Abstract
Single cell analysis of bacteria and subcellular protein localization dynamics has shown that bacteria have elaborate life cycles, cytoskeletal protein networks, and complex signal transduction pathways driven by localized proteins. The volume of multi-dimensional images generated in such experiments and the computation time required to detect, associate, and track cells and subcellular features pose considerable challenges, especially for high-throughput experiments. Therefore, there is a need for a versatile, computationally efficient image analysis tool capable of extracting the desired relationships from images in a meaningful and unbiased way. Here we present MicrobeJ, a plug-in for the open-source platform ImageJ. MicrobeJ provides a comprehensive framework to process images derived from a wide variety of microscopy experiments with special emphasis on large image sets. It performs the most common intensity and morphology measurements as well as customized detection of poles, septa, fluorescent foci, and organelles, determines their sub-cellular localization with sub-pixel resolution, and tracks them over time. Because a dynamic link is maintained between the images, measurements, and all data representations derived from them, the editor and suite of advanced data presentation tools facilitates the image analysis process and provides a robust way to verify the accuracy and veracity of the data.
The bottleneck in the ability to perform automated, quantitative analysis of cell shape, behavior, and fluorescence patterns of high throughput experiments is that analysis tools for extracting unbiased data efficiently from these image sets have lagged behind the technology used to collect them. This is especially true for bacterial cells, which pose additional challenges due to their size, and the ability to distinguish them from background particles due to their low contrast and irregular morphologies. In addition, bacterial cells often have external features, such as flagella, adhesive organelles, and pili that are difficult to readily detect, measure and associate with cells. In order to accomplish these tasks, and obtain information about cellular features such as poles, external organelles, sub-cellular localization patterns (mid-cell or polar), track these features over time, and relate the features to each other in meaningful ways, biologists either need to use specifically tailored analysis tools, be user savvy at applying individual algorithms, or have programming skills to extract data from images.
Currently, the available software tools for image analysis of bacterial cells (Supplementary Table 1) offer analysis methods that typically address some, but not all of these needs, and perform best within their specialized application areas 1–9. The vast majority of these tools fall into MATLAB or ImageJ based solutions. Many microbiologists are familiar with the MATLAB-based program MicrobeTracker6, which has been available for many years to perform quantitative analysis of bacterial cells. MicrobeTracker allows detailed analysis of bacterial cells, including subpixel resolution contours, quantitative measurements of fluorescence, cell segmentation, and subpixel localization of fluorescent foci. The current stand-alone implementation of MicrobeTracker, Oufti8, has improved its ability to detect confluent cells, non-rod shaped morphologies, handle large datasets, perform automated image analysis, and provides powerful tools for data analysis and plotting with an easier to use graphical user interface (GUI). However, while Oufti provides the ability to change the input values used by the segmentation algorithms, understanding the meaning of these values and their effect on the resulting output requires good knowledge of signal treatment. Because MATLAB has not been developed to deal with images per se, many common features available for working with image sets such as working with multiple images, selecting a region of interest (ROI), applying filters, and changing look-up tables (LUT) available in ImageJ-based programs are more difficult to accomplish in Oufti.
ImageJ-based7 or Icy-based10 solutions, such as Coli-Inspector2 or BactImAS4 provide an integrated and user-friendly approach to analyze spatio-temporal localization patterns and/or intensity of fluorescent protein fusions in time-lapse movies, but are specific to certain tasks and/or organisms. Coli-Inspector, for instance, is based on ObjectJ, which provides an integrated platform that dynamically links user-defined objects, such as cell axes, cell diameters, or constriction sites across images with their corresponding properties. With ObjectJ the user can easily interact with the images, the user-defined objects, and the associated results, and has access to interactive methods and visual inspection tools for quick elimination of artifacts. However, ObjectJ and similar tools offer a limited level of automation or rely on manual detection to delineate each cell on each micrograph.
Here we describe MicrobeJ, an ImageJ plugin, offering tools dedicated to address the challenges posed by automatically detecting and analyzing the wide variety of morphologies of bacterial cells and their associated organelles. This plugin was optimized to handle high throughput time-lapse microscopy experiments and deal with the obstacles described above for image analysis. MicrobeJ has an interactive GUI, including drag and drop loading of images, settings and template files, icon access to frequently used functions, hover box guidance for icon functions and user-interactive boxes (Fig. 1 & Supplementary Video 1). The GUI facilitates interaction between image sets, data, and post-processing data representation outputs, allowing users to determine the best detection parameters and ensure accuracy. MicrobeJ can be used in conjunction with the extensive library of functions and plugins that have been developed over the years as practical image analysis tools for ImageJ. A comprehensive description of the software is available in the supplementary information.
