Abstract
Cell-autonomous immunity is essential for host organisms to defend themselves against invasive microbes. In vertebrates, both the adaptive and the innate branches of the immune system operate cell-autonomous defenses as key effector mechanisms that are induced by pro-inflammatory interferons. Interferons can activate cell-intrinsic host defenses in virtually any cell type ranging from professional phagocytes to mucosal epithelial cells. Much of this interferon-induced host resistance program is dependent on four families of interferon-inducible GTPases: the myxovirus resistance proteins (Mx), the immunity related GTPases (IRGs), the guanylate binding proteins (GBPs) and the very large interferon-inducible GTPases (VLIGs). These GTPase families provide host resistance to a variety of viral, bacterial and protozoan pathogens through the sequestration of microbial proteins, manipulation of vesicle trafficking, regulation of antimicrobial autophagy (xenophagy), execution of intracellular membranolytic pathways, and the activation of inflammasomes. This review discusses our current knowledge of the molecular function of interferon-inducible GTPases in providing host resistance as well as their role in the pathogenesis of autoinflammatory Crohn’s disease. While substantial advances were made in the recent past, few of the known functions of interferon-inducible GTPases have been explored in any depth and new functions await discovery. This review will therefore highlight key areas of future exploration that promise to advance our understanding of the role of interferon-inducible GTPases in human diseases.
Keywords: IRG, IRGM, GBP, inflammasome, Crohn’s disease
Graphical abstract

1. INTRODUCTION
From unicellular amoeba to the individual cells of multicellular plants and animals - all eukaryotic cells possess the inherent capacity to fight off microbial invaders. This ability of a single cell to control and possibly eliminate an infectious agent is called cell-autonomous immunity. Cell-autonomous immune responses encompass the detection of invading microbes and the subsequent execution of antimicrobial effector pathways. These effector pathways mediate the killing, expulsion or intracellular encapsulation and containment of invasive microbes [1]. The repertoire of cell-autonomous defense modules can be extensive and varies from cell type to cell type and between host species. Some cell-autonomous effector mechanisms such as the oxidative burst are ancient and found throughout the animal and plant kingdoms [2], and yet other responses evolved more recently and are accordingly less ubiquitous. Integrating many separate defense modules of distinct evolutionary histories into a functionally coherent cell-autonomous immune program requires the careful coordination of their activities to promote synergy and to avoid antagonistic effects.
One critical regulatory mechanism of cell-autonomous immunity in humans and other vertebrates is provided by the temporal and spatial control that pro-inflammatory cytokines exert over the expression of many host defense proteins. Chief amongst these cytokines are the 3 types of Interferons (IFNs) [3]. Engagement of type I, II and III IFN receptors initiates Janus activated kinase (JAK) signalling resulting in the phosphorylation and nuclear translocation of members of the Signal Transducers and Activators of Transcription (STAT) protein family. Different combinations of STATs form transcription factor complexes that promote the expression of IFN-inducible target genes which are defined by the presence of IFN-responsive promoter elements, most notably the Interferon-sensitive response element (ISRE) and the Interferon-gamma Activated Sequence (GAS) [4]. Typically, the most prominently induced proteins found in an IFN-primed cells are members of four families of IFN-inducible dynamin-like GTPases (Figure 1). These families are known as the myxovirus resistance proteins (Mx), the immunity related GTPases (IRGs), the guanylate binding proteins (GBPs) and the very large IFN-inducible GTPases (VLIGs).
Figure 1. Maximum-likelihood phylogeny based on conserved coding regions of interferon-stimulated GTPase family members in humans and mice.
Branches for larger clades have been collapsed. Major families, including MX (blue), IRG (orange), GBP (purple), and VLIG (green), are indicated, with human K-Ras included as an outgroup. Structures of representative MX, IRG, and GBP family members are shown at right (PDB: 3SZR, 1TQ6, 1F5N).
The first IFN-inducible GTPases to be discovered were mouse Mx1 and human GBP1 more than 30 years ago [5, 6]. The functional importance of mouse Mx1 became immediately obvious when Lindenmann, Haller, Staeheli and colleagues found that a non-functional allele of Mx1 was responsible for the susceptibility of many inbred strains of mice to influenza and influenza-like viruses [7–10]. While the Mx1 paradigm suggested a universal role for IFN-inducible GTPases in host resistance, only a small number of laboratories developed and maintained an active research program focused on the biology of IFN-inducible GTPases. More than a decade later interest in IFN-inducible GTPases reignited when IRG proteins were found to protect against protozoan and bacterial infections [11–14]. The association of a polymorphic allele of human IRGM with increased risk to develop autoinflammatory Crohn’s disease highlighted for the first time the importance of IFN-inducible GTPases in inflammation and human disease [15, 16]. This time around, IFN-inducible GTPases were not to be ignored, and a flurry of research efforts has since begun. Here, we will provide a critical overview of the field, describe our current understanding of the molecular function of IFN-inducible GTPases in host resistance and inflammation, as well as highlight discrepancies in reported findings and current challenges to future advances.
2. HOST RESISTANCE MEDIATED BY IFN-INDUCIBLE GTPASES
Mx proteins mediate a multi-facetted antiviral response
A detailed review covering Mx-mediated immunity has been published recently [17]. We will therefore provide only a brief description of our current molecular understanding of the antiviral activity of Mx proteins. Expression of Mx genes is exclusively induced by type I and type III IFNs [18]. The Mx genes can be divided into two lineages that arose from an ancestral gene duplication event. Humans possess a single representative of each lineages: MxA and MxB. Rodents, on other hand, lack MxB-like genes, and instead harbor two MxA-like paralogs encoding proteins Mx1 and Mx2. The two Mx gene lineages mediate resistance to distinct spectra of viruses. The MxA lineage mediates resistance to a broad spectrum of viruses including influenza viruses. Standard mouse inbred strains lack both functional Mx1 and Mx2 genes and are highly susceptible to a number of viral pathogens including the orthomyxovirus influenza virus [17]. Transgenic complementation of Mx1/Mx2-deficient inbred mice with functional copies of Mx1 restores resistance to influenza infections [19]. Similarly, transgenic expression of human MxA also establishes anti-flu immunity in mice, a finding that supports an important role for MxA in protection against influenza in humans [20]. Indeed, human MxA binds to influenza virus nucleoprotein (NP) and NP mutations present in 1918 and 2009 H1N1 pandemic strains convey resistance to MxA-dependent immunity [21]. In contrast, human MxB is ineffective against influenza but provides cell-autonomous resistance to human immunodeficiency virus type 1 (HIV-1) [22–24]. A recent analysis of the evolution of MxB proteins in primates suggests that MxB proteins mediate host defense not only against lentiviruses but also other pathogens, yet the identification of these presumably viral pathogens awaits future discovery [25].
MxA proteins restrict a broad spectrum of DNA and RNA viruses that include for example the hepadnavirus hepatitis B virus (HBV), bunyaviruses, picornaviruses and togaviruses [17]. The antiviral breadth of MxA proteins is likely due to the presence of several distinct conformation-dependent protein surfaces that engage in highly specific interactions with a variety of viral proteins such as Influenza NP and the bunyavirus N protein [26]. The evolutionary conserved tripartite structure of MxA proteins consists of a globular G domain at its N-terminus, the central bundle signalling element (BSE) and the C-terminal elongated stalk consisting of a four-helix bundle. The stalk comprises the middle domain and the GTPase effector domain [27, 28]. A disordered loop, L4, protrudes from the protein stalk and mediates most interactions of MxA proteins with viral proteins [17]. While these interactions between MxA and viral proteins have not yet been visualized, several lines of evidence indicate that GTP-bound MxA dimers or tetramers mediate the initial binding to viral proteins. Following this nucleation event, additional MxA proteins self-assemble into higher order ring-like structures. Molecular modelling suggests that stalk-stalk and BSE-stalk interactions mediate the assembly of MxA proteins into these ring-like complexes, with the G domains facing outwards [27, 28]. This conformational arrangement would allow for G domains of neighboring rings to interact with each other and to promote GTPase activation. In agreement with such a model, GTP hydrolysis by MxA proteins is oligomerization-dependent [28]. Although the precise mechanisms by which GTP hydrolysis propels antiviral activities of human MxA and murine Mx1 proteins are currently unclear, it seems likely that hydrolysis-driven conformational changes promote the sequestration for viral proteins or the disruption of viral ribonucleoprotein complexes (vRNP) [17].
Mx proteins can interact with their viral targets both in the cytoplasm and in the cell nucleus. Rodent Mx1 is a nuclear protein that inhibits transcription performed by viral RNA polymerases in the cell nucleus [29, 30]. Accordingly, rodent Mx1 only blocks viruses such as influenza that have a nuclear replication step [29]. Rodent Mx2 on the other hand is cytoplasmic and can also inhibit viruses that exclusively replicate inside the cytoplasm [31]. Both human Mx proteins are localized to the cytoplasm but exert their antiviral activities through apparently distinct mechanisms. MxA mediates its antiviral activity through the sequestration of viral proteins into perinuclear complexes, interference with nuclear translocation of incoming vRNP, and the inhibition of viral transcription and replication in the cytoplasm [17]. Human MxB restricts the propagation of lentiviruses by targeting the pre-integration complex (PIC) that is formed following the reverse transcription of the RNA genome of incoming HIV-1 nucleocapsid. Human MxB blocks the uncoating, the nuclear import, or the integrase function of PIC [22–24]. Only MxA but not MxB proteins depend on protein oligomerization and GTP hydrolysis for most of their antiviral activities, further indicating that the antiviral effector mechanisms of these two closely related proteins are distinct [17]. As more viral proteins and viral species targeted by Mx proteins are discovered, it is becoming increasingly apparent that Mx proteins provide cell-autonomous immunity to a broad spectrum of viruses and are likely to fulfill an important barrier function against zoonotic infections in humans.
Recurrent gene duplication and loss in the evolution of the IRG gene family
Like the Mx family, the IRG gene family likely arose in the common ancestor of chordates [32]. However, in contrast to the relatively stable copy number of Mx genes across vertebrate species, the IRG gene family has undergone dramatic episodes of gene duplication and loss. For example, zebrafish harbour 11 IRG genes, pufferfish possess only 2 and birds appear to have lost the IRG gene family entirely. The expansion and deletion of IRG genes is also apparent in mammals: whereas rodent genomes contain up to 20 IRG genes or more, humans possess only 2 IRG genes [32–34].
