Abstract
Transverse relaxation rate measurements in MAS solid-state NMR provide information about molecular motions occurring on nanoseconds-to-milliseconds (ns-ms) time scales. The measurement of heteronuclear (13C, 15N) relaxation rate constants in the presence of a spin-lock radio-frequency field (R1ρ relaxation) provides access to such motions, and an increasing number of studies involving R1ρ relaxation in proteins has been reported. However, two factors that influence the observed relaxation rate constants have so far been neglected, namely (i) the role of CSA/dipolar cross-correlated relaxation (CCR), and (ii) the impact of fast proton spin flips (i.e. proton spin diffusion and relaxation). We show that CSA/D CCR in R1ρ experiments is measurable, and that this cross-correlated relaxation rate constant depends on ns-ms motions, and can thus itself provide insight into dynamics. We find that proton spin-diffusion attenuates this cross-correlated relaxation, due to its decoupling effect on the doublet components. For measurements of dynamics, the use of R1ρ rate constants has practical advantages over the use of CCR rates, and the present manuscript reveals factors that have so far been disregarded and which are important for accurate measurements and interpretation.
Keywords: protein, rotating frame relaxation, numerical spin simulations, spin-diffusion, J-doublet
Introduction
The structure of a protein is determined by a large number of individually weak interactions, which constantly re-shuffle at ambient temperatures. This dynamic exchange between different conformers is often important for functions such as binding or enzymatic turnover. NMR spectroscopy is very well suited to provide direct atomic-level access to the amplitudes and time scales of such motions. Whereas solution-state NMR is a well-established tool for such studies, magic-angle spinning (MAS) solid-state NMR (ssNMR) is rapidly establishing in studies of biomolecular dynamics, complementing its solution-state counterpart in cases where limited solubility or large size hamper the application of solution-state methods. In addition to opening new fields of applications, ssNMR also has theoretical advantages for determining dynamics, compared to solution-state NMR. In particular, the absence of overall tumbling motion enables the study of motions without “blind windows” of time scales, i.e. over all time scales from picoseconds to seconds, in contrast to its solution-state counterpart, which is severely challenged on the nanosecond to several-microseconds time scale.1 In MAS ssNMR, motions can be studied through (i) their averaging effect on anisotropic interactions (in particular, the averaging of dipolar couplings and chemical-shift anisotropies), which report on the amplitude of motion seen by these interactions, and (ii) through the investigation of nuclear spin relaxation rate constants, which depend on amplitudes and time scales of motions. Many biological processes, such as binding, gating or enzymatic turnover, occur on time scales of hundreds of nanoseconds to milliseconds (ns-ms). Studying these time scales is, therefore, of particular relevance when attempting to decipher the molecular basis of these processes. In MAS ssNMR, motion on these time scales can be accessed by measurements of transverse relaxation rate constants, such as the decay of spin coherence, R2. However, quantitative measurements of transverse relaxation rates in ssNMR are generally hampered by the fact that spin coherences in solid samples decay through two mechanisms, (i) due to random field fluctuations (i.e., dynamics), which is what we are interested in herein, and (ii) due to evolution under the incompletely averaged anisotropic interactions, in particular due to 1H-1H dipolar couplings. The latter, termed dipolar dephasing, strongly depends on the presence of (multiple) dipolar couplings, and on the employed MAS frequency. Accessing the dynamics-induced component requires that coherent contributions to signal decay (in particular dipolar dephasing) are suppressed to negligible levels. Several reported studies have shown that heteronuclear ( 13C, 15N) R1ρ rate constants, i.e. the decay of Cx (Nx) in the presence of a spin-lock RF field, can be measured accurately without significant interfering effects from dipolar dephasing if (i) high MAS frequencies and/or (ii) extensive deuteration are applied.2–5 R1ρ experiments under such conditions have been reported so far for 15N and 13C’ sites in small model proteins (SH3, GB1, ubiquitin).2–7 R1ρ experiments in solids are particularly informative also when the RF field strength approaches the rotary resonance conditions, as has been shown for small molecules8,9 and proteins.4,6
As an alternative method to probe ns-ms motions, it has been proposed to measure cross-correlated relaxation (CCR) between 1H-15N dipolar/15N chemical-shift anisotropy (CSA) interactions in R2’-type experiments (i.e. in the absence of a spin-lock RF field). These latter experiments measure the difference of the apparent transverse decay rate constants of the two 15N scalar-coupling doublet components, i.e. the decay of Nx-[Hα] and Nx-[Hβ] components in a spin-echo experiment.10 Even though each individual rate constant itself contains contributions from dipolar dephasing (and can, therefore, not be interpreted quantitatively), the respective difference can be quantitatively interpreted in terms of dynamics on ns-ms time scales.11 This experiment suffers, however, from the fact that the R2’ decay rates in MAS ssNMR are large, because of the dipolar-dephasing contribution. As a consequence, detection of a small differential relaxation rate from two individually large relaxation rate constants corresponding to the two doublet components (R2’+ΓCSA/D and R2’-ΓCSA/D) is challenging. Furthermore, the rapid signal loss throughout the experiment, based on scalar-coupling transfers,10 results in relatively low detection sensitivity.
Here we investigate the effects of dipolar/CSA CCR in heteronuclear ( 13C, 15N) R1ρ experiments in MAS ssNMR. In solution-state NMR, such cross-correlated relaxation effects have been reported12,13, but have so far been neglected in MAS ssNMR. We use explicit numerical spin dynamics simulations, as well as experimental measurements, and show that CCR rate constants under spin-lock RF fields are measurable, and that they report on molecular motion. The CCR rate constant has a similar dependence on dynamics as the in-phase R1ρ rate constant, as we show with numerical simulations. We assume here that the CCR rate constant in R1ρ measurements is essentially free from coherent contributions, for several reasons. First, it has been convincingly shown that CCR rate constants, derived from the above-mentioned experiments with free evolution (R2’) are primarily reflecting dynamics, even though each of the two doublet components itself is not free from coherent contributions.14 In R1ρ- based experiments the situation can be assumed to be even more favorable, because the decay of each of the two components is much less prone to coherent decay.3
Furthermore, we show that experimentally determined R1ρ-derived CCR rate constants (referred to here as ) reveal the expected patterns of motions in a well-studied protein, and correlate with R1ρ rate constants, which have been shown to contain only minimal coherent contributions. However, both CCR and R1ρ contain contributions from stochastic proton flips: we provide evidence that proton spin-diffusion reduces the CCR rate constants due to averaging of the doublet components. Furthermore, proton spin diffusion also leads to an increase of the auto-relaxation (R1ρ) rate constants, in a manner that depends on the RF field strength. Our experimental data and simulations thus suggest that knowledge of the proton spin-diffusion rate is required for accurately determining dynamics, either through in-phase R1ρ measurements or CCR measurements under spin-lock. Finally, we also discuss pulse schemes to suppress the effects of CCR, and thus eliminate the bi-exponentiality of decay in in-phase R1ρ measurements.