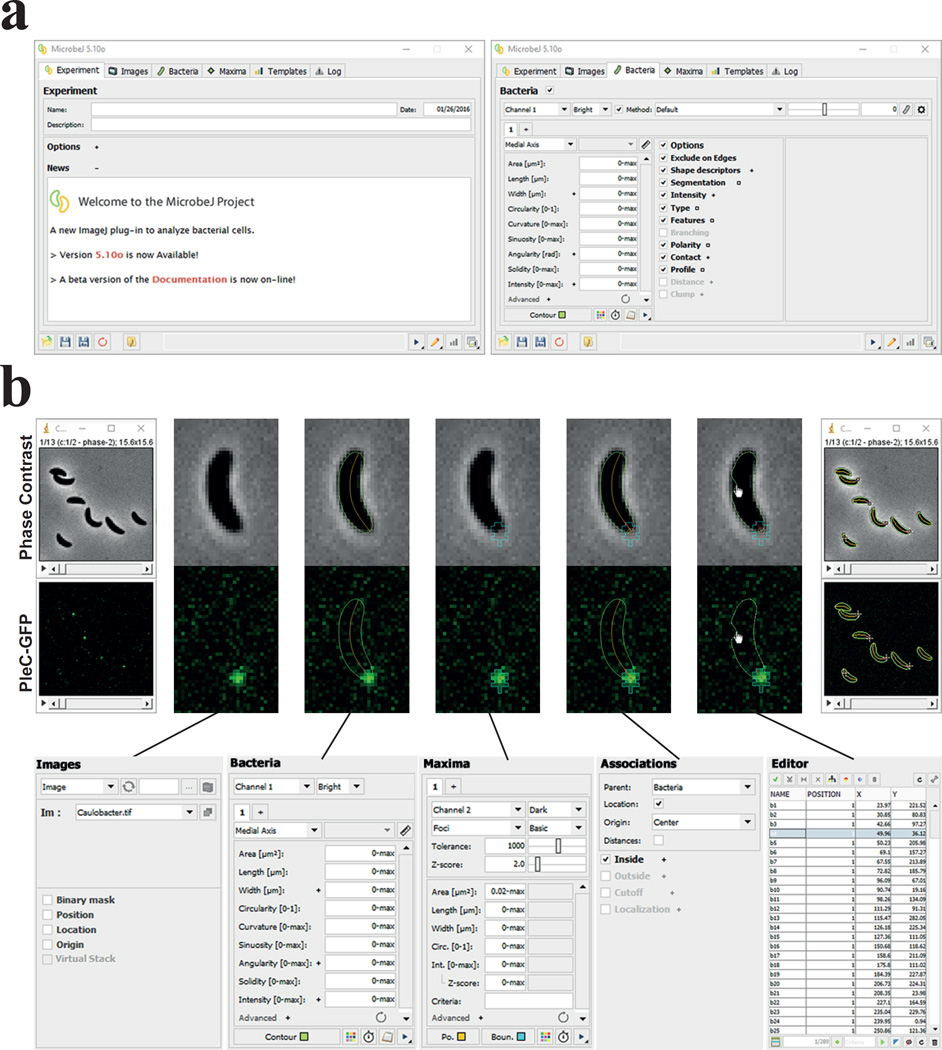
Figure 1. The MicrobeJ GUI and workflow.