The dramatic expansion of the IRG gene family in several vertebrate species is best explained by the action of strong selective forces imposed by the host interactions with pathogens. Host adaptions resulting in improved host resistance are often counteracted by microbial adaptations, which in turn force the host to respond with new adaptations, setting of a continuing process of cyclical adaptation events by both host and microbe. The need to constantly adapt and evolve to survive the molecular “arms race” with an ever-evolving opposing organism is defined as a Red Queen genetic conflict [35]. This conflict is beautifully illustrated by discrete interactions between the murine IRG resistance system and a repertoire of virulence factors encoded by T. gondii to evade IRG immunity. These virulence factors consist of a family of polymorphic rhoptry secretory kinases that have been amplified in the genome of the parasite [36, 37]. On the host side, the IRG resistance system has also undergone gene expansion and increased its allelic variation. This polymorphic complexity is most apparent in widtype-derived mouse lines and it has been shown that at least one IRG haplotype found in wild-derived mouse strains blocks the function of a T. gondii rhoptry kinase complex that inactivates the IRG resistance system in common mouse laboratory strains [38].
While a dramatic expansion of the IRG gene family is seen in rodents, many other vertebrate species display a collapse of the IRG resistance system. The partial or complete loss of the IRG system in birds and other vertebrate species could be explained by a lack of a selective pressure, or alternatively suggests that IRG resistance system may impose fitness costs upon hosts. The association of human IRGM gene variants with the development of inflammatory diseases hints at such evolutionary costs, which may have contributed to the loss or reduction of the IRG resistance system in many vertebrate species [15, 16].
A rodent IRG resistance system targets pathogen-containing vacuoles for destruction
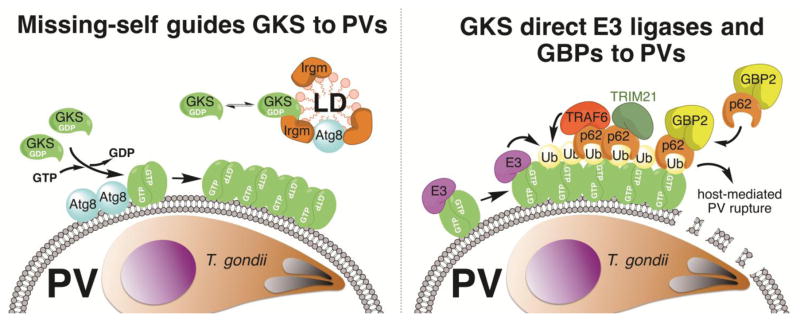
The genome of the standard laboratory mouse strain C57BL/6 contains 16 IRG genes and 4 annotated IRG pseudogenes [33, 38]. The IRG genes are organized in two adjacent clusters on mouse chromosomes 11 and a single cluster on chromosome 18. In most cases a single exon contains the entire open reading frame (ORF) encoding IRG proteins with a typical molecular weight of 47 kDa. However, a subset of 4 ‘tandem’ IRG proteins arose through the fusion of 2 ancestral IRG genes into a single mRNA that is spliced and translated into one protein with a molecular weight of 94 kDa, twice that of canonical IRG proteins [38]. All mouse IRG proteins can be placed into 2 subfamilies defined by sequence and functional differences: the GKS ‘effector’ proteins and the regulatory Irgm proteins [33, 39]. GKS proteins include the Irga, Irgb, Irgc, and Irgd groups and are defined by the presence of a canonical GxxxxGKS motif in the P-loop of the GTP-binding site. GKS proteins with the exception of Irgc are strongly induced by IFN receptor signalling. In uninfected, IFN-primed cells GKS proteins are nearly exclusively found in the GDP-bound form and reside in the cytosol or associate transiently with cell organelles such as the endoplasmic reticulum (ER) [39]. Once a cell is infected with IRG-susceptible pathogens such as T. gondii or Chlamydia trachomatis the pathogen-containing vacuole (PV) surrounding these pathogens becomes decorated with GTP-bound GKS proteins [39–42]. GTP binding and the resulting oligomerization of GKS proteins are essential for the association of GKS proteins with PV membranes [39, 41]. The murine Irgm proteins Irgm1, Irgm2 and Irgm3 on the other hand harbour the non-canonical GxxxxGMS P-loop sequence and are therefore sometimes referred to as ‘GMS proteins’ [33, 39]. Following their induction by IFNs, murine GDP-bound Irgm proteins stably reside on most if not all cell organelles including mitochondria, the ER, the Golgi apparatus and lipid droplets (LDs). Here, the regulatory Irgm proteins prevent off-target activation of GKS proteins and thereby guard self-structures against GKS binding [41]. Irgm proteins are largely absent from PVs, which enables the activation and binding of GKS proteins to PVs through a missing-self mechanism (Figure 2), conceptually similar to the binding of complement factors to extracellular bacteria that lack complement regulatory proteins on their surfaces [41, 43].
Figure 2. Targeting of GKS proteins and GBPs to T. gondii-containing parasitophorous vacuoles (PVs) in mouse cells.
The GKS class of IRG proteins is guided towards PV membranes through a missing-self principle. Whereas Irgm1 and Irgm3 proteins residing on “self” lipid droplets (LDs) block GKS protein activation, the absence of these Irgm proteins from PVs results in the docking of GTP-bound GKS dimers to PV membranes. Lipidated members of the Atg8 family of ubiquitin-like proteins further promote GKS association with PVs (left panel). GKS proteins promote the recruitment of ‘pioneering’ ubiquitin E3 ligases (E3) and p62-interacting E3 ligases (TRAF6, TRIM21) to PVs, which promote the decoration of PVs with ubiquitin. GKS proteins themselves are possible ubiquitination substrates, as depicted in this figure, but further studies are required to test this hypothesis. The ubiquitin-binding protein p62 also escorts GBP2 to PVs (right panel). Additional p62-independent mechanisms of GBP recruitment exist (not shown).
The delivery of GKS proteins to PVs has only been unambiguously demonstrated for three pathogens so far: the protozoan T. gondii, the bacterium C. trachomatis and, more recently, the microsporidian Encephalitozoon cuniculi [40, 42, 44]. In agreement with the missing-self mechanism by which GKS proteins assemble on Irgm-deficient structures [41], PVs formed by all 3 of these pathogens are largely devoid of Irgm proteins. That observation prompts the question as to why Irgm proteins are absent from these PVs. PVs are initially formed through the invagination of the plasma membrane during the process of microbial host cell invasion. The presence of Irgm proteins on host plasma membranes and phagocytic cups is controversial. While staining with anti-Irgm1 antibody originally indicated that Irgm1 localizes to phagocytic cups and early phagosomes engulfing Listeria monocytogenes or Mycobacterium bovis bacillus Calmette-Guérin (BCG) [13, 45], a subsequent study demonstrated that the same anti-Irgm1 antibody used in these earlier studies bound non-specifically to bacteria. Careful reexamination of the subcellular localization of Irgm1 with a set of new reagents argued against the presence of Irgm1 on Listeria- or Mycobacteria-containing phagosomes [46]. Nevertheless, Irgm proteins other than Irgm1 or alternative inhibitors of GKS protein activation could be present on phagosomal membranes. It has been proposed that pathogens entering cells through conventional phagocytosis would maintain these GKS activation inhibitors on their surrounding vacuolar membranes and therefore remain GKS-resistant [44]. Unique non-phagocytic entry mechanisms used by T. gondii, C. trachomatis and E. cuniculi, on the other hand, would circumvent the accumulation of Irgm proteins or alternative GKS inhibitors on PVs, thus allowing the recruitment of GKS proteins to these PVs. Such a model implies that GKS-susceptible pathogens derive some sort of fitness advantage from dwelling within an Irgm-devoid vacuolar compartment. In support of this idea, it has been proposed that Irgm1 associated with pathogen-containing phagosomes could promote the rapid fusion of these phagosomes with bactericidal lysosomes [13, 45]. Accordingly, the absence of Irgm proteins from PVs could provide an initial advantage to microbes such as T. gondii or C. trachomatis. However, the advantage would be short-lived, as Irgm-devoid PVs attract GKS proteins that execute the rupture of PVs. To further refine this model, we propose that the absence of Irgm proteins from PVs is necessary but not sufficient for the directed delivery of GKS proteins to PVs, and that additional markers enriched on PVs, but also present on some host cell organelles, promote the deposition of GKS proteins on PVs. Several observations support the latter model. For instance, the depletion of Irgm proteins from host cells triggers dramatic mistargeting of GKS proteins to LDs, but only minimal mislocalization of GKS proteins to mitochondrial and peroxisomal microdomains [41]. Accordingly, we would argue that LDs contain high concentrations of yet to be defined molecules that stimulate GKS binding. Strong candidate molecules to fulfilll such a function are members of the ubiquitin-like Atg8 protein family, which includes microtubule-associated protein 1A/1B light chain 3 (LC3). Lipidated LC3 has been to shown to localize not only to the surface of LDs [47] but also to GKS-decorated PV membranes [48] (Figure 2). Importantly, the enzymatic machinery that catalyses the lipidation of LC3 and other Atg8 proteins boosts the association of GKS proteins with T. gondii and C. trachomatis PVs independently of its role in the execution of canonical autophagy [48–51]. Future studies should determine which members of the Atg8 protein family assist in GKS recruitment and determine the underlying molecular mechanism. We propose the simple yet attractive hypothesis that Atg8-decorated membranes – either directly or indirectly - induce the exchange of GDP for GTP in GKS proteins resulting in GKS dimer formation and increased avidity for membranes decorated with lipidated Atg8 (Figure 2). In support of this hypothesis we demonstrated previously that a GTP-locked mutant of the GKS protein Irgb10, but not wildtype Irgb10, targets PVs with high efficiency in host cells lacking the Atg8 lipidation machinery [50], thus overcoming the requirement for lipidated Atg8.
The binding of GKS proteins to intracellular PVs and the binding of complement to extracellular bacteria share a common feature of targeting by missing-self mechanisms [43]. They also seem to share their main effector function: just as the formation of complement membrane attack complexes on the surface of extracellular bacteria results in bacterial lysis, the assembly of GKS proteins on PVs formed by T. gondii or C. trachomatis results in the lytic destruction of these vacuoles [42, 52–54]. GKS-decorated PV membranes vesiculate and ultimately rupture, thereby releasing T. gondii tachyzoites into the host cell cytosol where the parasite dies within a relatively short period of time [53, 54]. While the precise mechanism resulting in parasite death is unclear, some observations suggest that the parasite is stripped of its plasma membrane and that the denuded parasite is captured within autophagosomes that fuse with degradative lysosomes [54]. Following the destruction of the parasite some cell types undergo cell death that morphologically resembles necroptosis [53]; however, the molecular mechanism underlying this IRG-mediated host cell death is unknown. Whether IRG-dependent death of infected host cells occurs in vivo and whether IRG-dependent death alters the course of an infection remain open questions. The destruction of PVs requires GKS proteins to undergo GTP hydrolysis [53] suggesting that hydrolysis-driven conformational changes elicit mechanoenzymatic forces on the targeted PV membranes, similar to the dynamin-driven forces that result in vesicle scission during endocytosis [55]. Alternatively, GKS proteins work co-operatively with other host factors such as the GBPs and ubiquitin E3 ligases to induce PV lysis [52, 56]. Thus, while more detailed biochemical studies are required in order to clearly define the mechanism of IRG-mediated PV rupture, it represents an elegant and unique mechanism through which host immune effector proteins eradicate a subset of invading pathogens.
