Abstract
Importance
The present study identified potential genetic modifiers that may delay or accelerate age at onset of familial Alzheimer disease (AD) by examining age at onset in PSEN1 mutation carrier families, and further investigation of these modifiers may provide insight into the pathobiology of AD and potential therapeutic measures.
Objective
To identify genetic variants that modify age at onset of AD.
Design, Setting, And Participants
Using a subset of Caribbean Hispanic families that carry the PSEN1 p.G206A mutation, we performed a 2-stage genome study. The mutation carrier families from an ongoing genetic study served as a discovery set, and the cohort of those with LOAD served as a confirmation set. To identify candidate loci, we performed linkage analysis using 5 p.G206A carrier families (n = 56), and we also performed whole-exome association analysis using 31 p.G206A carriers from 26 families. To confirm the genetic modifiers identified from the p.G206A carrier families, we analyzed the GWAS data for 2888 elderly individuals with LOAD. All study participants were Caribbean Hispanics.
Main Outcomes and Measures
Age at onset of AD.
Results
Linkage analysis of AD identified the strongest linkage support at 4q35 (LOD [logarithm of odds] score, 3.69), and the GWAS of age at onset identified variants on 1p13.1, 2q13, 4q25, and 17p11. In the confirmation stage, genewise analysis identified SNX25, PDLIM3, and 3 SH3 domain genes (SORBS2, SH3RF3, and NPHP1) to be significantly associated with LOAD. Subsequent allelic association analysis confirmed SNX25, PDLIM3, and SORBS2 as genetic modifiers of age at onset of EOAD and LOAD and provided modest support for SH3RF3 and NPHP1.
Conclusions and Relevance
Our 2-stage analysis revealed that SNX25, PDLIM3, and SORBS2 may serve as genetic modifiers of age at onset in both EOAD and LOAD.
In 2001, we identified a p.Gly206Ala (g.44636G>C) variant in PSEN1 (OMIM 104311) in members of 8 Caribbean Hispanic families with early-onset Alzheimer disease (EOAD) from Puerto Rico.1 Subsequently, Arnold and colleagues2 also reported this mutation in additional individuals from Puerto Rico but not in other ethnic groups. The mean age at onset among mutation carriers was 55.6 years but was highly variable within families, ranging from 22 to 77 years. The presence of neither an APOE ε4 allele nor any antecedent environmental, health-related, or social factors could explain the differences in the age at onset among mutation carriers, leading us to suspect that other genes were involved in determining when, but not whether, p.G206A carriers develop AD.
Late-onset diseases, including AD,3,4 Huntington disease,5-7 and Parkinson disease,8,9 have been studied for genetic modifiers of age at onset in the presence of a major genetic risk factor (ie, pathogenic mutation), recognizing that there are likely genetic variants that contribute to phenotypic expression. Few studies10,11 have reported gene-gene interaction in AD because different individuals are likely to carry different genetic risk factors. Since there are approximately 185 known PSEN1 variants associated with AD and these variants are rare, most studies will not have sufficient statistical power.12
We conducted a 2-stage study on a set of families multiply affected by AD with at least one carrier of the p.G206A variant in the PSEN1 gene to identify genetic modifiers of age at onset among carriers. In the discovery stage, we used these genetically homogeneous families in which nearly all carriers of the PSEN1 mutation had p.G206A. Using a subset of carrier family members, we applied both genome-wide linkage and exome sequencing analyses (eFigure 1 in the Supplement). In the confirmation stage, we examined allelic association between age at onset and variants in the same candidate genes in a cohort of the same ancestry with late-onset AD (LOAD). We report that variants in SNX25, PDLIM3, SORBS2, and SH3RF3 were associated with variation in age at onset in families with a copy of PSEN1 p.G206A mutation and in LOAD. In addition, at least one variant in NPHP1 was associated with variation in age at onset of AD.
Methods
Discovery Cohort
We conducted a 2-stage study to identify genetic modifiers of age at onset of AD using multiple sets of data in which some were family based and others comprised unrelated individuals (eFigure 1 in the Supplement). The study protocol was approved by the Columbia University Institutional Review Board, and written informed consent was provided by all the participants. Using 56 family members from 5 families with EOAD with the PSEN1 p.G206A mutation, we performed linkage analysis to identify candidate loci. These families represent a subset of PSEN1 carrier families with a wide range of age at onset among affected individuals. Family members resided in the United States, Puerto Rico, Dominican Republic, and countries in South America. Although these families were selected because multiple members had EOAD (age at onset, ≤65 years), many of these families had some family members who had LOAD (age at onset, >65 years). In addition, we performed an exome sequencing experiment and found 31 carriers from 26 families who had multiple affected members and at least 2 who had the PSEN1 p.G206A mutation. Nine of the 31 individuals were also included in the linkage analysis. Given the small sample size of carrier families examined, the main goal of this discovery stage is to prioritize candidate loci.13
Confirmation Cohort
For the LOAD validation experiment, we examined 2888 Caribbean Hispanic elderly people who were noncarriers of the PSEN1 p.G206A mutation and were included in separate genome-wide association studies (GWAS) (eTable 1 in the Supplement).
Genetic Analysis
All persons were genotyped using the 6K Illumina linkage panel single-nucleotide polymorphism (SNP) chips (Illumina Inc). This SNP chip was designed to obtain information for linkage signals. Before analysis, we conducted the standard quality control checks, including checking of the reported family relationships using the software PREST,14 Hardy-Weinberg equilibrium, and genotyping rate. The details of the quality control procedures are presented elsewhere.15,16
Genome-wide Linkage Analysis
To identify regions that may contain variants that could modify age at onset, linkage analyses were performed using the 6K linkage panel SNP data from 56 individuals in 5 families. We examined AD and age at onset as phenotypes. We reasoned that variants that lead to early age at onset are likely to increase the risk of AD, whereas variants that delay age at onset may reduce the risk. For AD phenotype, single-point analysis assumed an autosomal dominant mode of inheritance and used the affecteds-only approach 17 in PSEUDOMARKER.18 For age-at-onset phenotype, we used age at onset for affected individuals and age at last examination for unaffected individuals. We then applied the multipoint variance component method to identify candidate loci as implemented in SOLAR statistical software.19 This variance component model was adjusted for sex, APOE ε4, and AD status.