Materials and Methods
Samples and NMR spectroscopy
In this study we investigate the cross-correlated relaxation between a heteronuclear CSA tensor (15N) and the 1H-15N dipolar coupling in amide moieties; our conclusion apply also to other 1H-X spin pairs, such as 1H-13C in backbone or side chain sites. We use u-[2H,15H]-labelled ubiquitin, crystallized with 2-methyl-2,4-pentane-diol (MPD)15, obtained by crystallization from a solution containing an effective H2O/D2O ratio of 60/40 (taking into account protons originating from MPD). Exchangeable sites, such as amides are protonated at a level of 60%, in an otherwise deuterated background. Protein samples were filled into 1.6 mm Agilent MAS rotors by ultracentrifugation using a dedicated filling device (in-house built). NMR experiments were performed on a 600 MHz Agilent VNMRS spectrometer equipped with a 1.6 mm MAS probe tuned to 1H,13C,15N frequencies, spinning at 38 kHz MAS frequency. The effective sample temperature, calibrated with internal standard using the bulk water frequency, was set to 300 K. Pulse sequences used in this study are shown in Figure S1 in the Supporting Information, and use 1H-15N out-and-back cross-polarization, and 1H detection. In particular, the sequence in Figure S1B was used for the CCR measurement, and (in-phase) R1ρ rate constants with and without 1H π pulse decoupling are shown in Figure S1A and C, respectively. Finally, the proton spin-diffusion experiment was performed with the sequence shown in Figure S1D. 15N spin-lock RF field frequencies were calibrated with a nutation experiment: after initial 1H-15N cross-polarization, a 5 ms spin-lock period was applied (to take into account possible detuning during a spin-lock, as this is relevant for a R1ρ experiment), followed by a 90 degree phase-shifted nutation pulse. If this pulse is a π/2 pulse, then the signal is zero. We determined the amplitude of this pulse by parametric incrementation. We find that two alternative calibration methods result in very similar calibrations: (i) a simple nutation pulse, applied at the beginning of the solvent-saturation period (see Figure S1), i.e. disregarding the spin-lock period, and (ii) the observation of the rotary resonance condition. For the latter, the signal intensity is monitored as a function of the 15N spin-lock RF field strength in an experiment that applies a 5 ms spin-lock just after the initial 1H-15N CP transfer (Figure S1A); a minimum is observed when the RF field matches the n=1 rotary-resonance condition νRF= νMAS. We find that the three methods provide essentially identical calibration results. Processing and analysis was done with nmrPipe16, and NMRView (OneMoon Scientific) as well as in-house written scripts in python language. Error estimates of rate constants were based on spectral noise estimates (3 times standard deviation of the signal in empty regions of the spectrum).
Numerical simulations
Simulations of the evolution of spin coherences were performed using in-house written programs using the GAMMA simulation library.17 The simulation approach includes two spins (1H, 15N), existing in two exchanging sub-systems, spanning a composite Liouville space.1,11,18,19 The simulation approach, as well as numerous results of similar kinds of simulations were reported recently.1 Briefly, the evolution of the appropriate coherences (Nx, 2NxHα, 2NxHβ, Hz) is followed over the course of a 15N spin-lock. The two exchanging sub-states differ in the orientations of the 1H-15N dipolar coupling tensor and 15N CSA tensor. The interaction tensor parameters were assumed as follows: 1H-15N dipolar coupling 22954 Hz (corresponding to 1.02 Å bond length), axially symmetric 15N tensor with Δσ=-170 ppm, i.e. σzz=-113 ppm (6780 Hz at 14.1 T B0 field strength). The 1H CSA parameters have negligible effect in this context. The time-domain data were fitted with python scripts in order to obtain relaxation rate constants, using mono- or bi-exponential functions as reported in the text.
Results and Discussion
We first aim at providing a quantitative understanding of the effects of CSA/D CCR in R1ρ experiments by numerical simulations. Although a very well-established theoretical framework for nuclear spin relaxation exists (Redfield theory20), this theory is not strictly valid in cases of slow motion, which are the focus of this paper. Recent systematic analyses have established that Redfield results may disagree with numerical simulations,1,7 even in cases where the actual relaxation is more than one order of magnitude slower than the dynamic event. In order to circumvent these problems we resort to numerical simulations for investigating the properties of R1ρ relaxation in rotating solids. The simulation approach is based on an explicit jump model of a 1H-15N moiety exchanging between two conformations. The employed composite Liouville-space approach has been used similarly before11,18,19.
Figure 1 shows an example of a simulated decay of Nx coherence for a two-site exchange, using powder averaging and MAS. The observed decay (red curve in Figure 1A) is clearly non-exponential, and the exponential fit (black) unsatisfactorily describes the decay. The non-exponentiality may be ascribed to two effects. The first is that the NMR interactions that lead to relaxation (dipolar coupling, CSA) are orientation-dependent, and therefore the relaxation rate constants are orientation-dependent, too. When summing all contributions from different crystallite orientations across the sample, as required when considering a sample with randomly oriented crystallites, the resulting relaxation is always multi-exponential, as has been described and reviewed earlier.1,21,22 If this is the only reason for non-monoexponential behavior, then individual crystallites (i.e. with a single orientation within the rotor-fixed frame) should show mono-exponential decays. However, we find that also when using single crystallites rather than a powder distribution the decay is bi-exponential (data not shown), pointing to an additional mechanism.
