Abstract
Cnidarians represent one of the few groups of venomous animals that lack a centralized venom transmission system. Instead, they are equipped with stinging capsules collectively known as nematocysts. Nematocysts vary in abundance and type across different tissues; however, the venom composition in most species remains unknown. Depending on the tissue type, the venom composition in sea anemones may be vital for predation, defense, or digestion. Using a tissue-specific RNA-seq approach, we characterize the venom assemblage in the tentacles, mesenterial filaments, and column for three species of sea anemone (Anemonia sulcata, Heteractis crispa, and Megalactis griffithsi). These taxa vary with regard to inferred venom potency, symbiont abundance, and nematocyst diversity. We show that there is significant variation in abundance of toxin-like genes across tissues and species. Although the cumulative toxin abundance for the column was consistently the lowest, contributions to the overall toxin assemblage varied considerably among tissues for different toxin types. Our gene ontology (GO) analyses also show sharp contrasts between conserved GO groups emerging from whole transcriptome analysis and tissue-specific expression among GO groups in our differential expression analysis. This study provides a framework for future characterization of tissue-specific venom and other functionally important genes in this lineage of simple bodied animals.
Keywords: gene ontology, tentacles, mesenterial filaments, column, toxins
Introduction
The majority of venomous taxa is equipped with a centralized venom gland or duct, from which they are able to transmit toxic peptides and other biomolecules into a target organism (Smith and Wheeler 2006; Fry et al. 2009; Castelin et al. 2012; Casewell et al. 2013). Cnidarians represent one of the few groups of venomous animals that lack a centralized venom transmission system: Their venom is transmitted at the cellular level using specialized stinging capsules called nematocysts. The nematocyst composition varies across tissues and is often attributed to tissue-specific functions (Mariscal 1974; Kass-Simon and Scappaticci 2002); however, the venom composition among the different tissue types in most species remains unknown. Within Cnidaria, sea anemones (Actiniaria) constitute the best studied group with regard to diversity of venom and cnidom (Mariscal 1974; Fautin 2009; Frazao et al. 2012; Reft and Daly 2012; Jouiaei et al. 2015), although this knowledge is limited taxonomically compared with other venomous lineages.
As toxin components can be differentially expressed across tissue types, the venom cocktail can be versatile throughout the polyp. Acrorhagi and acontia are both specialized tissues found exclusively in sea anemones, with venom components presumably used for intraspecific competition (Honma et al. 2005; Macrander, Brugler et al. 2015) and defense (Talvinen and Nevalainen 2002; Nevalainen et al. 2004), respectively. Tentacles have been the primary tissue from which toxin peptides have been isolated (Oliveira et al. 2012; Frazao et al. 2012) and are likely multifunctional to immobilize prey (i.e., neurotoxins) and/or repel potential predators by inducing pain (i.e., acid sensing ion channel targeting toxins). Mesenterial filaments of sea anemones also contain components of venom which aid in digestion, but may also be projected outside the anemone’s body for external digestion, competition, and defense (Fautin and Mariscal 1991; Basulto et al. 2006), similarly to the mesenterial filaments of corals, which are used in external digestion/competition (Lang 1973). Although there are no published studies which characterize toxins from the column in sea anemones, based on the function of this region, the venom assemblage here may be used exclusively for defense. Toxin assemblage may covary with nematocyst type and abundance; however, toxins have also been shown to be released from glandular cells (Moran et al. 2011) adding another layer of complexity to tissue-specific venom in sea anemones.
Next-generation sequencing has been a useful tool to investigate the diversity of components in sea anemone venom (Johansen et al. 2010; Rodriguez et al. 2012; Urbaravo et al. 2012; Macrander, Brugler et al. 2015). Of the better studied sea anemone toxin genes, voltage-gated sodium channel toxins (NaTxs) and type I and III voltage-gated potassium channel toxins (KTxs) appear to be specific to Actiniaria, with pore forming cytolysins found throughout Cnidaria (Frazao et al. 2012). Many of the poorly studied toxin candidates are members of large gene families and share a strong resemblance to their nonvenomous counterparts (Fry et al. 2009; Moran et al. 2013; Rachamim et al. 2014) or have been characterized in only one or two species (Frazao et al. 2012). Previous tissue-specific investigations focusing on venom assays of crude extracts or isolated proteins have found that nematocyst type and abundance do not always correlate with venom potency (Hessinger 1988; Nevalainen et al. 2004), that tissues thought to be devoid of nematocysts also have high concentrations of venom (Mathias et al. 1960), and that whole tissue extracts can contain more potent bioactive compounds when compared with isolated peptides (Hessinger and Lenhoff 1976). Tissue-specific transcriptome analyses provide an alternative path to elucidate the functional role of candidate toxin-genes or other synergistic proteins in a comparative context.
In this study, we use a combined RNA-seq and bioinformatic approach to characterize the venom composition across different tissues (tentacles, mesenterial filaments, and column) in three species of sea anemone: Anemonia sulcata (Actiniidae), Heteractis crispa (Stichodactylidae), and Megalactis griffithsi (Actinodendronidae) (fig. 1). These species vary considerably in their inferred venom potency, symbiotic associations, and morphology. Envenomation from M. griffithsi is extremely painful to humans (Auerbach 1997; JM and MD, personal observation). Although there appears to be no envenomation symptoms associated with A. sulcata and H. crispa following contact with the epidermis (personal observation). And no reported symptomology, despite A. sulcata being a focal taxa in venom research and the popularity of H. crispa in the aquarium trade. Although all of the focal taxa harbor symbionts, H. crispa is host to the widest reported variety of symbionts among our focal species (Dunn 1981; Fautin and Allen 1997; Marin et al 2004) and is an obligatory host for juvenile clownfishes (Huebner et al. 2012). Tissue-specific nematocyst assemblage and distribution also vary among these species, with A. sulcata and H. crispa having three types of nematocysts (spirocysts, basitrichs, and microbasic p-mastigophores: Schmidt 1972; Dunn 1981) and M. griffithisi having only two (spirocysts and basitrichs: Ardelean and Fautin 2004). Finally, A. sulcata is closely related to Anemonia viridis, which has one of the better characterized “venomes” among sea anemones (Kozlov and Grishin 2011; Frazao et al. 2012; Oliveira et al 2012; Nicosia et al. 2013); almost nothing is known about the venom of H. crispa or M. griffithsi. We predicted that we would find venom assemblages and other synergistic or functionally important genes conserved across tissue types, and that we would identify toxins and other differentially expressed genes that correlate with unique tissue-specific functions of the focal taxa or with their ecology, venom potency, nematocyst distribution, or evolutionary histories.
Fig. 1.—
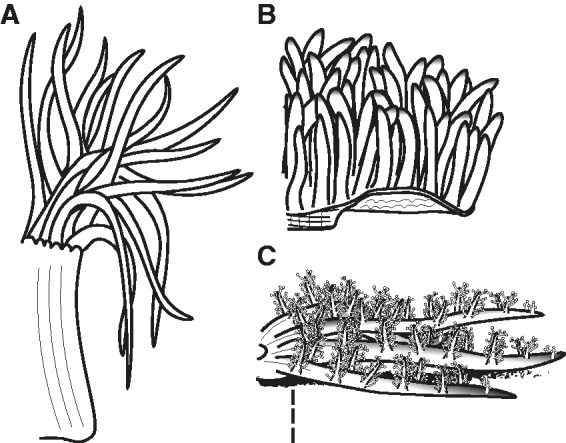
Tissues and focal taxa. Comparison of the general morphology of the three focal species (A) A. sulcata, (B) H. crispa, and (C) M. griffithsi. Tentacle morphology varies considerably across these species with long fusiform tentacles (A. sulcata), short filiform tentacles (H. crispa), and broad and highly branching tentacles with acrospheres (M. griffithsi). Unlike the many species of sea anemone that have their column exposed, our focal taxa have their column protected to some degree by either tentacles (A. sulcata), a broad oral disk (H. crispa), or buried in the sediment (M. griffithsi).
Materials and Methods
Specimens
For this study, a single specimen of A. sulcata was collected from Strangford Lough, Co. Down, Northern Ireland. A specimen of H. crispa, reportedly collected from Indonesia, was purchased from an online retailer (www.liveaquaria.com, last accessed 15 October 2013). A specimen of M. griffithsi, reportedly collected in the Indo-Pacific, was purchased from a commercial retailer (Seaside Tropical Fish; Huntington Beach, CA). The specimen of A. sulcata was placed in RNALater (QIAGEN) immediately after being collected. The other two specimens (H. crispa and M. griffithsi) were kept alive in the lab in aquaria filled with artificial sea water. Over a period of 2 weeks, specimens were placed in smaller acclimation tanks for 15 min, with the different tissues removed using tweezers and a scalpel, starting with tentacle, column, and mesenteric filaments last. Once the tissues were removed and flash frozen in liquid nitrogen, the specimens were returned to the original aquarium.
