Abstract
Long noncoding RNAs (lncRNAs), defined as non-translated transcripts greater than 200 nucleotides in length, are often differentially expressed throughout developmental stages, tissue types, and disease states. The identification, visualization, and suppression/overexpression of these sequences have revealed impacts on a wide range of biological processes, including epigenetic regulation. Biochemical investigations on select systems have revealed striking insight into the biological roles of lncRNAs and lncRNA:protein complexes, which in turn prompt even more unanswered questions. To begin, multiple protein- and RNA-centric technologies have been employed to isolate lncRNA:protein and lncRNA:chromatin complexes. LncRNA interactions with the multi-subunit protein complex PRC2, which acts as a transcriptional silencer, represent some of the few cases where the binding affinity, selectivity, and activity of a lncRNA:protein complex have been investigated. At the same time, recent reports of full-length lncRNA secondary structures suggest the formation of complex structures with multiple independent folding domains and pave the way for more detailed structural investigations and predictions of lncRNA three-dimensional structure. This review will provide an overview of the methods and progress made to date as well as highlight new methods that promise to further inform the molecular recognition, specificity, and function of lncRNAs.
Graphical Abstract
The “noncoding RNA revolution” (Cech, Stetiz) has revealed myriad functional RNA molecules with roles extending far beyond that of a messenger between DNA and protein.1 The world of noncoding RNAs (ncRNAs), or RNAs that are not usually translated to proteins, came to light in large part as a result of the Encyclopedia of DNA Elements (ENCODE) project.2 This consortium found that while up to 90% of the genome was transcribed only 1.2% was translated to protein. Furthermore, this large pool of untranslated transcripts demonstrated biochemical indices of function traditionally ascribed solely to proteins.3 Research exploring the biological activity of these ncRNA transcripts promptly grew. Among the many newly discovered functions of noncoding RNAs, several classes are now known to play critical roles in the regulation of gene expression1 as well as disease progression.4 NcRNAs are classified based on size, with small ncRNAs less than 200 n.t. and long noncoding RNAs (lncRNAs) greater than 200 n.t. Several small ncRNA classes, including microRNAs (miRNAs) and small-interfering RNAs (siRNA), regulate gene expression by forming partially complementary duplexes with mRNAs, which in turn promote mRNA degradation or inhibit mRNA translation into peptides.5–8 LncRNAs, on the other hand, have been found to exhibit a wide range of regulatory roles, including trafficking of proteins in the cytoplasm7,9 and epigenetic modulation in the nucleus,10 with the latter representing the most well studied function.11,12 Despite this rapid progress in lncRNA identification, the molecular characterization of most functional lncRNAs remains unexplored.
Perhaps the most complete lncRNA story is found in the Xist transcript. Xist is necessary for X-chromosome inactivation (Xci), which allows for even genetic dosage between female (XX) and male (XY) mammals.13 Xist coats the chromatin in cis and binds the polycomb repressive complex (PRC2), which imparts the repressive H3K27me3 mark.14 To date, the minimal RNA binding sequence of Xist (Repeat A, (RepA)), its binding affinity for PRC2 and PRC2 subunits, and its impact on methylase activity have been reported.15 Indeed, Xist represents one of the few lncRNAs that has been characterized from the organismal down to the biophysical level, yet precise details of the molecular interactions between Xist and PRC2 have yet to be determined.
While several excellent reviews have been published recently regarding the function of lncRNAs,12,16–18 few have focused on the molecular-level characterization of lncRNAs and lncRNA:protein complexes.19,20 While not exhaustive, this review will focus on select methods that have enabled recent successes in the biochemical characterization of mammalian lncRNAs, specifically those involved in human epigenetic regulation, as well as highlight new and traditional RNA methods that, when applied to lncRNAs, promise to facilitate a greater understanding of how the structure, function, and molecular interactions of these novel molecules impact their activity.
DISCOVERY OF FUNCTIONAL lncRNA TRANSCRIPTS
LncRNA sequences are defined as transcripts greater than 200 n.t., but many extend into the range of 1–5 kb. LncRNA, unlike mRNA, can vary in splicing, polyadenylation, cellular localization, and the transcribing polymerase.21 LncRNAs were first identified through transcriptome wide analysis using either tiled cDNA microarrays or high throughput sequencing.12,21,22 Importantly, the de novo identification of lncRNAs generally requires methods that allow for the enrichment of low abundance RNA sequences and that do not rely on the presence of a polyA tail or specific polymerase activity. The first and strongest indication of lncRNA functional relevance stemmed from the differential but specific expression levels observed in various developmental stages, tissue types, and disease states.16 To date, nearly 16,000 lncRNA sequences have been annotated in the GENCODE v23 database,23 while nearly 300 lncRNAs with verified function have been curated into the Long Noncoding RNA Database v2.0.24 Representative examples of these lncRNAs are highlighted in Table 1.
Table 1.
Examples of Mammalian lncRNA Sequences and Their Binding Partners
| shorthand | name | size (kb)a,b | binding partner(s) | known biological pathways | ref |
|---|---|---|---|---|---|
| Xist | Xi-specific transcripts | 17 | PRC2 | inactivates one X chromosome to ensure genetic equality between sexes | 25 |
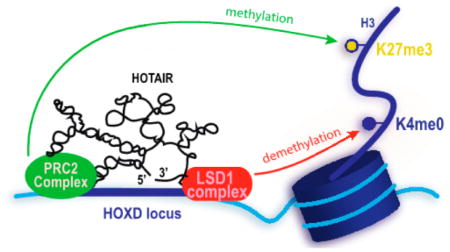
| HOTAIR | HOX transcript antisense intergenic RNA | 2.2 | LSD1; PRC2 | suppression of transcription through modification of histone 3 on the HOXD locus | 26 |
| MEG3 (GTL2) | maternally expressed gene 3 (gene trap locus 2) | 1.5–1.8 | PRC2 | potentially forms RNA-DNA triplex structures to regulate TGF-β genes | 27, |
| ANRIL | antisense noncoding RNA in the INK4 locus | 2.2 | PRC1; PRC2 | regulates histone modification in the CDKN2A/B locus | 29 |
| FENDRR | fetal-lethal noncoding developmental regulatory RNA | 2.6; 3.1 | PRC2; TrxG/MLL | binds Foxf1 gene promoter to regulate pathways controlling fate of ES cell differentiation | 30 |
| MALAT1 (NEAT2) | metastasis associated lung adenocarcinoma transcript 1 (nuclear-enriched abundant transcript 2) | 8.8 | SR proteins | recruits SR proteins, a family of splicing proteins, to nuclear speckles to control alternative splicing of pre-mRNA | 31 |
| NEAT1 (VINC) | nuclear paraspeckle assembly transcript 1 (virus inducible noncoding RNA) | 3.2; 22.7 | P54nrb; PSP1a | forms and maintains paraspeckle structure | 32 |
| PCAT1 | prostate cancer associated transcript 1 | 2 | PRC2 | post-transcriptionally silences BRCA2 | 33, 34 |
| SChLAP1 | second chromosome locus associated with prostate-1 | 1.3–1.7 | SWI/SNF | binds SWI/SNF complex, leading to aberrant gene expression | 35 |
| DANCR (ANCR) | differentiation antagonizing nonprotein coding RNA (antidifferentiation ncRNA) | 0.9 | PRC2 | promotes expression of IL6 and TNF-α at mRNA and protein levels in post-menopausal osteoporosis | 36, 37 |
| Lnc_bc060912 | 1.2 | PARP1; NPM | binds PARP1 and NPM1, DNA damage repair proteins, to suppress cell apoptosis | 38 | |
| Kcnq1ot1 | KCNQ1 overlapping transcript 1 | 91.5 | DNMT1 | imprints on multiple genes, leading to gene silencing | 39 |
| SRA | steroid receptor RNA activator | 0.87–0.92 | ER-α; SRC-1; Sharp; SLIRP; RNA helicases p68 and p72 | breast cancer tumorigenesis | 40 |
| PARTICLE | promoter of MAT2A-antisense radiation- induced circulating lncRNA | 1.4 | G9a; SUZ12 | implication in transcription regulation via triple-helix formation | 41 |
For lncRNA with <3 transcript isoforms, the transcripts are separated as such: ##; ##.
For lncRNA with ≥3 transcript isoforms, a range of transcript lengths are shown: ##-##.
CELLULAR LOCALIZATION AND FUNCTION
Cellular Localization
The localization of lncRNA transcripts further supports specific cellular functions and has been examined with traditional RNA detection methods including real-time quantitative polymerase chain reactions (RT-qPCR), Northern blotting, and RNA-fluorescent in situ hybridization (RNA-FISH).
RT-qPCR
The standard determination of lncRNA cellular localization relies upon cell fractionation and RT-qPCR. The first step, cell lysis, uses detergents that maintain segregation between the nuclear and cytoplasmic transcripts.42 Following separation, the transcripts are reverse transcribed and the cDNA submitted to RT-qPCR to quantitatively compare RNA levels in the separated cellular fractions. In a recent example, Kanduri and co-workers used these methods to determine the distribution of MEG3 lncRNA and found it to be almost exclusively expressed in the nuclear fraction.28 Compared to sequencing methods, RT-qPCR is limited to discrete sequences but can be highly quantitative even when performed on small amounts of sample.
