Abstract
Patients diagnosed with triple negative breast cancer (TNBC) have a high rate of tumor metastasis and a poor prognosis. The treatment option for these patients is currently chemotherapy, which results in very low response rates. Strategies that exploit the immune system for the treatment of cancer have now shown the ability to improve survival in several tumor types. Identifying potential targets for immune therapeutic interventions is an important step in developing novel treatments for TNBC. In this study, in-silico analysis of publicly available datasets and immunohistochemical analysis of primary and metastatic tumor biopsies from TNBC patients were conducted to evaluate the expression of the transcription factor brachyury, a driver of tumor metastasis and resistance and a target for cancer vaccine approaches, in TNBC. Analysis of breast cancer datasets demonstrated a predominant expression of brachyury mRNA in TNBC and in basal vs. luminal or HER2 molecular breast cancer subtypes. At the protein level, variable levels of brachyury expression were detected both in primary and metastatic TNBC lesions, and a strong association was observed between nuclear brachyury protein expression and the stage of disease, with nuclear brachyury being more predominant in metastatic vs. primary tumors. Survival analysis also demonstrated an association between high levels of brachyury in the primary tumor and poor prognosis. Two brachyury-targeting cancer vaccines are currently undergoing clinical evaluation; the data presented here provides rationale for utilizing brachyury-targeting immunotherapy approaches for the treatment of TNBC.
Keywords: brachyury, TNBC, tumor antigen, vaccine target
Introduction
Triple negative breast cancer (TNBC), which accounts for approximately 10-15% of all breast tumors, is an aggressive breast cancer subtype defined through biomarker analysis by the absence of estrogen (ER) and progesterone (PR) receptor expression and lack of epidermal growth factor receptor-2 (HER2) overexpression and/or amplification. Due to the absence of expression of these receptors, no effective therapy is currently available for TNBC patients, which typically present a high rate of tumor relapse compared to patients with other subtypes of breast cancer (Carey, et al. 2006; Dent, et al. 2007; Haffty, et al. 2006; O'Brien, et al. 2010). With the recent use of genome-wide expression analysis, breast cancer has also been classified into various molecular subtypes. The initial classification, currently known as the “intrinsic subtypes” and defined by the PAM50 assay, includes the luminal A, luminal B, HER2-enriched and basal-like groups (Perou, et al. 2000; Prat, et al. 2012). Tumors of the basal-like category are usually negative for ER, PR and HER2, and thus overlap with the biomarker-defined TNBC group (Metzger-Filho, et al. 2012). Several reports have now demonstrated that these tumors frequently exhibit features of tumor initiating cells (also designated as cancer stem cells), upregulation of mesenchymal markers and downregulation of epithelial markers, thus suggesting a possible link between the phenomenon of epithelial-mesenchymal transition (EMT) and TNBC (Gupta and Massague 2006; Jeong, et al. 2012; Sarrio, et al. 2008). EMT has been associated with a more invasive or metastatic tumor behavior and with the acquisition of resistance to a variety of anti-cancer therapies, including chemotherapy, radiation, small molecule-targeted therapies and immunotherapy (Creighton, et al. 2009; David, et al. 2016; Fernando, et al. 2016; Hamilton, et al. 2014; Huang, et al. 2013; Kalluri and Weinberg 2009; Larocca, et al. 2013; Thiery and Sleeman 2006).
Brachyury (encoded by the gene T) (Kispert, et al. 1995) is a transcription factor that induces EMT in human carcinoma cells (Fernando, et al. 2010), and whose expression has been reported to associate with poor prognosis in several human cancers including prostate (Pinto, et al. 2014), lung (Haro, et al. 2013), colon (Kilic, et al. 2011), and breast (Palena, et al. 2014b; Shao, et al. 2015), among others. Our group has previously reported on an increased proportion of brachyury-expressing breast carcinomas negative for ER and PR (Palena et al. 2014b), however, a limited sample number precluded the evaluation of brachyury in the TNBC subgroup. In the present study, the expression of brachyury was analyzed in primary and metastatic TNBC tumors utilizing both in-silico and immunohistochemical analyses. Increased frequency of samples positive for brachyury mRNA expression was observed in TNBC compared to other breast cancer subtypes. Furthermore, brachyury mRNA expression levels inversely correlated with the levels of mRNA encoding for ER-α (ESR1) and PR (PGR) receptors. Immunohistochemical analysis of primary and metastatic TNBC biopsies also demonstrated variable levels of brachyury protein expression in primary and metastatic TNBC lesions. Furthermore, a strong association was observed between nuclear brachyury expression and stage of disease, with nuclear brachyury being also predominant in metastatic vs. primary TNBC tumors. An association was also observed between high levels of brachyury in primary tumors and poor prognosis.
There are currently two brachyury-targeting cancer vaccines undergoing clinical evaluation (Heery, et al. 2015b); www.clinicaltrials.gov/ct2/show/NCT02179515). The data presented here provides rationale for utilizing brachyury-targeting immunotherapy approaches for the treatment of TNBC.
Methods
Tumor cell lines
Tumor cell lines were purchased from the American Tissue Culture Collection (ATCC), which uses STR analysis for identity verification. Cells were grown in DMEM medium (Corning) supplemented with 10% fetal bovine serum (Gemini Bio-Products), 1X antibiotic/antimycotic solution (Corning) and 4μg/mL recombinant human insulin (Gibco). Cell lines were used for experiments at the following passages from purchased stocks: MDA-MB-231, passage 6; MDA-MB-436, passage 4; Hs 578T, passage 6; BT-474, passage 2; ZR-75-1, passage 4; and MCF7, passage 1.
Patients and tissue collection
Seventy-seven patients with histologically diagnosed primary TNBC and ten patients with metastatic TNBC were enrolled in the Inter-Institutional Multidisciplinary BioBank of the Biomarker Discovery and Advanced Technologies (BioDAT) Laboratory, IRCCS San Raffaele Pisana, Rome, Italy. In collaboration with the Surgical and Pathology Departments of San Giovanni Addolorata Hospital and the Medical Oncology Unit of the “Tor Vergata” Clinical Center, Rome, Italy, primary breast and metastatic tumor tissue samples were collected at the time of surgery. Patient characteristics are provided in Table 1. Clinical follow up data was available from 58/77 patients with primary TNBC. Patients received the following treatment regimens: 30/58 received anthracycline-based therapy; 4/58 received non-anthracycline-based regimens (CMF); 5/58 received adjuvant radiotherapy alone; 8/58 did not receive treatment; not treatment data was available for the remaining 11/58 patients. Informed consent was obtained from each participating subject; the study was performed under the appropriate institutional ethics approvals and in accordance with the principles embodied in the Declaration of Helsinki.
Table 1.
Patient characteristics.
| Overall population (n=87) | Follow-up population (n=58) | |
|---|---|---|
| Primary breast cancer (n=77) | ||
| Age, y (Mean ± SD) | 63 ± 16 | 63 ± 16 |
| Menopausal status | ||
| Pre | 27 (35%) | 21 (36%) |
| Post | 50 (65%) | 37 (64%) |
| Pathological diagnosis | ||
| Infiltrating ductal carcinoma | 70 (91%) | 54 (93%) |
| Infiltrating lobular carcinoma | 5 (6%) | 2 (3.5%) |
| Others* | 2 (3%) | 2 (3.5%) |
| Grading | ||
| 1 | 0 (0%) | 0 (0%) |
| 2 | 0 (0%) | 0 (0%) |
| 3 | 77 (100%) | 58 (100%) |
| Stage | ||
| I | 28 (36%) | 21 (36%) |
| II | 27 (35%) | 24 (42%) |
| III | 20 (26%) | 13 (22%) |
| IV | 2 (3%) | 0 (0%) |
| p53 expression | ||
| Negative | 41 (53%) | 26 (45%) |
| Positive | 36 (47%) | 32 (55%) |
| Ki67 expression | ||
| Median (range) | 70 (40 – 90) | 56 (40 – 90) |
| Positive, n (%)** | 76 (99%) | 57 (98%) |
| Metastatic breast cancer (n=10) | ||
| Age, y (Mean ± SD) | 63 ± 17 | |
| Menopausal status | ||
| Pre | 4 (40%) | |
| Post | 6 (60%) | |
| Pathological diagnosis | ||
| Infiltrating ductal carcinoma | 9 (90%) | |
| Infiltrating lobular carcinoma | 1 (10%) | |
| p53 expression | ||
| Negative | 8 (80%) | |
| Positive | 2 (20%) | |
| Ki67 expression | ||
| Median (range) | 63 (50 – 80) | |
| Positive, n (%)** | 9 (90%) | |
Includes a case of comedocarcinoma and a metaplastic breast cancer.