Library Preparation, Sequencing, Cleanup, and Assembly
Total RNA was extracted following the RNeasy Mini Kit (QIAGEN) protocol. Several small (1.5–2 mm) ceramic beads (BioExpress) were added to each sample at the same step as Buffer RLT and the tissue was then macerated using a Mini-Beadbeater-8 (BioSpec Products) for 30 s at Homogenize. Following 30 s of homogenization, the tube was examined to determine whether the tissue was homogenized and then repeated as necessary. For each tissue, the RNA extractions were quantified on the Qubit 2.0 (Life Technologies) and RNA integrity number (RIN) values determined on the RNA BioAnalyzer RNA chip (Agilent Technologies). The Nucleic Acid Shared Resource-Illumina Core (The Ohio State University, Columbus, OH) carried out the first strand synthesis, library construction, and paired-end 100 base sequencing. Illumina sequencing libraries were constructed from total RNA using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina). The final libraries were quantified using the Qubit 2.0 (Life Technologies), and qualified on High Sensitivity DNA BioAnalyzer chip (Agilent Technologies). Libraries were pooled and sequenced on a single Illumina HiSeq 2500 Paired-End 100-bp High Output flow cell lane. Raw sequence data were demultiplexed by barcode and used in downstream analyses.
As initial error-correction steps can have a significant impact on the final transcriptome assembly (MacManes and Eisen 2013), alternative assemblies were explored by cleaning raw reads using both the ErrorCorrectReads.pl (ECR) script, which is built into the ALLPATHS-LG pipeline (Gnerre et al. 2011), and/or the program Trimmomatic (Bolger et al. 2014). Our initial survey used both the ECR script and Trimmomatic. The ECR script examines stacked raw reads along 24-mer associated segments and corrects sequences that occur at a number below a necessary threshold. Previously this has been accessible only through the ALPATHS-LG pipeline, but has recently been made available for genome and transcriptome assemblies (Gnerre et al. 2011). The program Trimmomatic is more widely used, and provides an alternative (and in this case additional) error correction step by removing adapters, leading and trailing low quality bases (< 3), reads below 36 bases in length, and the trailing portions of reads with an average quality score of 15 in a four-base sliding window. Our initial clean-up approach using both methods was then compared with a second approach which used only Trimmomatic (with the same parameters). As the ECR script is reportedly much more conservative than other error correcting programs (MacManes and Eisen 2013), the comparison allowed us to determine whether the additional step was necessary for all of our transcriptomes.
For both approaches, transcriptomes were assembled with default parameters in the de novo assembly program Trinity (Grabherr et al. 2011). This was done for each tissue-specific transcriptome (tentacles, mesenterial filaments, and column) in addition to a “combined” transcriptome for each species. Transcriptome completeness was determined using CEGMA (Core Eukaryotic Genes Mapping Approach: Parra et al. 2007) to assay the presence of 248 conserved eukaryotic proteins on iPlant (Goff et al. 2011). We compared the ECR- and non-ECR-prepared transcriptomes after assembly to evaluate the impact of the cleanup step on overall completeness and also conducted a reciprocal BLAST (Basic Local Alignment Search Tool) search of known sea anemone venom genes to determine whether the alternative cleanup strategies would result in a different number of candidate toxin genes. All assemblies and subsequent analyses were done on the OBCP (Ohio Biodiversity Conservation Partnership) cluster (4 × Intel Xeon E5-4650L-8 cores 16 threads, 256 GB RAM) unless otherwise specified.
Candidate Toxin Gene Identification
Using toxin genes from sea anemones and other venomous lineages in the ToxProt data set (http://www.uniprot.org/program/Toxins, last accessed 15 June 2015) as query sequences, we conducted a tBLASTn search (E-value < 10.0 and matching length > 60%) in order to identify toxin gene candidates from the combined transcriptomes for each focal taxa. Sea anemone toxin candidates were visually inspected for premature stop codons and the placement of key cysteine amino acid residues characteristic of the different venom types (Kozlov and Grishin 2012). Using a custom Python script all of the sea anemone toxin-candidate transcripts were trimmed to include only the open-reading frame (ORF) identified by Transdecoder (http://transdecoder.github.io). A MAFFT translation alignment (Katoh 2013) was done in Geneious for each of the focal sequence sets of with the L-INS-I algorithm, BLOSUM62 scoring matrix, a gap open penalty of 1.53, and offset value of 0.123. Toxin gene candidates were visually inspected to determine whether structurally and/or potentially functionally import amino acid residues were aligned. For the better studied sea anemone toxins (cytolysins, NaTx, and types I III KTx), we mapped the raw reads back onto the transcripts to identify any misassembled toxin transcripts that may result in unidentified toxin-like genes (Macrander, Broe et al. 2015).
Tissue-Specific Expression of Toxin Genes
Relative expression levels for each of the tissue-specific transcriptomes were calculated using the program RSEM (Li and Dewey 2011) within the Trinity programming suite (Grabherr et al. 2011) by mapping raw reads from each of the tissue-specific libraries back to the combined transcriptome. RSEM provides, among other calculations, measurements of expression as raw counts, transcripts per million (TPM), and fragments per kilobase of transcript per million (FPKM). TPM can be used to compare gene expression levels between libraries as it adjusts for differences in the library sizes and skewed expression of transcripts (Wagner et al. 2012). Although this value is specific to the focal transcriptome and does not lend itself to comparisons without a relative context, this approach avoids biases encountered when using raw read counts or FPKM (Robinson and Oshlack 2010; Li and Dewey 2011; Nakasugi et al. 2013). We also checked for biases in expression level by comparing TPM values for unmodified transcripts with those trimmed down to the toxin ORF. This allowed us to determine whether regions flanking transcripts were overinflating TPM values by incorporating incorrect raw reads on the focal transcript (Yang and Smith 2013). Using the TPM values, we evaluated overall venom composition among the different tissue types to determine whether any gene families were expressed proportionately higher than across all tissue types and species.
Differential Gene Expression and GO Analysis
Differential expression (DE) calculations for tissue-specific transcriptomes were determined using RSEM outputs in combination with the EdgeR bioconductor package (Robinson et al. 2010) in the Trinity programming suite. This approach uses TMM-normalized FPKM values to determine absolute, rather than relative measures of abundance for transcripts from each tissue-specific library (Robinson and Oshlack 2010). This allowed us to identify transcripts with significant deviations based on n-fold changes from expected genes within clusters of transcripts. To make inferences about the implications of the DE analysis, the transcripts were subjected to a BLAST search against the UniProt database. Using custom Python scripts and GO information for the top BLAST hit for each transcript, we averaged DE values derived from the counts matrix across all GO groups. The upregulated GO groups and the DE values were used in the program REVIGO (Supek et al. 2011) to visualize DE among these functional groups across tissues.
For our transcriptome-level GO analyses, we used two approaches to compare functional groups between tissue types for each species. First, we used the more common BLAST2GO approach (Conesa et al. 2005), focusing on the more highly expressed transcripts for each tissue. An initial screening of each transcriptome reduced the query sequences to those transcripts with a TPM value in the top 10% for each tissue-specific transcriptome. These transcripts were then used to perform a BLASTX search against the UniProtKB database (downloaded February 15, 2015). The top BLAST hits (E-value cutoff of 0.0001) for each transcript were then evaluated further in the BLAST2GO program where the GO groupings were mapped. As an alternative GO analysis, we used the Trinotate pipeline (http://trinotate.github.io/, last accessed May 12, 2015) in combination with custom Python scripts to account for different GO levels and an overabundance of GO groupings based on information within the UniProtKB BLAST results. Using the same BLASTX parameters (E-value 0.0001) against the UniProt database, we queried each tissue-specific transcriptome. We divided TPM values equally across all associated GO terms identified in each sequence. The GO groups were then arranged based on semantic similarities using the program REVIGO (Supek et al. 2011). The major difference between these two approaches is the hierarchical level of GO being reported (Level 2: BLAST2GO; any level: Trinotate and script) and how the expression values (TPM) are being used (Top 10% TPM only: BLAST2GO; scaling based on number of GO groups within each domain: Trinotate and script).