(a) The MicrobeJ GUI: This interface contains several tabs that allow users to set up an experiment (Experiment), select or load a set of images (Images), define parameters for bacterial cell detection and analysis (Bacteria), define parameters for fluorescent foci detection and analysis (Maxima), define and use templates (Templates), and track any errors that might arise during analysis (Log). (b) To start, the user can simply drag and drop an image file or stack from a disk onto the MicrobeJ GUI (1), here a stack of images with C. crescentus cells expressing the polarly localized PleC-GFP. Once the images are loaded and selected, the user can test the bacterial cell detection settings. At any time, the user can also select dedicated options such as cell segmentation (Segmentation), fluorescence intensity measurements (Intensity), cellular type detection (Type), features detection (Feature), (Branching), assign poles polarity based on any particle’s property (Polarity), measure number of particles within a defined distance of cell (Contact), or display cell profile along chosen axis (Profile) (2). To detect fluorescent foci, the user can test the maxima detection settings and further refine using another set of dedicated exclusion filters. The user can also select dedicated options such as the sub-pixel resolution (Gaussian Fit, not shown) or fluorescence intensity measurement (Intensity, not shown) (3). When both bacterial cells and fluorescent foci are detected, the user can associate them hierarchically using the association panel (4). At that point the user can either manually add, edit or delete any particles detected or missed during the detection process, as in this example of a cell contour being re-adjusted using the active handles distributed along the polygon (5), or run the analysis on the stack of images (6) and obtain the raw data (see Supplementary Fig. 4).
Bacterial cell detection can be achieved from either phase contrast or fluorescence images. Using conventional thresholding techniques, MicrobeJ detects particles with virtually any type of morphology, and then generates subpixel resolution contours and extracts medial axes (Fig. 2a). Branched cells can be detected by selecting the “Branching” option, whereas other shapes are automatically detected, with the ability to reject inappropriate particles based on their properties and attributes. Medial axes allow the determination of the geometrical and topological properties of the particle shape, such as its length, width, sinuosity, curvature and angularity, and establishment of the spatial information relative to the sub-cellular coordinate system (see below). Both the accuracy and the precision of MicrobeJ measurements are in accordance with those reported by other cell sub-pixel contour detection methods8,9 (Supplementary Fig. 1, Supplementary Table 1, see Supplementary Information).
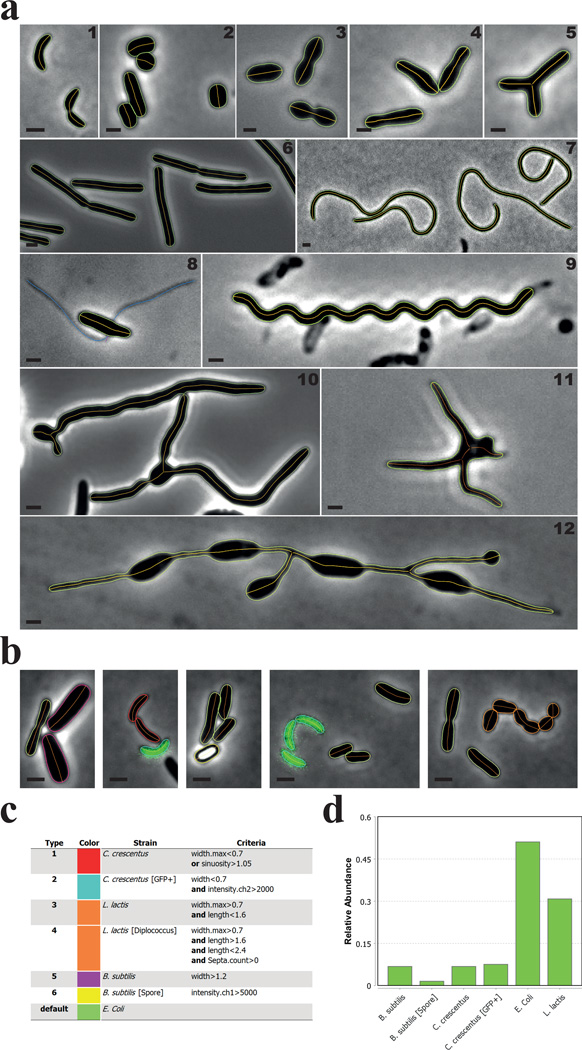
Figure 2. MicrobeJ can detect various cell morphologies.