To prioritize candidate genes under the linkage peak at 4q35.1, the top linkage peak, we screened candidate genes using CANDID, version 1.1.20 Higher weights were given to logarithm of odds (LOD) score (weight, 2) and conservation (weight, 2) compared with literature (which included the keywords Alzheimer disease, neurodegeneration, early onset, and beta amyloid) (weight, 1) and gene expression (weight, 1).
Exome Sequencing
Whole-exome capture libraries were constructed from DNA from whole blood of the samples after sample shearing, end repair, phosphorylation, and ligation to bar-coded sequencing adaptors (eFigure 2 in the Supplement). The ligated DNA was subjected to exonic hybrid capture using the Nimble exome capture array (http://www.nimblegen.com/seqcapez/). Samples were multiplexed and sequenced on multiple Illumina HiSeq flow cells for a mean target exome coverage of 80× to generate paired-end reads of 90 base pairs.
Read Mapping, Variant Calling, and Downstream Bioinformatics Analyses
Prealignment quality control of the read data to identify failed runs and lanes was performed using the R BioConductor package: ShortRead (http://www.bioconductor.org/). Sequence alignment was performed using the aligner BWA. Polymerase chain reaction duplicates (reads) were removed using Picard tools. Base quality recalibration and realignment around indels were performed using the Genome Analysis Toolkit (https://www.broadinstitute.org/gatk/). Multisample variant calling and quality control of the call were then performed using the Genome Analysis Toolkit. On the basis of the hypothesis that the mutation underlying this rarer form of EOAD was not present in the general population, SNPs identified in the 1000 Human Genomes (HG) project (version 2010-11; http://www.1000genomes.org) or in dbSNP (build 134; http://www.ncbi.nlm.nih.gov/projects/SNP) were removed. Exonic coding variants were identified and annotated by ANNOVAR (http://annovar.openbioinformatics.org/en/latest/). Non-exonic and synonymous variants were filtered from the variant list using predictions from SIFT (Sorting Intolerant From Tolerant) software, version 4.0 (http://sift.jcvi.org).
Genome-wide Association Analysis
Using 31 individuals with exome sequencing data, we examined candidate genes under the top linkage peak and the rest of the exome by applying a mixed linear model adjusting for sex, APOE genotype, affection status, and kinship coefficient. The kinship coefficient matrix using the R functions21 was included in the model to take into account relatedness among family members.
Replication Using the LOAD Hispanic GWAS Data Set: Genewise Analysis
To determine whether candidate genes identified from the EOAD cohort were similarly associated with age at onset of AD in the LOAD cohort of the same ethnic background, we performed gene wise association analysis using family-based SKAT software (FAMSKAT),22 adjusting for sex, AD status, and APOE ε4. This approach was necessary because (1) multiple variants within one gene had modest effects on age at onset and (2) most of the variants identified from exome sequencing were absent in the GWAS data of Caribbean Hispanics.
Imputation of the LOAD Hispanic GWAS Data Set
Imputations of 5 genes (SNX25, PDLIM3, SORBS2, SH3RF3, NPHP1) were performed based on cosmopolitan phased haplotypes of 1000 HG, version 2010–11 (data freeze, 2012-03-04 haplotypes; http://csg.sph.umich.edu//abecasis/MaCH/download/1000G.2012-03-14.html). PLINK, version 1.07 (http://pngu.mgh.harvard.edu/∼purcell/plink/) was used for imputation. Before imputing, a number of filters were implemented in the Hispanic GWAS genotypic data by removing markers that had a minor allele frequency less than 1%, a Hardy-Weinberg equilibrium P <10−6, Hispanic GWAS SNP alleles mismatched with those of 1000 HG project and not present in the 1000 HG panel, and flipping of any SNP when appropriate to the forward strand. A total of 348 924 SNPs for 2 regions of chromosome 2 (175 288 SNPs, approximately 14 Mbp) and chromosome 4 (173 636 SNPs, approximately 10 Mbp) were imputed. For single-variant and single-trait association with imputed genotypes, 2 additional filters were implemented: minor allele frequency greater than 1% and an information score greater than 0.8 (a quality score from the imputation), which reduced the analysis to 2636 variants within the 3 genes.
Association Analysis and Meta-analysis
We applied the same multivariate linear mixed model that adjusted for sex, AD status, and APOE ε421 as above to test the allelic association between age at onset and variants in LOAD. We then conducted a meta-analysis to assess whether the SNPs in the 3 candidate genes were significantly associated with the LOAD GWAS data sets. For this purpose, we estimated the meta-analysis of P values from 4 studies as implemented in the METAL (http://csg.sph.umich.edu//abecasis/metal/), which estimates a single summary P value across 3 data sets. An overall z statistic and P value were calculated while taking into account the number of individuals examined in each study.
Results
Study Participants
Table 1 details the 3 sets of Caribbean Hispanics investigated: (1) families who have multiple affected members and at least one carrying the PSEN1 p.G206A mutation were analyzed using linkage analysis; (2) carriers of the PSEN1 p.G206A mutation from families who have multiple affected members were analyzed using exome sequencing; and (3) 3 confirmatory GWAS data sets of Caribbean Hispanics primarily composed of LOAD were studied,15,23 which were analyzed using a linear mixed model to assess allelic association. All 3 data sets had both family members and unrelated individuals, and appropriate statistical methods were applied to account for nonindependence.