Results
Sequencing, Cleanup, and Assembly
Our tissue-specific transcriptomes ranged from 33,993,938 to 73,285,750 paired-end reads depending on tissue and taxon (table 1). Although the less conservative approach using only Trimmomatic kept approximately 96–99% of the raw reads for each library, the more conservative ECR script in combination with Trimmomatic reduced the number of raw reads incorporated into the tissue-specific assemblies substantially, retaining 86–91% of the reads for A. sulcata, 27–86% for H. crispa, and 59–84% for M. griffithsi. The H. crispa column transcriptome could not be assembled without an initial pass through the ECR script (table 1). Reasons for the difficulty of assembling the H. crispa column tissue-specific transcriptome remain unclear: Closer inspection of the Trimmomatic-cleaned data revealed no outliers or errors when compared with any other tissue in the FastQC analysis (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed 15 December 2013). Because there was no indication that the column transcriptomes needed to be excluded from further analyses due to quality concerns, we included them in our transcriptome assemblies and downstream analyses.
Table 1.
Summary of Sequencing, Cleanup, and Assembly of Tissue-Specific and Combined Transcriptomes
| Species | Sequencinga |
Trimmomatic |
Trinity |
CEGMA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | Yield (Mbases) | # PE Reads (2 × 100 bp) | F and R Recovered | F Only | R Only | Dropped | Isoforms | Genes | N50 | Complete (%) |
| Anemonia sulcata | ||||||||||
| Tentacle | 8,793 | 43,966,062 | 42,158,521 | 1,440,580 | 242,354 | 124,607 | 116,940 | 90,709 | 1,167 | 75 |
| EC | 40,114,944 | 32,725,211 | 3,534,322 | 2,282,523 | 1,572,888 | 54,365 | 39,503 | 1,262 | 77.02 | |
| Column | 10,480 | 52,402,099 | 49,231,820 | 2,561,688 | 385,801 | 222,790 | 134,535 | 105,984 | 1,052 | 90.73 |
| EC | 47,615,852 | 36,988,265 | 4,813,831 | 3,185,361 | 2,628,395 | 55,868 | 41,679 | 1,242 | 81.42 | |
| Filament | 14,657 | 73,285,750 | 69,599,682 | 3,027,316 | 411,698 | 247,054 | 167,752 | 140,339 | 789 | 88.31 |
| EC | 63,044,097 | 51,425,852 | 5,771,906 | 3,334,536 | 2,511,803 | 45,421 | 35,879 | 953 | 59.27 | |
| Combined | 33,930 | 169,653,911 | 160,990,023 | 7,029,584 | 1,039,853 | 594,451 | 303,773 | 245,737 | 1,186 | 94.76 |
| Heteractis crispa | ||||||||||
| Tentacle | 8,054 | 40,269,872 | 38,950,621 | 1,071,964 | 170,250 | 77,037 | 186,284 | 142,194 | 1,227 | 93.55 |
| EC | 34,831,282 | 28,422,326 | 2,916,966 | 2,165,659 | 1,326,331 | 112,900 | 85,627 | 1,162 | 85.05 | |
| Column | 11,629 | 58,142,549 | 55,456,896 | 2,220,420 | 318,314 | 146,919 | — | — | — | — |
| EC | 15,862,609 | 12,892,450 | 1,480,601 | 919,042 | 570,516 | 103,035 | 97,215 | 289 | 15.73 | |
| Filament | 6,975 | 34,877,218 | 33,252,993 | 1,315,400 | 207,390 | 101,435 | 307,498 | 275,492 | 421 | 55.25 |
| EC | 23,826,103 | 19,561,405 | 1,915,171 | 1,502,125 | 847,402 | 67,733 | 56,039 | 711 | 43.95 | |
| Combined | 26,658 | 133,289,639 | 127,660,510 | 4,607,784 | 695,954 | 325,391 | 655,116 | 581,957 | 545 | 94.34 |
| Megalactis griffithsi | ||||||||||
| T. Lobe | 7,201 | 33,993,938 | 31,743,659 | 2,008,599 | 147,255 | 94,425 | 208,940 | 146,020 | 1,119 | 88.71 |
| EC | 27,928,980 | 22,883,168 | 2,916,204 | 1,288,046 | 841,562 | 141,408 | 103,182 | 1,123 | 83.87 | |
| Column | 8,303 | 36,006,836 | 33,419,868 | 2,330,576 | 162,784 | 93,608 | 572,734 | 547,297 | 268 | 19.76 |
| EC | 21,270,705 | 17,482,032 | 2,312,185 | 849,156 | 627,332 | 85,311 | 93,406 | 590 | 17.75 | |
| Filament | 6,799 | 41,516,836 | 39,947,259 | 1,336,863 | 151,386 | 81,328 | 197,441 | 157,660 | 1,144 | 88.71 |
| EC | 35,093,477 | 29,602,445 | 2,759,483 | 1,633,100 | 1,098,449 | 93,566 | 76,840 | 1,157 | 87.50 | |
| Combined | 22,303 | 111,517,610 | 105,110,786 | 5,676,038 | 461,425 | 269,361 | 1,200,511 | 1,073,369 | 392 | 97.18 |
Note.—F, Forward; R, Reverse.
Values represent the number of paired end reads (2 × 100 bp) at each step, EC indicates that the raw reads were subjected to the errorcorrect.pl script from the ALLPATHS-LG pipeline (Gnerre et al. 2011).
The transcriptomes varied in number of trinity components and N50 values (table 1). Completeness scores determined in CEGMA were consistently high for the combined transcriptomes (> 94.34%), with the values for the tissue-specific transcriptomes varying among and within taxa (table 1). Of the tissue-specific transcriptome assemblies, the columns of H. crispa and M. griffithsi had the lowest CEGMA completeness scores (17.75% and 15.73%, respectively), which is especially notable compared with that of the column of A. sulcata (81.42%). The transcriptomes that had been cleaned up with the ECR script had a longer N50 and fewer isoforms and contigs (parameters that may indicate a better assembly); however, their CEGMA scores were consistently lower (table 1) than those of transcriptomes cleaned using only Trimmomatic. We therefore opted to use the less conservative Trimmomatic-only approach in our assemblies and analyses to avoid biases in our GO analyses and abundance estimates (Vijay et al. 2013; MacManes and Eisen 2013).
Candidate Toxin Gene Diversity and Tissue-Specific Assemblage
We identified 7 type I KTxs, 90 type III KTxs, 25 NaTxs, and 10 cytolysins (table 2) in the transcriptomes of our focal taxa. After mapping raw reads onto candidate toxin genes, the number of hidden candidate toxin genes for the type I KTxs and cytolysins were low, whereas there were considerably more toxin candidates for the type III KTx and NaTx (table 5). Due to the high abundance of type I KTx domains in nontoxin peptides, we considered a gene to be a candidate type I KTx toxin when it had a positive BLAST hit against previously characterized type I KTxs (Yamaguchi et al. 2010; Orts et al. 2013). The candidate type I KTxs exhibited a greater rate of amino acid substitution when compared with other type I KTxs previously characterized. In addition, our study identified a transcript from M. griffithsi containing a tandem duplication of the type I KTx domain (supplementary fig. S1, Supplementary Material online). The number of type III KTxs we recover accords with inferences from previous expressed sequence tag and bioinformatic approaches that hypothesize that this is a large gene family (Kozlov and Grishin 2011). Although cytolysins are reported to have multiple gene copies (Wang et al. 2008), we found many fewer cytolysins when compared with previous studies (table 2).
Table 2.