A rodent-adapted Chlamydia species and T. gondii strains actively evade IRG immunity
Considering the potency of the IRG resistance system, it comes as no surprise that pathogens have evolved counter-immune strategies. Two examples of IRG immune evasion have been reported so far. The first example resulted from studies on the pathogenesis of Chlamydia infections in mouse models. Infections of mice with the human pathogen C. trachomatis are rapidly cleared in an IFNγ-dependent manner [57]. A forward genetics approach in mice identified IRG genes as the critical IFNγ-inducible host resistance factors that promote C. trachomatis killing in vivo and in cell culture models [14, 58]. In contrast, the rodent-adapted pathogen Chlamydia muridarum is able to resist much of this IFNγ-orchestrated defense system [57]. This long-standing observation was finally explained when it was discovered that C. muridarum evades IRG-mediated immunity by blocking GKS protein accumulation at its PV [40]. Recent advances in the manipulation of Chlamydia genomes should pave the way towards the identification of the C. muridarum virulence factors that are responsible for immune evasion, and ultimately lead to a molecular understanding of the mechanism by which C. muridarum interferes with the IRG resistance system.
Following the report on active immune evasion of the IRG system by C. muridarum, it was shown that certain strains of T. gondii also escaped from IRG-mediated immunity. Different clonal lineages of T. gondii are commonly classified as ‘avirulent’ or ‘virulent’ based on the infectious dose required to induce lethality in standard inbred strains of mice [59]. Avirulence is conveyed by the IRG defense system [11, 60], as GKS proteins are able to efficiently load onto avirulent T. gondii PVs but not virulent T. gondii PVs [61, 62]. Virulent T. gondii strains interfere with loading through the secreted rhoptry kinase ROP16 and ROP18 that block deposition of GKS proteins onto their PV membranes. These rhoptry kinases do so by phosphorylating highly conserved threonine residues in the switch I region of the nucleotide binding site of several GKS proteins, thereby preventing GKS protein dimerization and PV membrane binding. Additional secreted rhoptry proteins facilitate phosphorylation by docking to GKS proteins and maintaining them in a conformation permissive for phosphorylation [61, 63–67]. Some wild-derived mouse strains have compensated for the actions of ROP18-expressing ‘virulent’ T. gondii strains, through expression of a highly polymorphic version of one of the aforementioned tandem GKS proteins, namely Irgb2-b1. IFNγ-inducible Irgb2-b1 acts as decoy for the ROP18 kinase complex, thereby preventing the phosphorylation and the resultant activation of canonical GKS proteins [38]. Thus, the evolutionary arms race between IRG-mediated immunity and T. gondii-employed immune evasion strategies appears to have played a major role in shaping the remarkable degree of polymorphic variation found in IRG genes amongst wild-derived mouse strains.
Human IRGM promotes autophagy and cell-autonomous immunity to bacterial infections
Controlling GKS protein activation and subcellular localization has emerged as a key role for IRGM proteins in rodents. However, IRGM proteins fulfilll additional important functions in host defense that are GKS-independent. At least some of these GKS-independent functions appear to be conserved between mice and humans.
Remarkably, the human genome lacks the large array of GKS proteins found in mice, with the only GKS protein present in the humans being IRGC. This gene is expressed only in testes and presumably does not play a large role in immunity [33]. Humans also possess a single gene encoding for the IRGM protein, which is expressed broadly in all cell types and tissue examined so far, albeit at very low levels. Its expression is augmented when cells are primed with lipopolysaccharide (LPS), hinting at a role for human IRGM in host defense [33, 34, 68, 69].
The human IRGM ORF is truncated at its 5’ and 3’ ends and encodes for a protein with a predicted molecular weight of 20 kDa. Due to its truncations and lack of IFN-inducible promoter elements, IRGM was initially classified as non-functional [33]. However, the association of the IRGM gene locus with susceptibility to Crohn’s disease and Mycobacterium tuberculosis (Mtb) infections argued against human IRGM as being a pseudogene [15, 16, 70].
The first functional role described for human IRGM was in regulating autophagic processing of Mycobacteria-containing phagosomes in macrophages [12]. This conclusion was based on studies in which IRGM expression was decreased by siRNA knockdown, leading to a reduction in IFNγ-induced autophagy in human macrophage cell lines that coincided with a decrease in autophagic maturation of M. bovis BCG-containing phagosomes. This role in controlling autophagic elimination of bacteria in host cells was buoyed by other studies showing decreased restriction of adherent-invasive Escherichia coli (AIEC) [71] and Salmonella typhimurium [72] when IRGM expression is similarly reduced. An underlying mechanism emerged only recently, and appears to involve a direct interaction of IRGM with core autophagy proteins Beclin, ATG16L1, and NOD2 to regulate assembly of the autophagosome [68]. Interaction of IRGM with the autophagic core is enhanced by ubiquitination of the protein [68]. Direct regulation of autophagy may well be the central function of IRGM that ultimately instructs its important roles in immunity. Nevertheless, the broader implications of IRGM-mediated autophagy beyond bacterial clearance are not yet clear, given that autophagy modulates multiple processes that ultimately instruct immunity and inflammation.
The question of whether murine Irgm proteins control autophagic processes, similar to human IRGM, remains unresolved. The Deretic lab initially reported that overexpression of mouse Irgm1 led to a boost in the number of autophagosomes and that siRNA-mediated interference with Irgm1 expression decreased the number of autophagosomes in IFNγ-primed RAW 264.7 cells, a commonly used mouse macrophage cell line [12, 73]. In contrast to these earlier reports it was later shown IFNγ priming promoted autophagic induction independently of Irgm1 in primary macrophages [74]. Future studies need to address these discrepancies and also carefully assess the roles of the paralogous Irgm2 and Irgm3 proteins in both autophagic induction and flux. Determining which of the mouse Irgm orthologs act as functional homologs of human IRGM is important in order to develop appropriate mouse models to study the role of Irgm proteins in the pathogenesis of infectious and autoinflammatory diseases.
Human IRGM and mouse Irgm1 proteins modulate mitochondrial function
Mitochondria are believed to be the principal source of radical oxygen species (ROS) required for autophagy induction [75]. In agreement with IRGM’s role as an orchestrator of autophagy, one study suggested an important role for human IRGM in regulating mitochondrial biogenesis and in controlling mitochondrial ROS production [69]. It found that a substantial portion of the cellular pool of human IRGM localizes to the inner mitochondrial membrane and associates with cardiolipin. An IRGM knockdown alters the morphology of the cellular mitochondrial pool, shifting it to a more fused and to a less punctate morphology. The converse experiment of overexpressing IRGM (specifically the IRGMd isoform) has the opposite effect in reducing fused and increasing punctate mitochondria. Together, these results suggest that IRGM can promote mitochondrial fission. This role in promoting fission appears to have ramifications for at least two broader cellular functions. First, overexpression of the IRGMd isoform, which leads to increased mitochondrial fission, also causes a decrease in mitochondrial membrane polarization, and a coincidental increase in apoptotic cell death. Second, when fission is reduced with IRGM knockdown, or similarly with knockdowns of mitochondrial fission factors Drp1 or Fis1, there is a decrease in mitochondrial ROS production and autophagosome formation in the cell. These results suggest that increased ROS production during mitochondrial fission may promote autophagy, presenting one mechanism through which IRGM may influence autophagy of bacterial phagosomes or cytosolic bacteria indirectly [12]. It is currently not clear how this function may be distinct from the recently defined, and more direct, role for IRGM in autophagosome assembly [68].
Limited studies of murine Irgm1 have yielded similar results: (a) Irgm1 localizes to mitochondria [76] (b) Irgm1-deficient fibroblasts display a more fused mitochondrial pool [76] and (c) overexpression of Irgm1 in fibroblasts leads to a more punctate mitochondrial pool [77]. The effect of overexpression is dependent on a functional GTPase domain in the protein, as well as the presence of an amphipathic helix and palmitoylation domain that mediate mitochondrial membrane association [77]. While it may seem more than coincidental that human IRGM and murine Irgm1 impact mitochondrial dynamics similarly, the mechanism(s) through which each protein does so are currently unknown.
Absence of Irgm1 causes lymphopenia in infected animals and broad susceptibility to many pathogens
The dramatic changes of the mitochondrial network of Irgm1-deficient cells are expected to also change the energy metabolism of the cell. Alterations of mitochondrial metabolism would have various pleiotropic effects. In support of a role for Irgm1 in energy metabolism, homozygous Irgm1 knockout mice tend to be runty and show signs of severe atrophy in multiple tissues (unpublished results). Altered mitochondrial function may also account for the striking observation that proliferating T cells rapidly succumb to a poorly characterized form of cell death, if they lack expression of Irgm1 [78, 79]. This effect is cell-autonomous and can be observed both in tissue culture and in vivo. Accordingly, Irgm1-deficient mice infected with Mycobacterium avium, T. gondii or lymphocytic choriomeningitis virus develop severe lymphopenia and ultimately succumb to these infections, while wildtype mice survive [79]. The infection-induced collapse of the adaptive immune system may contribute to the universal susceptibility of Irgm1−/− mice to pathogens that also include S. typhimurium, L. monocytogenes and Mtb in addition to the pathogens already mentioned [13, 60, 80].