RNA-FISH
RNA-FISH uses nucleic acid probes to fluorescently track the localization of a target transcript inside a cell through sequence-specific hybridization. In general, a fluorescently tagged short RNA construct (10–50 n.t.) with unique complementarity to the target sequence is added to chemically fixed cells.43 Ideally, the probe sequence selectively binds to the transcript of interest, allowing for direct imaging of the target. Improved sensitivity now enables the visualization of a target transcript at the single-molecule level while limiting the off-target effects. Furthermore, with the use of dozens of fluorescently labeled probes that effectively tile the target sequence, one can achieve significant enrichment over background/off-target complementation.44,45 Both cytoplasmic and nuclear-localizing lncRNAs, including Xist, NEAT1, and MALAT1, have been studied using RNA-FISH.46
Recently, Lee and co-workers used three-dimensional (3D) stochastic optical reconstruction microscopy (STORM), a high-resolution microscopy method, in conjunction with RNA-FISH to study the colocalization of PRC2 to Xist (Figure 1).47 In this two-color 3D STORM experiment, PRC2 was probed using an enhancer of zeste homologue 2 (EZH2)-antibody, and Xist was probed using RNA-FISH. Spectral overlap confirmed the expected colocalization, and over 75% of the end-to-end distances between the PRC2 and Xist fluorescent signals were within 100 nm. The resolution of STORM combined with the sequence-specific probing enabled by RNA-FISH will continue to generate crucial insight into the function of lncRNA and lncRNA:protein complexes.
Figure 1.

Nebulous microscopy image of Xist RNA using traditional technology in MEF cells by RNA FISH (A) as compared to 3D-STORM resolution of the same cells (B). Reprinted (adapted or reprinted in part) from ref 47 with permission. Copyright 2015 National Academy of Sciences.
Encoded Fluorescent Tagging
Similar to fluorescent protein tagging methods, RNA transcripts can be encoded with non-native sequences for fluorescence detection, though generation of the fluorescent signal usually requires a secondary step.48,49 Perhaps the most common method, MS2 tagging, takes advantage of the high affinity of the MS2 viral coat protein to the stem loop repeat MS2 aptamer sequence.50 When the MS2 protein is conjugated to a fluorescent protein, selective binding of the conjugate to the MS2 aptamer sequence results in fluorescent labeling. Visualization of NEAT1 lncRNA, for example, was achieved through incorporation of the MS2 aptamer downstream of the lncRNA.51 Cotransfection of the NEAT1-MS2 aptamer construct and an enhanced yellow fluorescent protein (EYFP)-MS2 protein construct effectively allowed in situ fluorescent tagging of the NEAT1 lncRNA.
Jaffrey and co-workers developed an alternative small molecule based method that relies on interactions between an RNA aptamer sequence (Spinach)52 and the cognate small molecule, 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), a derivative of the native green fluorescent protein (GFP) chromophore. In this method, the Spinach aptamer is encoded downstream of the target RNA sequence. Rather than requiring coexpression of a reporter protein, as in the case of MS2, the cell-permeable DFHBI molecule fluoresces upon binding the Spinach aptamer. The Jaffrey lab has continued to improve the stability and quantum yield of these RNA reporter complexes,53 and in the future these constructs may offer advantages in lncRNA detection over the bulky protein-based labeling approaches. Although powerful, encoded tags can interfere with native RNA folding, which is an important consideration when using these methods.
Cellular Function
Functional studies of lncRNAs generally begin with knock-down and overexpression methods, both in cell culture and animal models, though knockout/knockin methods using CRISPR-Cas9 technology have also been employed.
RNA Knockdown and Rescue
RNA interference (RNAi) methods such as small interfering RNA (siRNA) and short hairpin RNA (shRNA) allow for loss-of-function studies in cell culture or mouse models and are often the first experiment performed to investigate lncRNA function. RNAi protocols typically use transcripts of ~20–40 n.t. that are complementary to the target transcript. Upon target binding, the newly formed duplexes are degraded through existing cellular machinery.54 For instance, Chinnaiyan and co-workers developed siRNA and shRNA sequences toward a novel prostate-cancer associated lncRNA, second chromosome locus associated with prostate-1 (SChLAP1; also called LINC00913).35 Addition of siRNA or shRNA reduced invasion in cell culture and metastasis in mouse models, respectively. As a complement to knock-down studies, “rescue” experiments are some of the most compelling confirmations of function. After successful inhibition with siRNA, reintroduction of an siRNA-resistant target transcript should lead to recovery of lncRNA function and restore the native phenotype. Because the siRNA developed was specific for one SChLAP1 isoform, the same culture could be complemented with a second native but siRNA resistant SChLAP1 isoform. As expected, the invasive phenotype was restored, confirming the impact of SChLAP1 on this pathway.
Though RNAi has been crucial to the study of many lncRNA functions, it is important to note that lncRNA-targeted RNAi methods can be hindered by several factors, including the often low levels of native lncRNA expression,55 the nuclear localization of many lncRNAs,46 and the more structured nature of lncRNAs compared to mRNAs.56 As a result, several siRNA sequences are often screened before identifying a unique transcript for effective lncRNA knockdown. The use of chemically modified oligonucleotides has addressed some of these limitations. Corey and co-workers, for example, recently developed ssRNA sequences with chemical modifications to the backbone that facilitate nuclear localization and cotranscriptional silencing of ncRNA.57 Antisense oligonucleotides (ASOs) are synthetic nucleic acid derivatives that, while often less effective than siRNA, are more stable to degradation and more easily access nuclear RNA sequences.58 For example, concerns over the efficiency of nuclear siRNA targeting led Jeang and co-workers to use ASOs complementary to NEAT1 to confirm the decrease in nuclear paraspeckle formation previously observed with siRNA.59 The impact of cellular environments on siRNA must also be taken into account. For example, several authors have performed knockdown on MALAT1 using siRNA and shRNA in different cell lines and identified distinct functional pathways of MALAT1 activity.60–62 These results indicate that either siRNA and shRNA knockdown are not analogous methods or that the function of MALAT1, and potentially other lncRNAs, varies greatly between cell lines.
CRISPR-Cas9
The CRISPR-Cas9 system improves upon the shortcomings inherent to RNAi.63 Bacteria naturally harbor the CRISPR-Cas9 system as a means of adaptive immunity, and scientists have engineered this variable immune system to cleave directed DNA sites in systems that range from isolated plasmid64 to human cells.65 Whereas RNAi uses transcript complementarity to target transcribed RNA, leaving the potential for incomplete targeting and missed transcripts, the CRISPR-Cas9 system uses gene complementarity to bind and excise the target gene.64–66 One potential drawback to this technique, however, is that the removed genomic DNA may also play a regulatory or structural role. In CRISPR-Cas9 studies on Haunt lncRNA, Shen and co-workers utilized a combined knockout/knockin strategy to systematically delete regions of the Haunt gene without changing the amount of bulk genomic DNA.67 These studies revealed opposing functions of the Haunt DNA sequence and the lncRNA transcript, clearly demonstrating the importance of adequate controls and the power of the CRISPR-Cas9 system to study lncRNA function.
IDENTIFICATION OF lncRNA BINDING PARTNERS
As previously discussed, lncRNA can perform explicit biological functions, e.g., epigenetic regulation, when bound to a protein or protein complex (Table 1). In many cases, knockdown and related methods give insight into pathways that might be influenced by lncRNA transcripts, allowing researchers to infer which proteins might be involved. On the basis of these insights, protein-specific immunoprecipitation (IP) methods and RNA pull-down assays can be pursued. Methods that specifically focus on chromatin-bound lncRNAs are discussed in Methods for Identifying and Characterizing lncRNAs and lncRNA:Protein Complexes and Their Influence on Chromatin.
RIP(-Seq)
Antibody-based RNA immunoprecipitation (RIP) methods isolate RNA:protein complexes from cell culture.68,69 This well-established method isolates the protein of interest by exposing freshly harvested cell lysates to immobilized antibodies (Figure 2). While not always necessary, cross-linking is often employed prior to lysis to ensure coprecipitation of the RNA of interest. Following isolation, the cross-links can be reversed to allow for RNA isolation and downstream analyses such as RT-qPCR. In addition, other protein binding partners can be confirmed through Western blot analysis or identified de novo through proteomics. In early studies of HOTAIR, for example, antibodies to both EZH2, a member of the PRC2 complex, and lysine-specific demethylase 1A (LSD1) were used to IP HOTAIR.10 In the event that other RNA sequences may be bound to the protein target, the isolated RNA can be submitted to microarray or sequencing analysis for binding partner identification on a transcriptome wide scale.68 Using this method, Cech and co-workers were able to evaluate the promiscuity of the PRC2 complex for RNA binding via RIP-seq.70,71 Numerous combinations of these capture-and-characterize methods and the utility of Next Generation Sequencing (NGS) can be found in recent reviews.72–76
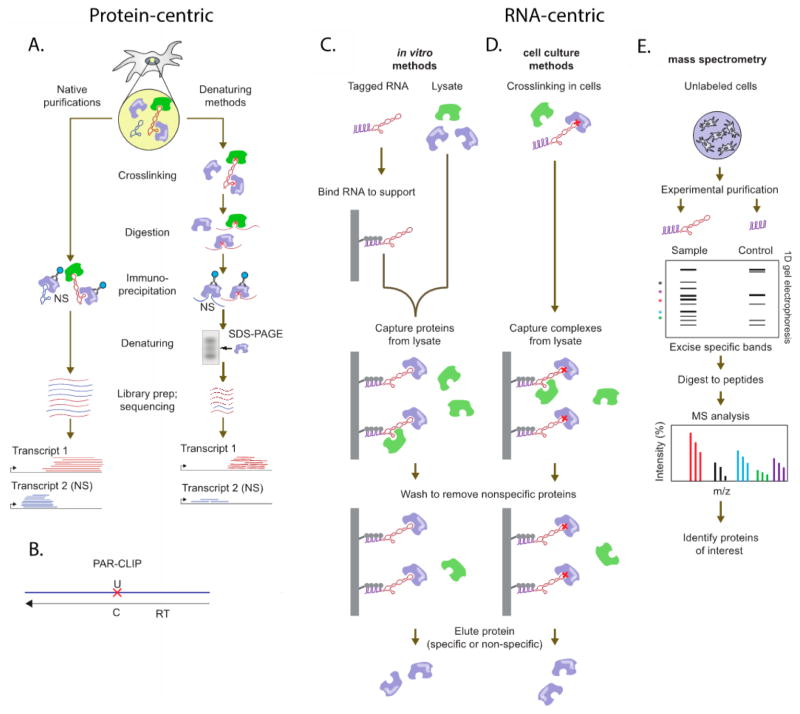
Figure 2.