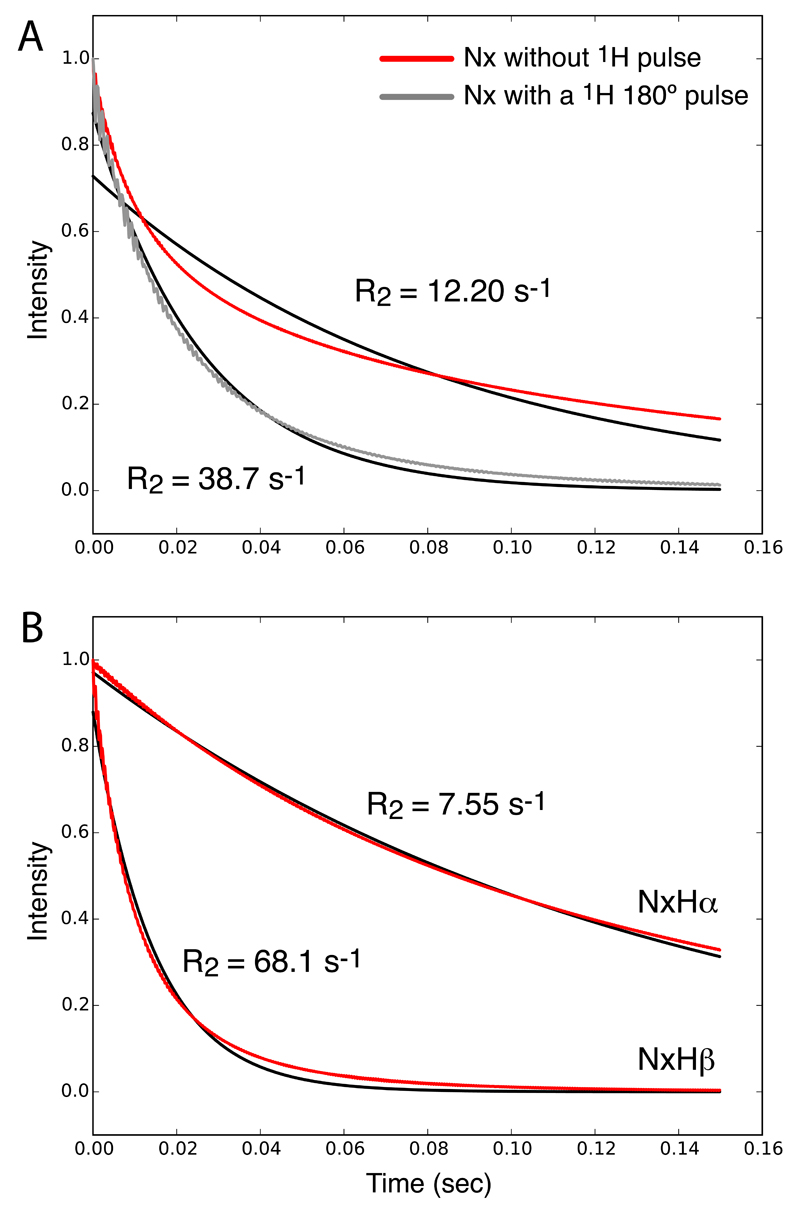
Figure 1.
Simulated evolution of 15N spin coherences under a spin-lock RF field. (A) Time evolution of in-phase Nx coherence (red) and a single-exponential fit, which clearly results in poor agreement with the decay. For the simulation shown in grey, a 1H π pulse was inserted in the center of the spin-lock period. An exponential fit is shown in black. (B) Simulation of the doublet components, Nx-[Hα], Nx-[Hβ] (red), and exponential fits (black). The jump angle was 20 degrees and the jump rate constant 30000 s-1.
This second possible origin of non-exponential decay is the presence of cross-correlated relaxation due to 15N-CSA/1H-15N dipolar-coupling interference. When observing the two components of the initial Nx coherence, namely the coherences Nx-[Hα] and Nx-[Hβ], the decays are nearly exponential when integrated over a powder (Figure 1B). If one considers individual crystallites then the decay of the two coherences are indeed perfectly exponential (Figure S2C,D). One can readily show that the bi-exponential decay of Nx is due to interference of CSA and dipolar mechanisms: removing either of the two mechanisms (i.e. setting the jump angle for one of the two interactions to zero, or, equivalently, setting the interaction strength to zero) leads to identical relaxation of the two components, and mono-exponential decay of Nx (data not shown). Furthermore, if a 1H π pulse is inserted, thus inverting the sign of the dipolar interaction, the near-monoexponential decay is restored, as shown in Figure 1A (grey line). This is expected, as the effect of the cross-correlated relaxation is eliminated.
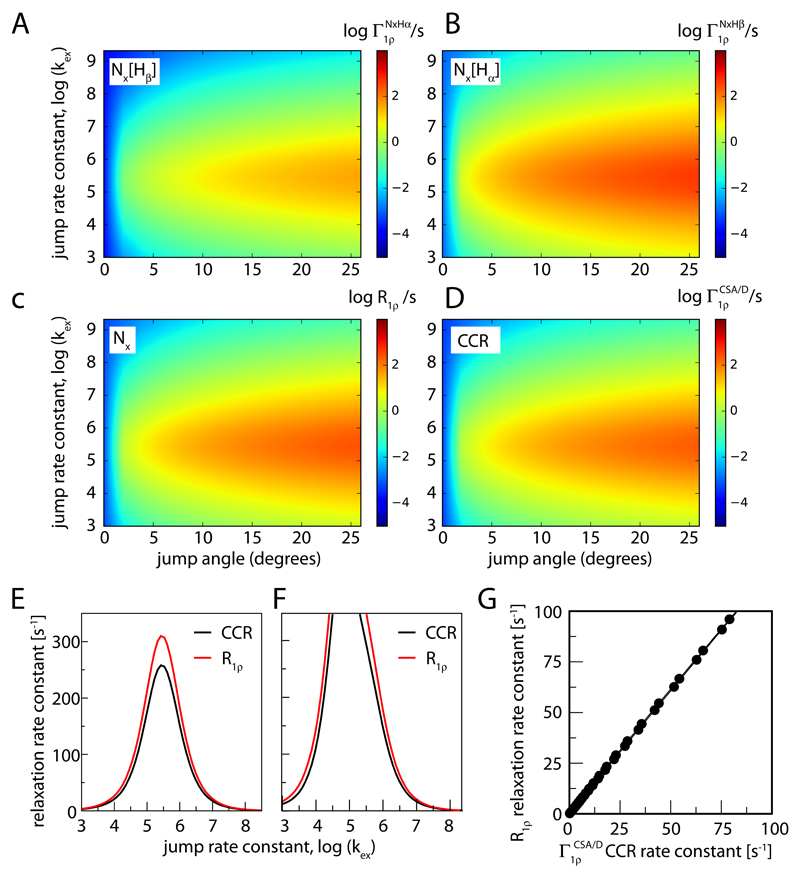
Numerical simulations also allow determining the difference of the rate constants of the two components, i.e. the CSA/D CCR rate constant in the R1ρ experiment, which we term here . Figure 2 establishes that the differential relaxation rate constant depends on the amplitude and time scale of the motion, as expected for a cross-correlated relaxation, and on the difference of RF field strength and MAS frequency, again as expected.1,23 It exhibits a maximum when the motion is on the time scale of microseconds to tens of microseconds, in a manner that depends on the spin-lock RF field strength (Figure 2 E, F).
Figure 2.