Table 1. Demographic and Clinical Characteristics of Sequenced Participants and Family Membersa.
| Characteristic | Genotyped (n = 56 [5 Families]) | Sequenced (n = 31 [26 Families]) | Combined GWAS (N = 2888) |
|---|---|---|---|
| Affection status | |||
| Affected | 21 (37.5) | 26 (83.9) | 1473 (51.0) |
| Unaffected | 32 (57.1) | 5 (16.1) | 1408 (48.8) |
| Unknown | 3 (5.4) | 0 | 6 (0.2) |
| Sex, male:female | 19:37 (33.9:66.1) | 12:19 (38.7:61.3) | 970:1918 (33.6:66.4) |
| Age at onset or age at last examination, mean (SD), y | 57.2 (7.0) | 57.9 (9.9) | 74.0 (10.0) |
| Educational level, mean (SD), y | 10.0 (4.6) | 9.1 (5.2) | 5.7 (4.7) |
| APOE frequency | |||
| E4 | 23 (20.5) | 19 (30.6) | 1238 (23.0) |
| E3 | 85 (75.9) | 43 (69.4) | 3849 (71.5) |
| E2 | 4 (3.6) | 0 (0.0) | 293 (5.5) |
Abbreviation: GWAS, genome-wide association study.
Data are presented as number (percentage) of participants unless otherwise indicated. The combined GWAS data set has a total of 2324 unrelated individuals (2321 families and 3 individuals who married into the families).
Linkage Analysis Set
Of 56 family members included in the linkage analysis, 21 (37.5%) were affected, and 32 (57.1%) were unaffected. A total of 37 (66.1%) of the family members were women, the mean (SD) age at onset of AD was 57.2 (7.0) years (range, 43.0-73.0 years), and the mean educational level was 10.0 years. The overall allele frequency of APOE ε4 was 20.5%.
Exome Sequencing Set
Of 31 PSEN1 p.G206A mutation carriers from 26 families who were included in the exome sequencing experiment, 26 (83.9%) of the mutation carriers had been clinically diagnosed as having AD. The other persons were unaffected carriers. Nineteen participants (61.3%) were women, the mean (SD) age was 57.9 (9.9) years (range, 44.0-77.0 years), and the mean educational level was 9.1 years. Most mutation carriers reported Puerto Rico as the country of origin, whereas the others were from the Dominican Republic.
Caribbean Hispanic GWAS Set
Among 2888 individuals in the confirmation set, 1473 (51.0%) of the individuals were affected with AD, and 1918 (66.4%) were women. Most affected individuals had LOAD, and their mean age at onset was 74.0 years (range, 30.0-105.0 years). The proportion of APOE ε4 carriers (23.0%) was comparable to those with EOAD family members.
Genome-wide Linkage Analysis of Families Carrying the PSEN1 p.G206A Mutation
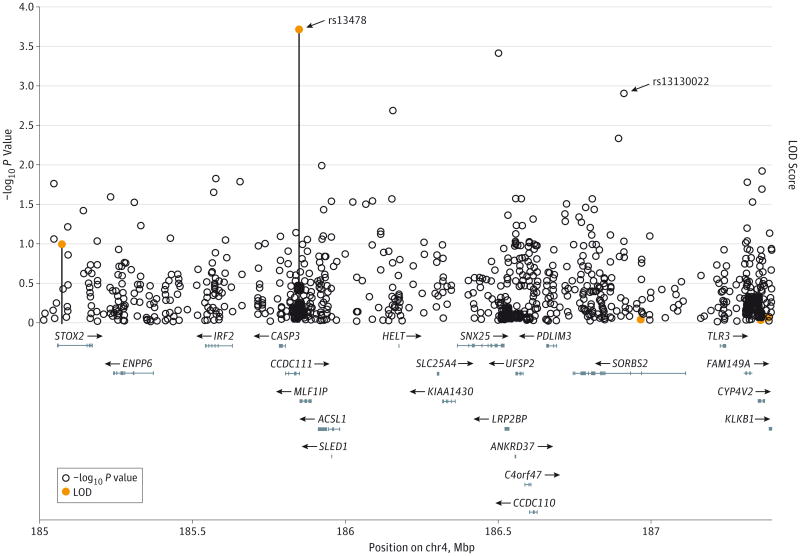
For the purpose of gene discovery, we conducted genome-wide linkage analyses of AD and age at onset using 5 multiplex families with variable age at onset. The linkage analysis of AD identified the strongest linkage support for rs13478 at 4q35.1 (LOD score, 3.69), and 3 additional SNPs (rs1024026, rs1983503, rs2036912) from the 6K linkage panel were suggestive (Figure). This locus included multiple genes in linkage disequilibrium, including C4orf41, STOX2, MLF1IP, SORBS2, and FAM149A. Our subsequent informatics analysis based on the CANDID algorithm20 identified 3 top candidate genes: SLC25A4, SORBS2, and KIAA1430. On the other hand, the linkage analysis of age at onset that adjusted for sex, AD status, and APOE ε4 did not identify any SNPs with suggestive or significant LOD scores. An additional model that included the presence or absence of the PSEN1 p.G206A variant in addition to sex, AD status, and APOE ε4 yielded the highest LOD score of 0.75.
Figure. Results From Linkage and Association Analysis for 4q35.
The bars represent logarithm of odds (LOD) scores from single-point linkage analysis, and points without the bar represent −log10 P values from association analysis. Approximate locations for relevant genes as well as physical locations on chromosome 4 (chr4) are presented at the bottom of the figure.
Targeted and Genome-wide Association Analysis of PSEN1 p.G206A Carriers
Using 31 PSEN1 p.G206Amutation carriers, we performed targeted association analysis with the genes located under the linkage peak at 4q35.1. In addition, we performed genome-wide association analysis of age at onset in the rest of the exome by applying a multivariate linear mixed model that adjusted for sex, AD status, APOE ε4, and kinship coefficient.21 The linear mixed model was applied to take into account 5 individuals who were related to others included in the exome sequencing experiment. Under the linkage peak, several SNPs provided modest support for association with age at onset, but we focused on SORBS2 because of the strong support from the informatics analysis and the linkage analysis. The effect of the minor allele of rs13130022 in SORBS2 was associated with a delay in age at onset by 11.1 years (see β coefficients in Table 2). In the remaining exome, the strongest signal for age at onset was observed at rs906815 in NPHP1 at 2q13 (P = 4.51× 10−6). Specifically, individuals carrying the rare A allele for rs906815 had an age at onset approximately 11.7 years earlier for having one copy of the A allele compared with those who do not, thereby leading to nearly a difference of 2 decades in age at onset (ie, 44 vs 64.17). In addition, the effect variants in SNX25 and SH3RF3 were associated with a delay in age at onset by 8.8 and 9.3 years, respectively, whereas the effect variant in PDLIM3 was associated with an earlier onset by 12 years. In addition, rs696662 on 1p13.1 (P = 6.8 × 10−6), rs906815 on 2q13 (P =4.5 × 10−6), and rs3026115 on 17p13.2 (P = 7.8 × 10−5) had SNPs with strong support for allelic association.