RNA- and protein-centric immunoprecipitation assays. (A) Protein-centric pull-down to identify RNA–protein interactions, using either native or denaturing methods. After immunoprecipitation, transcripts are separated and submitted for sequencing. (B) A schematic of PAR-CLIP, where the red X indicated the point mutation inserted during reverse transcription (RT). (C) In vitro methods of RNA-centric pull-downs to identify RNA-binding proteins (D) cell culture methods of RNA-centric pull-downs (E) RNA:protein complexes from (C) and (D) are separated, and proteins are submitted for proteomics. Reprinted (adapted or reprinted in part) with permission from ref 77. Copyright 2015 Nature.
PAR-CLIP
Photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) can reduce the number of false positives in traditional RIP experiments by photo-cross-linking newly synthesized RNA directly to protein binding partners that are in close proximity.78,79 First, cells are treated with photoreactive thioribonucleoside analogues. These analogues are incorporated into newly transcribed RNA, and exposure to UV light induces cross-linking of the nucleosides to nearby cysteine residues on the protein surfaces. Following IP, the RNA and protein can be separated through proteinase digestion, and the RNA fraction is reverse-transcribed into cDNA before being submitted to sequencing. This method not only allows the discovery of novel protein-bound RNA transcripts but also identifies the protein-binding domain of the RNA through the detection of distinct RT-induced mutations in the cDNA caused by the unnatural ribonucleosides. As with all cross-linking and IP techniques, robust background controls and statistical analyses must be performed.79 PAR-CLIP has been employed by Reinberg and co-workers to identify lncRNA binding partners of the jumonji family ARID-containing protein 2 (JARID2),80 which was discovered to be an adjunct component in PRC2 pathways.81,82 Through PAR-CLIP with a JARID2-specific antibody, Reinberg and co-workers identified 53 lncRNAs that were bound to both JARID2 and EZH2.
Several factors must be considered when performing RIP, and numerous variations of RIPs and CLIPs have been developed within the past five years and are well-described in recent reviews.83,84 For example, while cross-linking can help to capture low affinity or transient interactions, it is also prone to false positives.83 Non-native RNA:protein interactions can be formed in vitro after the cell lysate has been collected.85 The generally low expression levels of lncRNA may lead to masking of these sequences by more abundant RNAs, particularly given the logarithmic amplification of PCR.86 Single-molecule sequencing (SMS) can be used to avoid interference from abundant RNAs of all sizes as well as other amplification-induced biases.87 SMS covers a broad range of transcript concentrations without the need for PCR. Recently, Williams and co-workers used this technique to identify novel estrogen-regulated lncRNAs in breast cancer.88
RNA-Centric Methods
RNA pull downs can be performed both in vitro and in cell culture and can isolate a wide array of binding partners that may or may not appear in a protein-specific IP.69 For example, purified and biotinylated HOTAIR was immobilized on resin and exposed to HeLa cell lysates. HOTAIR was found to bind EZH2 and SUZ12, two subunits of the PRC2 complex, as well as the LSD1 complex.10 By immobilizing a series of HOTAIR deletion mutants, the fragments of HOTAIR critical to protein binding were identified. Cell culture pull downs often employ an encoded MS2-aptamer sequence that can be immobilized on an MS2-coat protein surface.86
BIOPHYSICAL CHARACTERIZATION OF lncRNA:PROTEIN COMPLEXES
Following identification of lncRNAs and lncRNA:protein complexes, a series of in vitro techniques can be utilized for further biophysical characterization. Many studies begin with the determination of binding affinity and selectivity of lncRNA and cognate protein binding partners. The determination of the structural components of these interactions, however, has been investigated in far fewer cases, though novel techniques used in the characterization of other long RNA sequences such as viral RNAs should facilitate these studies.
EMSA
Electrophoretic mobility shift assay (EMSA), also known as a gel shift assay or band shift assay, is an affinity-based technique that can be used to quantitatively or qualitatively study RNA-protein interactions based on the size difference between apo and protein-bound states of RNA.97 One can quantify the bound versus unbound RNA by comparing the RNA-protein band versus the apo RNA band, respectively. Specificity can be evaluated through the addition of unlabeled control RNA, which would not be expected to impact complex formation, as well as through the addition of unlabeled RNA target, which should displace the labeled RNA in a dose-dependent manner. Lee and co-workers, for example, used this method to test specificity of PRC2 for RepA, the minimal binding domain of Xist.14 While unlabeled RepA led to displacement of the observed complex, competition assays with the antisense sequence and scrambled sequences showed little to no disruption in RepA:PRC2 binding, consistent with a specific complex.
Both the Cech70 and the Lee89 laboratories pursued quantitative analysis of HOTAIR:PRC2 interactions and found dissociation constants ranging from 75 to 100 nM between PRC2 complexes and the 5′-end domain of HOTAIR. At the same time, significant differences in the selectivity of PRC2 for HOTAIR and other lncRNAs were observed, with the Cech lab concluding that PRC2 binding affinity was strongly dependent on the length rather than the sequence of the RNA. These discrepancies led the two laboratories to collaborate on a careful comparison between the two studies,71 and the researchers found that the RNA:PRC2 EMSA assay was highly sensitive to changes in experimental conditions. In addition, PRC2 was found to have multiple lncRNA binding partners, each with similar affinity; however, PRC2 did not interact with all lncRNAs or mRNAs of similar sized transcripts, leading both groups to define PRC2:lncRNA interactions as promiscuous but specific.
Filter-Binding Assay
In addition to EMSAs, more traditional filter-binding assays can be employed to determine binding affinities of RNA:protein complexes. Radiolabeled RNA is incubated with the protein of interest and is washed over a nitrocellulose filter paper. RNA that complexes with protein remains on the nitrocellulose, and the bound versus unbound fraction can be measured via scintillation counter. Lee and co-workers utilized this method not only to confirm binding constants of lncRNA:PRC2 complexes but also to study enzymatic activity.89 It was expected that RNA sequences with lower binding affinity to PRC2 would lower the histone methyltransferase (HMTase) activity. Methylation rates were measured by tracking the transfer of tritiated methyl groups from S-adenosyl methionine (SAM) onto recombinant histone H3 via a filter-binding assay. The rate of catalysis was thus directly proportional to the radioactivity of H3. The authors discovered that regulation by an additional PRC2-association protein, JARID2, was necessary to allow histone methylation, which is consistent with the reduction of the binding affinity of PRC2 for lncRNA in the presence of JARID2.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) allows for the study of RNA-protein interactions in a label-free, dynamic environment by measuring changes in the refractive index (RI) on the surface.90,98 Though SPR requires higher molecular weight particles to measure the RI, this may not be an issue for the study of large transcripts such as lncRNA and their oft multiprotein binding partners.98 For example, PARTICLE, or promoter of MAT2A-antisense radiation-induced circulating lncRNA, is located within the MAT2A gene.41 Knockdown of PARTICLE in sham-irradiated cells caused overexpression of MAT2A, indicating an epigenetic regulatory role for PARTICLE. Anastasov and co-workers examined the PARTICLE:MAT2A complex using SPR. Their results show an inhibitory DNA:RNA triple helix formation between the MAT2A gene and the PARTICLE lncRNA. These effects were only seen at low doses.
The use of additional methods may be required to accurately characterize lncRNA:protein complexes (Table 2), including fluorescence anisotropy techniques96 and ITC,99 which would provide quantitative evaluation of binding affinities. The large size of lncRNA and associated protein complexes may serve as an advantage for fluorescence anisotropy, which depends upon changes in tumbling rate.96 Novel methodologies to determine the specificity of RNA binding are under active investigation.100 For example, specificity for another promiscuous RNA-binding protein, RNase P protein subunit C5, has been studied using high-throughput sequencing kinetics approach (HITS-KIN), which allows simultaneous monitoring of thousands of RNA sequences. Methods such as these along with studies of the impact of lncRNA on enzymatic activity will be crucial to future characterizations of the lncRNA:protein interaction.
Table 2.