Simulated relaxation rate constants of a 15N spin due to its CSA tensor and 1H-15N dipolar coupling, within the two-site jump model. Shown are rate constants for the individual doublet components (A, B), the average rate constant R1ρ (C) and the CSA/D CCR rate constant (D), as a function of the jump angle and jump rate constant. (E) shows R1ρ and CCR rate constants for a constant jump angle (20°) as a function of the jump rate constant, assuming a MAS frequency of 40 kHz and a RF field strength of 12 kHz. (F) uses parameters as in (E), but with an RF field strength of 32 kHz. (G) Correlation of R1ρ and CCR rate constants, assuming a jump angle of 5° and a MAS frequency of 40 kHz and a RF field strength of 12 kHz. The jump angle can be related to the often-used order parameter S2 as S2 = 1/4(3cos2(θ) + 1) .34 According to this equation, jump angles of 5, 10, 15, 20, 25 degrees, for example, correspond to S2= 0.994, 0.977, 0.95, 0.91, 0.866, respectively.
For completeness and illustration purposes, we also provide the analytical expression for the CCR rate constant under spin-lock, , in the framework of Redfield theory (which is only approximately valid1):
| (1) |
Here, δNH is the anisotropy of the dipolar coupling tensor, δCSA is the chemical-shift anisotropy and P2(cos(θ)) is the second Legendre polynomial involving the angle between the principal axes of the dipolar and CSA interactions, θ . It is interesting to confront this equation to the expression of the R1ρ relaxation rate constant due to the dipolar interaction and CSA, which is given as:1,23
where
| (2) |
Comparison of (1) and (2) reveals that the two relaxation rate constants provide similar information. In this work we are interested primarily in motions slower than hundreds of nanoseconds (because in this range and R1ρ are largest, as shown below); in this regime one can neglect the J(ωN) term; furthermore, R1 is very small compared to R1ρ. Neglecting thus the J(ωN) terms one can see that the two rate constants depend on the same spectral density functions (see Equations 1 and 2). We thus expect that the two rate constants are correlated, an observation that is indeed confirmed both experimentally and through simulations (discussed further below, Figures 2G and 3C). In the remainder of this manuscript we do not use analytical calculations but use exclusively numerical simulations, because this allows to take into account additional factors, such as relaxation of the proton spin due to external mechanisms, as described below. By using numerical simulations we also circumvent the fact that Redfield theory is outside its range of validity for slow motions (microseconds).1
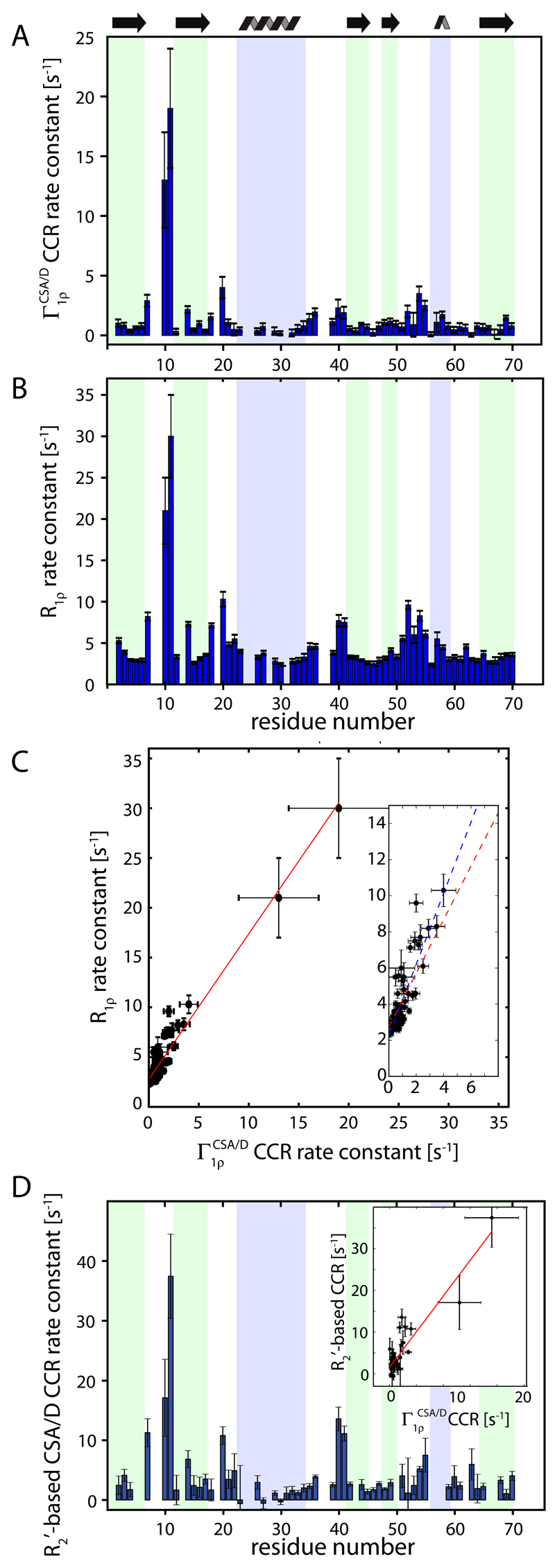
Figure 3.
Experimental observation of CSA/D CCR in 15N spin-lock relaxation experiments in deuterated, 60% back-exchanged ubiquitin, obtained at a MAS frequency of 38 kHz. (A) Differential relaxation rate constants as a function of the residue number, determined from the relaxation rate constant of the two doublet components. (B) R1ρ relaxation rate, i.e. obtained from the average of the two rate constants. In (C), a correlation of the two rate constants is shown. The red line corresponds to a correlation obtained over all available residues, with a slope of 1.48 and an intercept of 2.74 s-1. The insert focuses on the data points at low rate constants. The blue line has a slope of 1.96 s-1. Panel (D) shows previously published27 CSA/D CCR relaxation rate constants, obtained with a spin-echo (delay – π pulse – delay) sequence instead of a spin-lock, at the same B0 field strength. The insert shows the correlation of these data with the R1ρ-based CCR rate constants of panel (A).