Sea Anemone Toxin Tissue-Specific Diversity and Highest TPM Values
|
Anemonia sulcata |
Heteractis crispa |
Megalactis griffithsi |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | C | F | T | N | C | F | T | N | C | F | T | |
| ASIC toxins | 0 | — | — | — | 2 | 7 | 70 | 271 | 0 | — | — | — |
| Acrorhagins | 4 | 26 | 37 | 21 | 1 | 0.31 | 5 | 15 | 2 | 0 | 13.79 | 7.44 |
| AETX-like | 2 | 50 | 21 | 48 | 0 | — | — | — | 0 | — | — | — |
| Metalloproteases | 90+ | 271 | 265 | 157 | 40+ | 3 | 43 | 230 | 30+ | 2 | 87 | 15 |
| Class 9a (KTx) | 1 | 344 | 637 | 1726 | 0 | — | — | — | 0 | — | — | — |
| Type II Cytolysins | 3 | 2 | 0.19 | 6 | 6 | 3 | 19 | 986 | 1 | 0 | 3 | 0.03 |
| MACPFa | 6 | 3 | 0.2 | 2 | 7 | 0.38 | 4 | 4 | 0 | — | — | — |
| NaTx | 23 | 931 | 109 | 130 | 1 | 22 | 0 | 0 | 2 | 6 | 2803 | 32 |
| EGF-like | 5 | 7 | 124 | 34 | 2 | 1 | 9 | 26 | 0 | — | — | — |
| Type I KTxs | 2 | 36 | 103 | 89 | 5 | 0.13 | 12 | 1031 | 3 | 0 | 10 | 0 |
| PI/Type II KTxs | 39 | 333 | 471 | 426 | 37 | 6 | 402 | 1789 | 54 | 3 | 185 | 579 |
| Type III KTxs | 34 | 95 | 814 | 1663 | 14 | 29 | 55 | 499 | 42 | 0.17 | 329 | 751 |
| Type V KTxs | 2 | 64 | 9 | 47 | 3 | 0 | 3 | 4 | 0 | — | — | — |
| PLA2 | 17 | 33 | 108 | 74 | 19 | 0.28 | 53 | 142 | 9 | 0.09 | 162 | 70 |
Note.—N, number of candidate toxin genes; maximum TPM value for a single transcript in each of the tissue-specific transcriptomes: C, column; F, filament; T, tentacle/tentacular lobe. The highest TPM values are highlighted in bold. The better studied toxin genes are underlined. —indicates no candidate toxin identified. For a full list of candidate toxins, visit the associated bitbucket site (https://bitbucket.org/JasonMacrander/anemone_tissue/).
Only partial sequence recovered from transcripts.
Table 3.
Most Highly Expressed Transcripts with All the ToxProt Data BLAST Hits
| Transcript_id | TPM | ToxPROT | Group | Gene Name | E-Value |
|---|---|---|---|---|---|
| Anemonia suclata | |||||
| Column | |||||
| comp207685_c1_seq2 | 931 | P01533 | Anemone | Neurotoxin-1 | 6.00E-26 |
| comp196092_c0_seq1 | 777 | Q8AY75 | Snake | Calglandulin | 2.00E-34 |
| comp203492_c1_seq3 | 484 | Q9TXD8 | Spider | Venom peptide isomerase heavy chain | 6.00E-32 |
| comp203768_c0_seq4 | 357 | A6MFK8 | Snake | Venom prothrombin activator vestarin-D2 | 1.00E-42 |
| comp192322_c0_seq1 | 344 | P86467 | Anemone | Toxin Bcg III 23.41 | 1.00E-07 |
| comp203839_c0_seq1 | 333 | C0HJF3 | Anemone | Analgesic polypeptide HC3 | 8.00E-14 |
| comp213984_c0_seq4 | 332 | P0DKM7 | Snail | Turripeptide Lol9.1 | 2.00E-11 |
| comp177042_c0_seq1 | 310 | Q9TXD8 | Spider | Venom peptide isomerase heavy chain | 3.00E-29 |
| comp146633_c0_seq1 | 281 | M5B4R7 | Spider | Translationally controlled tumor protein | 4.00E-12 |
| comp205430_c0_seq1 | 271 | K7Z9Q9 | Anemone | Nematocyte expressed protein 6 | 6.00E-40 |
| Filament | |||||
| comp213984_c0_seq4 | 1433 | P0DKM7 | Snail | Turripeptide Lol9.1 | 2.00E-11 |
| comp197800_c0_seq1 | 814 | P11494 | Anemone | Antihypertensive protein BDS-1 | 2.00E-08 |
| comp177042_c0_seq1 | 751 | Q9TXD8 | Spider | Venom peptide isomerase heavy chain | 3.00E-29 |
| comp192322_c0_seq1 | 673 | P86467 | Anemone | Toxin Bcg III 23.41 | 1.00E-07 |
| comp196092_c0_seq1 | 656 | Q8AY75 | Snake | Calglandulin | 2.00E-34 |
| comp203768_c0_seq4 | 532 | A6MFK8 | Snake | Venom prothrombin activator vestarin-D2 | 1.00E-42 |
| comp208862_c2_seq12 | 471 | P10280 | Anemone | Kunitz-type proteinase inhibitor 5 II | 1.00E-32 |
| comp203492_c1_seq1 | 432 | Q9TXD8 | Spider | Venom peptide isomerase heavy chain | 2.00E-31 |
| comp146633_c0_seq1 | 423 | M5B4R7 | Spider | Translationally controlled tumor protein | 4.00E-12 |
| comp197800_c0_seq2 | 401 | P11494 | Anemone | Antihypertensive protein BDS-1 | 2.00E-08 |
| Tentacle | |||||
| comp192322_c0_seq1 | 1726 | P86467 | Anemone | Toxin Bcg III 23.41 | 1.00E-07 |
| comp205526_c0_seq2 | 1663 | P11494 | Anemone | Antihypertensive protein BDS-1 | 2.00E-23 |
| comp196092_c0_seq1 | 1250 | Q8AY75 | Snake | Calglandulin | 2.00E-34 |
| comp213984_c0_seq4 | 550 | P0DKM7 | Snail | Turripeptide Lol9.1 | 2.00E-11 |
| comp204748_c0_seq1 | 484 | B2DCR8 | Cephalopod | SE-cephalotoxin | 4.00E-04 |
| comp197800_c0_seq1 | 433 | P11494 | Anemone | Antihypertensive protein BDS-1 | 2.00E-08 |
| comp208862_c2_seq12 | 425 | P10280 | Anemone | Kunitz-type proteinase inhibitor 5 II | 1.00E-32 |
| comp208862_c2_seq5 | 346 | P10280 | Anemone | Kunitz-type proteinase inhibitor 5 II | 1.00E-32 |
| comp177042_c0_seq1 | 346 | Q9TXD8 | Spider | Venom peptide isomerase heavy chain | 3.00E-29 |
| comp203768_c0_seq4 | 323 | A6MFK8 | Snake | Venom prothrombin activator vestarin-D2 | 1.00E-42 |
| Heteractis crispa | |||||
| Column | |||||
| comp241739_c0_seq1 | 2563 | P0CI21 | Snail | Augerpeptide hhe53 | 8.00E-05 |
| comp312716_c1_seq1 | 40 | J3SFJ3 | Snake | Translationally controlled tumor protein | 4.00E-13 |
| comp245722_c0_seq1 | 30 | P69930 | Anemone | Peptide toxin Am-2 | 8.00E-52 |
| comp241777_c0_seq1 | 22 | P69928 | Anemone | Peptide toxin Am-3 | 7.00E-45 |
| comp299535_c0_seq1 | 14 | B2DCR8 | Cephalopod | SE-cephalotoxin | 3.00E-07 |
| comp186523_c0_seq1 | 13 | Q2XXL4 | Lizard | Vespryn | 6.00E-05 |
| comp275702_c0_seq1 | 12 | B2DCR8 | Cephalopod | SE-cephalotoxin | 3.00E-06 |
| comp301568_c0_seq1 | 11 | P81236 | Snake | Basic phospholipase A2 acanthin-1 | 2.00E-06 |
| comp312689_c0_seq1 | 11 | B6EWW8 | Snake | Snake venom 5'-nucleotidase | 2.00E-25 |
| comp312679_c0_seq1 | 11 | B6EWW8 | Snake | Snake venom 5'-nucleotidase | 9.00E-10 |
| Filament | |||||
| comp312716_c1_seq1 | 5695 | J3SFJ3 | Snake | Translationally controlled tumor protein | 4.00E-13 |
| comp286957_c0_seq1 | 1413 | Q8AY75 | Snake | Calglandulin | 8.00E-09 |
| comp297877_c0_seq1 | 547 | Q8AY75 | Snake | Calglandulin | 5.00E-12 |
| comp241739_c0_seq1 | 460 | P0CI21 | Snail | Augerpeptide hhe53 | 8.00E-05 |
| comp301796_c0_seq1 | 402 | P24541 | Snake | Kunitz-type serine protease inhibitor | 5.00E-13 |
| comp174862_c0_seq1 | 346 | P0DKM8 | Snail | Turripeptide Ici9.1 | 3.00E-10 |
| comp297655_c0_seq1 | 299 | Q8AY75 | Snake | Calglandulin | 4.00E-11 |
| comp175426_c0_seq1 | 219 | A9YME1 | Wasp | Venom allergen 5 | 2.00E-16 |
| comp298440_c1_seq3 | 110 | A6MFK8 | Snake | Venom prothrombin activator vestarin-D2 | 1.00E-42 |