(a) Representative phase contrast images of (1) C. crescentus cells, (2) Streptomyces venezuelae cells, (3) Streptococcus pneumoniae cells, (4) Rhodopseudomonas palustris cells, (5) Agrobacterium tumefaciens branched mutant cells, (6) Bacillus subtilis cells, (7) Cephalexin-treated C. crescentus cells, (8) Asticcacaulis biprosthecum cells, (9) 2 week old, viable, late stationary phase-adapted C. crescentus cells, (10) Vegetative S. venezuelae cells, (11) long-stalked morphotype Prosthecomicrobium hirshii cells, and (12) Rhodomicrobium sp. cells. The bacterial cell contours (green) and their corresponding medial axes (orange) are shown. A. biprosthecum stalks, detected and associated with the cell using the filament detection option, are shown in blue (8). Scale bar = 1µm. (b–d) In this experiment, C. crescentus CB15 cells (red contours), C. crescentus cells expressing a cytoplasmic GFP (mini-Tn7(Gm)gfp) (cyan contours), B. subtilis vegetative cells (magenta contours), B. subtilis spores (yellow contours), Lactococcus lactis cells (orange contours), and Escherichia coli MG1655 cells (green contours) were mixed together and imaged on an agarose pad. Cellular types were defined using the morphological and signal properties based on the specified criteria (c). The relative abundance of each type is shown (d).
For each particle, MicrobeJ provides a data structure that contains properties or complementary information, based on user-selected options, in the form of fields that can be accessed using an intuitive object-oriented semantic (Supplementary Table 2). For instance, the average and the minimal value of width measured along the medial axis of a particle can be obtained respectively using the semantics ‘SHAPE.width.mean’ and ‘SHAPE.width.min’. Using this terminology, particle properties can be used to define cellular types based on user-defined criteria. To demonstrate this, we mixed cells of various species with different morphologies or refractive index and used morphological and signal properties to define 6 different cellular types to determine their relative abundance (Fig. 2b&c, Supplementary Fig. 2).
Options are available for processes such as feature detection (constriction, fluorescent septa, or fluorescent patches) and fluorescence or width profile extraction. Using the selected options, MicrobeJ computes the specific properties and selected outputs for each particle during the detection process. Particle properties can be used to define cellular types (Fig. 2b&c), cell polarity, or temporal events. To illustrate those functionalities, we analyzed time-lapse images of Caulobacter crescentus cells harboring FtsZ-YFP and tracked the respective localization of FtsZ and the cell constriction during the cell cycle (Fig. 3a, Supplementary Video 2&3). In order to extract the time delay between these events and cell separation, we defined two temporal events. Temporal events consist of user-defined criteria designed to evaluate the particle properties over the time course of the experiment. When the particle meets the criteria of a specific event, the absolute and relative timing are recorded in the results table. In this example, we determined two temporal events based on the first appearance of a FtsZ-YFP fluorescent septum and a constriction, and from within MicrobeJ’s results interface, plotted the distribution of their timing before division for all the cell time courses detected in this experiments (Fig. 3b).
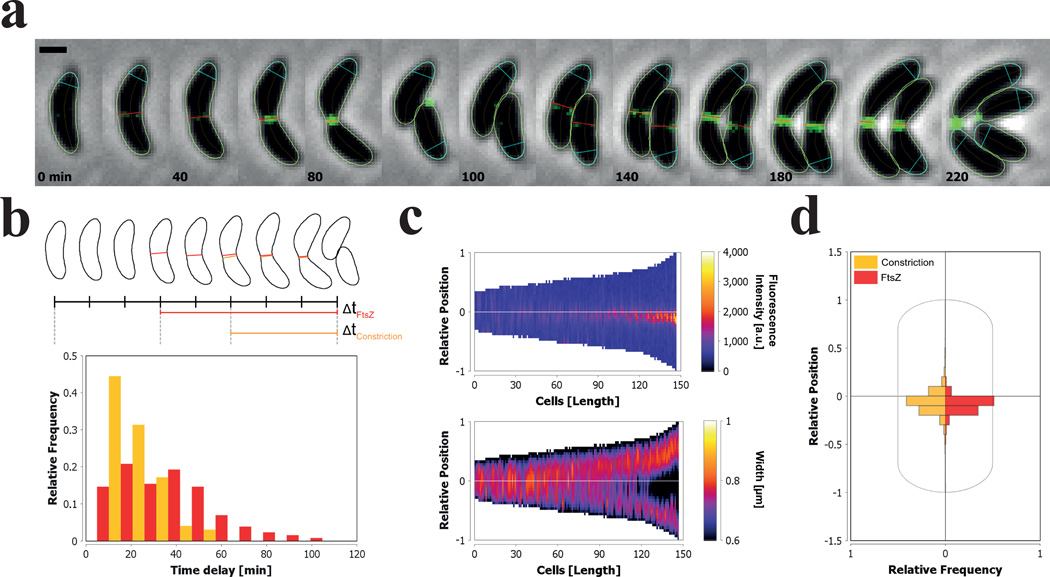
Figure 3. Detection and quantification of the localization of FtsZ-YFP and the constriction site during the cell cycle of C. crescentus.