Experimental detection of CSA/D cross-correlated relaxation
In order to experimentally verify the effects of cross-correlated relaxation in R1ρ experiments, we prepared a sample of 2H,15N labelled microcrystalline ubiquitin. The protein was crystallized in a buffer that results in a re-protonation of exchangeable sites, such as the amide sites that we are interested in here, at a level of 60%. In such highly deuterated samples, narrow 1H line widths can be achieved, and the simplicity of the spin system, that can be approximated as a two-spin system as far as 15N relaxation is concerned, also allows for straightforward analysis of relaxation data.24–26 Of particular relevance here, narrow 15N lines are obtained even without 1H decoupling, and therefore one can separately detect the decay of the doublet components. Figure 3 shows such 15N R1ρ data for the 15N doublet components of amide sites in ubiquitin, obtained at a MAS frequency of 38 kHz, and a 15N RF field strength of 12 kHz. For the majority of residues the decay rate constants of the two components are detectably different. The differential relaxation rate constant of the two components, , varies across the protein sequence, ranging from close to zero to about 20 s-1 for different residues (Figure 3A). Residues located in secondary structure elements have small values, below 1 s-1 in most cases. Largest values of , exceeding 10 s-1, are observed for residues G10 and K11 in the β1-β2 loop, which has been demonstrated before to undergo nanosecond motion in the microcrystals we are studying. 27,28 Cross-correlated relaxation rates are furthermore enhanced for residues located in the loop connecting β2 and ubiquitin’s α-helix, the α-β3 loop and the loop connecting β4 to the short 310 helix. These loop regions have also elevated R1ρ rate constants (Figure 3B). Figure 3C shows the correlation of residue-wise and R1ρ rate constants. Because the majority of residues are located in secondary structure elements, residue-wise variations are generally small. Basically, only residues located in loop regions have significantly enhanced and R1ρ rate constants. This limits the precision with which the correlation can be determined. The slope of the correlation is 1.48 when taking into account all residues (red line) and 1.96 when neglecting the two data points with largest rate constants. These experimental data unequivocally establish that the correlation has a non-zero offset: while R1ρ rate constants exceed 2 s-1 for all residues, many residues have a close-to-zero CSA/D cross-correlated relaxation rate constant. This finding cannot be understood based on the simulations in Figure 2. To understand this offset, one needs to take into account partial proton self-decoupling, as shown in the following section.
For completeness, we have also shown previously determined CSA/D CCR rate constants, determined with a spin-echo element instead of a spin-lock period. The drawback of the former experiment is that the decay with free evolution (R2’) is significantly faster than decay under spin-lock (in ubiquitin by up to one order of magnitude). Consequently, it is more difficult to determine small CCR rates from the difference of the two comparably large decay rate constants corresponding to the two doublet components. Qualitatively, the three observables highlight the same residues undergoing slow motions.
It shall The correlation of the rate constant
Proton spin diffusion and its effect on heteronuclear transverse relaxation
Longitudinal relaxation of the proton spin, or spin diffusion (i.e. magnetization exchange between protons) leads to interconversion of the spin states, α and β. Accordingly, the states Nx[Hα] and Nx[Hβ] inter-convert, and this would result in (partial) self-decoupling of these two 15N doublet components. In the context of cross-correlated relaxation, such self-decoupling is an important parameter, because it is expected to reduce the CCR rate constant.29 In this section we study the consequences of proton longitudinal relaxation or spin diffusion on heteronuclear relaxation and cross-correlated relaxation, using numerical simulations. At difference to the simulations shown in Figure 2, we added an ad hoc random field contribution acting on the proton spin. The effect of this random field is to relax the proton spin, and therefore interchange spin states H α and Hβ. In principle one might also attempt to explicitly simulate the proton spin diffusion or relaxation by using a large spin system with multiple protons. In practice, however, such simulations are prohibitively long if the spin system is larger than about 4-5 spins, and the accuracy of the proton evolution likely is not better than in our ad hoc external relaxation approach.
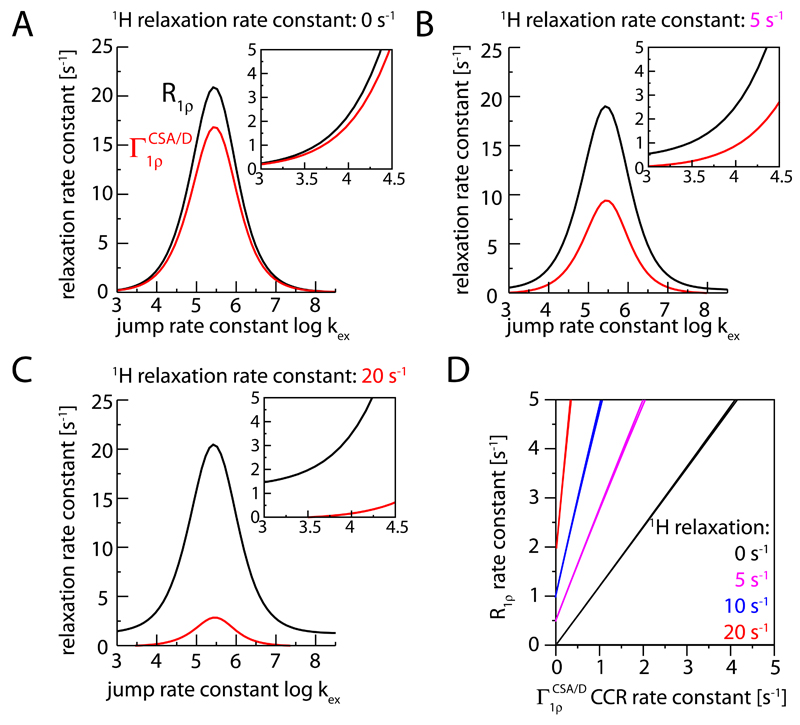
Figure 4 shows the results of numerical simulations of the heteronuclear R1ρ experiment in the presence of this additional proton relaxation, simulated here for the case of small-amplitude motion of the HN bond. Simulations with larger-amplitude motions are shown in Figure S3. These simulations establish that proton longitudinal relaxation induced by external relaxation mechanisms indeed reduces the CCR rate constant. The reduction of the rate constant depends on the rate constant of the proton relaxation, as can be seen by comparison the red curves shown in panels (A) to (C). This reduction of can be understood by the averaging of the two components. Figure 4 also shows that the 1H spin flips have an effect on the R1ρ rate constants. For the range of R1ρ rate constants that are observed experimentally, i.e. in the range of R1ρ of a few s-1 to tens of s-1 at most, the proton relaxation leads to an increase in R1ρ. The increase of R1ρ depends on the RF field strength and the MAS frequency (Figure S4). The origin of this increased R1ρ is the fact that stochastic proton spin flips counteract the MAS-induced recoupling of the dipolar interaction, as discussed in more detail in the Supporting Information. Taken together, the simulations of Figure 4 establish that the effect of the additional 1H spin relaxation is (i) a reduction of the CCR rate constant, and (ii) an increase of the R1ρ rate constant. The decreased CCR and increased R1ρ rate constants change the slope of the correlation of the two rate constants, and result in an offset of the two rate constants (Figure 4D). It is remarkable that this situation is qualitatively found in the experimental data, shown in Figure 3C, which reproduce an offset of R1ρ rate constants. It shall be noted that Figure 4 assumes a single amplitude and varies the time scale of the motion, a situation that is not likely to apply to the range of amide sites across a protein. Nonetheless, qualitatively the behavior observed in Figure 4 applies irrespectively of the precise motional model.
Figure 4.