| comp302180_c0_seq1 | 101 | Q8AY75 | Snake | Calglandulin | 1.00E-31 |
| Tentacle | |||||
| comp312716_c1_seq1 | 7514 | J3SFJ3 | Snake | Translationally controlled tumor protein | 4.00E-13 |
| comp301796_c0_seq1 | 1786 | P24541 | Snake | Kunitz-type serine protease inhibitor | 5.00E-13 |
| comp276054_c0_seq1 | 986 | Q86FQ0 | Anemone | Cytolysin Src-1 | 7.0E-127 |
| comp308440_c1_seq6 | 416 | Q8AY75 | Snake | Calglandulin | 4.00E-33 |
| comp312783_c0_seq1 | 287 | Q7M4I3 | Bee | Venom protease | 5.00E-42 |
| comp310460_c1_seq12 | 271 | C0HJB1 | Anemone | Toxin pi-PMTX-Pcr1a | 2.00E-10 |
| comp293159_c1_seq1 | 230 | K7Z9Q9 | Anemone | Nematocyte expressed protein 6 | 4.00E-41 |
| comp295958_c0_seq2 | 212 | P0CV91 | Snake | Peroxiredoxin-4 | 2.00E-14 |
| comp286957_c0_seq1 | 209 | Q8AY75 | Snake | Calglandulin | 8.00E-09 |
| comp298467_c1_seq1 | 193 | Q58L93 | Snake | Venom prothrombin activator porpharin-D | 1.00E-30 |
| Megalactis griffithsi | |||||
| Column | |||||
| comp1412551_c0_seq1 | 253 | P0CI21 | Snail | Augerpeptide hhe53 | 1.00E-04 |
| comp1220254_c0_seq1 | 19 | M5B4R7 | Spider | Translationally controlled tumor protein | 3.00E-12 |
| comp1418434_c0_seq1 | 16 | B2DCR8 | Cephalopod | SE-cephalotoxin | 1.00E-04 |
| comp1436556_c1_seq1 | 15 | W4VS99 | Spider | Neprilysin-1 | 2.00E-59 |
| comp1436580_c0_seq2 | 15 | P81383 | Snake | L-amino-acid oxidase | 3.00E-05 |
| comp1436525_c0_seq1 | 14 | B6EWW8 | Snake | Snake venom 5'-nucleotidase | 1.00E-09 |
| comp1436582_c0_seq1 | 14 | A8QL52 | Snake | l-amino-acid oxidase | 2.00E-16 |
| comp1436570_c0_seq2 | 8 | B6EWW8 | Snake | Snake venom 5'-nucleotidase | 2.00E-25 |
| comp1436570_c0_seq3 | 8 | B6EWW8 | Snake | Snake venom 5'-nucleotidase | 2.00E-25 |
| comp1220254_c0_seq1 | 8 | A0A024BTN9 | Snake | l-amino-acid oxidase | 8.00E-04 |
| Filament | |||||
| comp1220254_c0_seq1 | 10171 | M5B4R7 | Spider | Translationally controlled tumor protein | 3.00E-12 |
| comp1418434_c0_seq1 | 3712 | B2DCR8 | Cephalopod | SE-cephalotoxin | 1.00E-04 |
| comp1220094_c0_seq1 | 2803 | Q9NJQ2 | Anemone | Neurotoxin Ae I | 7.00E-21 |
| comp1220065_c0_seq1 | 1284 | Q8AY75 | Snake | Calglandulin | 1.00E-34 |
| comp1426766_c0_seq1 | 978 | B2DCR8 | Cephalopod | SE-cephalotoxin | 2.00E-05 |
| comp1430871_c1_seq3 | 395 | J3S9D9 | Snake | Reticulocalbin-2 | 9.00E-35 |
| comp1433029_c0_seq3 | 329 | P69930 | Anemone | Peptide toxin Am-2 | 1.00E-07 |
| comp1433029_c0_seq11 | 284 | P69930 | Anemone | Peptide toxin Am-2 | 7.00E-04 |
| comp1398541_c0_seq2 | 185 | Q8T3S7 | Spider | Kunitz-type serine protease inhibitor | 3.00E-18 |
| comp1435359_c2_seq1 | 162 | Q8WS88 | Anemone | Phospholipase A2 | 2.00E-34 |
| Tentacle | |||||
| comp1220254_c0_seq1 | 4030 | M5B4R7 | Spider | Translationally controlled tumor protein | 3.00E-12 |
| comp1220065_c0_seq1 | 882 | Q8AY75 | Snake | Calglandulin | 1.00E-34 |
| comp1433029_c0_seq3 | 751 | P69930 | Anemone | Peptide toxin Am-2 | 1.00E-07 |
| comp1420730_c0_seq1 | 579 | Q6T6S5 | Snake | Kunitz-type serine protease inhibitor bitisilin-2 | 8.00E-12 |
| comp1410836_c0_seq1 | 397 | Q8AY75 | Snake | Calglandulin | 1.00E-34 |
| comp1433029_c0_seq8 | 369 | P69930 | Anemone | Peptide toxin Am-2 | 1.00E-07 |
| comp1433029_c0_seq9 | 339 | P69930 | Anemone | Peptide toxin Am-2 | 7.00E-10 |
| comp1433029_c0_seq6 | 240 | P69930 | Anemone | Peptide toxin Am-2 | 1.00E-07 |
| comp1223905_c1_seq1 | 226 | Q8AY75 | Snake | Calglandulin | 2.00E-16 |
| comp1223905_c0_seq1 | 219 | Q8AY75 | Snake | Calglandulin | 3.00E-10 |
Note.—Transcripts are expressed as TPM; bold BLAST hits have an identified signaling region. Sequences with an E-value >0.001 were not included in our analysis. Repeat hits are highlighted in red and those from anemones are in bold. For a full list of candidate toxins, visit the associated bitbucket site (https://bitbucket.org/JasonMacrander/anemone_tissue/).
In addition to members of the well-studied toxin families, we also identified candidates belonging to the acid sensing ion channel toxins (ASIC) (Rodriguez et al. 2014), acrorhagins (Honma et al. 2005), AETX-like toxins (Shiomi et al. 1997), class 9a KTx (Zaharenko et al. 2008), metalloproteases (Moran et al. 2013), membrane-attack complex/perforin (MACPF) toxin gene fragments (Nagai et al. 2002; Oshiro et al. 2004), EGF-like toxins (Shiomi et al. 2003; Honma et al. 2008), protease inhibitors/type II KTxs (Schweitz et al. 1995; Honma et al. 2008; Kozlov et al. 2009), type V KTxs (Orts et al. 2013), and Phospholipase A2 genes (Razpotnik et al. 2010; Romero et al. 2010). Of these, the metalloproteases and protease inhibitors/type II KTxs had the highest number of transcripts across all of the focal species (table 2). Both of these are members of large gene families in which the majority of constituents have nonvenomous functions, making it difficult to discern venomous toxin genes using sequence data alone. Sequence alignments and functional information for all of the identified toxin gene candidates are described in more detail in the supplementary material, Supplementary Material online.
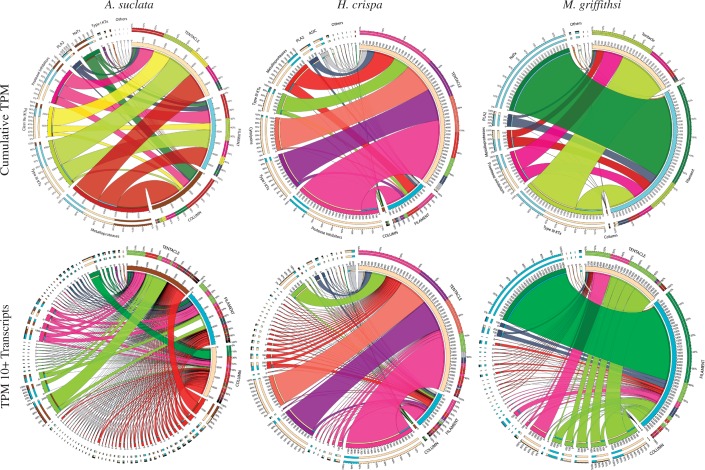
The tissue-specific toxin assemblages varied among the different tissue types. The most abundant toxin gene candidates were from the type III KTx, protease inhibitor/type II KTx, and metalloprotease gene families (fig. 2). In A. sulcata, the cumulative TPM for candidate toxin gene families were relatively consistent across tissue types, decreasing in their overall contribution in the mesenterial filaments and column, respectively (fig. 2). For each of the most abundant toxin gene families (protease inhibitors/type II KTxs, type I KTxs, and cytolysins), a single transcript was expressed at high levels in H. crispa. When compared with other focal taxa, H. crispa had the highest cumulative TPM (fig. 2) and single transcript TPM (table 2) in the tentacle. Megalactis griffithsi had the highest cumulative TPM values in the filament, with the largest TPM values from a single NaTx gene candidate (fig. 2). The second highest cumulative TPM values were from the type III KTxs and protease inhibitors/type II KTxs in the tentacles. Overall, the toxin assembly was predictable with regard a handful of toxin gene families consistently expressed at high levels, yet each species venom assemblage exhibited substantial variation within each tissue type.