(a–d) All the representations depicted here are created from within the Results Interface of MicrobeJ. (a) Sequence of representative phase contrast and fluorescent overlay images showing several round of division from a single C. crescentus cell expressing FtsZ-YFP. Time points are indicated for each time frame (Scale bar = 1 µm). The bacterial cell contours (green), their corresponding medial axes (orange), the position of fluorescent septum (red) and the position of the constriction (orange) are shown. Septa were detected using the intensity profile of pixels from the fluorescent image along the medial axis of the particle. Constrictions were detected using the width profile measured along the medial axis of the particle. The orientation of the medial axis of each cell, marked by the position of the pole polygon (cyan), is determined based on the position of the constriction relative to the cell center. (b) Distribution of the time delay between the first appearances of the fluorescence septum (red), the constriction (orange), and cell division (n=352). (c) Demographic representation of the FtsZ-YFP fluorescence intensities and the cellular widths measured along the medial axis of the cells. Cells are sorted according to their length. The Y-axis of each demograph represents the relative position along the cell body, where 0 represents mid-cell and 1 or −1 the cell poles. The 1 pole is the pole marked by the pole polygon shown in (a). (d) The distribution of the localization of the septa (red) and the constriction (orange) relative to the cell center (n=1521).
By default, the polarity of each particle is attributed randomly. However, cell polarity can be defined, based on user-defined criteria, with any geometrical and signal property (such as the width, the curvature, or the fluorescence intensity) or the presence or absence of fluorescent foci or cellular features (such as constrictions or septa). The division plane in C. crescentus cells is asymmetrically positioned between the old-pole (‘stalked’) and new-pole (‘swarmer’)6. Consequently, in the example of Figure 3, we used the relative position of the constriction site as a criterion to define the longitudinal polarity and extracted the fluorescence and the width profiles along the medial axes of the cells. The pixel intensity and the width are measured along the medial axis using the specified polarity and can be used to generate an oriented map of profiles also called a demograph (Fig. 3c). Finally, we measured the sub-cellular localization of the septa and the constrictions, and plotted their distribution relative to the center of the cell using sub-cellular localization charts (Fig. 3d).
To accommodate diverse applications requiring sub-cellular localization depiction, MicrobeJ provides four types of relative coordinate systems (Supplementary Fig. 3a, see Supplementary Information). Each coordinate system provides specific coordinates, such as the relative position along the medial axis of the parent particle or the angle relative to the medial axis of the parent particle. These properties can be used to study the spatial distribution or the dynamics of fluorescent foci inside their parent particle using dedicated subcellular representations such as charts (Supplementary Fig. 3b) or heatmaps (Supplementary Fig. 3c). Also, when fluorescent foci or features are detected and associated with bacterial cells, the sub-cellular localization, the localization type, and specific information (such as the number of foci per cell and their average fluorescence intensity per cell), are automatically computed for each associated particle.
Depending on image quality, most users can simply use the software’s default attribute values to detect bacterial cells with good results. In addition, MicrobeJ permits direct user interaction with the active image(s) during the detection process to optimize cell and subcellular particle detection parameters. Users can systematically change and test the settings, which may vary from experiment to experiment, prior to applying them to a full set of images for final analysis. In addition, the settings can be saved and applied to future image sets.
To improve the accuracy of detection in the final analysis, MicrobeJ has a comprehensive vetting system. The user can easily define a set of specific attributes referred to as ‘morphology’, including but not limited to area, length, width, circularity, curvature, sinuosity, angularity, solidity, and signal intensity, designed to evaluate the particles in the active image. Attribute values are displayed in the image, adjacent to each detected particle. Using the attributes of particles in the image, the user can constrain the attributes in the user interface in order to exclude inappropriate particles or include missed particles upon reanalysis. This strategy can also be used to exclude certain particles, for example filamentous cells, from subsequent analysis. Rejected particles or group of particles can be automatically segmented using the segmentation option. During the segmentation process, a set of methods are applied in parallel on the image in order to segment particles. Each method consists of specific filters (see Supplementary Information) or user-defined ImageJ macros. For each rejected particle, the method maximizing the number of particles that meet the specified morphology attributes is chosen.