Ki67 positivity defined as ≥20
Immunohistochemical detection of brachyury
Sections of formalin-fixed, paraffin-embedded tissues were evaluated for brachyury expression by using a rabbit monoclonal anti-brachyury antibody (MAb 54-1) at a 1:500 dilution (Hamilton, et al. 2015). Staining was performed on the Ventana BenchMark XT automated staining platform with the UltraView Universal DAB Detection Kit (Roche) according to manufacturer's instructions. Two pathologists independently evaluated the tumor and normal tissue samples in a blinded, randomized manner. For each slide, three to five random fields were evaluated; for each field, the percentage of positive tumor cells was calculated as: [(number of positive tumor cells/total number of tumor cells) x 100]. Nuclear, cytoplasmic and total brachyury staining were independently scored, with brachyury being observed either in the nucleus, the cytosol or both compartments of the tumor cells. For calculation of total brachyury expression (Table 2), the percentage of tumor cells with mutually exclusive cytosolic or nuclear staining was added; tumor cells showing brachyury in both compartments were scored only once. The relative staining intensity was scored as weak (+) for pale brown intensity, moderate (++) for intermediate brown intensity, and strong (+++) for intense, dark brown immunoprecipitate. Immunoreactivity index was calculated by multiplying the percentage of positive cells by the staining intensity. For normal tissues, the percentage of reactivity was individually evaluated for each cell type and calculated as: [(number of positive cells/total number of cells of the same type) x 100].
Table 2.
Immunohistochemical detection of brachyury in biopsies obtained from primary lesions of TNBC patients.
| Nuclear brachyury | Cytoplasmic brachyury | Total brachyury | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pt | Stage | Ki67 | p53 | Pos (%) | SI | Index | Pos (%) | SI | Index | Pos (%) | SI | Index |
| 1 | I | 90 | NEG | 100 | +++ | 300 | 70 | ++ | 140 | 100 | +++ | 300 |
| 2 | I | 25 | POS | 100 | +++ | 300 | 100 | ++ | 200 | 100 | +++ | 300 |
| 3 | I | 35 | POS | 70 | +++ | 210 | 100 | ++ | 200 | 100 | +++ | 300 |
| 4 | I | 60 | POS | 60 | +++ | 180 | 80 | ++ | 160 | 90 | +++ | 270 |
| 5 | I | 80 | NEG | 90 | +++ | 270 | 10 | + | 10 | 90 | +++ | 270 |
| 6 | I | 60 | NEG | 20 | ++ | 40 | 80 | +++ | 240 | 80 | +++ | 240 |
| 7 | I | 80 | NEG | 80 | ++ | 160 | 50 | + | 50 | 80 | ++ | 160 |
| 8 | I | 90 | POS | 20 | + | 20 | 80 | + | 80 | 80 | + | 80 |
| 9 | I | 90 | POS | 30 | + | 30 | 80 | ++ | 160 | 80 | ++ | 160 |
| 10 | I | 40 | NEG | 70 | ++ | 140 | 10 | + | 10 | 70 | ++ | 140 |
| 11 | I | 70 | POS | 50 | +++ | 150 | 70 | + | 70 | 70 | +++ | 210 |
| 12 | I | 20 | POS | 40 | +++ | 120 | 60 | ++ | 120 | 70 | +++ | 210 |
| 13 | I | 70 | NEG | 20 | + | 20 | 70 | ++ | 140 | 70 | ++ | 140 |
| 14 | I | 20 | NEG | 70 | +++ | 210 | 70 | + | 70 | 70 | +++ | 210 |
| 15 | I | 35 | POS | 60 | +++ | 180 | 20 | + | 20 | 60 | +++ | 180 |
| 16 | I | 40 | POS | 60 | ++ | 120 | 40 | + | 40 | 60 | ++ | 120 |
| 17 | I | 60 | POS | 20 | + | 20 | 50 | + | 50 | 50 | + | 50 |
| 18 | I | 80 | NEG | 20 | ++ | 40 | 50 | + | 50 | 50 | ++ | 100 |
| 19 | I | 90 | POS | 40 | ++ | 80 | 40 | + | 40 | 40 | ++ | 80 |
| 20 | I | 50 | NEG | 40 | ++ | 80 | 10 | + | 10 | 40 | ++ | 80 |
| 21 | I | 25 | NEG | 10 | + | 10 | 40 | + | 40 | 40 | + | 40 |
| 22 | I | 40 | NEG | 25 | ++ | 50 | 10 | + | 10 | 30 | + | 60 |
| 23 | I | 40 | NEG | 15 | ++ | 30 | 20 | + | 20 | 25 | + | 50 |
| 24 | I | 70 | POS | 15 | + | 15 | 15 | + | 15 | 20 | + | 20 |
| 25 | I | 80 | POS | 20 | + | 20 | 10 | + | 10 | 20 | + | 20 |
| 26 | I | 40 | POS | 20 | ++ | 40 | 0 | - | 0 | 20 | ++ | 40 |
| 27 | I | 70 | NEG | 10 | ++ | 20 | 10 | + | 10 | 15 | ++ | 30 |
| 28 | I | 90 | POS | 0 | - | 0 | 0 | - | 0 | 0 | - | 0 |
| 29 | IIA | 25 | POS | 100 | ++ | 200 | 10 | + | 10 | 100 | ++ | 200 |
| 30 | IIA | 90 | POS | 80 | +++ | 240 | 80 | ++ | 160 | 100 | +++ | 300 |
| 31 | IIA | 60 | POS | 20 | +++ | 60 | 100 | ++ | 200 | 100 | +++ | 300 |
| 32 | IIA | 90 | POS | 80 | +++ | 240 | 100 | +++ | 300 | 100 | +++ | 300 |
| 33 | IIA | 40 | POS | 100 | +++ | 300 | 10 | ++ | 20 | 100 | +++ | 300 |
| 34 | IIA | 45 | POS | 30 | + | 30 | 100 | ++ | 200 | 100 | ++ | 200 |
| 35 | IIA | 85 | POS | 90 | +++ | 270 | 20 | + | 20 | 90 | +++ | 270 |
| 36 | IIA | 90 | POS | 80 | +++ | 240 | 60 | + | 60 | 90 | +++ | 270 |
| 37 | IIA | 90 | NEG | 70 | +++ | 210 | 80 | + | 80 | 80 | +++ | 240 |
| 38 | IIA | 25 | NEG | 80 | +++ | 240 | 10 | + | 10 | 80 | +++ | 240 |
| 39 | IIA | 70 | POS | 30 | ++ | 60 | 80 | + | 80 | 80 | ++ | 160 |
| 40 | IIA | 40 | POS | 80 | ++ | 160 | 50 | ++ | 100 | 80 | ++ | 160 |
| 41 | IIA | 90 | NEG | 70 | +++ | 210 | 20 | ++ | 40 | 70 | +++ | 210 |
| 42 | IIA | 35 | POS | 50 | ++ | 100 | 30 | + | 30 | 50 | ++ | 100 |
| 43 | IIA | 60 | NEG | 25 | ++ | 50 | 10 | + | 10 | 30 | ++ | 60 |
| 44 | IIA | 70 | POS | 10 | + | 10 | 0 | - | 0 | 10 | + | 10 |
| 45 | IIA | 60 | POS | 10 | + | 10 | 0 | - | 0 | 10 | + | 10 |
| 46 | IIA | 90 | NEG | 10 | + | 10 | 0 | - | 0 | 10 | + | 10 |
| 47 | IIA | 25 | NEG | 5 | + | 5 | 0 | - | 0 | 5 | + | 5 |
| 48 | IIB | 35 | POS | 75 | ++ | 150 | 100 | + | 100 | 100 | ++ | 200 |
| 49 | IIB | 20 | POS | 20 | ++ | 40 | 80 | + | 80 | 80 | + | 160 |
| 50 | IIB | 60 | NEG | 40 | +++ | 120 | 70 | ++ | 140 | 70 | +++ | 210 |
| 51 | IIB | 80 | NEG | 70 | +++ | 210 | 30 | + | 30 | 70 | +++ | 210 |
| 52 | IIB | 90 | NEG | 50 | ++ | 100 | 50 | + | 50 | 60 | ++ | 120 |
| 53 | IIB | 80 | NEG | 60 | ++ | 120 | 20 | + | 20 | 60 | ++ | 120 |
| 54 | IIB | 90 | POS | 30 | +++ | 90 | 20 | ++ | 40 | 40 | +++ | 120 |
| 55 | IIB | 90 | NEG | 30 | + | 30 | 30 | + | 30 | 30 | + | 30 |
| 56 | IIIA | 90 | POS | 80 | +++ | 240 | 100 | + | 100 | 100 | +++ | 300 |
| 57 | IIIA | 90 | NEG | 100 | +++ | 300 | 0 | - | 0 | 100 | +++ | 300 |
| 58 | IIIA | 10 | NEG | 90 | +++ | 270 | 50 | ++ | 100 | 90 | +++ | 270 |
| 59 | IIIA | 30 | NEG | 90 | + | 90 | 30 | + | 30 | 90 | + | 90 |
| 60 | IIIA | 60 | NEG | 80 | ++ | 160 | 60 | ++ | 120 | 90 | ++ | 180 |
| 61 | IIIA | 80 | NEG | 70 | +++ | 210 | 60 | + | 60 | 80 | +++ | 240 |
| 62 | IIIA | 70 | NEG | 70 | +++ | 210 | 10 | + | 10 | 70 | +++ | 210 |