Fig. 2.—
Relative expression of toxin genes across tissues. CIRCOS plots with line thickness depicting both cumulative TPM values (top row) and transcript-specific TPM values (bottom row) for each toxin gene family across focal species and tissues. The tissue of origin is noted in capital letters, with the different toxin genes color coded across all plots. The transcript-specific plots (bottom row) show only transcripts with a TPM value >10 in at least one tissue type.
Expanding our BLAST queries to include nonsea anemone venom sequences identified toxin-like sequences that shared high sequence similarities to genes that encode toxins in snakes, spiders, snails, wasps, bees, cephalopods, or lizards (table 3). Several of the candidate toxin genes identified from nonsea anemone venomous taxa have high TPM values and are members of large gene families (e.g., peptidase S1, calmodulin, and translationally controlled tumor protein [TCTP]) or contain toxin-like domains or characteristics (e.g., EGF-like domain or cysteine rich regions) (table 3). Although our previous next-generation sequencing investigation into type I KTx diversity included only transcripts with a single Shk-like domain (Macrander, Brugler et al. 2015), in these species and tissues, we find dozens of transcripts that contained multiple type I KTx domains within a single transcript, often alongside other candidate metalloprotease toxin genes. We did not find any of the 167 nonactiniarian cnidarian toxin sequences in the transcriptomes of our focal taxa. This corroborated previous investigations into lineage-specific venom compositions among cnidarians (Rachamim et al. 2014).
DE and GO Analyses
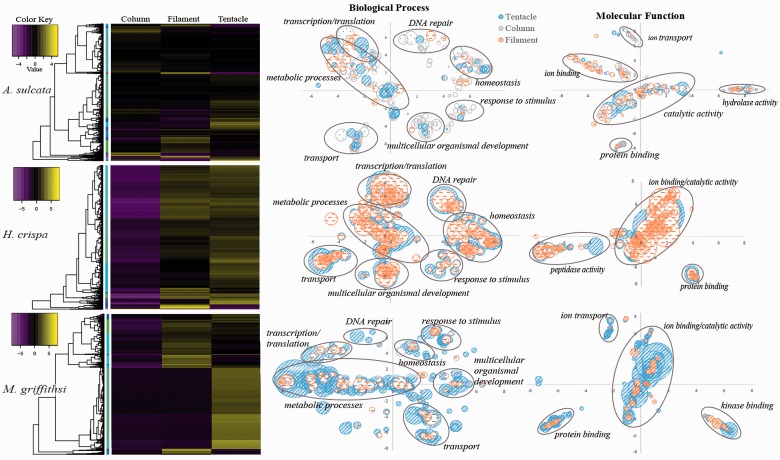
The EdgeR bioconductor package in the Trinity pipeline identified thousands of transcripts that were differentially expressed across tissue types. In A. sulcata, 7,302 transcripts showed significant (P < 0.001) DE with at least a 4-fold change across tissues. For H. crispa and M. griffithsi the threshold for significance was greater, likely due to the low expression levels in the column, with 3,533 transcripts being significant (P < 1.0 e-6) at a 12-fold change in H. crispa and 9,135 transcripts being significant (P < 1.0 e-4) at a 12-fold change in M. griffithsi. Of the differentially expressed transcripts, 4,484 (A. sulcata), 1,880 (H. crispa), and 5,065 (M. griffithsi) returned a positive result for the UniProt BLAST search to identify GO terms. To visualize the potential functional role of the differentially expressed genes that had a positive hit for a GO term, values from the DE matrix were averaged across GO groups, with multiple BLAST hits represented by the more abundant GO terms (fig. 3). Although the placement of similar GO groups is relatively consistent across plots, the manner in which REVIGO (Supek et al. 2011) constructs X, Y coordinates prevents absolute comparisons across species. The final BLAST2GO analysis provided GO information for just a small fraction of the original transcriptome across all tissue types and species, analyzing between 10% and 35% of the most highly expressed transcripts, with the actual GO analysis comprising 1–4% of the original assembly (table 4). Despite this small portion of the transcriptome being evaluated, we observed similar GO assemblages across each tissue type and taxa (fig. 4). The combination of transcriptome level GO analyses not incorporating gene expression information (but see Young et al. 2010) and genomic resources being limited for cnidarians and other lineages outside Bilateria (see Dunn et al. 2015) limits inferences that can be made for these types of investigations.
Fig. 3.—
DE analysis across tissue types. Heat maps depicting differentially expressed genes for A. sulcata, H. crispa, and M. griffithsi, with adjacent trees depicting the transcript clustering analysis. Plots depict upregulated transcripts and identified GO groups based on BLAST searches, with bubble size representing the proportion of each GO group (corrected for repeated GO groups).
Table 4.
Tissue-Specific BLAST2GO Analyses
| Species | Tissue | N | L | GO-Slim | BP | MF | CC |
|---|---|---|---|---|---|---|---|
| Anemonia sulcata | Column | 16,774 | 1,121 | 5,071 | 33,738 | 7,994 | 17,681 |
| Filament | 13,252 | 933 | 4,890 | 33,136 | 7,644 | 16,374 | |
| Tentacle | 11,693 | 1,146 | 4,194 | 27,757 | 6,619 | 14,502 | |
| Heteractis crispa | Column | 10,303 | 511 | 981 | 5,520 | 1,704 | 2,860 |
| Filament | 30,737 | 833 | 6,885 | 42,633 | 10,269 | 22,578 | |
| Tentacle | 18,620 | 1,083 | 4,157 | 26,878 | 6,386 | 13,837 | |
| Megalactis griffithsi | Column | 57,057 | 362 | 5,533 | 22,252 | 9,261 | 10,848 |
| Filament | 22,247 | 1,223 | 4,736 | 28,831 | 7,496 | 15,538 | |
| Tentacle | 20,884 | 892 | 5,298 | 30,364 | 6,922 | 16,477 |
Note.—For the gene ontology analysis, all values are the occurrence of gene ontology groupings in each of the GO domains. BP, biological process; MF, molecular function; CC, cellular component; N, number of sequences which were in the top 10% of the more highly expressed transcripts; L, average length of transcripts; GO-Slim, number of sequences processed.
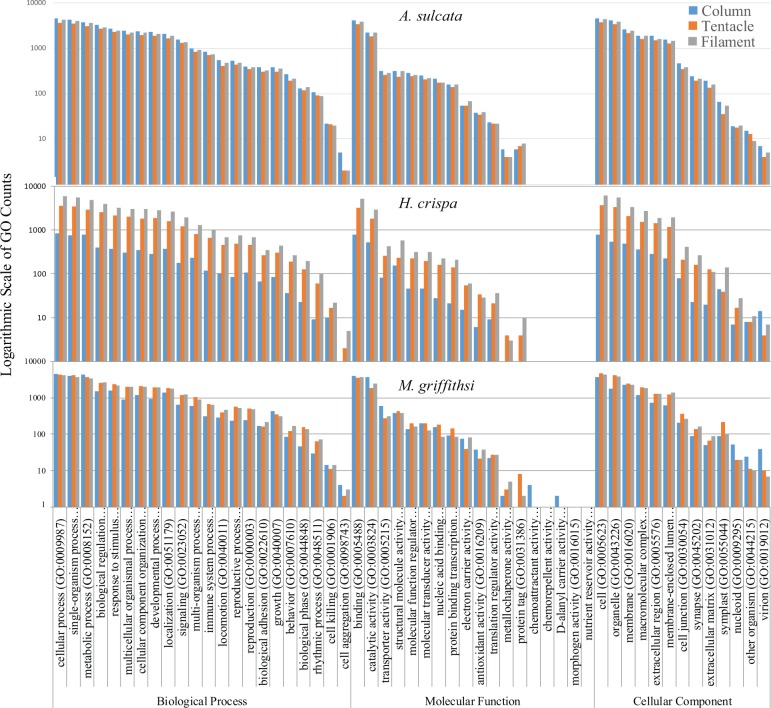
Fig. 4.—
BLAST2GO GO analysis. Species-level comparison of level 2 GO classifications of transcripts from each tissue-specific transcriptome. Counts are from the most highly expressed transcripts (top 10%, based on TPM values).