Table 2. Summary of Linkage, Allelic, and Genewise Association Analyses in EOAD and LOAD.
| Gene | EOAD Exome Sequencing–Allelic Association | LOAD-GWAS | ||||
|---|---|---|---|---|---|---|
| SNP | Location, bp | β | P Value | No. of SNPsa | P Value for Genewise Analysis | |
| Linkage Signal | ||||||
| 4q35 | ||||||
| STOX2 | rs28411264 | 184 908 447 | 13.01 | .03851 | 12 | .83 |
| ENPP6 | rs13147555 | 185 074 686 | −6.06 | .03024 | 28 | .002 |
| MLF1IP | rs1401359 | 185 640 216 | −5.11 | .09064 | 58 | 7.22 × 10−6 |
| SNX25 | rs11730401 | 186 268 129 | 8.75 | .0004 | 17 | 1.26 × 10−15 |
| PDLIM3 | rs28522047 | 186 446 018 | −11.97 | .1218 | 13 | 1.17 ×10−11 |
| SORBS2 | rs13130022 | 186 678 161 | 11.11 | .0013 | 143 | 3.80 × 10−6 |
| FAM149A | rs2276920 | 187 078 938 | 7.11 | .01671 | 14 | .04 |
| Association Signal | ||||||
| 2q13 | ||||||
| SH3RF3 | rs6542814 | 109 928 460 | 9.30 | .001 | 157 | 2.70 × 10−11 |
| NPHP1 | rs906815 | 110 942 496 | −11.69 | 4.51 × 10−6 | 12 | 5.47 × 10−5 |
Abbreviations: bp, base pair; EOAD, early-onset Alzheimer disease; GWAS, genome-wide association study; LOAD, late-onset Alzheimer disease; SNP, single-nucleotide polymorphism.
The SNP with the strongest P value from the exome sequencing experiment is given.
To be conservative, we performed one additional analysis of age at onset of AD in which we restricted the analysis to affected individuals only. This analysis yielded comparable results as the survival analysis approach. For example, rs906815 in NPHP1 had a P value of 4.13 × 10−6, and other SNPs that were significant in the earlier analysis remained statistically significant, but the P values were somewhat weakened (eTable 2 in the Supplement).
Replication in Individuals With LOAD: Genewise Analysis
To determine whether the variants discovered from the PSEN1 p.G206A carrier families were likely to be associated with variation in age at onset in LOAD, we first conducted genewise association analysis using FAMSKAT22 because the Hispanic GWAS data sets did not include the variants identified from exome sequencing of the discovery set. We confirmed that SNX25 (P = 1.26 × 10−15), PDLIM3 (P = 1.17 × 10−11), SORBS2 (P = 3.8 × 10−6), NPHP1 (P = 5.47 × 10−5), and SH3RF3 (P = 2.7 × 10−11) were associated with age at onset in Caribbean Hispanics with LOAD, further supporting that these genes may be involved functionally in altering phenotypic expression in AD.
Replication in Individuals With LOAD: SNP-wise Analysis
To further explore whether allelic association persists within those candidate genes in LOAD, we examined SNPs from the 3 GWAS data sets of Caribbean Hispanic ancestry, focusing on the SNPs in the same linkage disequilibrium block as the original discovery variants. Because the GWAS data sets did not have genotypes for many of the variants discovered from exome sequencing, we performed imputation using PLINK24 and then used both genotype and imputed SNPs for allelic association. We limited the use of imputed SNPs to those with an information score greater than 0.8 by PLINK analysis. As shown in eFigure 3 in the Supplement, pairwise D′ values across the chromosomal regions encompassing the discovery variants in candidate loci were high and were comparable across the 3 GWAS data sets.
Table 3 indicates that multiple genotyped or imputed data in the candidate genes were associated with age at onset in LOAD. In this confirmation analysis, 14 SNPs in SNX25 were associated with age at onset at P < .05, and variants were associated with 1 to 6 years of difference in age at onset in LOAD. For PDLIM3, 3 SNPs were associated with variation in age at onset and were associated with a difference of 2.6 to 3.6 years in mean age at onset. For SORBS2, 11 of 1246 SNPs or indels flanking the original discovery variant were significantly associated with age at onset of LOAD at P < .05, and the variants were associated with 1 to 4.6 years of mean age difference in onset. It is of interest to note that, for SORBS2, the same allele was associated with age at onset in all 3 GWAS data sets, and the allele frequency for the associated SNP was consistent across data sets, ranging from 0.032 to 0.222 with a median of 0.194. For SH3RF3, 5 SNPs were associated with age at onset at P < .05 in LOAD, and the difference in mean age at onset ranged from 1 to 2.9 years. For NPHP1, only one SNP was associated with age at onset.
Table 3. Meta-analysis in Late-Onset Alzheimer Disease and the Difference in Mean Age at Onset by Genotype Among Caribbean Hispanics.