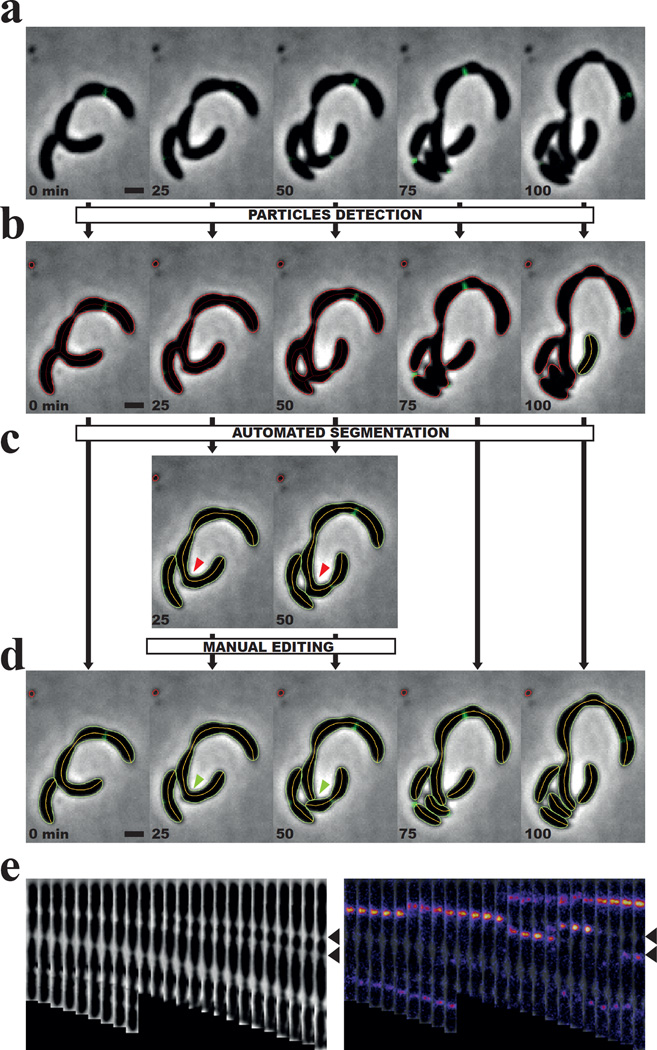
To illustrate the segmentation capabilities of MicrobeJ, we analyzed a micro-colony of C. crescentus cells harboring defects in cell division by time-lapse microscopy (Fig. 4a, Supplementary Video 4&5). Defects in cell division can lead to poorly defined septa or randomly localized division sites, both of which make the segmentation process more challenging (Fig. 4a&b). In this example, we optimized attribute values and used the default set of segmentation methods (see Supplementary Information) in order to segment the rejected particles (Fig. 4c). Error and imprecision in the contour detection or particle segmentation are sometimes observed when particles are in close proximity to each other or when there is poor resolution. In that case, a manual correction can be performed in order to add, edit, or delete any particles detected or missed during the automated detection process. In our example, we manually separated unsegmented cells (Fig. 4d & Supplementary Video 4&5) and then generated the montage of straightened phase contrast and fluorescence images (Fig. 4e).
Figure 4. The automated and manual segmentation processes.
(a–d) Sequence of representative phase contrast images showing several rounds of division from a group of C. crescentus cells harboring division defects. Time points are indicated for each time frame (Scale bar = 1 µm). (b–d) The particle contours computed by MicrobeJ at different steps of the segmentation process: before the automated segmentation process (b), after the automated segmentation process (c), and after the manual correction (d). Rejected particles are shown in red, while accepted particles are shown in green. (c) For clarity, the phase contrast images and their respective particle contours that did not require manual correction, were hidden. The red arrows highlight the cell that needed manual segmentation. (d) The green arrows highlight the result of the manual segmentation. (e) A montage of straightened phase contrast images (left panel) and fluorescence overlays showing the dynamic localization of FtsZ-YFP in a filamentous cell over time. Pictures were taken every 5 min. Black arrows highlight the two constrictions where FtsZ-YFP localizes alternatively until the cell divides.