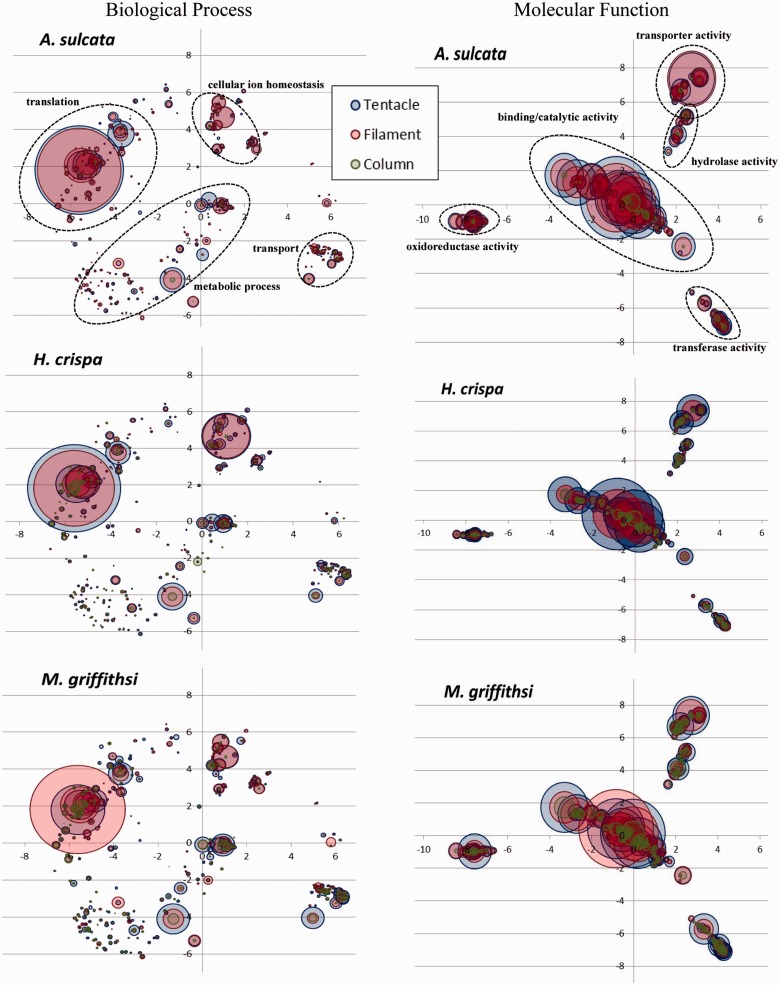
For all species, the column consistently had the lowest TPM values associated with the different GO groups. The tentacle had higher TPM values per GO groupings for H. crispa and A. sulcata, whereas the filament GO groups had higher TPM values in M. griffithsi (fig. 5). In the biological process domain, the translation [GO: 0006412], oxidation–reduction process [GO: 0055114], regulation of transcription, DNA-templated [GO: 0006355], and cellular iron ion homeostasis [GO: 0006879] GO groups consistently had the highest average TPM values. In the molecular function domain, the oxidoreductase activity [GO: 0016491], sequence-specific DNA binding [GO: 0043565], and the metal ion binding [GO: 0046872] consistently had the highest average TPM values. Ultimately our combined GO analyses complemented one another, with the hierarchical categorization in BLAST2GO depicting consistent associations among GO groups, whereas the combined Trinotate and TPM analyses highlighted where the variation occurred within these GO groups across tissues regardless of their hierarchical position.
Fig. 5.—
Trinotate GO analysis. Semantic similarity plots of average TPM per GO group for the Biological Process and Molecular Function GO domains. Labels correspond to the highest average TPM GO groups are shown in A. sulcata.
Discussion
Toxin Gene Diversity
We identified 603 of new candidate toxin genes with high sequence similarity to known toxins from sea anemones (table 2) and an additional 7,038 candidate toxins resembling toxins from other venomous lineages (table 5). Although some of these are expressed at high levels in the tentacles and previously characterized as toxins (fig. 2), many belong to nonvenomous gene families (e.g., metalloproteases, protease inhibitors, and PLA2s) that remain poorly characterized in sea anemones. Among the better studied toxins, the NaTxs and type III KTxs appear to be exclusively found in sea anemones (Rachamim et al. 2014) and share a unique cysteine arrangement not found in any other short venomous peptide (Diochot et al. 2004; Zaharenko et al. 2008; Peigneur et al. 2012). Although they are both found in all tissue types, the type III KTx are consistently found at higher TPM values in the tentacles, whereas NaTx occur at higher levels in the column and filaments for A. sulcata and M. griffithsi, respectively (fig. 2). Additionally, the tissue-specific expression for each Type III KTxs transcript varies considerably for A. sulcata and M. griffithsi: The most highly expressed transcript is not consistent across tissues. Because some type III KTx are capable of targeting both voltage-gated sodium and potassium channels (Nesher et al. 2014), changes in gene expression across tissue types may be influenced by how the gene family evolves (Kuhn & Beyer 2014), with selection taking place across tissues. Contrasting function and evolutionary history of the most abundant toxins for each tissue type may aid in understanding how gene copies take on tissue-specific toxin roles in sea anemones. By lacking a single point of envenomation sea anemones lend themselves to investigate this highly debated topic in venom research from a perspective not permissible in other taxa. Whether the observed patterns of localized expression or the high number of candidate genes correspond to functionally divergent gene copies is unknown, but the taxonomic restriction and target site variability make NaTx and Type III KTxs ideal candidates for future work on gene duplication and neofunctionalization (see Jouiaei et al. 2015). Because the venom of sea anemones is not localized in a venom gland, it is especially difficult to interpret the function of genes that belong to gene families with both venomous and nonvenomous functions. Differences in expression level across tissues that have different functions may provide insight into which transcripts in these gene families may exhibit a toxin like-behavior. In many venomous lineages, metalloproteases and protease inhibitors are both venomous and nonvenomous (Fry et al. 2009); these are expressed at high levels across several tissue types and species in our focal taxa. In sea anemones, metalloproteases (specifically NEP-6 and NEP-14) have been identified within nematocysts (Moran et al. 2013; Rachamim et al. 2014) and are hypothesized to have a dual role, being a weak toxin that targets voltage-gated potassium channels and a synergistic venomous protein that degrades extracellular matrix proteins (Galea et al. 2014). Protease inhibitors are weak venoms, targeting voltage-gated potassium channels, but are much more effective protease inhibitors (Schweitz et al. 1995; Honma et al. 2008). In other venomous taxa, comparative context (Calvete 2013) can help differentiate gene copies whose products differ in function, but we see nothing in the sequence of the candidate gene transcripts that allow us to distinguish them from known “venom” genes from these gene families (supplementary figs. S5 and S6, Supplementary Material online).
Table 5.
Hidden Gene Copies within Each Transcriptome
|
Anemonia sulcata |
Heteractis crispa |
Megalactis griffithsi |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| G | I | N | G | I | N | G | I | N | |
| Type I KTx | 1 | 1 | 2 | 4 | 4 | 5 | 2 | 2 | 3 |
| Type III KTx | 18 | 29 | 34 | 11 | 13 | 14 | 5 | 10 | 42 |
| Cytolysins | 2 | 2 | 3 | 3 | 5 | 6 | 1 | 1 | 1 |
| NaTx | 7 | 9 | 23 | 1 | 1 | 1 | 1 | 1 | 2 |
Note.—G , Trinity “genes”; I, Trinity transcripts; N, Total toxin candidates, including “hidden” gene copies.
By expanding beyond toxin genes previously characterized in sea anemones, we were able to identify several transcripts that may be components of sea anemone venom that share similar toxin-like protein domains. Our initial BLAST searches recovered transcripts that appeared to be a type I KTx, however, upon closer examination it was revealed that a cysteine arrangement found in the type I KTx is also a prevalent protein domain in several other proteins. These transcripts are likely members of the large astacin protein family (Pan et al. 1998, Galea et al. 2014) which contain a type I KTx domain. The type I KTxs have been used in pharmaceutical studies to combat autoimmune diseases (Harnett and Harnett 2008; Chi et al. 2012), with subsequent pharmacological potential for this toxin gene family being investigated by modifying the original type I KTx peptide (Pennington et al. 1996; Pennington et al. 2012). Nontoxic domains with this motif, including members of the matrix metalloprotease gene family found in mammals, are also capable of modifying voltage-gated ion channels (Rangaraju et al. 2010). Many of the transcripts identified in our analysis with the type I KTx domain share high sequence similarity with metalloproteases previously identified in nematocysts (Moran et al. 2013; Rachamim et al. 2014), with individual transcripts expressed at low levels across tissue types (fig. 2).