Common Biophysical Techniques That Have Been Used for lncRNA, Longer RNA Transcripts or Could Be Applied in Future Studiesa
| technique | shorthand | values solved | pros | cons | RNA examples | ref |
|---|---|---|---|---|---|---|
| electrophoretic mobility shift assay | EMSA | Kd | low-cost equipment | often requires P-32 radiolabel | HOTAIRb; Xistb | 71 |
| filter-binding assay | Kd | straight-forward method; efficient | requires radiolabeling (P-32); nonspecific binding to filter; aggregation of materials not related to native binding event | HOTAIRb | 89 | |
| surface plasmon resonance | SPR | Kd | label-free | target immobilization; cannot report on 2+ state binding model | PARTICLEb | 90 |
| isothermal titration calorimetry | ITC | n, Kd, ΔH, ΔG, ΔS | label-free; kinetic and thermodynamic information; can report on 2+ state systems | high concentrations of materials | HIV-1 TARc; purine riboswitchc; 16S rRNAc | 91–93 |
| Förster resonance energy transfer | FRET | n, Kd | relatively cheap; can measure distance relationship; sensitive | requires FRET pairs that do not interfere with RNA structure; depends on conformational change and may miss static binding events. | rRNAc, tRNAc | 94 |
| fluorescence polarization/anisotropy | FP/FA | n, Kd, aggregation, folding dynamics | no energy transfer requirement; can be HTP | requires fluorescent labeling; binding must significantly change the size of the fluorophore-labeled molecule | Tetrahymena Group I Intron Ribozymec | 95, 96 |
HTP = high-throughput; n = Hill’s coefficient; Kd = dissociation constant; ΔH = change in enthalpy; ΔS = change in entropy; ΔG = change in Gibb’s free energy.
Long ncRNA example.
Other ncRNA example.
Investigations of lncRNA Secondary Structure
The secondary structure of RNA refers to the arrangement of nucleotides in terms of loops and base-pairings. The differences in flexibility and dynamics of nucleotides that are base-paired compared to unpaired nucleotides render the latter more reactive to chemical or enzymatic probing methods. These relative reactivities lend valuable insight into the secondary structure of the RNA, providing parameters for the prediction of secondary structure and laying the groundwork for the prediction of tertiary interactions and protein binding domains.
Chemical and Enzymatic Probing
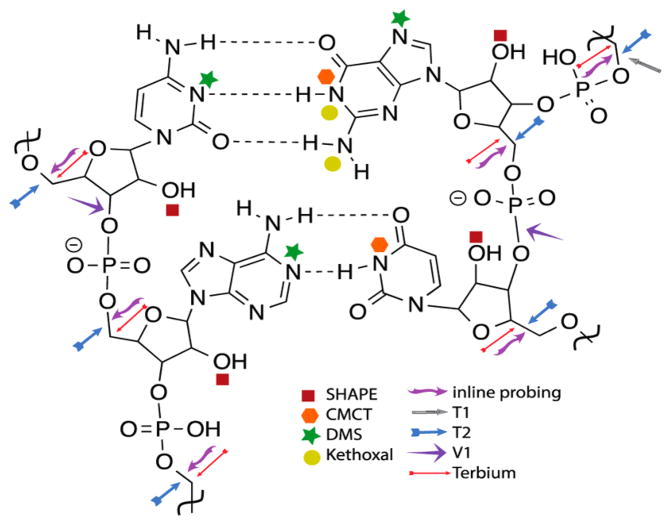
Chemical probing is the addition of reactive small molecules that target the base, sugar, or backbone of RNA molecules. Because of the high rigidity and structure of the A-form helix, Watson-Crick base-pairs are “protected” from reacting with the chemical probe, whereas highly mobile, single-stranded regions will react more often (Figure 3). These reactions can be analyzed though primer extension, reverse transcription, and cDNA sequencing via gel electrophoresis, respectively. Enzymatic probing utilizes RNases rather than small molecules, and important structural insights can be gained from the differential reactivity of RNases101 (Figure 3).
Figure 3.
Schematic mapping where various chemical probing materials target on nucleobases and backbone. Reprinted (adapted or reprinted in part) with permission from ref 102. Copyright 2015 Elsevier.
The size of lncRNA sequences renders structural determination particularly difficult; to date, only two secondary structures have been characterized. In the first example, Sanbonmatsu and co-workers reported the secondary structure of the steroid receptor RNA activator (SRA).103 The secondary structure of this 0.87 kb lncRNA was determined using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), in-line probing, dimethyl sulfate (DMS), and RNase V1 (enzymatic) probing. More recently, the Pyle lab published the secondary structure analysis of the lncRNA HOTAIR using SHAPE, DMS, and terbium (Tb).104 This work determined four independent folding domains within the 2.2 kb lncRNA by comparing the secondary structure data of overlapping fragments to the analysis of the full-length HOTAIR transcript using the shotgun secondary structure method (discussed below) (Figure 4).
Figure 4.
Secondary structure of human HOTAIR Domain 1 (1–530 n.t.; GI: 383286742). Pyle and co-workers performed chemical probing on the full-length HOTAIR transcript. Domain 1 contains the binding-domain of PRC2. Secondary structure rendered in VARNA.105
Improvements in secondary structure probing are ongoing. Weeks and co-workers recently reported a new SHAPE method, SHAPE-MaP (mutational profile), that can be used to predict the secondary structure of longer and more complex transcripts by detecting reduced enzyme processivity.106 This method takes advantage of the inherent ability of reverse transcriptase enzymes to either incorporate noncomplementary nucleotides or create deletions at the site of a SHAPE adduct. Additionally, Weeks and co-workers developed RNA interaction groups by mutational profiling (RING-MaP), a technique to explore correlations between clusters of chemically modified nucleobases, enabling the identification of through-space global interactions.107 Das and colleagues developed a technique named Mutate-and-Map (M2), to further explore the secondary structure of RNA.108 Within M2, each nucleotide is systematically mutated, and the resulting change in susceptibility of its base-pair to chemical adduction is measured and incorporated into the calculation. Chang and Kool have collaborated on the design and application of SHAPE probes that function in cell culture.109 Comparison of the SHAPE profiles for the 5S rRNA in vitro and in cell culture revealed that residues of the 5S that normally bind with ribosomal proteins were less accessible in cell culture, as expected. This method should allow for probing of lncRNA secondary structure in a biological context and reveal pivotal bases involved in RNA–protein interactions.
Shotgun Secondary Structure
To identify possible independently folding domains of HOTAIR, Pyle and coworkers used shotgun secondary structure (3S) fragment analysis.110 Following fragmentation, independent folding domains were determined by comparing the full-length probing map with the fragment probing maps. The analysis showed that HOTAIR is divided into four independent domains, of which the 5′-end domain and the 3′-end domain are known to interact with the PRC2 and the LSD1/REST/co-REST complex, respectively.10,70,71,89
Finally, both chemical and enzymatic probing can be used in the presence and absence of the RNA protein binding partner(s). This would not only determine the protein-binding region but could also give insight into the RNA structure rearrangement from apo to bound forms. It is also important to note that chemical and enzymatic probing methods result in structural predictions rather than absolute characterization. The probing experiments are often performed in parallel to provide a stronger model of the true base-pairings for a given structure. The solved secondary structures are thus a probability of base-pairs, a further reflection of the dynamic structure of RNA.111,112
Computational Prediction
To produce a secondary structure, data from chemical probing and enzymatic cleavage experiments are run through a secondary structure prediction program based on free energy minimization and the RNA base-pairing energetic parameters determined by Turner and coworkers113,114 These programs, such as Mfold115 or RNAs-tructure,116 attempt to reconcile the experimental data with free energy calculations to produce the most accurate secondary structure possible. While different programs specialize in particular chemical probing methods, all of them focus on increased sensitivity and positive predictive values (PPV) and are continually improving.
Investigations of lncRNA Three-Dimensional Structure and Tertiary Interactions
RNA three-dimensional structure is highly dependent upon backbone flexibility and/or rigidity, interhelical contacts, and coaxial stacking as well as less common motifs such as triple-helix formation or G-quadruplexes. While little is currently known about the three-dimensional structure of lncRNAs, the secondary structure probing of HOTAIR revealed significant and diverse structure, with more than half of the nucleotides found in base-pairs along with the presence of several higher order junctions.104 These results, along with the known and specific protein interactions, are consistent with HOTAIR having a complex 3D structure, though such complexity may not be true of all lncRNAs.
To facilitate studies of native lncRNA 3D structure and tertiary contacts, the Pyle lab recently reported a clear protocol for the in vitro transcription and native purification of lncRNAs.117 Nondenaturing purification avoids non-native buffer conditions and high temperatures that would cause the lncRNA to melt and reanneal, which can result in non-native folding. Sedimentation velocity analytical ultracentrifugation (SV-AUC) analyses support the importance of native purification as the formation of multiple lncRNA conformations were observed upon exposure to denaturing conditions.
X-ray Diffraction and Cryo-Electron Microscopy
X-ray crystallography has been a staple of structural biology for decades. For large, dynamic, and flexible structures such as RNA, however, obtaining an atomic-resolution structure is difficult, as evidenced by the 20:1 ratio in protein to RNA structures deposited in the Protein Data Bank.118 While the largest ribosome structures in the PDB contain as many as 5070 nucleotides119 (PDB: 4UG0), the highly structured RNA–protein complex samples only a limited range of conformations after its initial folding, and lncRNAs are expected to be more conformationally dynamic in comparison. Furthermore, the well-ordered structure of the ribosome is an exception: generally, as the unit cell size increases and becomes more complex, the resolution decreases substantially. The only non-ribosomal RNA that approaches the length of a lncRNA is the Bluetongue viral RNA (BTV) at 1906 nucleotides.120 At 10 Å resolution, the overall topology of a standard A-form helix is the only discernible structural feature (PDB: 1H1K). An additional concern with X-ray diffraction is that of structural artifacts: as the crystal itself forms, it is possible to induce structures that favor the formation of a stable crystal lattice and/or the optimization of crystal contacts but are not representative of the most biologically relevant structure. This is less of a concern for solution-phase techniques such as NMR and SAXS/SANS, which allow RNA to sample all possible conformations of a given structure and will be discussed later.