Normal T cell function can be restored in Irgm1-deficient T cells through the additional deletion of Irgm3. Proliferating Irgm1/Irgm3-deficient T cells are protected against cell death that occurs in proliferating Irgm1-deficient T cells [78]. However, Irgm1/Irgm3-deficient mice and cells remain highly susceptible to C. trachomatis infections, indicating that the susceptibility of the single knockout mice to C. trachomatis is not the result of a defective T cell response but rather due to a failure of the cell-autonomous immune response to control intracellular replication of C. trachomatis [14, 40, 78]. The situation is fundamentally different for S. typhimurium and Mycobacteria. Here, the deletion of Irgm3 in an Irgm1−/− mouse background ameliorated the decreased resistance of Irgm1-deficient mice [81]. Therefore, the increased susceptibility of Irgm1−/− mice to Salmonella and mycobacterial infections is best explained with infection-induced lymphopenia rather than a defect in cell-autonomous macrophage immunity. The mechanism by which removal of Irgm3 reveres Irgm1−/− phenotypes remains a mystery and needs to be explored in the future. Another important avenue of future research is to determine whether human IRGM also protects proliferating human T cells from death, and whether T cell death in Irgm1- or human IRGM-deficient cells is the result of mitochondrial dysfunction.
The VLIG gene family is pseudogenized in humans
VLIG genes are exclusively found in vertebrates and absent from other chordates [32]. Similar to the IRG genes, the number of VLIG genes varies greatly between vertebrate taxa: in zebrafish the VLIG family consists of 21 genes and 16 pseudogenes but the chicken genome contains only a single representative. Several vertebrate species including humans appear to have only one or two pseudogenized copies of VLIG [32, 33]. However, the story of human IRGM – once classified as a pseudogene [33] – serves as a cautionary tale and a more detailed analysis of human VLIG expression is warranted to determine whether or not truncated VLIG protein is expressed in humans. Indeed, truncated VLIG proteins are expressed in at least one vertebrate species, namely zebrafish [32]. The molecular weight of the full-length version of canonical VLIG protein as expressed for instance in mice is approximately 280 kDa, making VLIG the largest known GTPase. Because VLIGs are strongly induced by IFNs in mice and in zebrafish [32, 82], these proteins likely fulfill an important function in host defense. However, no functional studies in any organism have been reported up to this point.
GBPs provide immunity to viral, bacterial and protozoan infections
GBPs are a family of 65–73 kDa GTPases that were first isolated as highly expressed proteins in murine and human cells stimulated with IFNγ [6, 83]. Although classified as part of the dynamin superfamily, they are unique members as their primary sequences share little homology with other dynamin proteins [84]. While other GTPases contain a canonical (N/T)(K/Q)xD motif in their G domain, GBPs rather contain a TLRD or TVRD sequence for GTP binding [85]. As typical for dynamin-like proteins GBPs comprise a C-terminal α-helical regulatory domain, an assembly domain, a middle domain, and a GTPase effector domain (GED) [86, 87]. GBPs have the ability to oligomerize in a nucleotide dependent manner and bind GTP in a concentration dependent manner [87, 88].
Like Mx and IRG family members, GBPs likely arose in the ancestor of chordates. GBPs have undergone dynamic evolution in the vertebrate lineage and functional gene copies appear to be present in most vertebrate genomes surveyed including mice and humans [32, 33]. Since their discovery, seven orthologs and one pseudogene have been identified in humans. These human GBP genes are located within one gene cluster on chromosome 1 [89]. In contrast, mice possess eleven genes in addition to two pseudogenes. These murine GBPs are contained within two gene clusters on chromosome 3 and chromosome 5 [85]. Murine and human GBPs share a high degree of homology and, as expected, the most conserved region among GBP proteins is found within the N-terminal G domains [89].
GBPs function in a range of pathways including those involved in cellular proliferation, angiogenesis, and immunity [90, 91]. Although they were discovered in the early 1980s, their role in host defense had not been addressed until the 1990s when human GBP1 (hGBP1) was shown to control infections of vesicular stomatitis virus (VSV) and encephalomyocarditis virus (EMCV) when overexpressed in cell lines [92]. Murine GBP2 (mGBP2) was later discovered to similarly restrict these pathogens [93] and a splice variant of hGBP3 was shown to mediate anti-influenza activity through the repression of the viral polymerase complex [94]. While the loss of GTP binding in hGBP1 and mGBP2 mutant variants abrogated their ability to attenuate viral production of EMCV, these mutations had no effect on the restriction of VSV replication suggesting that distinct GBP restriction mechanisms act specifically upon subsets of viruses [92, 93]. More recently human GBP5 was found to restrict HIV-1 and other retroviruses by interfering with the processing of the viral envelope glycoprotein. It was shown that he antiviral activity of human GBP5 required isoprenylation but not protein oligomerization or GTPases activity [95]. While all of these studies demonstrated that several GBPs could exert some control over viral infections, their role in restriction is relatively weak compared to the antiviral properties of Mx proteins [17].
GBPs were first found to control bacterial or protozoan infections in studies of L. monocytogenes and T. gondii [11, 60]. Mice infected with these pathogens express high levels of numerous GBPs. As described for IRGs, mGBP1 and mGBP2 localized to T. gondii PVs but failed to associate with PVs formed by virulent T. gondii strains [96]. Human GBP1 and GBP2 were later reported to associate with intracellular C. trachomatis and to inhibit bacterial growth when overexpressed [97, 98]. However, we have failed to confirm these findings [99] and propose to reexamine the role for human GBPs in host resistance to C. trachomatis. Nonetheless, these early studies uncovered the importance of the GBP protein family for a range of infections. However, the specific function for individual GBPs remained unclear. Using siRNA knockdown, the role of the entire murine GBP family was examined during L. monocytogenes and M. bovis BCG infections. While many of the GBPs were required for restriction, their relative importance varied with some only providing very minimal protection against these pathogens [100]. In depth analyses demonstrated that mGBP7 regulated the production of ROS through its interactions with the subunits of NADPH oxidase 2 (Nox2) [100]. However, a subsequent study failed to confirm these findings [56]. Co-immunoprecipitation assays also showed mGBP7 bound to the autophagy protein, Atg4b. A different set of proteins that included the ubiquitin binding protein p62 was reported to interact with mGBP1 [100]. The specificities of these protein interactions have yet to be explained.
Disparate responses to different pathogens have also been observed during in vivo analyses. Mice lacking the cluster of Gbps located on chromosome 3 (Gbpchr3−/−), which includes Gbp1, Gbp2, Gbp3, Gbp5 and Gbp7 exhibited decreased survival in response to T. gondii infections [56], which was also observed in Gbp2−/− mice alone [101]. However, during infections of L. monocytogenes, Gbp2−/− mice showed equivalent survival rates to wildtype mice [101], while Gbp1−/− mice displayed significant weight loss and increased L. monocytogenes burden [100]. Together these results suggest a model where different GBPs have variations in either target selection or effector mechanisms during antimicrobial responses. This model is further supported by the recent finding that specific subsets of mGBPs cluster together as supramolecular complexes in vesicle-like structure (VLS) [102]. These clusters may organize mGBPs into groups, which share synergistic or cooperative functions.
GBPs associate with PV membranes and the plasma membrane of T. gondii
An important property of GBPs that contributes to their role in restriction is their ability to target PVs. Similar to IRGs, GBPs colocalize with membranous vesicles shed from PVs suggesting they have a role in the disruption of these structures [56, 103]. In uninfected cells stimulated with IFNγ, GBPs have been found to associate with VLS throughout the cytoplasm [102, 104]. Upon pathogen infection of murine cells, however, the distribution of these proteins changes as they relocate to the sites of infection. While GBPs have minimal variations in structure, the ability of specific members to target PVs can differ dependent on the PV-resident pathogen. Using immunofluorescence assays, mGBP1, mGBP7, and mGBP10 were observed localizing to PVs in macrophages infected with L. monocytogenes or M. bovis BCG, but mGBP3 did not target these structures [100]. In contrast, mGBP3 did associate with T. gondii PVs, which were in turn devoid of mGBP5 [96]. Furthermore, mGBP2 may be unique in its ability to enter the PV space and associate directly with the plasma membrane of the parasite [102]. These data suggest that specific host or pathogen features modulate the targeting of a subset of mouse GBPs although the specifics of these phenotypes have yet to be explained.
The CaaX motif is a C-terminal signal for prenylation, which mediates protein interactions with cellular membranes. GBP1, GBP2, and GBP5 are the only GBPs in both the murine and human families that contain CaaX motifs and these GBPs can be found localizing to various organelles and vesicles [85, 105]. The CaaX motif of mGBP1, however, inherently lacks isoprenylation leading to a cytoplasmic distribution of the protein [104, 106]. While the CaaX motif is required for targeting of individual GBPs to intracellular endomembranes, it is dispensable for their localization to PVs [104, 105]. Mutant forms of mGBP1, mGBP2, and mGBP5 lacking the cysteine residue for prenylation targeted T. gondii vacuoles similar to wildtype proteins [107]. Because unprenylated GBPs can target PVs, alternative lipidation moieties or domains, such as the polybasic region of hGBP1, may mediate membrane interactions [96, 105]. The heterodimerization of GBPs as well as interactions with additional host proteins could promote the localization of unprenylated proteins to PVs [50, 52, 56, 61, 105].
Both GTP binding and GTPase activity are critical for the docking of GBPs to PVs. Mutant mGBP2 and mGBP1 defective in GTP binding failed to target T. gondii or C. trachomatis PVs while GTP-locked mutants targeted PVs with the same efficiency as wildtype proteins [41, 107, 108]. Control over mGBP2 activation regulates its immune function: the Rab guanine dissociation inhibitor (GDI)-α specifically interacts with mGBP2 and maintains mGBP2 in its GDP-bound state. The absence of RabGDI-α results in increased targeting of GTP-bound mGBP2 to T. gondii PVs and in enhanced host resistance [109]. GTP binding promotes GBP homodimerization as well as heterodimerization. In human cells, it was shown that different types of GBP dimers display differential subcellular localization patterns. Homodimers of unprenylated hGBP3 and hGBP4 presented as cytoplasmic or nuclear staining, respectively. Heterodimers of hGBP3 or hGBP4 with prenylated hGBP1, hGBP2, and hGBP5, however, localized to vesicles, the nucleus, or the Golgi, characteristic of the corresponding homodimers of hGBP1, hGBP2, and hGBP5. These data suggest that a hierarchy exists where prenylated proteins such as hGBP2 dictate the targeting of unprenylated GBPs [105].
The GBP and IRG resistance systems functionally interact in mouse cells. A first hint at their functional relationship came from the observation that mGBP2 proteins localize to aggregates of GKS IRGs that form in the cytoplasm of cells lacking Irgm1 or Irgm3 or both genes concomitantly [110]. Further highlighting a link between IRG and GBP systems, targeting of mGBP2 to T. gondii and C. trachomatis PVs is dramatically reduced in Irgm1/Irgm3 double knockout cells [41]. Recent work provides a model to account for these observations; it was shown that the IRG system mediates the recruitment of ubiquitin E3 ligases to T. gondii and C. trachomatis PVs, which results in the attachment of polyubiquitin to PVs and the consequential recruitment of the ubiquitin-binding protein p62 (Figure 2). Because p62 exists together with mGBP2 in cytoplasmic complexes, p62 then escorts mGBP2 to ubiquitinated PVs [52].