| 63 | IIIA | 70 | NEG | 40 | + | 40 | 60 | + | 60 | 60 | + | 60 |
| 64 | IIIA | 50 | NEG | 50 | ++ | 100 | 10 | + | 10 | 50 | ++ | 100 |
| 65 | IIIA | 60 | NEG | 40 | + | 40 | 15 | + | 15 | 45 | + | 45 |
| 66 | IIIB | 95 | POS | 40 | +++ | 120 | 100 | + | 100 | 100 | ++ | 200 |
| 67 | IIIB | 35 | NEG | 100 | +++ | 300 | 50 | ++ | 100 | 100 | +++ | 300 |
| 68 | IIIB | 70 | NEG | 100 | +++ | 300 | 30 | + | 30 | 100 | ++ | 200 |
| 69 | IIIB | 80 | NEG | 100 | +++ | 300 | 10 | + | 10 | 100 | +++ | 300 |
| 70 | IIIB | 80 | NEG | 90 | +++ | 270 | 20 | + | 20 | 90 | +++ | 270 |
| 71 | IIIB | 40 | POS | 80 | +++ | 240 | 40 | + | 40 | 80 | +++ | 240 |
| 72 | IIIB | 50 | POS | 60 | ++ | 120 | 60 | ++ | 120 | 80 | ++ | 160 |
| 73 | IIIB | 80 | NEG | 70 | ++ | 210 | 70 | + | 70 | 80 | +++ | 240 |
| 74 | IIIB | 90 | NEG | 50 | +++ | 150 | 50 | + | 50 | 50 | +++ | 150 |
| 75 | IIIB | 90 | POS | 30 | + | 30 | 30 | + | 30 | 40 | + | 40 |
| 76 | IV | 90 | NEG | 30 | ++ | 60 | 40 | + | 40 | 50 | ++ | 100 |
| 77 | IV | 80 | NEG | 15 | +++ | 45 | 10 | + | 10 | 10 | +++ | 30 |
SI: Staining Intensity; NEG: negative; POS: positive.
In-silico analysis of the TCGA dataset
Relative expression levels of indicated mRNAs were assessed utilizing the TCGA dataset containing data from 1026 breast carcinoma patients (http://cancergenome.nih.gov). For the analysis, breast cancer samples were subdivided into three groups according to the level of brachyury (T) expression: 881/1026 samples with no detectable brachyury expression were classified as negative (Neg), while the remaining 145 samples were ranked and subdivided into “brachyury-high” (High, 73/1026) and “brachyury-low” (Low, 72/1026) groups based on an arbitrary cutoff set at the median value for the 145 samples. The level of expression of mRNA encoding for ER-α (ESR1), ER-β (ESR2), PR (PGR) and HER2 (ERBB2) were evaluated in each group. A subset of tumors in the database (n=513) for which data was available regarding the four intrinsic subtypes using the PAM50 discriminator assay (Parker, et al. 2009) was further assessed for brachyury expression (Cancer Genome Atlas 2012). Samples also were subdivided into subgroups according to ER, PR and HER2 status for comparison of the levels of mRNAs encoding for brachyury, various EMT-transcription factors, and the chemokine IL-8. Samples were classified as triple positive (ER+, PR+, HER2+), TNBC (ER−, PR− and HER2−), and non-TNBC, the latter corresponding to tumors that were at least positive for one marker (ER, PR or HER2). All data were analyzed utilizing the Nexus Expression 3 analysis software package (BioDiscovery).
Immunofluorescence
Tumor cells were cultured in 96-well black, clear-bottom plates (Greiner Bio-One). Following fixation with 3% paraformaldehyde (Electron Microscopy sciences), permeabilization with 0.05% TritonX, and blockade in 1X PBS supplemented with 1% BSA and 10% goat serum, cells were stained using antibodies reactive against vimentin (Dako), fibronectin, ZO1 (BD Biosciences), and brachyury (MAb 54-1), and Alexa Fluor 488 anti-mouse or anti-rabbit secondary antibodies. Nuclei were stained using DAPI (Thermo Fisher Scientific), and images were acquired using a Celigo S Cell Imaging Cytometer (Nexcelom Bioscience). For silencing of brachyury expression, control and brachyury-targeting ON-TARGETplus SMARTpool siRNAs were purchased from Dharmacon and used according to the manufacturer's instructions (GE Lifesciences). Cells were incubated for 72 hours in antibiotic-free medium prior to use for analysis of various markers by immunofluorescence.
Quantitative real-time PCR
Total RNA was prepared using the PureLink RNA Mini Kit (Thermo Fisher Scientific) and reverse-transcribed with the XLAScript cDNA MasterMix (WordWide Life Sciences). The resulting cDNA (10ng) was amplified in triplicate with the following TaqMan human gene expression assays (Life Technologies): T (brachyury) (Hs00610080_m1) and GAPDH (4326317E) using a 7300 Applied Biosystems instrument. Expression of brachyury relative to GAPDH was calculated as 2-(Ct(GAPDH) – Ct(target gene)).
Statistical methods
The unpaired two-sample Student's t-test was used for comparison of mean expression levels of indicated transcripts in samples in the TCGA dataset. For survival analysis, samples for which clinical follow up was available (n=58) were assigned into a low vs. high brachyury group, based on an arbitrary cutoff of 240 for the overall brachyury reactivity index, set at the 75th percentile for the overall population (n=87). A Kaplan-Meier analysis was used to evaluate the association between brachyury expression and relapse-free survival. Cox Proportional-Hazards regression was used to adjust for potential confounding factors such as stage of disease and p53 expression. All statistical tests were two-sided.
Results
Brachyury mRNA expression in TNBC
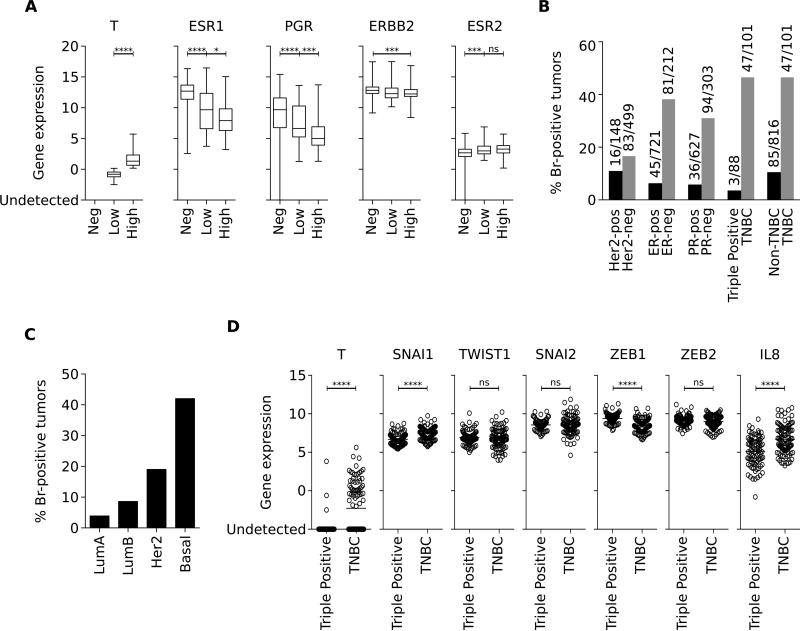
Samples (n=1026) from the TCGA breast cancer database were subdivided according to the level of brachyury (T) mRNA expression into brachyury negative (n=881), low (n=72) or high (n=73) groups, and subsequently analyzed for the level of expression of ER, PR and HER2 receptor-encoding mRNAs. The most significant associations were observed with ER-α and PR-encoding mRNAs (ESR1 and PGR, respectively), which demonstrated a marked decrease of expression with increasing levels of brachyury (Figure 1A). This reverse association, however, was not seen in relation to HER2 or ER-β encoding mRNAs (ERBB2 and ESR2, respectively, Figure 1A). Further analysis of samples subdivided according to the HER2, ER and PR status demonstrated a predominant expression of brachyury mRNA among ER− vs. ER+ (38% vs. 6%), PR− vs. PR+ (31% vs. 6%), TNBC vs. triple positive (47% vs. 3%), or TNBC vs. non-TNBC (47% vs. 10%) samples. Unlike with ER and PR, no difference was observed regarding the percentage of brachyury positive tumors subdivided based on their HER2 expression (Figure 1B). Brachyury mRNA expression was also evaluated in samples from the same dataset stratified based on the POM50 gene signature (Cancer Genome Atlas 2012; Parker et al. 2009). As shown in Figure 1C, brachyury mRNA was detectable in 9/232 (3.9%) Luminal A, 11/128 (8.6%) Luminal B, 11/58 (19.0%) HER2-enriched, and 40/95 (42.1%) basal breast carcinoma samples.