The effect of proton relaxation on 15N transverse relaxation parameters, from numerical simulations with an ad hoc added random field fluctuation leading to proton relaxation. Shown are the simulated relaxation rate constants of Nx (i.e. the relaxation with a 1H π pulse in the center of the spin-lock relaxation period), R1ρ, and the cross-correlated relaxation rate constant, , obtained from the differences of relaxation of the two doublet components. Different rate constants of the 1H relaxation are assumed: 0 s-1 (A), 5 s-1 (B) and 20 s-1 (C); R1ρ and rate constants are shown in black and red, respectively. Panel (D) correlates the R1ρ and relaxation rate constants for different 1H relaxation rate constants. The jump angle was set to 5 degrees, corresponding to an order parameter of S2=0.994. More simulations with a large motional amplitude are shown in Figure S3.
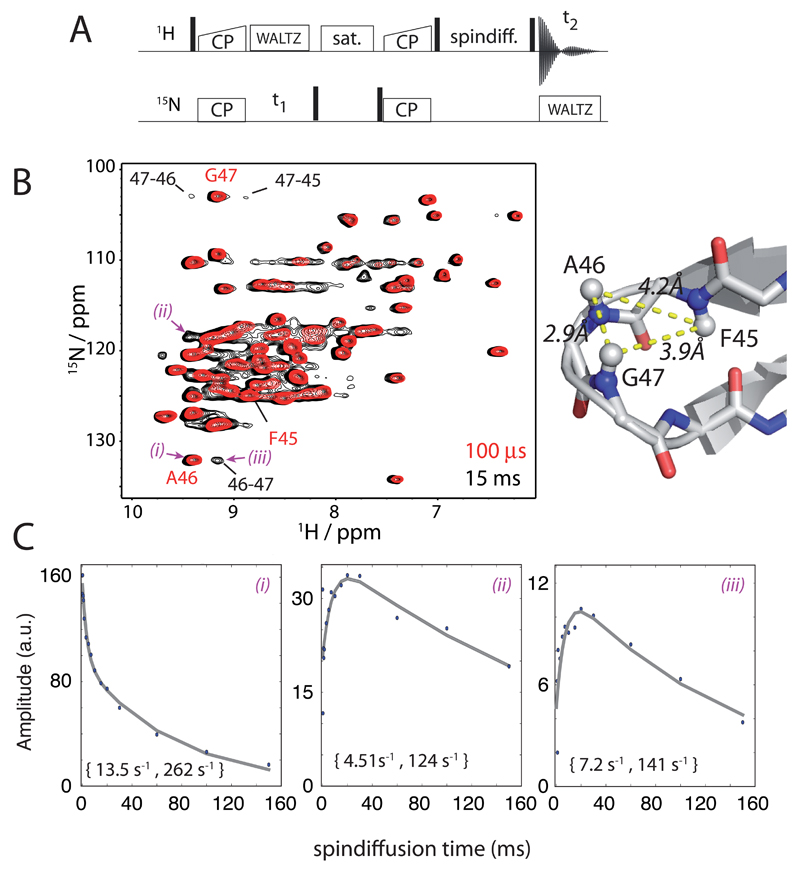
The amount by which the CCR rate is reduced depends on the rate at which the 1H spin flip (Figure 4). We therefore need to estimate which spin diffusion rate constants are realistic in our experimental setting, i.e. at a MAS frequencies of 38 kHz and with the deuteration level we employed (60% back-exchange). Figure 5 shows experimental measurements of spin-diffusion between amide protons. By comparison of the spectra obtained with very short mixing time (100 µs, red) and 15 ms mixing (black), one can observe that there are additional peaks pointing to 1H-1H spin diffusion. Figure 5B highlights a few cases of well-resolved cross-peaks, corresponding to spin diffusion in a β-turn region (residues 45-47); for the cases of these cross-peaks, the corresponding amide sites are within ~4.2 Å at most. Figure 5C shows the time evolution of representative diagonal- and cross-peaks. The diagonal peaks show non-monoexponential decay, which we ascribe to contributions of spin-diffusion/cross-relaxation (fast component) and relaxation of the magnetization, presumably at the most dynamic sites, or through exchange with solvent. When fitting these curves with bi-exponential functions, respective rate constants are several tens per second for the fast rate (about 30-40% amplitude) and about 5-10 s -1 (60-70% amplitude), respectively. If attempting to describe the decay with a single rate constant, approximated by the time point where the signal decay to 1/e ~37%, apparent rate constants of 20 s-1 are obtained. The observable cross-peaks show a buildup within tens of milliseconds, similar to the fast rate observed in the diagonal-peak decay curves. Taken together, these data allow one to estimate that proton relaxation occurs at a rate of approximately 10-20 s -1. (We note that the spin-diffusion rate is expected to decrease at higher MAS frequency and higher deuteration levels. A very recent report, which became available during the revision of the present manuscript,30 used higher MAS frequencies (60 kHz), but lower deuteration (100% back-protonation of exchangeable sites), and values of about 10-20 s-1 were reported for the majority of the residues. While this is in rather good agreement with the present estimates, no apparent multi-exponential decay has been reported there, nor did the authors report on spin-diffusion cross-peaks.)
Figure 5.
Experimental investigation of proton relaxation and spin diffusion in the deuterated, 60% back-exchanged ubiquitin sample at 38 kHz MAS (14.1 T). (A) Pulse sequence used to detect 1H relaxation/spin diffusion. (B) Overlay of two spectra obtained with different mixing times as indicated. Selected amide peaks, and the corresponding spin-diffusion cross-peaks are indicated, and the distances between these sites are indicated on the right. (C) Representative time traces observed at peak positions corresponding to amide sites (diagonal peaks), and two spin-diffusion cross-peaks, obtained at spectral positions at which intensity is observed only with a mixing time. The grey lines correspond to bi-exponential fits, and the fitted rate constants are indicated in brackets. The identity of the peaks for which the graphs are shown in C is indicated in the spectrum with (i)-(iii).
Using this estimate of 1H relaxation, one can simulate its effect on measured and R1ρ rate constants, and possibly correct for them. It has been proposed in solution-state NMR that 1H-1H NOE (i.e. relaxation-induced spin-diffusion) can be taken into account when analyzing cross-correlated relaxation,31 using an analytical approach. In principle, such an approach could also be applied in solids. However, the effect of the proton spin flips on the 15N relaxation depends on the RF field and the MAS frequency, and an appropriate formalism is currently not developed. However, one may resort to numerical simulations, such as the ones we use here, to evaluate the effects of proton spin flips on heteronuclear R1ρ or CCR rate constants. Experimentally determined proton decay rate constants, or at least global estimates, may be used to this end.
We illustrate this with the data shown in Figure 4. For example, assume that the experimentally determined R1ρ rate constant is 3 s-1, i.e. including the effects of proton relaxation. The actual R1ρ rate constant, corrected for a 1H relaxation rate constant of 20 s-1 (10 s-1), is 1.8 s-1 (2.6 s-1), i.e. the excess decay induced by the 1H relaxation is of the order of 0.4 to 1.2 s-1. In principle, one can take into account these effects, and use the corrected values when fitting motional models to transverse relaxation data.