Interestingly, GKS targeting to PVs also depends on GBPs, as Gbpchr3−/− cells exhibit a partial decrease in targeting of Irga6, Irgb6 and Irgb10 to T. gondii and C. trachomatis PVs [50, 56]. Further work demonstrated that absence of mGBP2 was sufficient to decrease PV targeting of Irga6 but not Irgb6 and that the absence of mGBP1 led to decreased targeting of Irgb6 but not Irga6 [103, 109], suggesting functional interactions between specific pairs of mGBPs and GKS proteins. To probe for these interactions, co-immunoprecipitation assays were performed using a pan-specific antibody against Gbpchr3 proteins. These studies found an interaction between mGBPs and Irgb6 [56], but the specific GBP interacting with Irgb6 was not defined.
Together these observations suggest a hierarchy amongst IRGs and mGBPs, and that Irgm1/Irgm3-dependent delivery of GKS proteins to T. gondii and C. trachomatis PVs initiates the process of loading PVs with antimicrobial proteins. The initial deposition of GKS proteins triggers the attachment of ubiquitin resulting in the recruitment of GBPs complexed with ubiquitin-binding proteins. Interactions between specific mGBPs and GKS provide a feed-forward mechanism that enhances the efficiency with which GKS and mGBP proteins associate with PVs. While the importance of the IRG system in controlling the deposition of mouse GBPs on T. gondii and C. trachomatis PVs is evident, it is currently unclear how human cells direct GBPs towards their intracellular microbial targets and what these targets are. Because GKS proteins are absent from human cells, we anticipate that the subcellular location of GBPs or its regulation differ between mice and humans. Indeed, our recent findings suggest that T. gondii and C. trachomatis PVs are not targeted by GBPs in human cells [99].
3. IFN-INDUCIBLE GTPASES AS REGULATORS OF INFLAMMATION
Human IRGM alleles associate with increased risks for Crohn’s disease
Much interest in human IRGM has been triggered by its identification as a Crohn’s disease susceptibility locus through genome wide association studies [15, 16]. Several independent studies have identified multiple IRGM gene variants that enhance susceptibility to Crohn’s disease. These variants not only increase the risk of developing Crohn’s disease, but also increase the severity of disease, including ileal involvement [111], fistulating behavior [112], and need for surgery [113]. The associations with Crohn’s disease have been strong among many populations of European origin [111, 112, 114–117], while much weaker in Asian populations [118]. A meta-analysis has indicated that some IRGM gene variants are more tightly associated with susceptibility than others [118].
Most of the IRGM variants occur in non-coding regions of the gene and are thought to affect expression of the protein. For instance, a deletion polymorphism 20.1 kb upstream of the human IRGM gene correlates with decreased expression of IRGM in cultured cells, as well as impaired induction of autophagy and clearance of Salmonella typhimurium [119]. The same allele has been associated with changes in IRGM splicing [34, 119], and thus may also affect the spectrum of IRGM splice variants that are produced in cells. Another IRGM variant (rs11747270) is located 280 bp upstream from the beginning of the fourth exon of IRGM, close to a splice acceptor site, and consequently, may also affect expression of IRGM spliced forms [120]. The IRGM SNP variant, rs9637876, has been significantly associated with increased levels of expression and contributes to protection from intracellular pathogen M. tuberculosis [70]. Yet another IRGM variant, rs10065172, leads to loss of recognition and regulation by miR-196 microRNAs, which bind to and decrease expression of only the wild-type, protective form of IRGM [121]. The overall effect of this IRGM variant, therefore, is to increase expression, although it is important to point out that this differential regulation does not occur in Paneth cells, which may be a cell that is subject to IRGM-mediated regulation (see below). In summary, mounting evidence suggests that IRGM gene variants may either increase or decrease IRGM expression in a cell-specific manner (Table 1). It is possible that maintaining proper IRGM protein levels is imperative, with fluctuations in a positive or negative direction being equally deleterious.
Table1.
List of polymorphisms in human IRGM associated with changes in IRGM expression and human disease susceptibilities in comparison to phenotypes observed in Irgm1-deficient mice.
| Gene Deletion or SNP | Gene Position | Impact on Gene Expression | Crohn’s Disease (human) or Experimental Colitis (mice) | Resistance to Mtb in Humans or Mice in vivo | Bacterial Killing in vitro | Ref | |
|---|---|---|---|---|---|---|---|
IRGM
|
20.1 kb in LD with rs13361189 | −4299 |
|
|
[119] | ||
| rs9637876 | −261 |
|
|
|
[15, 70] | ||
| rs10065172 | +313 |
|
|
[121] | |||
| rs4958847 | +11866 |
|
[15] | ||||
|
Irgm1 |
Irgm1 gene deletion | protein coding region |
|
|
|
|
[13, 76, 80] |
Despite much work to date establishing the Crohn’s disease association, there is little known regarding how IRGM regulates inflammatory homeostasis in the intestine. It is likely to be linked to the emerging role for IRGM in coordinating autophagic responses, as already discussed. Nevertheless, studies in mice have shown that absence of Irgm1 in mice leads to autophagic dysregulation and impairments in a range of immune responses ranging from decreased control of bacterial pathogens in macrophages, to altered T cell homeostasis, enhanced inflammatory cytokine production, and Paneth cell dysfunction [76]. While these mouse studies should be interpreted carefully given differences between mouse and human IRGMs, they illustrate the potentially pleiotropic effects of altered expression of human IRGM. In light of this, as much information as possible should be gleaned from mouse models to infer human IRGM intestinal function, while also undertaking novel approaches to studying the role of human IRGM in the human intestine.
Loss of function in murine Irgm1 increases risk for intestinal inflammation
The role for human IRGM in Crohn’s disease described above has prompted examination of the impact of mouse Irgm proteins on intestinal inflammation, which to date has included a single study examining mice with a complete knockout of Irgm1 [76]. While Irgm1−/− mice display little or no basal intestinal inflammation, when they are acutely exposed to dextran sodium sulphate (DSS), a standard chemical inducer of experimental colitis in mice [122]), they display increases in both ileal and colonic inflammation over those seen in wildtype mice. The ileal inflammation is notable, as this is typically not a component of DSS-induced intestinal inflammation in most susceptible mouse models, while ileitis is, nevertheless, a frequent component of human Crohn’s disease. Accompanying the histologic inflammation in DSS-treated Irgm1−/− mice are enhanced weight loss, colonic shortening, intestinal bleeding, and loss of stool consistency.
The mechanism that underlies the diverse Irgm1−/− phenotypes is currently not clear. Irgm1 is expressed in a variety of cells including intestinal epithelial cells (unpublished results), macrophages [123], and T cells [79]; thus, deficient function in any of those cell types may contribute to the observed phenotypes. In the published study [76] there is clear evidence of altered epithelial responses in Irgm1−/− mice that may or may not reflect a primary dysfunction in the epithelium. Most striking are Paneth cell abnormalities including hyperplasia, ectopic placement along the villi, and altered secretory granule morphology. Further, there are decreased production of Paneth cell secretory products including lysozyme and certain α-defensins. A decrease in Paneth cell function would likely lead to a decreased capacity to maintain homeostasis with the bacterial microbiota in the intestine that could contribute to inflammation. It is striking that this Paneth cell phenotype parallels closely that in autophagy-deficient mice [124] (e.g. Atg16L1 hypomorphic mice [125, 126]), suggesting that altered autophagic responses noted in Irgm1-deficient cells could ultimately instruct the enhanced inflammatory responses in Irgm1−/− mice. An additional phenotype apparent in Irgm1−/− mice is the accumulation of swollen or altered mitochondria in ileal enterocytes. This may reflect the role for Irgm1 in mitochondrial dynamics, as discussed above, or it may be a secondary manifestation of altered autophagy. Thus, there are distinct challenges in examining this model in the future that include determining the cellular origin of inflammation, pinpointing the underlying biochemical defect, and assessing the potential impact of the intestinal flora on the development of inflammation, as specific floral components drive inflammation in autophagy-deficient mice [127].
GBPs are positive regulators of canonical and noncanonical inflammasome activation
Altered function of human IRGM and loss of function in mouse Irgm1 clearly correlate with increased inflammation, yet the cellular and molecular mechanisms responsible for IRGM-linked inflammation are largely obscure, as discussed above. A more refined picture is emerging for the role of GBPs in controlling inflammation. Here, it is evident that GBPs control the activation of inflammasomes, although controversy exists regarding the precise role for GBPs in this activation process.
A function for GBPs in inflammasome activation and the execution of pyroptosis was initially described in studies that demonstrated that overexpression of GBP5 promoted caspase-1-dependent cell death in response to S. typhimurium infections [128]. A later study by the MacMicking lab reported a marked reduction in NLRP3-dependent activities in Gbp5− − macrophages infected with S. typhimurium or treated with potassium efflux agonists [129]. However, using an independently produced Gbp5−/− mouse line, the labs of Broz and Kanneganti failed to confirm these original findings [130, 131]. While different priming conditions could underlie these discrepant results, we propose that strain-specific alleles of Gbp genes adjacent to Gbp5 modify the Gbp5−/− phenotype: the Gbp5−/− mouse published by the MacMicking lab was generated using 129-derived embryonic stem (ES) cells; Gbp5−/− mice were then backcrossed extensively to C57BL/6 mice effectively generating a C57BL/6 mouse carrying a congenic interval of 129-derived DNA that contains the Gbp5−/− locus and the 129 alleles of the adjacent Gbp1, Gbp2, Gbp3 and Gbp7 genes. Contrarily, the Gbp5−/− mice used by the Broz and Kanneganti labs carry C57BL/6 alleles of Gbp1, Gbp2, Gbp3 and Gbp7. These genetic differences in the Gbp gene cluster are likely functionally relevant considering that expression levels of mGBP proteins vary dramatically between different inbred strains of mice [132, 133]. Future studies will need to revisit the specific role for GBP5 in NLRP3 inflammasome activation and determine whether its function is partly redundant with that of other members of the GBP protein family.