One potential solution to structural studies of lncRNAs using X-ray crystallography is to obtain X-ray diffraction structures of the independent folding domains, such as those in HOTAIR mentioned above, and use these structural fragments to reconstruct the full length structure by cryo-EM. Cryo-EM is capable of capturing conformational changes and dynamic interactions for structures that are too large for NMR yet too dynamic for crystallography.121,122 While this would not provide a perfect rendering of the lncRNA structure, significant insight into global interactions would be possible, and it follows the trends in methodology used to elucidate the structures of other large biomolecules.119,123,124
At the same time, X-ray crystallography has not been without success in lncRNA. Steitz and co-workers successfully crystallized the 3′-end of MALAT1 along with a portion of its A-rich 3′-UTR (PDB: 4PLX).125 These results yield valuable insight into the local lncRNA structure and are consistent with the stabilization of MALAT1 against degradation as a potential mechanism for nuclear accumulation.
NMR Spectroscopy
Solution state nuclear magnetic resonance (NMR) spectroscopy is limited by the size of lncRNA due to both severe spectral overlap and poor resolution from increased tumbling times of large systems.126 At the same time, the advantage of NMR lies in the potential to obtain atomic resolution ensembles in solution state conditions that are more biologically relevant than the crystal-lattice conditions needed for X-ray diffraction. Additionally, recent advances in selective labeling techniques promise to help circumvent spectral overlap in long RNA samples.127
While a lncRNA structure via NMR has not yet been reported, Summers, Telesnitsky, and co-workers recently reported the longest RNA transcript structure (155 n.t.) solved by 2D-NMR methods, namely, the HIV-1 5′-UTR.128 The universal issue of spectral overlap was avoided by separately deuterium-doping segments of the HIV-1 core encapsidation signal (rCES), which encompasses the shorter fragments of the HIV-1 5′-UTR, and then annealing the 3′-end with a nonlabeled 5′-end (and vice versa) using rapid denaturation and snap cooling. This allowed for unambiguous assignment by utilizing the nuclear Overhauser effects (NOEs) to obtain sequence dependent correlations and ultimately the identification of long-range interhelical relationships. This work was able to resolve discrepancies between nonstructural studies (phylogenetic, biochemical, nucleotide reactivity, and mutagenesis) and the many NMR structure studies performed on shorter segments of the HIV-1 5′-UTR.129
Small Angle Scattering
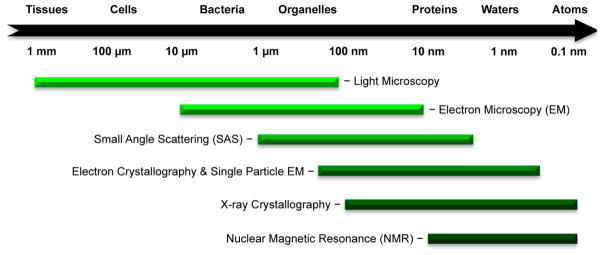
Small-angle scattering (SAS) provides the user with direct structural information about the size, shape, organization, and flexibility of a biopolymer and can utilize both X-ray and neutron scattering (SAXS and SANS, respectively).130 This method also uses solution phase sample data collection, similar to conventional biomolecular NMR. Contrary to NMR, however, SAS does not have a size limit nor does it require large concentrations of sample. It is important to note that the resolution of SAS is significantly lower than that of NMR or X-ray diffraction, and the method does not provide atomic-level detail (Figure 5).
Figure 5.
A scale showing the different ranges of electromagnetic dispersion for various structural methods.
For large structures such as lncRNAs, however, SAS might be the only reasonable method for directly acquiring structural data, as evidenced by the characterization of large viral RNA structures. For example, Rein, Wand and co-workers were able to resolve the topological map of the HIV-1 Rev response element (RRE; 233 n.t.) using SAXS, revealing an asymmetric A-shaped structure.131 Musier-Forsyth and co-workers were able to characterize the 3D structure of the 5′-UTR by combining SAXS topological maps of three ~100 nucleotide fragments with previously reported NMR data. This analysis revealed tertiary interactions consistent with several proposed functions of the 5′-UTR, including tRNA mimicry.132
Small-angle neutron scattering (SANS) can be used to resolve distinct classes of biomacromolecules, including nucleic acids and proteins, based on differences in the bound-atom neutron scattering lengths within each species. More specifically, proteins scatter in the range of (1–5) × 10−12 cm, while nucleic acids scatter in the range of (8–12) × 10−12 cm.133 Despite the inherent low resolution of SAS, this fundamental characteristic of neutron scattering may make it possible to discretely study the conformation of a lncRNA molecule in the context of any number of protein binding partners.
While no studies of SAS on lncRNAs have been reported to date, it is expected that the methods described here will be directly applicable and will provide numerous insights into RNA dynamics, folding, binding, and activity. SAS may further serve as a suitable “first pass” structural assessment of a de novo complex that can be refined by obtaining high-resolution structures of individual fragments.
Computational Modeling
As with secondary structure, computational modeling can provide insight into the 3D structure of RNA.113,114,134 Indeed, ab initio methods based only on primary sequence have improved to take into account not only canonical and noncanonical base-pairing but also some pseudoknots and unpaired regions. In 2007, Das and Baker presented the Rosetta-based fragment-based structure prediction algorithm for automatic de novo prediction of RNA 3D structures,135 which evolved into the Fragment Assembly of RNA with Full Atom Refinement (FARFAR) algorithm now available on the Rosetta Online Server that Includes Everyone (ROSIE).136,137 Predicted RNA structures can be inserted into a molecular dynamics (MD) simulation using force fields such as AMBER138 or CHARMM139 in order to further sample their conformational space.140 MD simulations predict multiple thermodynamically allowed structural conformations for a given sequence, which can lend insight into the dynamic flexibility, and potentially the mode of action, of an RNA structure.
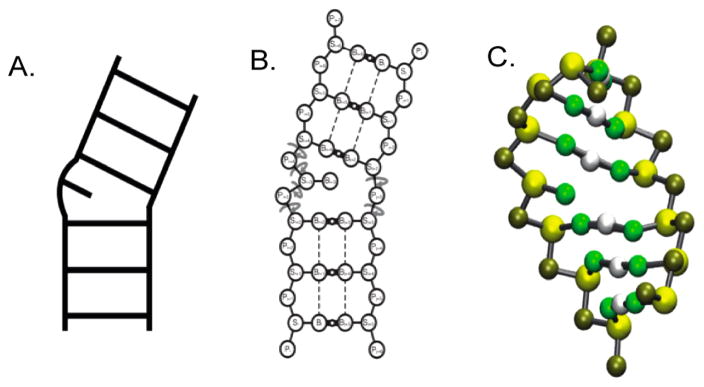
Though powerful tools, both computational modeling and MD are still limited to shorter RNA sequences based on current computing power. Coarse-grain modeling overcomes the time needed to run full-length, all-atom MD simulations while still allowing the exploration of topologically allowed space. Restrictions are placed to limit the degrees of freedom within the structure, and pseudoatoms are used in lieu of full structural elements to simplify the calculation. Caution must be taken when choosing a program, however, as most coarse-grain modeling programs lack parameters unique to RNA structure. The topological mapping of RNA (TOPRNA) program,141 on the other hand, was parametrized using RNA structure databases such as the Nucleic Acid Data Base (NBD).142 TOPRNA reduces the secondary structure of a given transcript to four pseudoatoms corresponding to the phosphate, ribose, nucleobases, and hydrogen bonds between paired nucleobases (omitted in unpaired bases; Figure 6) and uses the CHARMM force field to explore topological 3D space within the coarse grain parameters. This program focuses on connectivity and sterics for a given transcript, ignoring noncovalent interactions. TOPRNA successfully reproduced NMR residual dipolar couplings (RDCs) of tRNA-PHE, confirming that topological restrictions are a major factor in predicting RNA 3D structure. High-resolution RNA (HiRE-RNA) coarse-grain modeling incorporates seven pseudoatoms per nucleotide rather than four.143,144 This extra information allows for stacking and base-pairing terms, which are ignored in TOPRNA, but has a slower processing time.
Figure 6.
Conversion of known secondary structure (A) into one of four possible identities: phosphate, ribose, nucleobase, hydrogen bond (B). Three-dimensional assignment in the coarse-grain modeling program TOPRNA: gold, phosphate; yellow, ribose; green, nucleobase; white, hydrogen bond (C). Reprinted (adapted or reprinted in part) with permission from ref 141. Copyright 2014 Oxford University Press.
The ability to predict topologically allowed 3D structures from a secondary structure might lend valuable insight into the stability and accessibility of lncRNA domains. As parametrizations and computer processing improve, we can expect to see more detailed representations of RNA and access to longer constructs.
METHODS FOR IDENTIFYING AND CHARACTERIZING lncRNAs AND lncRNA:PROTEIN COMPLEXES AND THEIR INFLUENCE ON CHROMATIN
LncRNA sequences that interact with chromatin, chromatin-binding proteins, and/or genomic DNA are increasingly appreciated as regulators of gene expression.145–147 Several recent methods have led to insight into the specific interactions of lncRNA with chromatin and the influence of lncRNA on chromatin structure.