Figure 1. Brachyury mRNA upregulation in TNBC and basal breast cancers.
(A) Analysis of expression of indicated transcripts in the breast cancer TCGA dataset, according to the level of brachyury (T) mRNA expression. Percent of tumors in the TCGA database that are positive for brachyury (T) mRNA expression in samples classified based on their hormone receptor expression (B) or according to the molecular classification (C). Analysis of expression of indicated transcripts in breast cancer samples of the TCGA database, classified as triple positive vs. TNBC, in accordance with their expression of hormone receptors (D). Statistics were calculated using an unpaired t- test, [* p<0.05, *** p<0.001, **** p<0.0001, ns: not significant].
In addition to brachyury, the TCGA dataset was used to interrogate the expression of various transcription factors known to induce EMT. While brachyury (T) mRNA was predominantly expressed in TNBC (47/101) vs. triple positive (3/88) breast carcinoma samples, only a slight increase of SNAI1, a decrease of ZEB1, and no changes in the mRNA levels of TWIST1, SNAI2, or ZEB2 were observed in TNBC vs. triple positive breast tumors (Figure 1D). Interestingly, we also noted a significant increase in IL-8 mRNA levels in TNBC vs. triple positive breast tumors (Figure 1E), a result in support of our previous studies demonstrating a positive association between brachyury and the IL-8 axis in breast cancer (Fernando, et al. 2011).
Brachyury protein in primary and metastatic TNBC
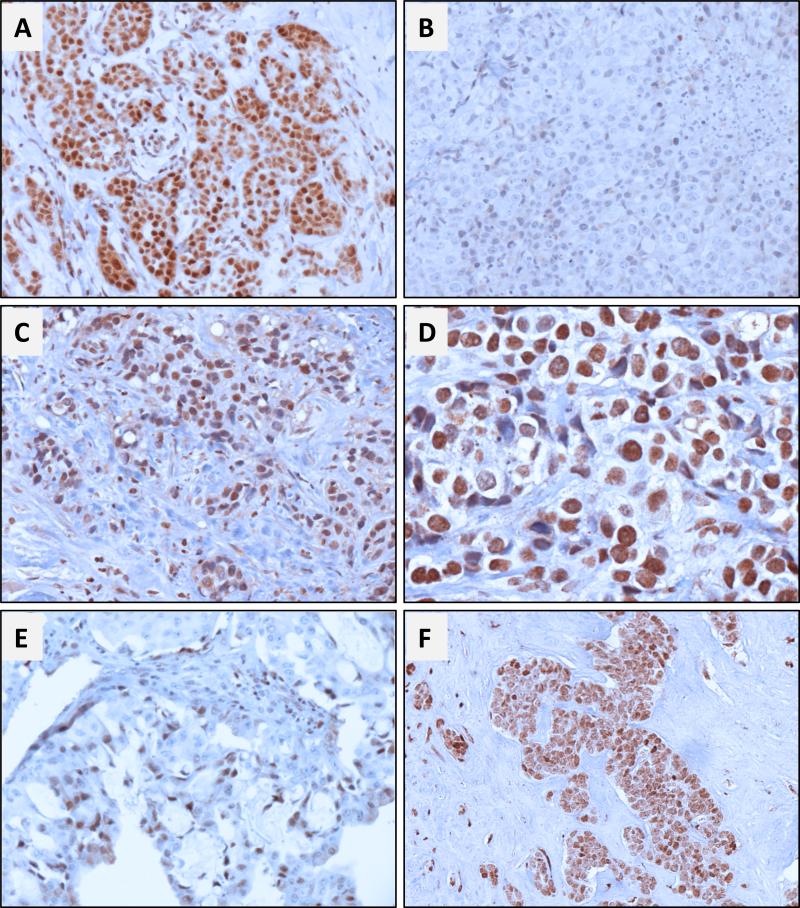
Brachyury protein expression was assessed by IHC in 87 tumor tissues from patients diagnosed with TNBC by using a rabbit monoclonal anti-brachyury antibody (MAb 54-1) previously described (Hamilton et al. 2015). Among 77 primary TNBC tumors, 71/77 (92%) showed some level of brachyury expression in >10% of the cancer cells, either in the nucleus or the cytoplasmic compartment (Table 2). As previously reported with other types of carcinomas, expression of brachyury was highly variable among primary tumors, with the percentage of brachyury positive cells ranging from 10% to 100% and the intensity of staining varying between (+) and (+++). Representative images of two primary TNBC cases positive vs. negative for brachyury expression are shown in Figures 2A and B, respectively. In addition to primary tumors, brachyury expression was evaluated in metastatic lesions from 10 patients with TNBC (Table 3 and representative images in Figures 2 C-F). Brachyury was observed in 100% of metastasis, the majority of cases showing high intensity staining for brachyury (+++) in 40% to 100% of the tumor cells.
Figure 2. Immunohistochemical analysis of brachyury expression in primary and metastatic TNBC.
Representative photomicrographs of brachyury staining on primary TNBC cases are reported on panels A and B. (A) Stage IIIB T2N3aM0 primary IDC (patient 71) showing positive nuclear and cytoplasmic staining in 80% and 40% of cancer cells, respectively. (B) Stage I T1cN0M0 primary IDC (patient 28) showing negative (<5%) nuclear and cytoplasmic staining in cancer cells. (C) Pleural metastatic lesion (patient 84) showing positive nuclear and cytoplasmic staining in 80% and 20% of cancer cells, respectively, and (D) detail of brachyury staining at higher magnification (400X). (E) and (F) represent bone (patient 86) and lymph node (patient 78) metastasis with positive nuclear (60% and 100%, respectively) and cytoplasmic (70% and 100%, respectively) brachyury expression in cancer cells. Magnification: 200X (except panel D: 400X).
Table 3.
Immunohistochemical detection of brachyury in biopsies obtained from metastatic lesions of TNBC patients.