Despite the uncertainty surrounding the role of GBP5 in NLRP3 inflammasome activation, studies using Gbpchr3−/− mice lacking the entire cluster of Gbp genes on chromosome 3 have firmly established a functional link between GBPs and the activation of the canonical NLRP3 and AIM2 as well as the noncanonical Caspase-11 inflammasomes. Here, mGBP2 emerged as a critical activator of AIM2 and Caspase-11 inflammasomes [130, 131, 134–136]. To explain how GBPs can promote inflammasome activation, three models have been proposed. The first two models both suggest that GBP-mediated membranolytic activities make pathogen-associated molecular patterns (PAMPs) such as LPS or bacterial DNA more readily available to cytosolic inflammasomes. One model suggests that mGBP2 promotes the lysis of PV membranes, thereby expelling S. typhimurium from its vacuole into the host cell cytosol where it is detected by Caspase-11 [131] (Figure 3). However, mGBPs appeared to play no role in the lysis of Legionella-containing vacuoles but were nonetheless required for the activation of Caspase-11-dependent pyroptosis in response to Legionella pneumophila infections [136]. The second model proposes that mGBP2 binds to cytosolic bacteria and lyses it, thereby releasing PAMPs directly into the cytosol [130, 135] (Figure 3). The latter model is in agreement with recent findings demonstrating the mGBP2 can associate with the parasite plasma membrane of T. gondii, and that mGBP2 eventually localizes to the cytoplasm of lysed, dead parasites [102]. As discussed earlier for IRG-mediated lysis, the mechanism by which GBP-dependent lysis is achieved is currently unknown.
Figure 3. Distinct models to account for the role of GBPs in inflammasome activation.
Three non-mutually exclusive models provide a conceptual framework for the function of GBPs in inflammasome activation: 1) GBPs promote the lysis of S. typhimurium-containing vacuoles thereby releasing the pathogen into the host cell cytosol; 2) GBPs bind to and lyse cytosolic F. novicida prompting the spillage of bacterial DNA into the host cell cytosol; 3) GBPs accelerate the kinetics of caspase-11 and/or caspase-1 oligomerization.
Several lines of evidence point towards a third model by which GBPs provide additional non-lytic functions in inflammasome activation. Injection of LPS into the host cell cytosol by different experimental methods universally triggers Caspase-11 activation that is partly GBP-dependent [136]. GBPs are not essential but rather fulfill an auxiliary role in Caspase-11 activation. Therefore, the defect of Gbpchr3−/− macrophages in noncanonical inflammasome activation is dependent on the type of LPS species and concentration used, and the duration of LPS treatment [131, 136]. Similarly, GBPs are not required but rather enhance the kinetics of both canonical and noncanonical inflammasome activation in macrophages infected with C. muridarum [134]. Interactions of GBPs with inflammasome components and their clustering through GBP oligomerization, as demonstrated for the interaction between GBP5 and NLRP3 [129], likely accelerate inflammasome assembly and drive fast-kinetics inflammasome activation [134] (Figure 3).
The three models that have been proposed to account for the role of GBPs in inflammasome activation are not mutually exclusive. The specific roles of individual GBPs in inflammasome activation need to be defined more carefully in the future. It seems quite reasonable to hypothesize that a subset of GBPs promote bacteriolysis and the release of PAMPs while another subset of GBPs promote inflammasome assembly. Linking these two processes through some shared components may further improve the efficiency of pathogen detection by the inflammasome system.
4. PERSPECTIVES
Ironically, many IFN-inducible GTPases appear to predate the emergence of interferons themselves. Homologs of IRG, GBP and Mx family members are observed in the distantly related cephalochordates, while VLIG genes likely arose in the ancestor of vertebrates [32]. Given the ubiquity of cell-autonomous immune defenses across the tree of life, it is perhaps not surprising that such factors arose and have persisted throughout metazoan evolution. Despite their age, however, IFN-inducible GTPase families have undergone dramatic episodes of gene duplication and loss.
Cases of gene loss may be explained by a lack of sustained selective pressure resulting in eventual pseudogenization. However, it is also possible that gene loss events could be driven by natural selection. Genetic links between IRGM and Crohn’s disease suggest a fitness cost to this activity in the form of autoinflammatory disorders [15, 16]. On the other hand, gene duplications and the resulting expansion of the repertoire of IFN-inducible GTPase may equip the host with the ability to recognize and target a more diverse array of pathogens. Functional studies of GBPs in particular support the notion that specific family members recognize and respond to specific types of pathogens or PVs [96, 100, 102, 136]. Not mutually exclusive, a larger repertoire of these antimicrobial GTPase may increase the number of effector functions directed at a microbial target and allow for the organization of GTPases into separate supramolecular complexes with distinct functional organization [102]. Thirdly, some members of GTPase families may take on decoy functions to overcome microbial immune evasion mechanism, as demonstrated for Irgb2-b1 [38]. Currently, a detailed understanding of the specific function of individual members of the respective GTPase families is lacking and will be a major focus of future research efforts; this is particularly true for the GBP family.
In addition to whole-gene duplication events, IFN-inducible GTPase function has evolved at the level of amino acid substitutions. Recent studies have demonstrated that recurrent positive selection – the spread of new beneficial alleles – has shaped the function of several cell-autonomous immunity factors during evolutionary “arms races” with pathogens [35, 137–139]. Studies by Mitchell et al. recently identified signatures of positive selection in MxA and MxB among anthropoid primates [25, 140]. In the case of MxA, the authors observed that a cluster of rapidly evolving sites in the unstructured loop L4 mediate specificity against influenza and Thogoto virus, pinpointing this region as a critical target specificity domain in MxA. The consequences for rapid evolution on antiviral activity of other Mx family members remain to be determined. Similar evolutionary studies could provide important insights on the contribution of other GTPase families to host range and immunity. Intriguingly, the human GBP4 and GBP7 gene loci contain sequences derived from the Neandertal and Denisovan genomes at enriched frequencies, suggesting positive selection of archaic GBP alleles during human evolution [141].
Reverse and forward mouse genetics approaches have demonstrated that IFN-inducible GTPases are critical mediators of host defense in vivo against viral, bacterial and protozoan pathogens, and it seems likely that these proteins also play a role in immunity to fungal pathogens – although this hypothesis awaits experimental testing. The association of an allelic variant of human IRGM with altered susceptibility to tuberculosis clearly establishes the relevance of IRGM-dependent immunity in human resistance to infectious diseases [70]. Whether and how genetic variants or polymorphisms of additional IFN-inducible GTPases affect the occurrence and outcome of human infections remains to be determined. We can also expect that many pathogens – including those highly adapted to the human host – have evolved strategies to block or circumvent the function of many IFN-inducible GTPases. This notion is already supported by a few examples but many more can be expected to exist. Discovery of these immune evasion strategies and the dissection of their underlying molecular mechanisms provide untapped opportunities to develop novel antimicrobial treatment strategies in the future.
HIGHLIGHTS.
IFN-inducible GTPases mediate host resistance to viral, bacterial and protozoan infections
IFN-inducible GTPases localize to intracellular microbes and solicit defense pathways
Guanylate binding proteins control inflammasome activation
Acknowledgments
This work was supported by American Heart Association predoctoral award 12PRE10440003 (to DP), by a Veterans Administration Merit Review Grant (to GAT), and by National Institute Health grant R01AI57831(to GAT) and R01AI103197 (to JC).
ABBREVIATIONS
- IFN
interferon
- IRG
immunity related GTPase
- GBP
guanylate binding protein
- VLIG
very large IFN-inducible GTPases
- ISRE
Interferon-sensitive response element
- GAS
Interferon-gamma Activated Sequence
- Mx
myxovirus resistance protein
- HIV-1
human immunodeficiency virus type 1
- HBV
hepatitis B virus
- NP
nucleoprotein
- JAK
Janus activated kinase
- STAT
Signal Transducers and Activators of Transcription
- BSE
bundle signalling element
- AIEC
adherent-invasive Escherichia coli
- PIC
pre-integration complex
- ORF
open reading frame
- ER
endoplasmic reticulum
- LD
lipid droplet
- PV
pathogen-containing vacuole
- LC3
microtubule-associated protein 1A/1B light chain 3
- LPS
lipopolysaccharide
- GED
GTPase effector domain
- VSV
vesicular stomatitis virus
- EMCV
encephalomyocarditis virus
- Nox2
NADPH oxidase 2
- BCG
bacillus Calmette-Guérin
- GDI
guanine dissociation inhibitor
- PAMPs
pathogen-associated molecular patterns
- DSS
dextran sodium sulphate
- ROS
radical oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Randow F, MacMicking JD, James LC. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science. 2013;340:701–6. doi: 10.1126/science.1233028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawahara T, Quinn MT, Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol. 2007;7:109. doi: 10.1186/1471-2148-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 5.Horisberger MA, Staeheli P, Haller O. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc Natl Acad Sci U S A. 1983;80:1910–4. doi: 10.1073/pnas.80.7.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng YS, Colonno RJ, Yin FH. Interferon induction of fibroblast proteins with guanylate binding activity. J Biol Chem. 1983;258:7746–50. [PubMed] [Google Scholar]
- 7.Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44:147–58. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- 8.Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Molecular and cellular biology. 1988;8:4518–23. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haller O, Acklin M, Staeheli P. Influenza virus resistance of wild mice: wild-type and mutant Mx alleles occur at comparable frequencies. J Interferon Res. 1987;7:647–56. doi: 10.1089/jir.1987.7.647. [DOI] [PubMed] [Google Scholar]
- 10.Lindenmann J. Resistance of mice to mouse-adapted influenza A virus. Virology. 1962;16:203–4. doi: 10.1016/0042-6822(62)90297-0. [DOI] [PubMed] [Google Scholar]
- 11.Taylor GA, Collazo CM, Yap GS, Nguyen K, Gregorio TA, Taylor LS, et al. Pathogen-specific loss of host resistance in mice lacking the IFN-gamma-inducible gene IGTP. Proc Natl Acad Sci U S A. 2000;97:751–5. doi: 10.1073/pnas.97.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 13.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–9. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc Natl Acad Sci U S A. 2006;103:14092–7. doi: 10.1073/pnas.0603338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller O, Staeheli P, Schwemmle M, Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015;23:154–63. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Holzinger D, Jorns C, Stertz S, Boisson-Dupuis S, Thimme R, Weidmann M, et al. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. Journal of virology. 2007;81:7776–85. doi: 10.1128/JVI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnheiter H, Skuntz S, Noteborn M, Chang S, Meier E. Transgenic mice with intracellular immunity to influenza virus. Cell. 1990;62:51–61. doi: 10.1016/0092-8674(90)90239-b. [DOI] [PubMed] [Google Scholar]