| SNP | Location, bp (HG19) | A1a | A2a | Freq1a | z Score | P Value | Directionb | S1c | S2c | S3c | Differenced |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNX25(4q35) | |||||||||||
| rs72706074 | 186 135 900 | G | A | 0.072 | 2.07 | .03821 | +−+ | I | I | I | 1.2 |
| rs72706075 | 186 136 511 | C | A | 0.071 | 2.07 | .03847 | +−+ | I | I | I | 1.0 |
| rs111645866 | 186 137 102 | A | C | 0.071 | 2.07 | .03847 | +−+ | I | I | I | 1.0 |
| rs72708003 | 186 143 673 | T | C | 0.043 | −2.40 | .01634 | −−− | I | I | I | −0.4 |
| rs73873407 | 186 157 838 | C | T | 0.019 | −2.25 | .02472 | −−− | I | I | I | −6.2 |
| rs73873412 | 186 167 895 | G | C | 0.035 | −2.42 | .01538 | ?−− | I | I | −2.1 | |
| rs73873414 | 186 169 190 | C | A | 0.035 | −2.42 | .01538 | ?−− | I | I | −2.1 | |
| rs73873415 | 186 171 421 | T | C | 0.035 | −2.42 | .01538 | ?−− | I | I | −2.2 | |
| rs73873416 | 186 174 202 | G | A | 0.034 | −2.15 | .03193 | ?−− | I | I | −2.1 | |
| rs10028004 | 186 177 378 | G | A | 0.021 | −2.06 | .03991 | −−? | I | I | −1.9 | |
| rs114698167 | 186 188 966 | A | C | 0.048 | 2.09 | .0368 | ??+ | I | 1.8 | ||
| rs72709980 | 186 207 628 | A | T | 0.034 | 2.50 | .01242 | +?? | I | 2.8 | ||
| rs140555723 | 186 241 606 | G | A | 0.042 | 2.54 | .01122 | +++ | I | I | I | 2.7 |
| rs1288550 | 186 259 737 | T | C | 0.034 | 2.32 | .02016 | +?+ | I | I | 2.4 | |
| PDLIM3(4q35) | |||||||||||
| rs1385580 | 186 414 852 | C | T | 0.036 | −2.33 | .02006 | −?? | I | −3.4 | ||
| rs143417134 | 186 415 740 | T | C | 0.010 | −2.34 | .01955 | −?? | I | −3.6 | ||
| rs79071575 | 186 442 445 | T | C | 0.027 | −2.35 | .01877 | −?? | I | −2.6 | ||
| SORBS2(4q35) | |||||||||||
| rs146271237 | 186 662 630 | C | T | 0.043 | −1.97 | .04856 | −−− | I | I | I | −4.6 |
| rs10009306 | 186 663 793 | C | T | 0.144 | −2.17 | .02995 | −−? | G | I | −1.0 | |
| rs7679506 | 186 664 542 | G | T | 0.153 | −2.55 | .01084 | −−− | G | G | G | −1.0 |
| rs1039239 | 186 667 811 | A | G | 0.169 | 2.09 | .03679 | +++ | I | I | I | 0.2 |
| rs13130022e | 186 678 161 | C | T | 0.213 | 1.90 | .05763 | +++ | I | I | I | 1.5 |
| rs1021678 | 186 678 768 | A | G | 0.204 | 2.30 | .02155 | +++ | I | I | I | 1.6 |
| rs12715855 | 186 687 554 | A | C | 0.203 | 2.19 | .02827 | +++ | I | I | I | 1.6 |
| 186687667 | 186 687 667 | TCA | T | 0.204 | 2.30 | .02155 | +++ | I | I | I | 1.6 |
| 186687668 | 186 687 668 | CACAT | C | 0.203 | 2.44 | .01451 | +++ | I | I | I | 1.7 |
| rs1039236 | 186 688 918 | T | C | 0.203 | 2.19 | .02827 | +++ | I | I | I | 1.6 |
| rs1039235 | 186 690 013 | A | T | 0.203 | 2.44 | .01451 | +++ | I | I | I | 1.7 |
| 186691125 | 186 691 125 | G | GC | 0.202 | 2.34 | .0193 | +++ | I | I | I | 1.7 |
| SH3RF3(2q13) | |||||||||||
| rs72939527 | 109 917 895 | T | C | 0.012 | 2.18 | .0295 | ?++ | I | I | 2.9 | |
| rs6542814e | 109 928 460 | .001 | 1.1 | ||||||||
| rs11679797 | 109 973 646 | G | T | 0.026 | 1.99 | .0462 | ?+? | I | 1.1 | ||
| rs13013000 | 109 982 224 | T | G | 0.034 | 2.16 | .0307 | ?+? | I | 0.8 | ||
| rs11674377 | 109 984 093 | A | G | 0.034 | 2.16 | .0307 | ?+? | I | 1.1 | ||
| rs113309962 | 109 987 615 | T | C | 0.011 | −2.37 | .0179 | −?? | I | … | ||
| NPHP1(2q13) | |||||||||||
| rs17842680 | 110 906 930 | G | A | 0.041 | 1.97 | .04942 | +++ | I | I | I | 0.9 |
| rs906815e | 110 942 496 | A | G | 0.282 | 0.31 | .7547 | −++ | I | G | G | −0.9 |
Abbreviations: A1, allele 1; A2, allele 2; bp, base pair; Freq1, frequency of allele 1; G, genotype; HG19, human genome assembly 19; I, imputed; S1, GWAS dataset 1; S2, GWAS dataset 2; S3, GWAS dataset 3; SNP, single-nucleotide polymorphism.
Minor allele and its frequency.
Direction of the β coefficient in 3 Hispanic genome-wide association data sets; plus sign indicates age at onset increases with dosage of allele; minus sign, age at onset decreases; and question mark, not computed because quality of imputation is poor.
I and G in each Hispanic genome-wide data set, where S1 represents dataset 1, S2, dataset 2, and S3 dataset 3. Imputed SNPs with an information score greater than 0.8 by PLINK analysis were used; otherwise, imputed SNPs were dropped, representing blank cells.
Difference in mean age by genotype. When available, mean ages at onset of 2 homozygotes were used to compute the difference in mean onset. When a homozygous rare variant was absent, mean age at onset of heterozygous genotype was used.
The SNPs from the discovery.
Table 4 summarizes reanalysis of the 10 SNPs from the 2 candidate genes (SORBS2 and SH3RF3) using genotyped data. The rationale for genotyping these subsets of SNPs from Table 3 was that the information score for the imputed SNPs in those 2 genes did not reach the satisfactory quality control threshold of 0.8 according to our PLINK analysis; thus, the results from the meta-analysis based on smaller data sets are likely to be less reliable when compared with other SNPs with high information scores. When genotyped data were used, rs72939527 in SH3RF3 had the strongest support for allelic association (P = .007). The T allele in rs72939527 was associated with a 2.7-year earlier age at onset compared with the reference allele, whereas other variants did not reach statistical significance (P = .22-.47).