Once the final image analysis has been completed, MicrobeJ provides a results interface for fast data post-processing workflow and to evaluate the accuracy of the output. This interface combines information about the experiment, the detected particles, and complementary data based on user-selected options (Supplementary Fig. 4a). Although particle measurements can be saved as a .csv file or copied into other programs for subsequent data analysis, the user can easily produce data representations of common statistical analyses (Supplementary Fig. 4b), interactive charts (Supplementary Fig. 4c), fluorescence map profiles (Supplementary Fig. 4d), regression analyses (Supplementary Fig. 4e), or simple custom results tables directly from within the results interface. The major advantage of data post-processing in MicrobeJ is that each row of data for a cell or focus is dynamically linked to the image and to each type of data representation derived from it. Thus, users can rapidly display subsets of data easily across all data representations. Since one major challenge biologists face when using automated image analysis programs lies in determining the accuracy and veracity of the data, exploiting the dynamic link between the images, data, and data representations provides an efficient strategy to verify data points and exclude any from further analysis that appear to be artifacts. Finally, the user can save the list of the generated data representation outputs as a template that can be reused for future image analyses. The use of templates significantly speeds up the creation of frequently compared cellular parameter outputs, simplifying analysis and interpretation of data without specific training in image analysis or programming (Supplementary Video 1). Finally, a batch-processing interface allows consistent analysis across images and experiments (Supplementary Fig. 5). These features facilitate customizable, high-throughput analysis of large image datasets (Supplementary Information).
In summary, we have shown how MicrobeJ can be used to analyze the large amounts of data contained in multi-dimensional image sets. MicrobeJ introduces several new features and concepts for analyzing bacterial cells within a highly customizable user interface. Earlier versions of MicrobeJ were successfully used to analyze different types of bacterial cells, notably to quantify the sub-cellular localization patterns of a developmental regulator and morphogen in the genera Caulobacter and Asticcacaulis11, the spatial dynamics of a versatile surface transporter in Myxococcus xanthus12, as well as the dynamic subcellular localization of a respiratory complex in Escherichia coli13. MicrobeJ is supported by a detailed user’s guide and video tutorials with examples that are available on a dedicated website (http://www.indiana.edu/~microbej/), and will remain freely available and open-source. Additional functionalities will be added over time based on user feedback and new research needs, and the latest releases will be updated on-line. We expect that MicrobeJ will facilitate quantitative image analysis for both novices and experts in image analysis.
MATERIAL AND METHODS
Bacterial Strains, Plasmids, and Growth
C. crescentus, B. subtilis, A. biprosthecum, Rhodomicrobium sp, and P. hirshii were grown in PYE14 at 30°C. A. tumefaciens, S. venezuelae, L. lactis, were grown in LB15 at 30°C and E. coli was grown in LB15 at 37°C. M. xanthus were grown at 32°C in CYE16. S. pneumonia were grown at 37°C in THY17. Rhodopseudomonas palustris CGA009 was grown anaerobically in defined mineral medium (PM)18 supplemented with 10 mM succinate and incubated at 30°C with constant illumination from a 60 W incandescent light bulb.
Phase and fluorescence time-lapse imaging was performed on a Nikon Ti-E inverted microscope, equipped with a Plan Apo 60×, 1.40 NA, Oil, Ph3 DM objective and 1.5× magnifier. Images were acquired every 5 min, and fluorescent proteins were illuminated with a Lumencor Spectra × light engine equipped with excitation filters 470/24 (GFP), 510/25 (YFP) or 575/25 (mCherry), Chroma emission filters 510/40 (GFP), 545/30 (YFP), 530/60 (mCherry) and either a quad polychroic DAPI/FITC/Cy3/Cy5 or triple polychroic CFP/YFP/mCherry cube for Lumencor SpectraX. Images were acquired using an Andor iXon3 DU885 EM CCD camera driven by NIS Elements Advanced Research software (Nikon, Melville, NY)
Cultures from strain YB4667 CB15::pvan-ftsZ-yfp were grown in PYE medium at 30°C and induced for 2 hours with 0.5 mM vanillic acid to express FtsZ-YFP. Exponentially growing cells from this culture were spotted onto a 0.8 mm thick 1% agarose pad made with PYE medium containing 0.5 mM vanillic acid and timelapse images were acquired every 5 minutes from 16 different slide positions for 54 time points. For cell division inhibition, 30 µg/ml of cephalexin was added to the agarose pad during the imaging period.