| Nuclear brachyury | Cytoplasmic brachyury | Total brachyury | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pt | Site | Ki67 | p53 | Pos (%) | SI | Index | Pos (%) | SI | Index | Pos (%) | SI | Index |
| 78 | LN | 30 | NEG | 100 | +++ | 300 | 100 | + | 100 | 100 | +++ | 300 |
| 79 | LN | 90 | NEG | 90 | +++ | 270 | 50 | + | 50 | 100 | +++ | 300 |
| 80 | LN | 65 | POS | 80 | +++ | 240 | 0 | - | 0 | 80 | +++ | 240 |
| 81 | LN | 50 | NEG | 70 | +++ | 210 | 50 | + | 50 | 80 | +++ | 240 |
| 82 | LN | 70 | NEG | 80 | +++ | 240 | 0 | - | 0 | 80 | +++ | 240 |
| 84 | Dist | 90 | NEG | 80 | +++ | 240 | 20 | + | 20 | 80 | +++ | 240 |
| 86 | Dist | 60 | NEG | 60 | ++ | 120 | 70 | ++ | 140 | 80 | ++ | 160 |
| 83 | Dist | 50 | NEG | 60 | +++ | 180 | 40 | ++ | 80 | 70 | +++ | 210 |
| 87 | Dist | 10 | POS | 60 | +++ | 180 | 20 | + | 20 | 60 | +++ | 180 |
| 85 | Dist | 80 | NEG | 30 | ++ | 60 | 20 | + | 20 | 40 | ++ | 80 |
SI: Staining Intensity; NEG: negative; POS: positive. LN: lymph node; Dist: distant metastasis
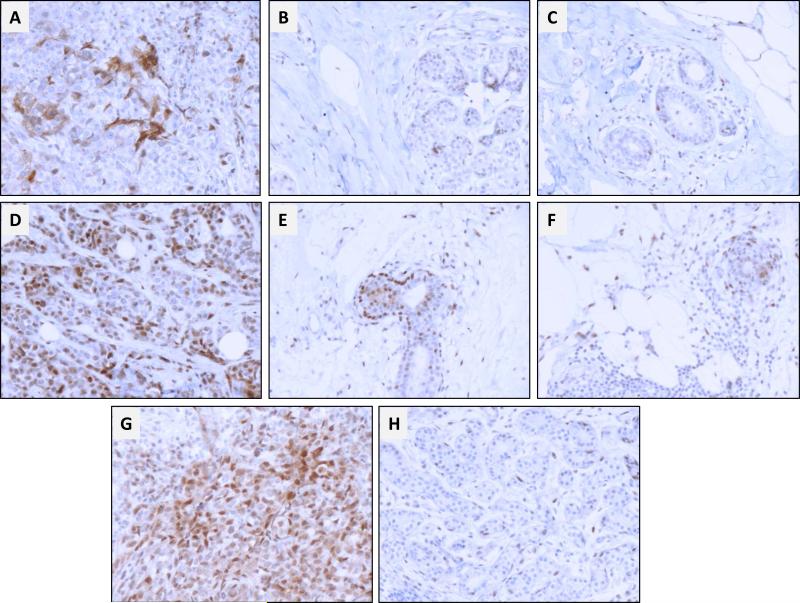
Expression of brachyury was also evaluated in breast tissues adjacent and distant to the tumor. As shown in Figure 3, a gradient of expression was observed with the highest degree of brachyury positivity detected in the tumor > adjacent breast > distant breast tissue. In the examples shown in Figures 3A and D, the primary tumor overall nuclear and cytoplasmic brachyury staining ranged from 20% to 80% of the cancer cells, while expression in breast tissues adjacent vs. distal to tumor corresponded to approximately 10% (Figure 3B and E) and <5% (Figure 3C and F) of the cells, respectively. Similarly, brachyury expression in the primary tumor shown in Figure 3G corresponded to 80% of the cells, while expression in histologically normal breast tissue from the same patient demonstrated <1% positivity for brachyury (Figure 3H). These results are in agreement with our previous study utilizing a different anti-brachyury monoclonal antibody (Palena et al. 2014b), where brachyury-positive cells were observed in the breast tissue adjacent (15/27 cases) but not distal to the tumor, while no expression was detected in 14 benign breast tissues, with the exception of two fibroadenoma cases in which focal expression of brachyury was observed. At present, it is unknown whether brachyury-expressing cells in the tissues adjacent to the tumor are cancer cells that have migrated towards the surrounding stroma, or correspond to normal stromal cells that upregulated brachyury in response to tumor-derived secreted factors.
Figure 3. Immunohistochemical analysis of brachyury expression in primary TNBC and normal breast tissues.
(A) Stage IIB T2N1aM0 primary tumor (patient 54) showing positive nuclear and cytoplasmic staining in 30% and 20% of cancer cells, respectively; (D) Stage IIA T2N0M0 primary tumor (patient 37) showing nuclear and cytoplasmic staining in 70% and 80% of cancer cells, respectively. Panels (B) and (E) correspond to brachyury staining in breast tissues adjacent to the tumor; panels (C) and (F) correspond to brachyury staining in breast tissues distant from the tumor. (G) Stage IIA T2N0M0 primary tumor (patient 30) showing both nuclear and cytoplasmic staining in 80% of cancer cells. (H) Brachyury staining in histologically normal breast tissue surrounding the tumor. Panel Magnification: 200X.
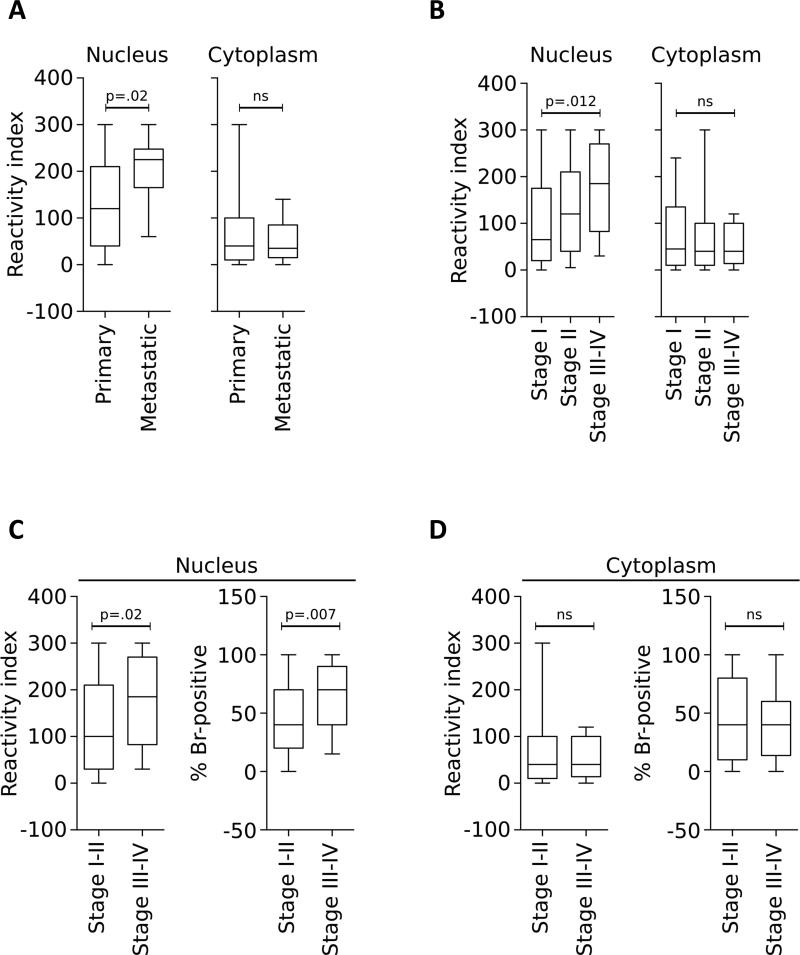
In agreement with previous studies, brachyury protein was observed either in the nucleus and/or the cytosol of the TNBC cells. A comparative analysis of brachyury expression demonstrated a significant increase in nuclear brachyury protein levels, as detected by an increased reactivity index, in metastatic vs. primary tumor samples (p=0.02, Figure 4A), an association that was not observed with cytoplasmic brachyury. A similar association between nuclear localization of brachyury and clinical stage of disease was also observed, with samples from higher tumor stages (III-IV) having higher nuclear brachyury reactivity index (p=0.012) than samples of stage I (Figure 4B). The same difference was observed when combined samples from tumors stages I-II were compared to those of stages III-IV, where nuclear expression of brachyury was significantly higher in tumors of advanced stages, both in terms of reactivity index (p=0.02) and % positive nuclei (p=0.007, Figure 4C), a correlation that was not observed with cytoplasmic brachyury expression (Figure 4D).
Figure 4. Prevalence of nuclear brachyury expression in metastatic and high-grade TNBC tumors.
Brachyury reactivity index observed in the nucleus or the cytoplasm of primary vs. metastatic (A) or TNBC samples according to stage (B-D). Samples were analyzed by IHC with an anti-brachyury monoclonal antibody. P values were calculated using an unpaired t-test, with values for panel B representing a comparison between Stage I and Stages III-IV. [ns: not significant].