A similar estimation for suggests, for example, that in the presence of a 1H relaxation with a rate constant of 10 s-1, a measured of 1 s-1 points to an actual rate constant of 3.6 s-1. Arguably, it is more difficult to correct for 1H relaxation effects, because the values of this rate constant are smaller than those for R1ρ, and 1H relaxation has a stronger effect on than on R1ρ.
It is noteworthy that such a correction appears more difficult for the CCR rate constant, which may become very small and thus error-prone, than for R1ρ. Of course, one may attempt to suppress the effects of proton spin flips even more, by using higher MAS frequencies and/or higher deuteration levels. Increasing MAS frequency will, however, not suppress proton magnetization decay which is actually due to dynamics or chemical exchange, such that it appears unlikely that one may suppress its effects completely.
Suppression of the effect of CSA/D CCR in R1ρ experiments
Finally, we note that the presence of CSA/D CCR might hamper accurate extraction of R1ρ rate constants. In solution-state NMR, schemes for suppression of the effect of CSA/D CCR have been reported.13,32 They generally require irradiation of 1H spins. Suppression of CCR in R1ρ experiments, even in solution-state NMR, is complicated by the fact that 1H RF fields applied during heteronuclear spin-lock may partially re-introduce the scalar coupling Hamiltonian in solution and solids,12,13 and it may also reintroduce the dipolar coupling in the solid state. In MAS ssNMR particular care has to be taken to avoid rotary resonance conditions, i.e. 1H multi-pulse or CW schemes which counteract the averaging brought about by MAS. To investigate these effects, we have tested two different suppression schemes, namely (i) a single 1H π pulse, applied in the center of the relaxation period and (ii) a train of 1H π pulses, spaced by 5 ms delays. Both schemes, or variants thereof, are used in solution-state NMR.12,13,33 Interestingly, we find that the multi-pulse scheme, even if the 1H pulse spacing (5 ms) is very long compared to the rotor period (~20-30 µs), the heteronuclear coherences decay significantly faster than in the absence of 1H pulses or with a single 1H π pulse (Figure S5). The use of a single 1H π pulse appears as a straightforward method to suppress the effects of CCR. We note, however, that even with a suppression of CCR, one does not expect mono-exponential decay, due to the inherent multi-exponential nature of decays in solids, outlined above.
Conclusions
We have shown here that CSA/D cross-correlated relaxation can be measured in MAS ssNMR of deuterated proteins. This relaxation rate constant reports on dynamics on nanosecond-to-microsecond time scales, as we illustrate with an application to microcrystalline ubiquitin. In many practical applications, however, the more easily accessible R1ρ rate constant of the decoupled line has advantages over the measurement of CSA/D CCR under spin-lock, in particular due to higher sensitivity and resolution in a decoupled vs a non-decoupled spectrum. We also provided evidence, by numerical simulations, that rapid 1H spin flips, as mediated by 1H-1H spin diffusion, reduces the CCR rate constant, and also contributes to the observed R1ρ rate constant. This finding also shows that for accurate quantitative analyses of R1ρ rate constants one should correct the measured rate constants for the contribution arising from proton relaxation. The present report of cross-correlated relaxation adds to the increasing arsenal of techniques to study protein dynamics on long time scales by MAS ssNMR, and stresses the importance of considering hitherto ignored effects when interpreting relaxation rate constants in ssNMR.
Supporting Information Description
Figures showing the pulse sequences used in this study; figures with additional simulations, and experimental data; extended discussion about the increase of R1ρ rate constants due to proton spin relaxation. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
This work was financially supported by the European Research Council (ERC-StG-311318 ProtDyn2Function) and the Commissariat a l’énergie atomique et aux énergies alternatives (CEA), the Centre National de la Recherche Scientifique (CNRS) and the Université Grenoble Alpes. This work used the platforms of the Grenoble Instruct Center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). We thank Matthias Ernst for stimulating discussions and for assistance with implementing the proton random field fluctuations in the GAMMA simulations.
References
- (1).Schanda P, Ernst M. Studying Dynamics by Magic-Angle Spinning Solid-State NMR Spectroscopy: Principles and Applications to Biomolecules. Prog Nucl Magn Reson Spectrosc. 2016;96:1–46. doi: 10.1016/j.pnmrs.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Krushelnitsky A, Zinkevich T, Reichert D, Chevelkov V, Reif B. Microsecond Time Scale Mobility in a Solid Protein as Studied by the 15N R1ρ Site-Specific NMR Relaxation Rates. J Am Chem Soc. 2010;132:11850–11853. doi: 10.1021/ja103582n. [DOI] [PubMed] [Google Scholar]
- (3).Lewandowski JR, Sass HJ, Grzesiek S, Blackledge M, Emsley L. Site-Specific Measurement of Slow Motions in Proteins. J Am Chem Soc. 2011;133:16762–16765. doi: 10.1021/ja206815h. [DOI] [PubMed] [Google Scholar]
- (4).Ma P, Haller JD, Zajakala J, Macek P, Sivertsen AC, Willbold D, Boisbouvier J, Schanda P. Probing Transient Conformational States of Proteins by Solid-State R1rho Relaxation-Dispersion NMR Spectroscopy. Angew Chemie - Int Ed. 2014;53:4312–4317. doi: 10.1002/anie.201311275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lamley JM, Lougher MJ, Sass H-J, Rogowski M, Grzesiek S, Lewandowski JR. Unraveling the Complexity of Protein Backbone Dynamics with Combined 13 C and 15 N Solid-state NMR Relaxation Measurements. Phys Chem Chem Phys. 2015;17:21997–22008. doi: 10.1039/c5cp03484a. [DOI] [PubMed] [Google Scholar]
