10.1016/s0022-3476(85)80291-2. [DOI] [PubMed] [Google Scholar]
- 28.Koh WJ, et al. Prevalence of gastroesophageal reflux disease in patients with nontuberculous mycobacterial lung disease. Chest. 2007;131:1825–1830. doi: 10.1378/chest.06-2280. [DOI] [PubMed] [Google Scholar]
- 29.Leopold PL, et al. Smoking is associated with shortened airway cilia. PLoS ONE. 2009;4:e8157. doi: 10.1371/journal.pone.0008157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster WM, Costa DL, Langenback EG. Ozone exposure alters tracheobronchial mucociliary function in humans. J. Appl. Phys. 1987;63:996–1002. doi: 10.1152/jappl.1987.63.3.996. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, et al. Triglyceride-rich lipoprotein lipolysis releases neutral and oxidized FFAs that induce endothelial cell inflammation. J. Lipid Res. 2009;50:204–213. doi: 10.1194/jlr.M700505-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickson RP, et al. Cell-associated bacteria in the human lung microbiome. Microbiome. 2014;2:28. doi: 10.1186/2049-2618-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twigg HL, et al. Comparison of whole acellular bronchoalveolar lavage to oral wash microbiomes Should acellular bronchoalveolar lavage be the standard? Ann. Am. Thorac. Soc. 2014;11:S72–S73. [Google Scholar]
- 36.Dickson RP, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann. Am. Thorac. Soc. 2015;12:821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui L, et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am. J. Respir. Crit. Care Med. 2015;191:932–942. doi: 10.1164/rccm.201409-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willner D, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am. J. Respir. Cell Mol. Biol. 2012;46:127–131. doi: 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q, Garrity GM, Tiedje JM, Cole J. R Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knights D, et al. Bayesian community-wide culture-independent microbial source tracking. Nature Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao G, et al. Identification of novel viruses using VirusHunter—an automated data analysis pipeline. PLoS ONE. 2013;8:e78470. doi: 10.1371/journal.pone.0078470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wikoff WR, et al. Pharmacometabolomics reveals racial differences in response to atenolol treatment. PLoS ONE. 2013;8:e57639. doi: 10.1371/journal.pone.0057639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fiehn O, et al. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 52.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Semple PL, et al. Regulatory T cells attenuate mycobacterial stasis in alveolar and blood-derived macrophages from patients with tuberculosis. Am. J. Respir. Crit. Care Med. 2013;187:1249–1258. doi: 10.1164/rccm.201210-1934OC. [DOI] [PubMed] [Google Scholar]
- 54.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 55.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 56.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.