Table 4. Reanalysis of SH3RF3 and SORBS2 in Late-Onset Alzheimer Disease Using Genotype Data and Difference in Age at Onset of Alzheimer Disease by Genotype.
| Chromosome | SNP | Location, bp (HG19) | A1 | No. of Genotyped Persons | Age-at-Onset Phenotype | Alzheimer Disease Phenotype | Onset Difference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β | Statistic | P Value | β | Statistic | P Value | ||||||
| SH3RF3a | |||||||||||
| 2 | rs72939527 | 109 917 895 | T | 1705 | −2.863 | −2.692 | .007 | 1.123 | 2.624 | .009 | −2.9 |
| 2 | rs6542814 | 109 928 460 | A | 1697 | 0.374 | 1.231 | .22 | 1.092 | 2.537 | .01 | −0.2 |
| 2 | rs11679797 | 109 973 646 | G | 1703 | −1.007 | −1.032 | .30 | 1.091 | 2.543 | .01 | −1.1 |
| 2 | rs13013000 | 109 982 224 | T | 1697 | −0.695 | −0.729 | .47 | 1.114 | 2.593 | .01 | −0.8 |
| 2 | rs11674377 | 109 984 093 | A | 1699 | −1.037 | −1.073 | .28 | 1.100 | 2.559 | .01 | −1.1 |
| 2 | rs17842680 | 110 906 930 | G | 1700 | 0.686 | 1.054 | .29 | 1.028 | 2.385 | .02 | 0.9 |
| 2 | rs906815 | 110 942 496 | A | 1702 | −0.329 | −1.015 | .31 | 1.116 | 2.596 | .01 | −0.2 |
| SORBS2 | |||||||||||
| 4 | rs10009306 | 186 663 793 | C | 1700 | 0.103 | 0.244 | .81 | 1.077 | 2.504 | .01 | 0.2 |
| 4 | rs1021678 | 186 678 768 | A | 1700 | 0.272 | 0.700 | .48 | 1.111 | 2.583 | .01 | 0.7 |
Abbreviations: A1, allele 1; bp, base pair; HG19, human genome assembly 19; SNP, single-nucleotide polymorphism.
rs113309962 (109987615bp) was homozygous.
Discussion
We focused on founder mutation PSEN1 p.G206A with high penetrance to identify genetic modifiers of age at onset of EOAD and LOAD and report that variants in SNX25, PDLIM3, SORBS2, SH3RF3, and NPHP1 may contribute to variation in age at onset in EOAD and LOAD. This study of a founder mutation allows identification of a second locus that may alter the effect of the highly penetrant, nonsynonymous PSEN1 p.G206A variant. This approach overcomes some of the limitations that are common in studies that examine gene-gene interaction in variants with modest to weak effect sizes. Furthermore, changes in age at onset by genotype among PSEN1 p.G206A carrier families ranged from 10 to 20 years, whereas those among individuals with LOAD ranged from 1 to 6.2 years. This finding suggests that the difference in age at onset by genotype is likely to be smaller in noncarriers compared with G206A carriers.
Familial EOAD is caused by mutations in 3 genes: APP, PSEN1, and PSEN2.25-28 Support clearly exists for genetic modifiers because age at onset varies widely for carriers of APP, PSEN2, and PSEN1. To date, only a few studies3,4,11 have reported modifiers. Investigators have attributed observed variable age at onset in PSEN2 to the APOE ε4 allele.3,4 For PSEN1, Vélez and colleagues11 conducted a pooling- and bootstrap-based GWAS to identify multiple modifiers. Among those, they observed that rs10173717 in NPHP1 may alter age at onset in PSEN1 p.Glu280Ala carriers. However, rs10173717 reported by Vélez et al11 was located 40.5 kilobases (kb) away from our own discovery signal rs906815 and 76.1 kb away from our GWAS signal rs17842680. The authors did not report the difference in age at onset by genotype.
In this study, 5 promising genetic modifiers were identified among families multiply affected by AD: SNX25, PDLIM3, SORBS2, SH3RF3, and NPHP1. SNX25 has been implicated in regulating endosome sorting and signaling.29,30 Given its potential role in regulating membrane protein trafficking excess levels of amyloid β in individuals with the PSEN1 p.G206A variant, this gene may be a biologically relevant modifier. This hypothesis is further supported by an earlier report31 that found that SNX3, a gene in the same family, was associated with AD, and SNX3 regulates and interacts with the retromer membrane. PDLIM3 is reported to be associated with diabetes and cardiomyopathy.32 Although there have been reports of insulin-related genes, little is known about the role of PDLIM3 in dementia to date. Interestingly, however, the PDZ domain is regulated by SNX27 retromer protein, and deficiencies in retromers have been associated with AD.33
The remaining 3 candidate genes encode proteins with a functional domain, SH3. These evolutionarily conserved domains are defined by sequence homology and are involved in protein-protein interaction.34,35 SH3 domains are ubiquitous intracellular protein modules; structural and functional studies34-37 reveal how they interact with their praline-rich ligands to promote the formation of specific protein aggregates. The SH3 domain is related to the WW and PTB domains37 and may interact with APP to modulate the degradation of APP.35 The SORBS2 gene—an SH3-binding domain gene located in 4q35.1 under the strongest linkage peak—transcribes a brain-specific splice variant known as nARGBP2.38 This variant was reported to influence the integrity off-actin in the dendritic spines of neurons. Cestra and colleagues39 observed that overexpression of nARGB2 in mice caused aggregation of f-actin bundles in dendritic spines and may influence phenotypic expression. Because alteration of synaptic shape may be associated with the biological progression of AD, variants in SORBS2 may influence variation in age at onset by affecting dendritic spine morphologic and subsequent neurodegenerative processes.