- 20.Pavlovic J, Arzet HA, Hefti HP, Frese M, Rost D, Ernst B, et al. Enhanced virus resistance of transgenic mice expressing the human MxA protein. Journal of virology. 1995;69:4506–10. doi: 10.1128/jvi.69.7.4506-4510.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manz B, Dornfeld D, Gotz V, Zell R, Zimmermann P, Haller O, et al. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog. 2013;9:e1003279. doi: 10.1371/journal.ppat.1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–6. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–62. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, et al. The Interferon-Inducible MxB Protein Inhibits HIV-1 Infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell PS, Young JM, Emerman M, Malik HS. Evolutionary Analyses Suggest a Function of MxB Immunity Proteins Beyond Lentivirus Restriction. PLoS Pathog. 2015;11:e1005304. doi: 10.1371/journal.ppat.1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell PS, Emerman M, Malik HS. An evolutionary perspective on the broad antiviral specificity of MxA. Curr Opin Microbiol. 2013;16:493–9. doi: 10.1016/j.mib.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao S, von der Malsburg A, Dick A, Faelber K, Schroder GF, Haller O, et al. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity. 2011;35:514–25. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–6. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 29.Pavlovic J, Haller O, Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. Journal of virology. 1992;66:2564–9. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krug RM, Shaw M, Broni B, Shapiro G, Haller O. Inhibition of influenza viral mRNA synthesis in cells expressing the interferon-induced Mx gene product. Journal of virology. 1985;56:201–6. doi: 10.1128/jvi.56.1.201-206.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zurcher T, Pavlovic J, Staeheli P. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology. 1992;187:796–800. doi: 10.1016/0042-6822(92)90481-4. [DOI] [PubMed] [Google Scholar]
- 32.Li G, Zhang J, Sun Y, Wang H, Wang Y. The evolutionarily dynamic IFN-inducible GTPase proteins play conserved immune functions in vertebrates and cephalochordates. Mol Biol Evol. 2009;26:1619–30. doi: 10.1093/molbev/msp074. [DOI] [PubMed] [Google Scholar]
- 33.Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, Ventura M, et al. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annual review of genetics. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 36.Behnke MS, Khan A, Wootton JC, Dubey JP, Tang K, Sibley LD. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A. 2011;108:9631–6. doi: 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reese ML, Zeiner GM, Saeij JP, Boothroyd JC, Boyle JP. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc Natl Acad Sci U S A. 2011;108:9625–30. doi: 10.1073/pnas.1015980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lilue J, Muller UB, Steinfeldt T, Howard JC. Reciprocal virulence and resistance polymorphism in the relationship between Toxoplasma gondii and the house mouse. Elife. 2013;2:e01298. doi: 10.7554/eLife.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, et al. Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J. 2008;27:2495–509. doi: 10.1038/emboj.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coers J, Bernstein-Hanley I, Grotsky D, Parvanova I, Howard JC, Taylor GA, et al. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J Immunol. 2008;180:6237–45. doi: 10.4049/jimmunol.180.9.6237. [DOI] [PubMed] [Google Scholar]
- 41.Haldar AK, Saka HA, Piro AS, Dunn JD, Henry SC, Taylor GA, et al. IRG and GBP host resistance factors target aberrant, "non-self" vacuoles characterized by the missing of "self" IRGM proteins. PLoS Pathog. 2013;9:e1003414. doi: 10.1371/journal.ppat.1003414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, et al. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coers J. Self and Non-self Discrimination of Intracellular Membranes by the Innate Immune System. PLoS Pathog. 2013;9:e1003538. doi: 10.1371/journal.ppat.1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Ferreira-da-Silva MF, Springer-Frauenhoff HM, Bohne W, Howard JC. Identification of the microsporidian Encephalitozoon cuniculi as a new target of the IFNgamma-inducible IRG resistance system. PLoS Pathog. 2014;10:e1004449. doi: 10.1371/journal.ppat.1004449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiwari S, Choi HP, Matsuzawa T, Pypaert M, MacMicking JD. Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat Immunol. 2009;10:907–17. doi: 10.1038/ni.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Springer HM, Schramm M, Taylor GA, Howard JC. Irgm1 (LRG-47), a regulator of cell-autonomous immunity, does not localize to mycobacterial or listerial phagosomes in IFN-gamma-induced mouse cells. J Immunol. 2013;191:1765–74. doi: 10.4049/jimmunol.1300641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, et al. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochemical and biophysical research communications. 2009;382:419–23. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 48.Choi J, Park S, Biering SB, Selleck E, Liu CY, Zhang X, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40:924–35. doi: 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Zeer MA, Al-Younes HM, Braun PR, Zerrahn J, Meyer TF. IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS One. 2009;4:e4588. doi: 10.1371/journal.pone.0004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haldar AK, Piro AS, Pilla DM, Yamamoto M, Coers J. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS One. 2014;9:e86684. doi: 10.1371/journal.pone.0086684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohshima J, Lee Y, Sasai M, Saitoh T, Su Ma J, Kamiyama N, et al. Role of mouse and human autophagy proteins in IFN-gamma-induced cell-autonomous responses against Toxoplasma gondii. J Immunol. 2014;192:3328–35. doi: 10.4049/jimmunol.1302822. [DOI] [PubMed] [Google Scholar]
- 52.Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, et al. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A. 2015;112:E5628–37. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao YO, Khaminets A, Hunn JP, Howard JC. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 2009;5:e1000288. doi: 10.1371/journal.ppat.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–71. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams M, Kim K. From membranes to organelles: emerging roles for dynamin-like proteins in diverse cellular processes. Eur J Cell Biol. 2014;93:267–77. doi: 10.1016/j.ejcb.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, et al. A Cluster of Interferon-gamma-Inducible p65 GTPases Plays a Critical Role in Host Defense against Toxoplasma gondii. Immunity. 2012 doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Perry LL, Su H, Feilzer K, Messer R, Hughes S, Whitmire W, et al. Differential sensitivity of distinct Chlamydia trachomatis isolates to IFN-gamma-mediated inhibition. J Immunol. 1999;162:3541–8. [PubMed] [Google Scholar]
- 58.Bernstein-Hanley I, Balsara ZR, Ulmer W, Coers J, Starnbach MN, Dietrich WF. Genetic analysis of susceptibility to Chlamydia trachomatis in mouse. Genes Immun. 2006;7:122–9. doi: 10.1038/sj.gene.6364285. [DOI] [PubMed] [Google Scholar]
- 59.Gazzinelli RT, Mendonca-Neto R, Lilue J, Howard J, Sher A. Innate resistance against Toxoplasma gondii: an evolutionary tale of mice, cats, and men. Cell Host Microbe. 2014;15:132–8. doi: 10.1016/j.chom.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, Woude GF, et al. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med. 2001;194:181–8. doi: 10.1084/jem.194.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khaminets A, Hunn JP, Konen-Waisman S, Zhao YO, Preukschat D, Coers J, et al. Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol. 2010;12:939–61. doi: 10.1111/j.1462-5822.2010.01443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y, Ferguson DJ, Wilson DC, Howard JC, Sibley LD, Yap GS. Virulent Toxoplasma gondii evade immunity-related GTPase-mediated parasite vacuole disruption within primed macrophages. J Immunol. 2009;182:3775–81. doi: 10.4049/jimmunol.0804190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niedelman W, Gold DA, Rosowski EE, Sprokholt JK, Lim D, Farid Arenas A, et al. The rhoptry proteins ROP18 and ROP5 mediate Toxoplasma gondii evasion of the murine, but not the human, interferon-gamma response. PLoS Pathog. 2012;8:e1002784. doi: 10.1371/journal.ppat.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim D, Gold DA, Julien L, Rosowski EE, Niedelman W, Yaffe MB, et al. Structure of the Toxoplasma gondii ROP18 kinase domain reveals a second ligand binding pocket required for acute virulence. J Biol Chem. 2013;288:34968–80. doi: 10.1074/jbc.M113.523266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, et al. Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe. 2010;8:484–95. doi: 10.1016/j.chom.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fleckenstein MC, Reese ML, Konen-Waisman S, Boothroyd JC, Howard JC, Steinfeldt T. A Toxoplasma gondii Pseudokinase Inhibits Host IRG Resistance Proteins. PLoS Biol. 2012;10:e1001358. doi: 10.1371/journal.pbio.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steinfeldt T, Konen-Waisman S, Tong L, Pawlowski N, Lamkemeyer T, Sibley LD, et al. Phosphorylation of mouse immunity-related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii. PLoS Biol. 2010;8:e1000576. doi: 10.1371/journal.pbio.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chauhan S, Mandell MA, Deretic V. IRGM Governs the Core Autophagy Machinery to Conduct Antimicrobial Defense. Mol Cell. 2015;58:507–21. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, et al. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol. 2010;12:1154–65. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Intemann CD, Thye T, Niemann S, Browne EN, Amanua Chinbuah M, Enimil A, et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 72.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008 doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 74.Matsuzawa T, Kim BH, Shenoy AR, Kamitani S, Miyake M, Macmicking JD. IFN-gamma Elicits Macrophage Autophagy via the p38 MAPK Signaling Pathway. J Immunol. 2012 doi: 10.4049/jimmunol.1102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell death and differentiation. 2015;22:377–88. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu B, Gulati AS, Cantillana V, Henry SC, Schmidt EA, Daniell X, et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. American journal of physiology Gastrointestinal and liver physiology. 2013;305:G573–84. doi: 10.1152/ajpgi.00071.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Henry SC, Schmidt EA, Fessler MB, Taylor GA. Palmitoylation of the immunity related GTPase, Irgm1: impact on membrane localization and ability to promote mitochondrial fission. PLoS One. 2014;9:e95021. doi: 10.1371/journal.pone.0095021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coers J, Gondek DC, Olive AJ, Rohlfing A, Taylor GA, Starnbach MN. Compensatory T Cell Responses in IRG-Deficient Mice Prevent Sustained Chlamydia trachomatis Infections. PLoS Pathog. 2011;7:e1001346. doi: 10.1371/journal.ppat.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng CG, Zheng L, Jankovic D, Bafica A, Cannons JL, Watford WT, et al. The immunity-related GTPase Irgm1 promotes the expansion of activated CD4+ T cell populations by preventing interferon-gamma-induced cell death. Nat Immunol. 2008;9:1279–87. doi: 10.1038/ni.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47) J Immunol. 2007;179:6963–72. doi: 10.4049/jimmunol.179.10.6963. [DOI] [PubMed] [Google Scholar]