ChIRP
A method developed by Chang and co-workers, chromatin isolation by RNA purification (ChIRP), can directly map lncRNA on the genomic scale.148,149 ChIRP enables pull-down of lncRNA:chromatin complexes by tiling the target lncRNA sequence with short biotinylated oligonucleotides. This method removes the need for known protein partners, pathways, or structure but also permits false positives, as colocalization does not directly imply cofunctionality. Recently, Kanduri and co-workers investigated how the maternally expressed gene 3 (MEG3) lncRNA associates with its target chromatin while bound to PRC2.28 The authors mapped the location of MEG3 binding using ChIRP-seq in conjunction with 4-SU photoactivatable in vivo cross-linking and discovered an overenriched GA-rich sequence motif in genomic regions associated with gene regulation. The authors used the Triplexator software150,151 to computationally predict a higher potential for triplex formation at these GA-rich regions when compared to nonenriched regions. Follow up EMSA assays confirmed the formation of a complex between the 20 nucleotide GA-rich ssRNA sequence within the 5′-end of MEG3 and the GA-rich dsDNA sequences found at the chromatin targets. While these results show the potential for triple-helix formation, it is not possible to know from these experiments if the same segment of RNA would be spatially available for triple-helix formation within the full-length MEG3 transcript.
Chromosome Conformation Capture
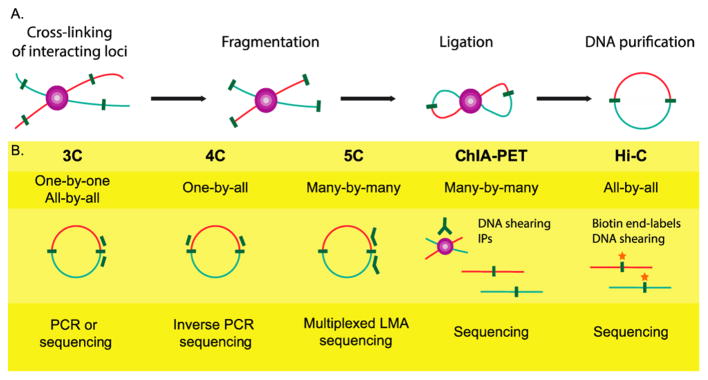
The study of large-scale chromosome structure has advanced significantly within the past decade.152–154 The development of chromosome confirmation capture (3C) and its subsequent advances (i.e., 4C,155 5C,156 Hi-C,157 ChIP-loop,155 ChIA-PET158) enables the studying of chromosome structure in both the presence and absence of other regulatory factors (Figure 7). Originally, 3C was developed to show cis-regulation of the chromosome between genes separated by many kilobases. In short, the chromatin is cross-linked and then exposed to digestion enzymes that remove any non-cross-linked DNA. Exposed DNA ends in close proximity are then ligated, and the newly generated sequences can be identified through PCR or sequencing following cross-link reversal. The successors of 3C allow for necessary prior knowledge of only one target sequence as opposed to both (4C); analysis of multiple junctions and contact points (5C); biotin-labeled ligation junction and specific purification (Hi-C); the combination of 3C and ChIP (ChIP-loop); and the removal of nonspecific interactions while also assessing de novo chromatin interactions (ChIA-PET). The method 3C has been adapted by Hoffman and Hu to study lncRNA:DNA complexation (R3C).159 In the R3C method, genome-bound ssRNA, in this case lncRNA Kcnq1ot1, is cross-linked to genomic DNA and then reverse transcribed with biotin-labeling. This cDNA is subjected to digestion via EcoRI and then ligated to the original target dsDNA. The biotin-labeled cDNA:dsDNA complex is pulled down using streptavidin beads and subjected to PCR to map the chimeric structure. This promising assay is able to map binding of lncRNA to specific loci, in this case the Kcnq1 promoter region.
Figure 7.
(A) The general steps of chromosome conformation capture (3C), from left to right: covalent cross-linking of overlapping chromatin regions; chemical or auditory fragmentation; ligation of cross-linked segments; cross-link reversal and DNA purification. (B) Expansive methods of 3C that allow for higher processivity and more spatial information. Reprinted (adapted or reprinted in part) with permission from ref 152. Copyright 2014 BioMed Central Ltd.
Future Directions
lncRNA transcripts and lncRNA:protein complexes play an undeniably essential role in epigenetic regulation, impacting a wide range of biological pathways from embryonic development to the progression of metastatic cancer, yet many details of lncRNA molecular function remain to be elucidated. While robust methods exist to identify the ever-increasing number of novel lncRNA transcripts and their specific protein partners, the order and regulation of these binding events in biological systems remain unknown. Biophysical characterization of these relationships will rely on continued efforts to standardize existing protocols and the application of innovative characterization methods. The impact of lncRNA:protein interactions on the localization and chemical activity of chromatin-modifying enzymes also merits continued investigation and may benefit from the use of synthetic nucleosomes as model systems.160,161 Arguably one of the most pressing and exciting areas of lncRNA research will involve structural elucidation, which has progressed impressively at the secondary structure level and will likely benefit from the application of coarse-grained predictions and the fitting of structural fragments to cryo-EM or small angle scattering envelopes. Furthermore, the impact of N6-methyladenosine (m6A) base modifications, discovered in lncRNA MALAT1 by Pan, He, and co-workers,162,163 may unlock yet another level of complexity as work in the m6 A field progresses. The application of these and other methods to lncRNA is expected to reveal striking insights into the molecular recognition, specificity, and function of these enigmatic molecules.
Another unexplored area of lncRNA research is the development of selective small molecule probes for lncRNA. While guiding principles for small molecule:RNA recognition are still being developed,164,165 protein-targeted small molecule ligands have been indispensible for both functional probing and therapeutic development, suggesting an enormous potential impact of lncRNA-targeted small molecules in the lncRNA field. The selective inhibition of discrete lncRNA:protein complexes, for example, would not only reveal the functional relevance of these interactions but may also allow the therapeutic potential of targeting lncRNA:protein interactions to be explored.
Acknowledgments
Funding
This work was supported by Duke University and the NIH Predoctoral Training Grant T32GM008555 in Biomolecular and Tissue Engineering to E.J.M.
We thank members of the lab for helpful suggestions and proofreading of the manuscript: A. Donlic, C. Eubanks, J. Forte, G. Kapral, B. Morgan, and N. Patwardhan. We also thank D. K. Merriman for helpful discussions, particularly pertaining to NMR, and J. Link for assistance with TOC graphic. Apologies are given to the many scientists whose great work could not be included within this manuscript due to space restriction.
ABBREVIATIONS
- 3C
chromosome confirmation capture
- 4C
circularized chromosome confirmation capture
- 4-SU
4-thiouridine
- 5C
carbon-copy chromosome confirmation capture
- 6-SG
6-thioguanosine
- AMBER
Assisted Model Building with Energy Refinement
- ANRIL
antisense noncoding RNA in the INK4 locus
- ARID
AT-rich interaction domain
- ASO
antisense oligonucleotide
- BTV
Bluetongue viral RNA
- Cas9
CRISPR associated protein 9
- cDNA
complementary DNA
- CHARMM
Chemistry at Harvard Macromolecular Mechanics
- ChIA-PET
chromatin interaction analysis by paired-end tag sequencing
- ChIP
chromatin immunoprecipitation
- ChIP-loop
ChIP-3C
- ChIRP
Chromatin isolation by RNA purification
- ChIRP-seq
ChIRP followed by high-throughput sequencing
- Co-REST
corepressor for REST
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- Cryo-EM
cryo-electron microscopy
- Cy5
cyanine dye 5
- DANCR
differentiation antagonizing nonprotein coding RNA
- DDX3
DEAD (Asp-Glu-Ala-Asp) box helicase 3
- DFHBI
3,5-difluoro-4-hydroxybenzylidene imidazolinone
- DIS
dimer initiation site
- DLS
dynamic light scattering
- DMS
dimethyl sulfate
- DNA
deoxyribonucleic Acid
- DNMT1
DNA (cytosine-5)-methyl-transferase 1
- dsDNA
double-stranded DNA
- EED
embryonic ectoderm development protein
- EMSA
electrophoretic mmobility shift assay
- ENCODE
Encyclopedia of DNA elements
- ER-α
estrogen receptor alpha
- EYFP
enhanced yellow fluorescent protein
- EZH2
enhancer of zeste homologue 2
- FARFAR
fragment assembly of RNA with Full Atom Refinement
- FARNA
fragment assembly of RNA
- FENDRR
fetal-lethal noncoding developmental regulatory RNA
- FP/FA
fluorescence polarization/anisotropy
- FRABASE
RNA fragments search engine and database
- FRET
Förster resonance energy transfer
- GFP
green fluorescent protein
- H3K27me3
trimethylation of histone 3 lysine 27
- H3K9me2
demethylation of histone 3 lysine 9
- hHotair
human HOTAIR
- HiRE-RNA
high resolution RNA
- HITS-KIN
high-throughput sequencing kinetics approach
- HIV 5′-UTR
human immunodeficiency virus 5′ untranslated region
- HMTase
histone methyltransferase
- HOTAIR
HOX transcript antisense intergenic RNA
- HTS
high-throughput screen
- IP
immunoprecipitation
- ITC
isothermal titration calorimetry
- JARID
Jumonji, AT-rich interaction domain
- JARID2
Jumonji, AT-rich interaction domain 2
- Kcnq1ot1
KCNQ1 overlapping transcript 1
- lncRNA
long noncoding RNA
- LSD1
lysine-specific demethylase 1A
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- MaP
mutational profile
- MBP
maltose binding protein
- MD
molecular dynamics
- MEG3
maternally expressed gene 3
- mHotair
mouse HOTAIR
- MLL
myeloid/lymphoid or mixed-lineage leukemia
- mRNA
messenger RNA
- MS2
male-specific bacteriophage 2
- NBD
nucleic data base
- ncRNA
noncoding ribonucleic acid
- NEAT1
nuclear enriched abundant transcript 1
- NMR
nuclear magnetic resonance
- NOESY
nuclear Overhauser effect spectroscopy
- NPM
nucleophosmin
- Nt
nucleotides
- P54nrb
54 kDa nuclear RNA- and DNA-binding protein
- PAR-CLIP
photoactivatable-ribonucleoside-enhanced cross-linking and immunoprecipitation
- PARP1
poly [APD-ribose] polymerase 1
- PBS
primer binding site
- PCAT1
prostate cancer associated transcript 1
- PCR
polymerase chain reaction
- PDB
Protein Data Bank
- PRC1
polycomb repressive complex 1
- PRC2
polycomb repressive complex 2
- PSP1a
Paraspeckle protein 1a