For precision assessment of MicrobeJ, Molecular Probes FluoSpheres carboxylate-modified microspheres (F8823), 1± 0.0480 µm lot #1761288 were spotted onto a 1% agarose pad made with deionized water and images were acquired for 30 ms using the same microscope, camera and objective as cells.
Supplementary Material
Acknowledgments
We thank all members of the Brun lab for many fruitful discussions. In particular, we thank Breah LaSarre and Courtney Ellison for useful feedback about MicrobeJ. We also thank Dave Kysela and Leon Espinosa for useful discussions and carefully reading the manuscript. We thank Susan Schlimpert, Nate Feirer, Christophe Grangeasse, Thierry Doan, Pam Brown and Breah LaSarre for kindly providing images from S. venezuelae cells, A. tumefaciens, S. pneumonia, B. subtilis, P. hirshii, and R. palustris, respectively. We thank Martin Thanbichler for the pvan-ftsZ-yfp carrying Caulobacter strain. This research was supported by National Institutes of Health grants GM51986 and GM113172, and by seed funding from the Indiana University Office of the Vice-President for Research. This project was supported, in part, by the Indiana Clinical and Translational Sciences Institute funded, in part by Grant Number UL1TR001108 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Footnotes
A.D. wrote the ImageJ plugin; A.D. and Y.V.B. planned the project; A.D. and E.M.Q. performed experiments; A.D., E.M.Q., and Y.V.B. analyzed the data; A.D., E.M.Q., and Y.V.B. wrote the paper.
REFERENCES
- 1.Homepage of ObjectJ. [Accessed: 29th September 2015]; Available at: https://sils.fnwi.uva.nl/bcb/objectj/index.html. [Google Scholar]
- 2.Vischer NOE, et al. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Dazzo FB, Glagoleva O, Yu B, Jain AK. CMEIAS: A Computer-Aided System for the Image Analysis of Bacterial Morphotypes in Microbial Communities. Microb. Ecol. 2001;41:173–194. doi: 10.1007/s002480000004. [DOI] [PubMed] [Google Scholar]
- 4.Mekterović I, Mekterović D, Maglica Z. BactImAS: a platform for processing and analysis of bacterial time-lapse microscopy movies. BMC Bioinformatics. 2014;15:251. doi: 10.1186/1471-2105-15-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christen B, et al. High-throughput identification of protein localization dependency networks. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4681–4686. doi: 10.1073/pnas.1000846107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paintdakhi A, et al. Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol. Microbiol. 2015 doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: Noise and Asymmetry in Bacterial Division Revealed by Computational Image Analysis at Sub-Pixel Resolution. PLOS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Chaumont F, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 11.Jiang C, Brown PJB, Ducret A, Brun1 YV. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature. 2014;506:489–493. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wartel M, et al. A Versatile Class of Cell Surface Directional Motors Gives Rise to Gliding Motility and Sporulation in Myxococcus xanthus. PLoS Biol. 2013;11:e1001728. doi: 10.1371/journal.pbio.1001728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alberge F, et al. Dynamic subcellular localization of a respiratory complex controls bacterial respiration. eLife. 2015;4 doi: 10.7554/eLife.05357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 15.Miller JH, Ippen K, Scaife JG, Beckwith JR. The promoter-operator region of the Lac operon of Escherichia coli. J. Mol. Biol. 1968;38:413–420. doi: 10.1016/0022-2836(68)90395-1. [DOI] [PubMed] [Google Scholar]
- 16.Bustamante VH, Martínez-Flores I, Vlamakis HC, Zusman DR. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol. Microbiol. 2004;53:1501–1513. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- 17.Todd EW, Hewitt LF. A new culture medium for the production of antigenic streptococcal hæmolysin. J. Pathol. Bacteriol. 1932;35:973–974. [Google Scholar]
- 18.Kim M-K, Harwood CS. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol. Lett. 1991;83:199–203. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.