Brachyury and TNBC prognosis
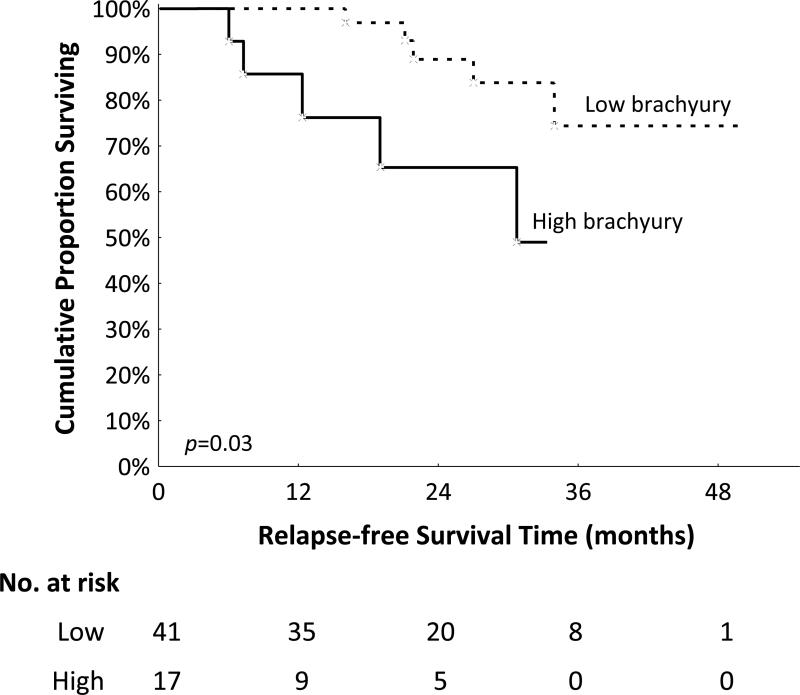
In order to evaluate a potential association of brachyury expression in primary TNBC with prognosis, samples for which clinical follow up was available (n=58) were assigned into a low vs. high brachyury group, based on an arbitrary cutoff of 240 for the overall brachyury reactivity index, set at the 75th percentile for the overall population (n=87). A Kaplan Meier estimate of survival showed that high brachyury expression in the primary tumor significantly associated with decreased survival (p=0.03, n=58, Long-rank test=2.17, Figure 5). To minimize the effect of clinical-pathological variables that might cause false positive association between brachyury and poor prognosis, a multivariate Cox Proportional-Hazards Regression survival analysis was conducted. It was found that high brachyury expression (reactivity index ≥240) significantly associates with low relapse-free survival (p=0.047, n=58, COXPH, HR=3.92, CI=1.02–15.1), compared to the low brachyury expression group (Table 4).
Figure 5. Brachyury expression and prognosis.
Kaplan-Meier estimates of recurrence-free survival in 58 cases of TNBC classified based on the brachyury expression level. The two groups (low vs. high brachyury) were defined based on the overall brachyury reactivity index set at a cutoff of 240, corresponding to the 75th percentile of expression in the overall population (n=87).
Table 4.
Multivariate Cox Proportional-Hazards Regression analysis of the predictive value of clinical-pathological variables and brachyury expression on relapse-free survival of TNBC patients.
| Recurrence |
|||||
|---|---|---|---|---|---|
| Variable | n | Yes | No | HR (CI) | p value |
| Stage of disease | |||||
| I | 21 | 2 (10%) | 19 (90%) | ||
| II | 24 | 4 (17%) | 20 (83%) | ||
| III | 13 | 4 (31%) | 9 (69%) | 1.83 (0.60 – 5.57) | 0.285 |
| p53 expression | |||||
| Negative | 26 | 4 (15%) | 22 (85%) | ||
| Positive | 32 | 6 (19%) | 26 (81%) | 1.34 (0.30 – 5.91) | 0.699 |
| Brachyury expression* | |||||
| Negative (<240) | 41 | 5 (12%) | 36 (88%) | ||
| Positive (≥240) | 17 | 5 (29%) | 12 (71%) | 3.92 (1.02 – 15.1) | 0.047 |
Categorized according to an arbitrary cutoff calculated as the 75th percentile of brachyury reactivity index for total brachyury staining in the overall population (n=87).
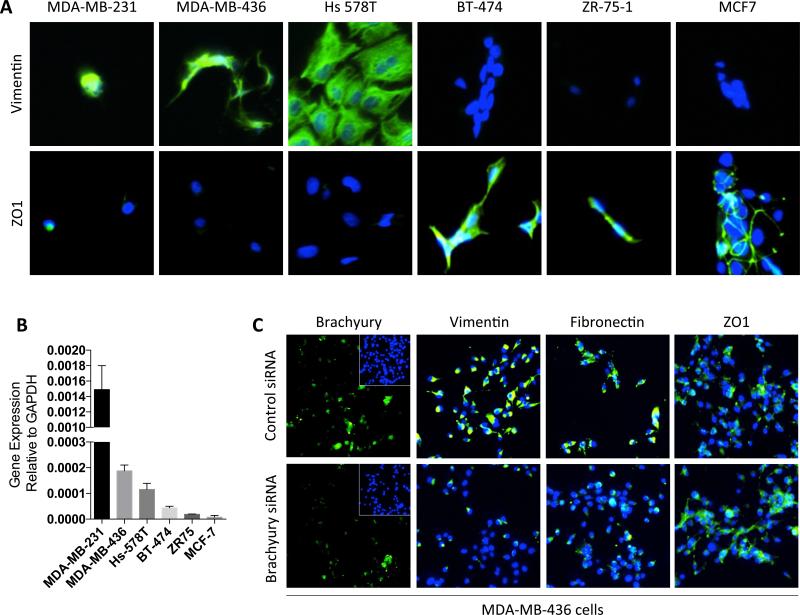
Brachyury expression in TNBC cell lines
Immunofluorescent staining of mesenchymal vimentin and epithelial ZO1 proteins was used for phenotypic confirmation of a panel of TNBC (MDA-MB-231, MDA-MB-436, and Hs 578T) and non-TNBC (BT-474, ZR 75-1, and MCF7) human breast carcinoma lines. As expected, strong expression of mesenchymal vimentin and little expression of epithelial ZO1 were observed with TNBC cells, while non-TNBC cells showed high amounts of ZO1 in the absence of vimentin expression (Figure 6A). When expression of brachyury mRNA was evaluated in each cell line, higher levels were observed in TNBC vs. non-TNBC cells (Figure 6B). The potential role of brachyury in tumor EMT was also investigated with MDA-MB-436 cells, where silencing of brachyury resulted in reduced expression of vimentin and fibronectin and increased expression of epithelial ZO1 (Figure 6C). These observations thus suggested that brachyury not only associates with, but is also required to maintain a mesenchymal-like phenotype in the MDA-MB-436 triple-negative breast cancer cell line.
Figure 6. Brachyury associates with mesenchymal features in human TNBC cell lines.
(A) Immunofluorescent detection of vimentin and ZO1 (green signal) in a panel of TNBC and non-TNBC cell lines. (B) Relative brachyury mRNA expression levels in the various cell lines. Error bars indicate the standard error of the mean of triplicate measurements. (C) Silencing brachyury expression in MDA-MB-436 cells is associated with a reduction in the expression of mesenchymal markers vimentin and fibronectin and increased expression of the epithelial marker ZO1 (green signal). Blue signal corresponds to DAPI staining of nuclei.
Discussion
Elucidation of novel, potential therapeutic targets for the treatment of TNBC remains an urgent need in the absence of current, effective treatment for this aggressive tumor type. Our group and others have previously described the selective expression of the transcription factor brachyury, an immunotherapy target and a driver of tumor EMT (Fernando et al. 2010; Hamilton, et al. 2013; Palena, et al. 2014a; Palena and Hamilton 2015; Palena, et al. 2007), in primary and metastatic lesions of invasive ductal carcinomas of the breast (Palena et al. 2014b; Shao et al. 2015). In the present study we expanded those previous observations and demonstrated for the first time a prevalent expression of brachyury mRNA and protein in TNBC compared to other breast cancer subtypes. Variable levels of brachyury protein were observed by IHC in approximately 90% of primary and 100% of metastatic TNBC lesions analyzed. Expression of brachyury, however, was highly variable among primary tumors, with the percentage of positive cells ranging from 10% to 100% and the intensity of staining varying between (+) and (+++). Based on the relevant role of brachyury in tumor dissemination and/or resistance to anti-cancer treatments, it is expected that a small fraction of brachyury-positive cells in a tumor mass could have a major impact in terms of clinical outcome. With this functional relevance of brachyury in mind, a tissue has been designated here as “brachyury positive” if as little as 10% of the cancer cells exhibited some level of staining.
It is important to point out that in addition to providing evidence of variable levels of brachyury expression in TNBC, the data presented here also indicate that some level of brachyury could be present in normal breast tissues adjacent or distal to the tumor. These results contrast with our previous observations with normal breast tissues from patients with benign conditions, where brachyury was not detected (Palena et al. 2014b). Two recently completed Phase I clinical trials of a yeast-brachyury vaccine (Heery et al. 2015b) or a MVA-brachyury-TRICOM vaccine (Heery, et al. 2015a) demonstrated no evidence of any autoimmune occurrence in the presence of measurable brachyury-specific CD4+ and CD8+ T-cell responses, including in normal testis and thyroid tissues that have been previously shown to express some level of the brachyury protein. The presence of brachyury in normal breast tissues surrounding TNBC, however, raises concern that an autoimmune event against those tissues could take place in vaccinated patients and warrants careful evaluation in future studies of brachyury-based vaccines.