- 81.Henry SC, Daniell XG, Burroughs AR, Indaram M, Howell DN, Coers J, et al. Balance of Irgm protein activities determines IFN-gamma-induced host defense. J Leukoc Biol. 2009;85:877–85. doi: 10.1189/jlb.1008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klamp T, Boehm U, Schenk D, Pfeffer K, Howard JC. A giant GTPase, very large inducible GTPase-1, is inducible by IFNs. J Immunol. 2003;171:1255–65. doi: 10.4049/jimmunol.171.3.1255. [DOI] [PubMed] [Google Scholar]
- 83.Gupta SL, Rubin BY, Holmes SL. Interferon action: induction of specific proteins in mouse and human cells by homologous interferons. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:4817–21. doi: 10.1073/pnas.76.10.4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nature reviews Molecular cell biology. 2004;5:133–47. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 85.Kresse A, Konermann C, Degrandi D, Beuter-Gunia C, Wuerthner J, Pfeffer K, et al. Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC genomics. 2008;9:158. doi: 10.1186/1471-2164-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng YS, Patterson CE, Staeheli P. Interferon-induced guanylate-binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Molecular and cellular biology. 1991;11:4717–25. doi: 10.1128/mcb.11.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–71. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 88.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–4. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 89.Olszewski MA, Gray J, Vestal DJ. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2006;26:328–52. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- 90.Guenzi E, Topolt K, Lubeseder-Martellato C, Jorg A, Naschberger E, Benelli R, et al. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 2003;22:3772–82. doi: 10.1093/emboj/cdg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guenzi E, Topolt K, Cornali E, Lubeseder-Martellato C, Jorg A, Matzen K, et al. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 2001;20:5568–77. doi: 10.1093/emboj/20.20.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson SL, Carton JM, Lou J, Xing L, Rubin BY. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 1999;256:8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- 93.Carter CC, Gorbacheva VY, Vestal DJ. Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2: differential requirement for wild-type GTP binding domain. Arch Virol. 2005;150:1213–20. doi: 10.1007/s00705-004-0489-2. [DOI] [PubMed] [Google Scholar]
- 94.Nordmann A, Wixler L, Boergeling Y, Wixler V, Ludwig S. A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication. FASEB J. 2012;26:1290–300. doi: 10.1096/fj.11-189886. [DOI] [PubMed] [Google Scholar]
- 95.Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Sturzel CM, et al. Guanylate Binding Protein (GBP) 5 Is an Interferon-Inducible Inhibitor of HIV-1 Infectivity. Cell Host Microbe. 2016;19:504–14. doi: 10.1016/j.chom.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 96.Degrandi D, Konermann C, Beuter-Gunia C, Kresse A, Wurthner J, Kurig S, et al. Extensive characterization of IFN-induced GTPases mGBP1 to mGBP10 involved in host defense. J Immunol. 2007;179:7729–40. doi: 10.4049/jimmunol.179.11.7729. [DOI] [PubMed] [Google Scholar]
- 97.Tietzel I, El-Haibi C, Carabeo RA. Human guanylate binding proteins potentiate the anti-chlamydia effects of interferon-gamma. PLoS One. 2009;4:e6499. doi: 10.1371/journal.pone.0006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Al-Zeer MA, Al-Younes HM, Lauster D, Abu Lubad M, Meyer TF. Autophagy restricts Chlamydia trachomatis growth in human macrophages via IFNG-inducible guanylate binding proteins. Autophagy. 2013;9:50–62. doi: 10.4161/auto.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnston AC, Piro A, Clough B, Siew M, Virreira Winter S, Coers J, et al. Human GBP1 does not localise to pathogen vacuoles but restricts Toxoplasma gondii. Cell Microbiol. 2016 doi: 10.1111/cmi.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–21. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 101.Degrandi D, Kravets E, Konermann C, Beuter-Gunia C, Klumpers V, Lahme S, et al. Murine guanylate binding protein 2 (mGBP2) controls Toxoplasma gondii replication. Proc Natl Acad Sci U S A. 2013;110:294–9. doi: 10.1073/pnas.1205635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kravets E, Degrandi D, Ma Q, Peulen TO, Klumpers V, Felekyan S, et al. Guanylate binding proteins (GBPs) directly attack via supramolecular complexes. Elife. 2016:5. doi: 10.7554/eLife.11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HWt, et al. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 2013;9:e1003320. doi: 10.1371/journal.ppat.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vestal DJ, Gorbacheva VY, Sen GC. Different subcellular localizations for the related interferon-induced GTPases, MuGBP-1 and MuGBP-2: implications for different functions? Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2000;20:991–1000. doi: 10.1089/10799900050198435. [DOI] [PubMed] [Google Scholar]
- 105.Britzen-Laurent N, Bauer M, Berton V, Fischer N, Syguda A, Reipschlager S, et al. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One. 2010;5:e14246. doi: 10.1371/journal.pone.0014246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stickney JT, Buss JE. Murine guanylate-binding protein: incomplete geranylgeranyl isoprenoid modification of an interferon-gamma-inducible guanosine triphosphate-binding protein. Molecular biology of the cell. 2000;11:2191–200. doi: 10.1091/mbc.11.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Virreira Winter S, Niedelman W, Jensen KD, Rosowski EE, Julien L, Spooner E, et al. Determinants of GBP recruitment to Toxoplasma gondii vacuoles and the parasitic factors that control it. PLoS One. 2011;6:e24434. doi: 10.1371/journal.pone.0024434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kravets E, Degrandi D, Weidtkamp-Peters S, Ries B, Konermann C, Felekyan S, et al. The GTPase activity of murine guanylate-binding protein 2 (mGBP2) controls the intracellular localization and recruitment to the parasitophorous vacuole of Toxoplasma gondii. J Biol Chem. 2012;287:27452–66. doi: 10.1074/jbc.M112.379636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ohshima J, Sasai M, Liu J, Yamashita K, Ma JS, Lee Y, et al. RabGDIalpha is a negative regulator of interferon-gamma-inducible GTPase-dependent cell-autonomous immunity to Toxoplasma gondii. Proc Natl Acad Sci U S A. 2015;112:E4581–90. doi: 10.1073/pnas.1510031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Traver MK, Henry SC, Cantillana V, Oliver T, Hunn JP, Howard JC, et al. Immunity-related gtpase M (IRGM) proteins influence the localization of guanylate-binding protein 2 (GBP2) by modulating macroautophagy. J Biol Chem. 2011 doi: 10.1074/jbc.M111.251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561–5. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 112.Latiano A, Palmieri O, Cucchiara S, Castro M, D'Inca R, Guariso G, et al. Polymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn's disease. Am J Gastroenterol. 2009;104:110–6. doi: 10.1038/ajg.2008.3. [DOI] [PubMed] [Google Scholar]
- 113.Sehgal R, Berg A, Polinski JI, Hegarty JP, Lin Z, McKenna KJ, et al. Mutations in IRGM are associated with more frequent need for surgery in patients with ileocolonic Crohn's disease. Diseases of the colon and rectum. 2012;55:115–21. doi: 10.1097/DCR.0b013e31823ccea8. [DOI] [PubMed] [Google Scholar]
- 114.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710–2. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–5. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 116.Weersma RK, Stokkers PCF, Cleynen I, Wolfkamp SCS, Henckaerts L, Schreiber S, et al. Confirmation of Multiple Crohn's Disease Susceptibility Loci in a Large Dutch-Belgian Cohort. American Journal of Gastroenterology. 2009;104:630–8. doi: 10.1038/ajg.2008.112. [DOI] [PubMed] [Google Scholar]
- 117.Palomino-Morales RJ, Oliver J, Gomez-Garcia M, Lopez-Nevot MA, Rodrigo L, Nieto A, et al. Association of ATG16L1 and IRGM genes polymorphisms with inflammatory bowel disease: a meta-analysis approach. Genes Immun. 2009;10:356–64. doi: 10.1038/gene.2009.25. [DOI] [PubMed] [Google Scholar]
- 118.Li Y, Feng ST, Yao Y, Yang L, Xing Y, Wang Y, et al. Correlation between IRGM genetic polymorphisms and Crohn's disease risk: a meta-analysis of case-control studies. Genetics and molecular research : GMR. 2014;13:10741–53. doi: 10.4238/2014.December.18.15. [DOI] [PubMed] [Google Scholar]
- 119.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bekpen C, Xavier RJ, Eichler EE. Human IRGM gene "to be or not to be". Semin Immunopathol. 2010;32:437–44. doi: 10.1007/s00281-010-0224-x. [DOI] [PubMed] [Google Scholar]
- 121.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 122.Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541–6. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 123.Sorace JM, Johnson RJ, Howard DL, Drysdale BE. Identification of an endotoxin and IFN-inducible cDNA: possible identification of a novel protein family. J Leukoc Biol. 1995;58:477–84. doi: 10.1002/jlb.58.4.477. [DOI] [PubMed] [Google Scholar]
- 124.Cadwell K, Patel KK, Komatsu M, Virgin HWt, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–2. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 127.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rupper AC, Cardelli JA. Induction of guanylate binding protein 5 by gamma interferon increases susceptibility to Salmonella enterica serovar Typhimurium-induced pyroptosis in RAW 264.7 cells. Infect Immun. 2008;76:2304–15. doi: 10.1128/IAI.01437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–5. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 130.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015 doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–70. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 132.Nguyen TT, Hu Y, Widney DP, Mar RA, Smith JB. Murine GBP-5, a new member of the murine guanylate-binding protein family, is coordinately regulated with other GBPs in vivo and in vitro. J Interferon Cytokine Res. 2002;22:899–909. doi: 10.1089/107999002760274926. [DOI] [PubMed] [Google Scholar]
- 133.Staeheli P, Prochazka M, Steigmeier PA, Haller O. Genetic control of interferon action: mouse strain distribution and inheritance of an induced protein with guanylate-binding property. Virology. 1984;137:135–42. doi: 10.1016/0042-6822(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 134.Finethy R, Jorgensen I, Haldar AK, de Zoete MR, Strowig T, Flavell RA, et al. Guanylate Binding Proteins enable rapid activation of canonical and noncanonical inflammasomes in Chlamydia-infected macrophages. Infect Immun. 2015 doi: 10.1128/IAI.00856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015 doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. 2014;111:6046–51. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–9. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102:2832–7. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barber MF, Elde NC. Nutritional immunity. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346:1362–6. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mitchell PS, Patzina C, Emerman M, Haller O, Malik HS, Kochs G. Evolution-guided identification of antiviral specificity determinants in the broadly acting interferon-induced innate immunity factor MxA. Cell Host Microbe. 2012;12:598–604. doi: 10.1016/j.chom.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016 doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]