- R3C
RNA 3C (chromosome confirmation capture)
- RDC
residual dipolar couplings
- RepA
repeat A transcript
- REST
RE1 silencing transcription factor
- RIP
RNA immunoprecipitation
- RNA
ribonucleic Acid
- RNA-FISH
RNA-fluorescent in situ hybridization
- RNAi
RNA interference
- RNase
ribonuclease
- ROSIE
Rosetta Online Server that Includes Everyone
- roX
RNA on X chromosome
- RRE
Rev response element
- rRNA
ribosomal RNA
- RT-qPCR
real-time quantitative polymerase chain reaction
- SAM
S-adenosyl methionine
- SANS
small-angle neutron scattering
- SAS
small-angle scattering
- SAXS
small-angle X-ray scattering
- SChLAP1
second chromosome locus associated with prostate 1
- SD
splice donor
- SEC
size exclusion chromatography
- SELEX
systematic evolution for ligands by exponential enrichment
- SHAPE
selective 2′-hydroxyl acylation analyzed by primer extension
- SHARP
SMRT/HDAC1-associated repressor protein
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- SLIRP
SRA stem-loop interacting RNA binding protein
- SMS
single-molecule sequencing
- SPR
surface plasmon resonance
- SR proteins
proteins containing long repeats of serine and arginine amino acid residues
- SRA
steroid receptor RNA activator
- SRC-1
steroic receptor coactivator 1
- ssRNA
single-stranded RNA
- STORM
stochastic optical reconstruction microscopy
- SUZ12
suppressor of zeste 12
- SV-AUC
sedimentation velocity analytical ultracentrifugation
- SWI/SNF
switch/sucrose non-fermentable complex
- TAR
trans-activation response element
- Tat
trans-activator of transcription
- Tb
terbium
- TLE
tRNA3Lys-like element
- TOPRNA
topological mapping of RNA program
- tracrRNA
transactivation crRNA
- tRNA
transfer RNA
- TrxG
trithorax group proteins
- Xci
X-chromosome inactivation
- Xi
inactivated X chromosome
- Xist
X-inactive specific transcript
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A Strategy for Probing the Function of Noncoding RNAs Finds a Repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 10.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 12.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 13.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, Visvikis A, Branlant C. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novikova IV, Hennelly SP, Sanbonmatsu KY. Tackling structures of long noncoding RNAs. Int J Mol Sci. 2013;14:23672–23684. doi: 10.3390/ijms141223672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 22.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. BioEssays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 23.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quek XC, Thomson DW, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME, Maag JLV. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 26.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 28.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 30.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh AL, Tuzova AV, Bolton EM, Lynch TH, Perry AS. Long noncoding RNAs and prostate carcinogenesis: the missing ‘linc’? Trends Mol Med. 2014;20:428–436. doi: 10.1016/j.molmed.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong X, Gu PC, Xu SZ, Lin XJ. Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci, Biotechnol, Biochem. 2015;79:732–737. doi: 10.1080/09168451.2014.998617. [DOI] [PubMed] [Google Scholar]
- 37.Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 38.Luo H, Sun Y, Wei G, Luo J, Yang X, Liu W, Guo M, Chen R. Functional Characterization of Long Noncoding RNA Lnc_bc060912 in Human Lung Carcinoma Cells. Biochemistry. 2015;54:2895–2902. doi: 10.1021/acs.biochem.5b00259. [DOI] [PubMed] [Google Scholar]
- 39.Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol. 2011;22:343–350. doi: 10.1016/j.semcdb.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 40.Leygue E. Steroid receptor RNA activator (SRA1): unusual bifaceted gene products with suspected relevance to breast cancer. Nucl Recept Signaling. 2007;4:e006. doi: 10.1621/nrs.05006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Leary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, Moertl S, Trau M, Atkinson MJ, Anastasov N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015;11:474–485. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: A two minute cell fractionation method. BMC Res Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of Single RNA Transcripts in Situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 44.Swanger SA, Bassell GJ, Gross C. High-resolution fluorescence in situ hybridization to detect mRNAs in neuronal compartments in vitro and in vivo. Methods Mol Biol. 2011;714:103–123. doi: 10.1007/978-1-61779-005-8_7. [DOI] [PubMed] [Google Scholar]
- 45.Dunagin M, Cabili M, Rinn J, Raj A. Visualization of lncRNA by Single-Molecule Fluorescence In Situ Hybridization. In: Nakagawa S, Hirose T, editors. Nuclear Bodies and Noncoding RNAs. Springer; New York: 2015. pp. 3–19. [DOI] [PubMed] [Google Scholar]
- 46.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sunwoo H, Wu JY, Lee JT. The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc Natl Acad Sci U S A. 2015;112:E4216–4225. doi: 10.1073/pnas.1503690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weil TT, Parton RM, Davis I. Making the message clear: visualizing mRNA localization. Trends Cell Biol. 2010;20:380–390. doi: 10.1016/j.tcb.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 50.LeCuyer KA, Behlen LS, Uhlenbeck OC. Mutagenesis of a stacking contact in the MS2 coat protein-RNA complex. EMBO J. 1996;15:6847–6853. [PMC free article] [PubMed] [Google Scholar]
- 51.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136:16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang K, Marran K, Valentine A, Hannon GJ. RNAi in Cultured Mammalian Cells Using Synthetic siRNAs. Cold Spring Harb Protoc. 2012;2012:957–961. doi: 10.1101/pdb.prot071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo KQ, Chang DC. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem Biophys Res Commun. 2004;318:303–310. doi: 10.1016/j.bbrc.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 57.Matsui M, Prakash TP, Corey DR. Transcriptional silencing by single-stranded RNAs targeting a noncoding RNA that overlaps a gene promoter. ACS Chem Biol. 2013;8:122–126. doi: 10.1021/cb300490j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sioud M. Overcoming the challenges of siRNA activation of innate immunity: design better therapeutic siRNAs. Methods Mol Biol. 2015;1218:301–319. doi: 10.1007/978-1-4939-1538-5_19. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4:e00596–12. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 62.Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Heidelberg, Ger ) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 63.Barrangou R, Birmingham A, Wiemann S, Beijersbergen RL, Hornung V, Smith A. Advances in CRISPR-Cas9 genome engineering: lessons learned from RNA interference. Nucleic Acids Res. 2015;43:3407–3419. doi: 10.1093/nar/gkv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Mali P, Kim-Kiselak C, Church G. CRISPR-Cas-mediated targeted genome editing in human cells. Methods Mol Biol. 2014;1114:245–267. doi: 10.1007/978-1-62703-761-7_16. [DOI] [PubMed] [Google Scholar]
- 66.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, Zhao Y, Shen B, Huang X, Zhu H, Orkin Stuart H, Shen X. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16:504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 68.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McHugh CA, Russell P, Guttman M. Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014;15:203. doi: 10.1186/gb4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cook KB, Hughes TR, Morris QD. High-throughput characterization of protein-RNA interactions. Briefings Funct Genomics. 2015;14:74–89. doi: 10.1093/bfgp/elu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 74.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: Methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta. 2016;1859:3–15. doi: 10.1016/j.bbagrm.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 77.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;551:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Friedersdorf MB, Keene JD. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014;15:R2. doi: 10.1186/gb-2014-15-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.König J, Zarnack K, Luscombe NM, Ule J. Protein–RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 84.Milek M, Wyler E, Landthaler M. Transcriptome-wide analysis of protein-RNA interactions using high-throughput sequencing. Semin Cell Dev Biol. 2012;23:206–212. doi: 10.1016/j.semcdb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 85.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gong C, Popp MWL, Maquat LE. Biochemical Analysis of Long Non-coding RNA-containing Ribonucleoprotein Complexes. Methods. 2012;58:88–93. doi: 10.1016/j.ymeth.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ozsolak F, Milos PM. Transcriptome profiling using single-molecule direct RNA sequencing. Methods Mol Biol. 2011;733:51–61. doi: 10.1007/978-1-61779-089-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jonsson P, Coarfa C, Mesmar F, Raz T, Rajapakshe K, Thompson JF, Gunaratne PH, Williams C. Single-molecule sequencing reveals estrogen-regulated clinically relevant lncRNAs in breast cancer. Mol Endocrinol. 2015;29:1634–1645. doi: 10.1210/me.2015-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katsamba PS, Park S, Laird-Offringa IA. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods. 2002;26:95–104. doi: 10.1016/S1046-2023(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 91.Suryawanshi H, Sabharwal H, Maiti S. Thermodynamics of peptide-RNA recognition: the binding of a Tat peptide to TAR RNA. J Phys Chem B. 2010;114:11155–11163. doi: 10.1021/jp1000545. [DOI] [PubMed] [Google Scholar]