SH3RF3 isan important paralog of SORBS2. Because paralogs arise from gene duplication within the same species and we found allelic association between age at onset and SORBS2 as well as SH3RF3, SH3RF3 is a biologically plausible gene. In the present data set, the rare homozygous genotype carriers had age at onset approximately 20 years earlier than those with the wild-type homozygous genotype under the model that adjusted for sex, AD, APOE ε4, and kinship coefficient. These results suggest that some individuals harbor a variant in SH3RF3 that may modify the effect of the PSEN1 p.G206A mutation on the disease phenotype. We note that the SH3 domain is found in proteins of signaling pathways regulating the cytoskeleton. They also regulate the activity state of adaptor proteins (which mediate specific protein-protein interactions that drive the formation of protein complexes) and other tyrosine kinases (which transfer a phosphate group from adenosine tri-phosphate to a protein in a cell, thereby functioning as an on-and-off switch). Thus, it is plausible that this gene may be involved in variable gene expression of related genes, which in turn modify the age at onset of disease.
NPHP1, another SH3 domain gene, encodes a nephrocystin-1 protein that may influence cell division and cell signaling localized to microtubule-based structures.40 This gene has been reported to be associated with retinitis pigmentosa, nephronophthisis, and neurodegeneration with brain iron accumulation 1. This disease is associated with dementia as part of the phenotypic spectrum, supporting biological relevance of this gene in the neurodegenerative process.
This study has some limitations. Age at onset of AD phenotype is inherently difficult to measure accurately because it is difficult to detect prodromal symptoms unless each individual is followed up longitudinally from preclinical stage to development of AD using sensitive biomarkers. Thus, reported age at onset is an approximation. In addition, the genetic variants identified from this study may be relatively common in Caribbean Hispanics but not in the general US populations. Thus, the identified variants themselves may have limited value in other ethnic cohorts. However, our earlier studies41-43 have found that when 2 cohorts of different ethnicity are studied, the same set of genes tends to be implicated, but the effect size associated with each variant differs somewhat. For example, the effect size for each SNP vary among Caribbean Hispanics, African Americans, and whites41,42; however, all 3 ethnic groups had SNPs that were significantly associated with AD within the gene. Similarly, SNPs in ABCA7 were associated with AD in African Americans and whites, but the effect sizes differed.43
Conclusions
This study of carriers of the PSEN1 p.G206A mutation has identified variants in SNX25, PDLIM3, and a family of SH3 domain genes (SORBS2, SH3RF3, NPHP1) that may modify age at onset of EOAD. Furthermore, variants in SORBS2 consistently were associated with delayed onset in EOAD and LOAD, although less profoundly so in LOAD. On the other hand, the discovery variant and flanking variants in SH3RF3 and NPHP1 were significantly associated with EOAD, but their effect size was modest with LOAD. One possible explanation for lack of association in the present data set may be that the effect size for SH3RF3 and NPHP1 may be relatively modest, and a larger study may be needed to detect significant association. The present findings are worthy of further investigation to understand the pathobiology of AD.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by grants R37-AG15473, P50-AG08702, and R01AG037212 from the National Institute on Aging, National Institutes of Health and grant R01-MH084995 from the National Institute of Mental Health.
Role of the Funder/Sponsor: Genentech contributed to the design, acquisition of sequencing and GWAS data, bioinformatics analysis of the sequence data, and review of the manuscript but did not play a role in management of the data, statistical genetics analysis, approval of the manuscript, and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Lee and Mayeux had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Lee, Cheng, Mayeux.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Lee, Mayeux.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Lee, Cheng, Vardarajan, Bhangale.
Obtained funding: Mayeux.
Administrative, technical, or material support: Lee, Lantigua, Reyes-Dumeyer, Ortmann, Behrens, Medrano, Jiménez-Velázquez, Mayeux.
Study supervision: Lee, Mayeux.
Supplemental content at jamaneurology.com
Conflict of Interest Disclosures: Drs Ortmann, Graham, Bhangale, and Behrens reported being full-time employees of Genentech, Inc. No other disclosures were reported.
Additional Contributions: Jessica Russo, BA, provided thorough examination of the candidate genes. The Columbia University team performed analyses of genotype-phenotype relations in the PSEN1 p.G206A carrier families and the GWAS of LOAD. Ms Russo was compensated for her work.
References
- 1.Athan ES, Williamson J, Ciappa A, et al. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. JAMA. 2001;286(18):2257–2263. doi: 10.1001/jama.286.18.2257. [DOI] [PubMed] [Google Scholar]
- 2.Arnold SE, Vega IE, Karlawish JH, et al. Frequency and clinicopathological characteristics of presenilin 1 Gly206Ala mutation in Puerto Rican Hispanics with dementia. J AIzheimers Dis. 2013;33(4):1089–1095. doi: 10.3233/JAD-2012-121570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchani EE, Bird TD, Steinbart EJ, et al. Evidence for three loci modifying age-at-onset of Alzheimer's disease in early-onset PSEN2 families. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(5):1031–1041. doi: 10.1002/ajmg.b.31072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wijsman EM, Daw EW, Yu X, et al. APOE and other loci affect age-at-onset in Alzheimer's disease families with PS2 mutation. Am J Med Genet B Neuropsychiatr Genet. 2005;132B(1):14–20. doi: 10.1002/ajmg.b.30087. [DOI] [PubMed] [Google Scholar]
- 5.Metzger S, Walter C, Riess O, et al. REGISTRY Investigators of the European Huntington's Disease Network. The V471A polymorphism in autophagy-related gene ATG7 modifies age at onset specifically in Italian Huntington disease patients. PLoS One. 2013;8(7):e68951. doi: 10.1371/journal.pone.0068951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrow M, Chua P, Churchyard A, Bradshaw JL, Chiu E, Georgiou-Karistianis N. Proximity to clinical onset influences motor and cognitive performance in presymptomatic Huntington disease gene carriers. Cogn Behav Neurol. 2006;19(4):208–216. doi: 10.1097/01.wnn.0000213914.64772.b6. [DOI] [PubMed] [Google Scholar]