Three cancer vaccine platforms against brachyury have been developed and two are currently undergoing Phase I/II clinical evaluation (Gabitzsch, et al. 2015; Hamilton et al. 2013; Heery et al. 2015a; Heery et al. 2015b). Brachyury-based vaccines have demonstrated in Phase I clinical studies the ability to elicit CD8+ and CD4+ brachyury-specific T-cell immune responses in the blood of patients, post-vaccination. These brachyury-specific T cells have been shown to produce IFN-γ, IL-2, TNF-α, and/or to express CD107a, a marker of lytic potential. Although some evidence of clinical response was observed, the small number of patients evaluated precluded the analysis of any potential association between the degree of antigen-specific immune response and clinical outcome. Further studies with larger number of patients will need to be conducted to investigate whether such association could be established. In a previous report we have shown that brachyury-specific T cells generated from the blood of cancer patients can lyse basal MDA-MB-231 cancer cells in an MHC-restricted manner (Palena et al. 2014b). Those experiments were conducted with unfractionated CD8+ T cells and it is expected that a higher degree of lysis would be observed with tetramer-isolated, brachyury-specific T cells.
Although the molecular classification of breast cancer into various subtypes has revealed a high degree of heterogeneity in this disease, the majority of TNBC cases (56-95%) can be molecularly categorized as basal-like (Prat et al. 2012). Tumors of this subtype are defined by the expression of genes typically present in normal breast basal/myoepithelial cells and are characterized by a high proliferation index, p53 mutations, and an aggressive course with frequent tumor relapse (Reis-Filho and Tutt 2008). The phenomenon of EMT, a phenotypic switch that allows epithelial cells to acquire motility, invasiveness, and resistance to cell death while exhibiting features characteristic of mesenchymal cells, is being postulated as a relevant mechanism that fosters progression towards metastatic disease (Kalluri and Weinberg 2009). In agreement, it is the aggressive basal-like group of breast cancer that commonly exhibits mesenchymal features, including expression of vimentin, smooth-muscle-actin (SMA) and N-cadherin in place of epithelial E-cadherin (Blick, et al. 2008; Sarrio et al. 2008).
The transcription factor brachyury has been previously shown to induce phenotypic changes in carcinoma cells reminiscent of an EMT, a phenomenon associated with tumor dissemination, metastasis and acquisition of resistance to a variety of anti-tumor therapies (Huang et al. 2013; Larocca et al. 2013). In this regard, we and others have shown that the presence of high levels of brachyury in the primary tumor can predict poor prognosis in a range of human carcinomas including lung (Haro et al. 2013), hepatocellular (Du, et al. 2014), GIST (Pinto, et al. 2015), prostate (Pinto et al. 2014), colorectal (Kilic et al. 2011; Sarkar, et al. 2012) and hormone receptor positive breast cancer (Palena et al. 2014b). In agreement, here we have observed that high levels of brachyury in primary TNBC tumors associate with poor survival. It is important to point out, however, that studies of the association between brachyury and patient survival were based on an arbitrary cutoff of ≥240 for the brachyury reactivity index. Thus, expression of brachyury in primary TNBC is proposed to be a bad prognostic indicator only for the fraction of tumors that express brachyury in at least 80% of the cancer cells and at high staining intensity (+++), which will result in reactivity index values ≥ 240.
In our previous studies we have conducted gain- and loss-of-function experiments to investigate the particular role of brachyury in breast carcinoma cells. Utilizing basal-like, hormone-receptor negative MDA-MB-436 cells, we have shown that silencing of brachyury is able to significantly reduce cell invasiveness and the tumor cells’ ability to form mammospheres in primary and secondary cultures, a measure of tumor stemness, relative to their control counterparts. The loss of brachyury was also shown to significantly sensitize basal-like MDA-MB-436 cells to the cytotoxic activity of docetaxel, thus demonstrating a potential role for brachyury in tumor invasiveness and resistance to cell death in TNBC (Palena et al. 2014b). Moreover, a recent report (Ben-Hamo, et al. 2014) has identified brachyury as a dominant gene in a network of genes that most significantly discriminate TNBC from non-TNBC samples. Here we have further demonstrated the prevalent expression of brachyury in a panel of TNBC human breast carcinoma cell lines, and validated its role as a driver of tumor EMT with MDA-MB-436 TNBC cells, where brachyury silencing markedly reduced the expression of mesenchymal vimentin and fibronectin while increasing the expression of epithelial ZO1.
In the present study, the expression of brachyury mRNA was shown to be predominant among TNBC vs. triple positive samples in the TCGA database, while that was not the case with other EMT drivers, including TWIST1, SNAI2 or ZEB2 mRNA. These results seemed to contradict the expected enrichment of EMT markers in TNBC samples. Although the reason for these observations is unknown at this time, we have previously observed that expression of TWIST1, SNAI1 and SNAI2 mRNA, for example, is detectable in normal tissues at levels comparable to those observed in tumors (Palena et al. 2014b; Roselli, et al. 2012). As the mRNA analysis includes not only tumor cells but also surrounding normal tissues, one possible explanation for the absence of difference of expression of the EMT markers between different tumor types could be the simultaneous detection in adjacent normal cells. Unlike TWIST1, SNAI1 and SNAI2, we have previously shown that expression of brachyury mRNA is undetectable in normal breast tissues (Palena et al. 2014b).
Among genes that were differentially expressed between TNBC and other tumors was IL-8, a chemokine that was previously associated with tumor stemness and EMT in breast cancer (Fernando et al. 2011; Ginestier, et al. 2010; Palena, et al. 2012). We have previously demonstrated that IL-8 is able to significantly upregulate brachyury expression in breast cancer cells at the transcriptional level, and that upregulation of brachyury leads to increased secretion of IL-8 and overexpression of IL-8 receptors, thus establishing an autocrine feed-forward loop that sustains the mesenchymal, resistant phenotype. We hypothesize that the presence of this autocrine regulatory loop in TNBC could drive the expression of brachyury and, potentially, the acquisition of mesenchymal and stem-like features by the tumor cells.
While there have been some discrepancies regarding the detection of brachyury protein in various types of carcinomas, we believe that these inconsistencies are due to the utilization of various antibodies possessing a range of affinity and specificity for the target protein brachyury. We and others have shown, for example, that brachyury can be detected in the nucleus and/or the cytosol of carcinoma cells, while its expression in chordomas is predominantly seen in the nucleus of the tumor cells (Hamilton et al. 2015; Miettinen, et al. 2015; Pinto et al. 2014). In agreement with those previous observations, here we demonstrated that brachyury protein can be detected either in the nucleus and/or the cytosol of TNBC cells, although their expression is somehow linked with a positive correlative trend observed between the nuclear and cytoplasmic reactivity index (r=0.236, p=0.04). While the biological relevance of nuclear vs. cytosolic brachyury localization in tumor cells remains unknown, it was interesting to observe in this study that nuclear (but not cytosolic) expression of brachyury associated with clinical stage of disease and was more predominant in metastatic vs. primary tumor tissues. This potentially indicates a role for brachyury in tumor progression when localized in the nucleus, where its transcriptional activity is expected to take place. Future studies are warranted to evaluate, in a larger cohort of TNBC patients, the potential association between nuclear vs. cytoplasmic brachyury expression and clinical outcome.
Altogether our observations demonstrate the potential of brachyury as a target for the treatment of early or metastatic TNBC, and provide rationale for utilizing a brachyury-targeting vaccine approach for treatment of this disease.
Acknowledgements
The authors would like to thank Dr. Jeffrey Schlom from the National Cancer Institute, NIH, for his invaluable input along the study. Some of the results published here are based upon data from the TCGA Research Network.
Financial Support: This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health and by research funding from the European Social Fund, under the Italian Ministry of Education, University and Research PON03PE_00146_1/10 BIBIOFAR (CUP B88F12000730005) to FG.
Footnotes
Authors contributions: DHH and CP carried out studies at the mRNA level and cell line-based assays, data analysis and wrote the manuscript. MR, PF, LC, FC, MT and FG, carried out the immunohistochemical analyses, performed data analysis and wrote the manuscript. CP and FG designed and supervised the study. All authors read and approved the final manuscript.