- (6).Lamley JM, Öster C, Stevens RA, Lewandowski JR. Intermolecular Interactions and Protein Dynamics by Solid-State NMR Spectroscopy. Angew Chemie - Int Ed. 2015;54:15374–15378. doi: 10.1002/anie.201509168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ma P, Xue Y, Coquelle N, Haller JD, Yuwen T, Ayala I, Willbold D, Colletier J, Skrynnikov NR. Observing the Overall Rocking Motion of a Protein in a Crystal. Nat Commun. 2015;6:1–24. doi: 10.1038/ncomms9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Quinn CM, McDermott AE. Quantifying Conformational Dynamics Using Solid-State R1ρ Experiments. J Magn Reson. 2012;222:1–7. doi: 10.1016/j.jmr.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Quinn CM, McDermott AE. Monitoring Conformational Dynamics with Solid-State R1ρ Experiments. J Biomol NMR. 2009;45:5–8. doi: 10.1007/s10858-009-9346-7. [DOI] [PubMed] [Google Scholar]
- (10).Chevelkov V, Diehl A, Reif B. Quantitative Measurement of Differential 15-N-H A/β T2 Relaxation Rates in a Perdeuterated Protein by MAS Solid-State NMR Spectroscopy. Magn Reson Chem. 2007;45:S156–S160. doi: 10.1002/mrc.2129. [DOI] [PubMed] [Google Scholar]
- (11).Skrynnikov N. Asymmetric Doublets in MAS NMR: Coherent and Incoherent Mechanisms. Magn Reson Chem. 2007;45:S161–S173. doi: 10.1002/mrc.2162. [DOI] [PubMed] [Google Scholar]
- (12).Massi F, Johnson E, Wang C, Rance M, Palmer AG. NMR R1ρ Rotating-Frame Relaxation with Weak Radio Frequency Fields. J Am Chem Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- (13).Korzhnev DM, Skrynnikov NR, Millet O, Torchia DA, Kay LE. An NMR Experiment for the Accurate Measurement of Heteronuclear Spin-Lock Relaxation Rates. J Am Chem Soc. 2002;124:10743–10753. doi: 10.1021/ja0204776. [DOI] [PubMed] [Google Scholar]
- (14).Chevelkov V, Reif B. TROSY Effects in MAS Solid-State NMR. Concepts Magn Reson Part A. 2008;32A:143–156. [Google Scholar]
- (15).Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand aJ. Assignments of Carbon NMR Resonances for Microcrystalline Ubiquitin. J Am Chem Soc. 2004;126:6720–6727. doi: 10.1021/ja030547o. [DOI] [PubMed] [Google Scholar]
- (16).Delaglio F, Grzesiek S, Vuister G, Zhu G, Pfeifer J, Bax A. NMRPIPE - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- (17).Smith S, Levante T, Meier B, Ernst R. Computer Simulations in Magnetic Resonance. An Object-Oriented Programming Approach. J Magn Reson. 1994;106:75–105. [Google Scholar]
- (18).Abergel D, Palmer AG. On the Use of the Stochastic Liouville Equation in Nuclear Magnetic Resonance: Application to R1ρ Relaxation in the Presence of Exchange. Concepts Magn Reson Part A Bridg Educ Res. 2003;19:134–148. [Google Scholar]
- (19).Kubo R. Stochastic Liouville Equations. J Math Phys. 1963;4:174–183. [Google Scholar]
- (20).Redfield AG. On the Theory of Relaxation Processes. IBM J Res Devel. 1957;1:19–31. [Google Scholar]
- (21).Torchia DA, Szabo A. Spin-Lattice Relaxation in Solids. J Magn Reson. 1982;49:107–121. [Google Scholar]
- (22).Giraud N, Blackledge M, Goldman M, Böckmann A, Lesage A, Penin F, Emsley L. Quantitative Analysis of Backbone Dynamics in a Crystalline Protein from Nitrogen-15 Spin-Lattice Relaxation. J Am Chem Soc. 2005;127:18190–18201. doi: 10.1021/ja055182h. [DOI] [PubMed] [Google Scholar]
- (23).Kurbanov R, Zinkevich T, Krushelnitsky A. The Nuclear Magnetic Resonance Relaxation Data Analysis in Solids: General R1/R1ρ Equations and the Model-Free Approach. J Chem Phys. 2011;135:184104. doi: 10.1063/1.3658383. [DOI] [PubMed] [Google Scholar]
- (24).Reif B. Ultra-High Resolution in MAS Solid-State NMR of Perdeuterated Proteins: Implications for Structure and Dynamics. J Magn Reson. 2012;216:1–12. doi: 10.1016/j.jmr.2011.12.017. [DOI] [PubMed] [Google Scholar]
- (25).Schanda P, Huber M, Boisbouvier J, Meier BH, Ernst M. Solid-State NMR Measurements of Asymmetric Dipolar Couplings Provide Insight into Protein Side-Chain Motion. Angew Chem Int Ed Engl. 2011;50:11005–11009. doi: 10.1002/anie.201103944. [DOI] [PubMed] [Google Scholar]
- (26).Knight MJ, Webber AL, Pell AJ, Guerry P, Barbet-Massin E, Bertini I, Felli IC, Gonnelli L, Pierattelli R, Emsley L, et al. Fast Resonance Assignment and Fold Determination of Human Superoxide Dismutase by High-Resolution Proton-Detected Solid-State MAS NMR Spectroscopy. Angew Chem Int Ed Engl. 2011;50:11697–11701. doi: 10.1002/anie.201106340. [DOI] [PubMed] [Google Scholar]
- (27).Schanda P, Meier BH, Emst M. Quantitative Analysis of Protein Backbone Dynamics in Microcrystalline Ubiquitin by Solid-State NMR Spectroscopy. J Am Chem Soc. 2010;132:15957–15967. doi: 10.1021/ja100726a. [DOI] [PubMed] [Google Scholar]
- (28).Haller JD, Schanda P. Amplitudes and Time Scales of Picosecond-to-Microsecond Motion in Proteins Studied by Solid-State NMR: A Critical Evaluation of Experimental Approaches and Application to Crystalline Ubiquitin. J Biomol NMR. 2013;57:263–280. doi: 10.1007/s10858-013-9787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hu K, Vögeli B, Clore GM. Interference between Transverse Cross-Correlated Relaxation and Longitudinal Relaxation Affects Apparent J-Coupling and Transverse Cross-Correlated Relaxation. Chem Phys Lett. 2006;423:123–125. [Google Scholar]
- (30).Smith AA, Testori E, Cadalbert R, Meier BH, Ernst M. Characterization of Fibril Dynamics on Three Timescales by Solid-State NMR. J Biomol NMR. 2016 doi: 10.1007/s10858-016-0047-8. [DOI] [PubMed] [Google Scholar]
- (31).Reif B, Diener A, Hennig M, Maurer M, Griesinger C. Cross-Correlated Relaxation for the Measurement of Angles between Tensorial Interactions. J Magn Reson. 2000;143:45–68. doi: 10.1006/jmre.1999.1980. [DOI] [PubMed] [Google Scholar]
- (32).Massi F, Johnson E, Wang C, Rance M, Palmer AG. NMR R1rho Rotating-Frame Relaxation with Weak Radio Frequency Fields. J Am Chem Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- (33).Palmer AG, Massi F. Characterization of the Dynamics of Biomacromolecules Using Rotating-Frame Spin Relaxation NMR Spectroscopy. Chem Rev. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- (34).Clore GM, Driscoll PC, Wingfield PT, Gronenborn AM. Analysis of the Backbone Dynamics of Interleukin-1 Beta Using Two-Dimensional Inverse Detected Heteronuclear 15N-1H NMR Spectroscopy. Biochemistry. 1990;29:7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.