- 92.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 93.Kaul M, Pilch DS. Thermodynamics of aminoglycoside-rRNA recognition: the binding of neomycin-class aminoglycosides to the A site of 16S rRNA. Biochemistry. 2002;41:7695–7706. doi: 10.1021/bi020130f. [DOI] [PubMed] [Google Scholar]
- 94.Munro JB, Vaiana A, Sanbonmatsu KY, Blanchard SC. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Minuesa G, Antczak C, Shum D, Radu C, Bhinder B, Li Y, Djaballah H, Kharas MG. A 1536-well fluorescence polarization assay to screen for modulators of the MUSASHI family of RNA-binding proteins. Comb Chem High Throughput Screening. 2014;17:596–609. doi: 10.2174/1386207317666140609122714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi X, Herschlag D. Fluorescence polarization anisotropy to measure RNA dynamics. Methods Enzymol. 2009;469:287–302. doi: 10.1016/S0076-6879(09)69014-5. [DOI] [PubMed] [Google Scholar]
- 97.Rio DC. 5′-End Labeling of RNA with [γ-32P]ATP and T4 Polynucleotide Kinase. Cold Spring Harb Protoc. 2014;2014:441–443. doi: 10.1101/pdb.prot080739. [DOI] [PubMed] [Google Scholar]
- 98.Di Primo C, Dausse E, Toulme JJ. Surface plasmon resonance investigation of RNA aptamer-RNA ligand interactions. Methods Mol Biol. 2011;764:279–300. doi: 10.1007/978-1-61779-188-8_19. [DOI] [PubMed] [Google Scholar]
- 99.Feig AL. Studying RNA-RNA and RNA-protein interactions by isothermal titration calorimetry. Methods Enzymol. 2009;468:409–422. doi: 10.1016/S0076-6879(09)68019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jankowsky E, Harris ME. Specificity and nonspecificity in RNA-protein interactions. Nat Rev Mol Cell Biol. 2015;16:533–544. doi: 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sloma MF, Mathews DH. Improving RNA secondary structure prediction with structure mapping data. Methods Enzymol. 2015;553:91–114. doi: 10.1016/bs.mie.2014.10.053. [DOI] [PubMed] [Google Scholar]
- 103.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Somarowthu S, Legiewicz M, Chillón I, Marcia M, Liu F, Pyle AM. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc. 2015;10:1643–1669. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Homan PJ, Favorov OV, Lavender CA, Kursun O, Ge X, Busan S, Dokholyan NV, Weeks KM. Single-molecule correlated chemical probing of RNA. Proc Natl Acad Sci U S A. 2014;111:13858–13863. doi: 10.1073/pnas.1407306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cordero P, Kladwang W, VanLang CC, Das R. The mutate-and-map protocol for inferring base pairs in structured RNA. Methods Mol Biol. 2014;1086:53–77. doi: 10.1007/978-1-62703-667-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. RNA SHAPE analysis in living cells. Nat Chem Biol. 2012;9:18–20. doi: 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Novikova IV, Dharap A, Hennelly SP, Sanbonmatsu KY. 3S: shotgun secondary structure determination of long non-coding RNAs. Methods. 2013;63:170–177. doi: 10.1016/j.ymeth.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 111.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 112.McGinnis JL, Dunkle JA, Cate JH, Weeks KM. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu M, McDowell JA, Turner DH. A Periodic Table of Tandem Mismatches in RNA. Biochemistry. 1995;34:3204–3211. doi: 10.1021/bi00010a009. [DOI] [PubMed] [Google Scholar]
- 115.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinf. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chillón I, Marcia M, Legiewicz M, Liu F, Somarowthu S, Pyle AM. Native Purification and Analysis of Long RNAs. Methods Enzymol. 2015;558:3–37. doi: 10.1016/bs.mie.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- 120.Diprose JM, Grimes JM, Sutton GC, Burroughs JN, Meyer A, Maan S, Mertens PP, Stuart DI. The core of bluetongue virus binds double-stranded RNA. J Virol. 2002;76:9533–9536. doi: 10.1128/JVI.76.18.9533-9536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct. 2002;31:303–319. doi: 10.1146/annurev.biophys.31.082901.134202. [DOI] [PubMed] [Google Scholar]
- 122.Lengyel J, Hnath E, Storms M, Wohlfarth T. Towards an integrative structural biology approach: combining Cryo-TEM, X-ray crystallography, and NMR. J Struct Funct Genomics. 2014;15:117–124. doi: 10.1007/s10969-014-9179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 125.Brown JA, Bulkley D, Wang J, Valenstein ML, Yario TA, Steitz TA, Steitz JA. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat Struct Mol Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Foster MP, McElroy CA, Amero CD. Solution NMR of large molecules and assemblies. Biochemistry. 2007;46:331–340. doi: 10.1021/bi0621314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu K, Heng X, Summers MF. Structural Determinants and Mechanism of HIV-1 Genome Packaging. J Mol Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Neylon C. Small angle neutron and X-ray scattering in structural biology: recent examples from the literature. Eur Biophys J. 2008;37:531–541. doi: 10.1007/s00249-008-0259-2. [DOI] [PubMed] [Google Scholar]
- 131.Fang X, Wang J, O’Carroll IP, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, Kjems J, Schwieters CD, Seifert S, Winans RE, Watts NR, Stahl SJ, Wingfield PT, Byrd RA, Le Grice SF, Rein A, Wang YX. An unusual topological structure of the HIV-1 Rev response element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jones CP, Cantara WA, Olson ED, Musier-Forsyth K. Small-angle X-ray scattering-derived structure of the HIV-1 5′ UTR reveals 3D tRNA mimicry. Proc Natl Acad Sci U S A. 2014;111:3395–3400. doi: 10.1073/pnas.1319658111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Harroun TA, Wignall J, Katsaras J. Introduction. In: Fitter JG, Gutberlet T, Katsaras J, editors. Neutron Scattering in Biology. 1. Springer-Verlag; Berlin Heidelberg: 2006. pp. 1–18. [Google Scholar]
- 134.Tinoco I, Jr, Uhlenbeck OC, Levine MD. Estimation of secondary structure in ribonucleic acids. Nature. 1971;230:362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- 135.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc Natl Acad Sci U S A. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lyskov S, Chou FC, Conchuir SO, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ, Das R. Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE) PLoS One. 2013;8:e63906. doi: 10.1371/journal.pone.0063906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yildirim I, Stern HA, Tubbs JD, Kennedy SD, Turner DH. Benchmarking AMBER Force Fields for RNA: Comparisons to NMR Spectra for Single-Stranded r(GACC) Are Improved by Revised χ Torsions. J Phys Chem B. 2011;115:9261–9270. doi: 10.1021/jp2016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Denning EJ, Priyakumar UD, Nilsson L, Mackerell AD. Impact of 2′-hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. J Comput Chem. 2011;32:1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zgarbová M, Otyepka M, Šponer J, Mládek A, Banáš P, Cheatham TE, Jurečka P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J Chem Theory Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mustoe AM, Brooks CL, 3rd, Al-Hashimi HM. Topological constraints are major determinants of tRNA tertiary structure and dynamics and provide basis for tertiary folding cooperativity. Nucleic Acids Res. 2014;42:11792–11804. doi: 10.1093/nar/gku807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cragnolini T, Laurin Y, Derreumaux P, Pasquali S. Coarse-Grained HiRE-RNA Model for ab Initio RNA Folding beyond Simple Molecules, Including Noncanonical and Multiple Base Pairings. J Chem Theory Comput. 2015;11:3510–3522. doi: 10.1021/acs.jctc.5b00200. [DOI] [PubMed] [Google Scholar]
- 144.Pasquali S, Derreumaux P. HiRE-RNA: a high resolution coarse-grained energy model for RNA. J Phys Chem B. 2010;114:11957–11966. doi: 10.1021/jp102497y. [DOI] [PubMed] [Google Scholar]
- 145.Bohmdorfer G, Wierzbicki AT. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roberts TC, Morris KV, Weinberg MS. Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics. 2014;9:13–20. doi: 10.4161/epi.26700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Visualized Exp. 2012;61:e3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res. 2012;22:1372–1381. doi: 10.1101/gr.130237.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Buske FA, Bauer DC, Mattick JS, Bailey TL. Triplex-Inspector: an analysis tool for triplex-mediated targeting of genomic loci. Bioinformatics. 2013;29:1895–1897. doi: 10.1093/bioinformatics/btt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenet Chromatin. 2014;7:25. doi: 10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 154.Lajoie BR, Dekker J, Kaplan N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods. 2015;72:65–75. doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 156.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 157.Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107:30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, Cui J, Wang G, Qian G, Higgins MJ, Fan X, Hoffman AR, Hu JF. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol. 2014;204:61–75. doi: 10.1083/jcb.201304152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Burg JM, Makhoul AT, Pemble CWt, Link JE, Heller FJ, McCafferty DG. A rationally-designed chimeric KDM1A/KDM1B histone demethylase tower domain deletion mutant retaining enzymatic activity. FEBS Lett. 2015;589:2340–2346. doi: 10.1016/j.febslet.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nguyen UT, Bittova L, Muller MM, Fierz B, David Y, Houck-Loomis B, Feng V, Dann GP, Muir TW. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T. N-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding. J Mol Biol. 2015 Sep 4; doi: 10.1016/j.jmb.2015.08.021. No. 0022-2836(15)00486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Disney MD, Yildirim I, Childs-Disney JL. Methods to enable the design of bioactive small molecules targeting RNA. Org Biomol Chem. 2014;12:1029–1039. doi: 10.1039/c3ob42023j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]