Competing interests: No competing interests to declare
References
- Ben-Hamo R, Gidoni M, Efroni S. PhenoNet: identification of key networks associated with disease phenotype. Bioinformatics. 2014;30:2399–2405. doi: 10.1093/bioinformatics/btu199. [DOI] [PubMed] [Google Scholar]
- Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, Thompson EW. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–642. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JM, Hamilton DH, Palena C. MUC1 upregulation promotes immune resistance in tumor cells undergoing brachyury-mediated epithelial-mesenchymal transition. Oncoimmunology. 2016;5:e1117738. doi: 10.1080/2162402X.2015.1117738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Du R, Wu S, Lv X, Fang H, Wu S, Kang J. Overexpression of brachyury contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33:105. doi: 10.1186/s13046-014-0105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando RI, Castillo MD, Litzinger M, Hamilton DH, Palena C. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 2011;71:5296–5306. doi: 10.1158/0008-5472.CAN-11-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando RI, Hamilton DH, Dominguez C, David JM, McCampbell KK, Palena C. IL-8 signaling is involved in resistance of lung carcinoma cells to erlotinib. Oncotarget. 2016 doi: 10.18632/oncotarget.9662. Epub online 2016/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando RI, Litzinger M, Trono P, Hamilton DH, Schlom J, Palena C. The T-box transcription factor Brachyury promotes epithelial-mesenchymal transition in human tumor cells. J Clin Invest. 2010;120:533–544. doi: 10.1172/JCI38379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabitzsch ES, Tsang KY, Palena C, David JM, Fantini M, Kwilas A, Rice AE, Latchman Y, Hodge JW, Gulley JL, et al. The generation and analyses of a novel combination of recombinant adenovirus vaccines targeting three tumor antigens as an immunotherapeutic. Oncotarget. 2015;6:31344–31359. doi: 10.18632/oncotarget.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, Wicinski J, Cabaud O, Charafe-Jauffret E, Birnbaum D, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120:485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–5657. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- Hamilton DH, Fernando RI, Schlom J, Palena C. Aberrant expression of the embryonic transcription factor brachyury in human tumors detected with a novel rabbit monoclonal antibody. Oncotarget. 2015;6:4853–4862. doi: 10.18632/oncotarget.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DH, Huang B, Fernando RI, Tsang KY, Palena C. WEE1 inhibition alleviates resistance to immune attack of tumor cells undergoing epithelialmesenchymal transition. Cancer Res. 2014;74:2510–2519. doi: 10.1158/0008-5472.CAN-13-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DH, Litzinger MT, Jales A, Huang B, Fernando RI, Hodge JW, Ardiani A, Apelian D, Schlom J, Palena C. Immunological targeting of tumor cells undergoing an epithelial-mesenchymal transition via a recombinant brachyury-yeast vaccine. Oncotarget. 2013;4:1777–1790. doi: 10.18632/oncotarget.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro A, Yano T, Kohno M, Yoshida T, Koga T, Okamoto T, Takenoyama M, Maehara Y. Expression of Brachyury gene is a significant prognostic factor for primary lung carcinoma. Ann Surg Oncol. 2013;20(Suppl 3):S509–516. doi: 10.1245/s10434-013-2914-9. [DOI] [PubMed] [Google Scholar]
- Heery CR, Donahue R, Lepone L, Grenga I, Richards J, Metenou S, Fernando RI, Dirmeier U, Singh H, Madan R, et al. Phase I, dose-escalation, clinical trial of MVA-Brachyury-TRICOM vaccine demonstrating safety and brachyury-specific T cell responses. Journal for ImmunoTherapy of Cancer. 2015a;3:1–2. [Google Scholar]
- Heery CR, Singh BH, Rauckhorst M, Marte JL, Donahue RN, Grenga I, Rodell TC, Dahut W, Arlen PM, Madan RA, et al. Phase I Trial of a Yeast-Based Therapeutic Cancer Vaccine (GI-6301) Targeting the Transcription Factor Brachyury. Cancer Immunol Res. 2015b;3:1248–1256. doi: 10.1158/2326-6066.CIR-15-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Cohen JR, Fernando RI, Hamilton DH, Litzinger MT, Hodge JW, Palena C. The embryonic transcription factor Brachyury blocks cell cycle progression and mediates tumor resistance to conventional antitumor therapies. Cell Death Dis. 2013;4:e682. doi: 10.1038/cddis.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Ryu YJ, An J, Lee Y, Kim A. Epithelial-mesenchymal transition in breast cancer correlates with high histological grade and triple-negative phenotype. Histopathology. 2012;60:E87–95. doi: 10.1111/j.1365-2559.2012.04195.x. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic N, Feldhaus S, Kilic E, Tennstedt P, Wicklein D, Wasielewski R, Viebahn C, Kreipe H, Schumacher U. Brachyury expression predicts poor prognosis at early stages of colorectal cancer. Eur J Cancer. 2011;47:1080–1085. doi: 10.1016/j.ejca.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyury is a tissue-specific transcription factor. EMBO J. 1995;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca C, Cohen JR, Fernando RI, Huang B, Hamilton DH, Palena C. An autocrine loop between TGF-beta1 and the transcription factor brachyury controls the transition of human carcinoma cells into a mesenchymal phenotype. Mol Cancer Ther. 2013;12:1805–1815. doi: 10.1158/1535-7163.MCT-12-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, Bradbury I, Bliss JM, Azim HA, Jr., Ellis P, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Wang Z, Lasota J, Heery C, Schlom J, Palena C. Nuclear Brachyury Expression Is Consistent in Chordoma, Common in Germ Cell Tumors and Small Cell Carcinomas, and Rare in Other Carcinomas and Sarcomas: An Immunohistochemical Study of 5229 Cases. Am J Surg Pathol. 2015;39:1305–1312. doi: 10.1097/PAS.0000000000000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KM, Cole SR, Tse CK, Perou CM, Carey LA, Foulkes WD, Dressler LG, Geradts J, Millikan RC. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res. 2010;16:6100–6110. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena C, Fernando RI, Hamilton DH. An immunotherapeutic intervention against tumor progression: Targeting a driver of the epithelial-to-mesenchymal transition. Oncoimmunology. 2014a;3:e27220. doi: 10.4161/onci.27220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena C, Hamilton DH. Immune Targeting of Tumor Epithelial-Mesenchymal Transition via Brachyury-Based Vaccines. Adv Cancer Res. 2015;128:69–93. doi: 10.1016/bs.acr.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena C, Hamilton DH, Fernando RI. Influence of IL-8 on the epithelialmesenchymal transition and the tumor microenvironment. Future Oncol. 2012;8:713–722. doi: 10.2217/fon.12.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palena C, Polev DE, Tsang KY, Fernando RI, Litzinger M, Krukovskaya LL, Baranova AV, Kozlov AP, Schlom J. The human T-box mesodermal transcription factor Brachyury is a candidate target for T-cell-mediated cancer immunotherapy. Clin Cancer Res. 2007;13:2471–2478. doi: 10.1158/1078-0432.CCR-06-2353. [DOI] [PubMed] [Google Scholar]
- Palena C, Roselli M, Litzinger MT, Ferroni P, Costarelli L, Spila A, Cavaliere F, Huang B, Fernando RI, Hamilton DH, et al. Overexpression of the EMT driver brachyury in breast carcinomas: association with poor prognosis. J Natl Cancer Inst. 2014b;106:dju054. doi: 10.1093/jnci/dju054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pinto F, Campanella NC, Abrahao-Machado LF, Scapulatempo-Neto C, de Oliveira AT, Brito MJ, Andrade RP, Guimaraes DP, Reis RM. The embryonic Brachyury transcription factor is a novel biomarker of GIST aggressiveness and poor survival. Gastric Cancer. 2015 doi: 10.1007/s10120-015-0505-0. [DOI] [PubMed] [Google Scholar]
- Pinto F, Pertega-Gomes N, Pereira MS, Vizcaino JR, Monteiro P, Henrique RM, Baltazar F, Andrade RP, Reis RM. T-box transcription factor brachyury is associated with prostate cancer progression and aggressiveness. Clin Cancer Res. 2014;20:4949–4961. doi: 10.1158/1078-0432.CCR-14-0421. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135:301–306. doi: 10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- Roselli M, Fernando RI, Guadagni F, Spila A, Alessandroni J, Palmirotta R, Costarelli L, Litzinger M, Hamilton DH, Huang B, et al. Brachyury, a driver of the epithelial-mesenchymal transition, is overexpressed in human lung tumors: an opportunity for novel interventions against lung cancer. Clin Cancer Res. 2012;18:3868–3879. doi: 10.1158/1078-0432.CCR-11-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Shields B, Davies ML, Muller J, Wakeman JA. BRACHYURY confers cancer stem cell characteristics on colorectal cancer cells. Int J Cancer. 2012;130:328–337. doi: 10.1002/ijc.26029. [DOI] [PubMed] [Google Scholar]
- Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- Shao C, Zhang J, Fu J, Ling F. The potential role of Brachyury in inducing epithelial-to-mesenchymal transition (EMT) and HIF-1alpha expression in breast cancer cells. Biochem Biophys Res Commun. 2015;467:1083–1089. doi: 10.1016/j.bbrc.2015.09.076. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]