- 7.Djoussé L, Knowlton B, Hayden MR, et al. Evidence for a modifier of onset age in Huntington disease linked to the HD gene in 4p16. Neurogenetics. 2004;5(2):109–114. doi: 10.1007/s10048-004-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thobois S, Ribeiro MJ, Lohmann E, et al. French Parkinson's Disease Genetics Study Group. Young-onset Parkinson disease with and without parkin gene mutations. Arch Neurol. 2003;60(5):713–718. doi: 10.1001/archneur.60.5.713. [DOI] [PubMed] [Google Scholar]
- 9.Foroud T, Uniacke SK, Liu L, et al. Parkinson Study Group. Heterozygosity for a mutation in the parkin gene leads to later onset Parkinson disease. Neurology. 2003;60(5):796–801. doi: 10.1212/01.wnl.0000049470.00180.07. [DOI] [PubMed] [Google Scholar]
- 10.Lalli MA, Garcia G, Madrigal L, et al. Exploratory data from complete genomes of familial Alzheimer disease age-at-onset outliers. Hum Mutat. 2012;33(12):1630–1634. doi: 10.1002/humu.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vélez JI, Chandrasekharappa SC, Henao E, et al. Pooling/bootstrap-based GWAS (pbGWAS) identifies new loci modifying the age of onset in PSEN1 p.Glu280Ala Alzheimer's disease. Mol Psychiatry. 2013;18(5):568–575. doi: 10.1038/mp.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33(9):1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terwilliger JD. On the resolution and feasibility of genome scanning approaches. Adv Genet. 2001;42:351–391. doi: 10.1016/s0065-2660(01)42032-3. [DOI] [PubMed] [Google Scholar]
- 14.Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54(2):99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Cheng R, Barral S, et al. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. 2011;68(3):320–328. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wijsman EM, Pankratz ND, Choi Y, et al. NIA-LOAD/NCRAD Family Study Group. Genome-wide association of familial late-onset Alzheimer's disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet. 2011;7(2):e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knapp M. Alternative test for linkage between two loci. Genet Epidemiol. 1998;15(5):511. doi: 10.1002/(SICI)1098-2272(1998)15:5<511::AID-GEPI5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Hiekkalinna T, Schäffer AA, Lambert B, Norrgrann P, Göring HH, Terwilliger JD. PSEUDOMARKER: a powerful program for joint linkage and/or linkage disequilibrium analysis on mixtures of singletons and related individuals. Hum Hered. 2011;71(4):256–266. doi: 10.1159/000329467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutz JE, Kraja AT, McLeod HL, Province MA. CANDID: a flexible method for priorityzing candidate genes for complex human traits. Genet Epidemiol. 2008;32(8):779–790. doi: 10.1002/gepi.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Therneau T, Atkinson E, Sinnwell J, Matsumoto M, Schaid D, McDonnell S. [Accessed June 15, 2015];Package ‘kinship2’. 2012 http://cran.r-project.org/web/packages/kinship2/kinship2.pdf.
- 22.Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet Epidemiol. 2013;37(2):196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Center for Biotechnology Information. [Accessed 2014];dbGaR. 2014 http://www.ncbi.nlm.nih.gov/
- 24.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St George-Hyslop PH, Petit A. Molecular biology and genetics of Alzheimer's disease. C R Biol. 2005;328(2):119–130. doi: 10.1016/j.crvi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Lendon CL, Ashall F, Goate AM. Exploring the etiology of Alzheimer disease using molecular genetics. JAMA. 1997;277(10):825–831. [PubMed] [Google Scholar]
- 27.Schellenberg GD. Genetic dissection of Alzheimer disease, a heterogeneous disorder. Proc Natl Acad Sci USA. 1995;92(19):8552–8559. doi: 10.1073/pnas.92.19.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardy J. The Alzheimer family of diseases. Proc Natl Acad Sci USA. 1997;94(6):2095–2097. doi: 10.1073/pnas.94.6.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Petralia RS, Kurushima H, et al. Arc/Arg3.1 regulates an endosomal pathway essential for activity-dependent β-amyloid generation. Cell. 2011;147(3):615–628. doi: 10.1016/j.cell.2011.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wassmer T, Attar N, Harterink M, et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell. 2009;17(1):110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vardarajan BN, Bruesegem SY, Harbour ME, et al. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol Aging. 2012;33(9):2231.e15–2231.e30. doi: 10.1016/j.neurobiolaging.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohno K, Masuda A. RNA pathologies in neurological disorders. In: Blass JP, editor. Neurochemical Mechanisms in Disease. New York, NY: Springer Science and Business Media; 2011. pp. 399–415. [Google Scholar]
- 33.Muhammad A, Flores I, Zhang H, et al. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Aβ accumulation. Proc Natl Acad Sci U S A. 2008;105(20):7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolis B, Borg JP, Straight S, Meyer D. The function of PTB domain proteins. Kidney Int. 1999;56(4):1230–1237. doi: 10.1046/j.1523-1755.1999.00700.x. [DOI] [PubMed] [Google Scholar]
- 35.Sudol M. From Src homology domains to other signaling modules: proposal of the “protein recognition code”. Oncogene. 1998;17(11 reviews):1469–1474. doi: 10.1038/sj.onc.1202182. [DOI] [PubMed] [Google Scholar]
- 36.Morton CJ, Campbell ID. SH3 domains. Curr Bbl. 1994;4(7):615–617. doi: 10.1016/s0960-9822(00)00134-2. [DOI] [PubMed] [Google Scholar]
- 37.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92(17):7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hand D, Eiden LE. Human sorbin is generated via splicing of an alternative transcript from the ArgBP2 gene locus. Peptides. 2005;26(7):1278–1282. doi: 10.1016/j.peptides.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Cestra G, Toomre D, Chang S, De Camilli P. The Abl/Arg substrate ArgBP2/nArgBP2 coordinates the function of multiple regulatory mechanisms converging on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102(5):1731–1736. doi: 10.1073/pnas.0409376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mollet G, Silbermann F, Delous M, Salomon R, Antignac C, Saunier S. Characterization of the nephrocystin/nephrocystin-4 complex and subcellular localization of nephrocystin-4 to primary cilia and centrosomes. Hum Mol Genet. 2005;14(5):645–656. doi: 10.1093/hmg/ddi061. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Cheng R, Schupf N, et al. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64(4):501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reitz C, Jun G, Naj A, et al. Alzheimer Disease Genetics Consortium. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ε4, and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309(14):1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.