Many transcripts resembling toxin components described from distantly related venomous lineages were expressed at high levels in the tentacles and filaments (table 3). Among these, transcripts resembling calglandulin and the TCTP were identified in the nematocyst proteome of Acropora digitifera (Gacesa et al. 2015). Of these, the TCTP is the only one that has been functionally characterized in Cnidaria (specifically Hydra vulgaris), where it is expressed exclusively in the column and is involved in cell proliferation (Yan et al. 2000), but has not been investigated for possible function in venom. A transcript resembling the TCTP was consistently expressed at high levels across all tissue types in our focal species (table 3) and also expressed at high levels previously in acrorhagi of A. elegantissima (see Macrander, Brugler, et al. 2015). Venomous TCTP toxins induce vascular permeability, resulting in an inflammatory response (Rattmann et al. 2008; Sade et al. 2012) which is likely due to their ability to target mast cells, releasing histamine that can ultimately cause pain, edema, and erythema (Mulenga and Azad 2005; Sade et al. 2012). Symptoms exhibited by TCTP toxins in noncnidarian lineages resemble that of thalassine, an uncharacterized component in sea anemone venom described early on in sea anemone venom research (Richet 1903a, 1903b, 1905). Extracts from tentacles of Actinia equina and A. sulcata were shown to have a compound which released histamine from mast cells (Jaques and Schachter 1954); however, the actual molecular component behind this was never identified (Turk and Kem 2009). Due to the high expression levels of the TCTP-like transcripts and symptoms exhibited by their venomous constituents, it is likely that this peptide is the toxic component in sea anemones that has evaded identification for over 100 years.
Tissue-Specific DE and Venom Assemblage
We present the first investigation into tissue-specific differential gene expression and venom composition in sea anemones. Previous tissue-specific transcriptomic investigations have used GO analyses to provide a snapshot of the functional role of expressed genes across tissues in cnidarians (Siebert et al. 2011; Sanders et al. 2014). These studies optimized their GO analyses to identify potential genes of interest by removing transcripts below specific lengths or expression levels. Even after reducing our transcriptomes to include only the most highly expressed transcripts for the BLAST2GO analysis, we observed very little variation in GO assemblage across species and tissues (fig. 4). The lower GO variation across tissue types may be attributed to our lack of implementing a threshold for transcript lengths. As many toxic components are short peptides, we did not want to limit our GO identification to transcripts larger than typical sea anemone venom components, ultimately this may have introduced biases as GO representation of short incomplete transcripts, overpowering any potential functional information from complete transcripts in this analysis. Additionally, we may have an overrepresentation of some of the more common GO groups by using only the Trimmomatic-cleaned transcriptomes in our analyses. However, the more conservative raw read cleaning step, which combined both the ECR script and the Trimmomatic, would have undersampled the transcriptomes, with the variation across tissues being an artifact of the final transcriptome assembly rather than functional importance of tissue types.
Our combined gene expression and GO analyses in Trinotate provided a unique perspective for highlighting subtle differences in each of the tissue-specific transcriptome GO groups (fig. 5). Our analyses identified several GO groups corresponding to physiological and functional roles throughout the polyp, with transcripts corresponding to GO groups associated with ion bonding, transport, oxidation–reduction, and homeostasis expressed at high levels in the tentacles (fig. 5). Genes involved with these processes are not only targets of several venomous peptides (Casewell et al. 2013), but are members of conserved gene families often recruited into venom arsenals (Fry et al. 2009). Transcripts in these (and other) GO groups identified in our analysis may be additional toxin-like transcripts or key functional genes in each of the tissue-specific transcriptomes.
The composition of toxin genes varied considerably across tissue type and species. Although it is unlikely that all of the toxin-like transcripts identified here are functionally venomous, the high abundance in the tentacle for some transcripts (fig. 2) may indicate that they are used in prey capture or defense. Proteins can acquire new functions when they are expressed in multiple tissues and may experience some shift in gene expression following gene duplication (Brawand et al. 2011; Kuhn & Beyer 2014; Assis and Bachtrog 2015). This phenomenon has been of recent interest in elucidating the origin and evolution of venom in snakes by contrasting the expression of venom genes in the venom gland and nonvenomous tissues (Hargreaves et al. 2014; Junqueira-de-Azevedo et al. 2015; Reyes-Velasco et al. 2015). This approach cannot be directly applied to sea anemones because all of their tissues contain venom. However, we expected that there would be some differences in the venom assemblage across these tissues based on their inferred function.
Among our focal taxa, the highest numbers of toxin genes were consistently metalloproteases, protease inhibitors, and type III KTxs (table 2). Cumulatively, these were also among the most abundant toxins across tissue types; however, the proportion of TPM for each transcript varied considerably across tissues (fig. 2). Venom in the column has not been characterized previously and we anticipated that these toxins would be less diverse and expressed at lower levels when compared with tentacles or filaments. The column did have a lower abundance of venom components in all species (fig. 2); however, the almost nonexistent expression levels for toxin genes in H. crispa and M. griffithsi were unexpected. For A. sulcata, individual transcripts from the metalloprotease and NaTx gene families made a large contribution to the overall column toxin assemblage. Among the focal taxa, the potential ecological role of these toxin-like transcripts may play in this tissue-specific venom composition (discussed below) indicates that these genes may be functionally venomous in a tissue that is typically characterized as being nonvenomous in nature. Although not evaluated here, tissue-specific toxin expression could vary over time and across environments (Dutertre et al. 2014; Sunagar et al. 2016). Future tissue-specific investigations in sea anemone venom composition would benefit from systematically monitoring changes in toxin gene expression during ecologically relevant interactions (i.e., in the presence of symbionts, predators, or prey) over longer time scales.
Species-Specific Insight
The majority of previously characterized sea anemone toxins has been isolated from Anemonia viridis, a closely related congener to A. sulcata that is often treated as its synonym (see Oliveira et al. 2012). Among the most abundant toxic components identified throughout the polyp (e.g., NaTx, type III KTx, and protease inhibitors) many of these have previously been characterized in Anemonia (Beress 2004; Oliveira et al. 2012). Although metalloproteases were found throughout the polyp (fig. 2), they were not previously isolated from the nematocysts of A. sulcata despite being found in the nematocysts of Nematostella, Hydra, and Aurelia (Moran et al. 2013; Rachamim et al. 2014). The class 9a type KTx in A. sulcata was the only poorly studied toxin gene candidate expressed at high levels that is not part of a large nonvenomous gene family. In other taxa, this short peptide (∼28 amino acids) targets the voltage-gated potassium ion channel (Zaharenko et al. 2008), as well as the acid sensing ion channel reversing inflammatory and acid-induced pain (Osmakov et al. 2013). The role of metalloproteases in sea anemone venom remains unclear. The column is regularly exposed in polyps of A. sulcata (Edmunds et al 1976), and if these toxin candidates are venomous, they may deter potential predators. If this pattern holds true for the other species with an exposed column, it may provide some functional insight which toxins deter predators. The combination of our tissue-specific and gene ontology (GO) analyses provides the necessary framework for future studies of sea anemone venom.
Within the family Stichodactylidae, there have been several species that have been the focus of sea anemone venom research, although H. crispa has been poorly studied compared with some of its congeners (see Oliveira et al. 2012). Previous studies on sea anemone venom that have focused on members of Stichodactylidae have revealed a diverse assemblage of toxins, some of which appear to be lineage specific (i.e., type II NaTx). Although we did not identify any of these toxins here (supplementary fig. S3, Supplementary Material online), the cumulative TPM for H. crispa was highest in the tentacles (fig. 2), with type I KTx and cytolysins making significant contributions to the venom assemblage in addition to other toxin gene candidates that are thought to behave synergistically (Chi et al. 2012). The diverse and apparently robust venom assemblage in the tentacles in H. crispa may be due to their ecological role as hosts to juvenile clownfishes (Huebner et al. 2012). Although there were no symbionts associated with our specimen at the time of RNA extraction, the high expression of toxin genes could be a fixed trait due to coevolutionary processes that have occurred in this lineage. Continually upregulated venom genes in the tentacles may provide ongoing predatory defense and shelter for juvenile clownfishes, which are protected from anemone stings through behavioral modifications (Fautin 1991).
Despite their highly potent venom on humans and popularity in the aquarium trade, this study is the first to characterize the venom composition of any actinodendronid. Due to the severity of symptoms following envenomation in M. griffithsi, we anticipated that the tentacles would have high expression levels of sea anemone toxins known to induce apoptosis, hemolytic activity, or other cell membrane disruptions. Although we identified the types of toxins that are associated with these symptoms, they were expressed at low levels compared with other toxins when considering cumulative TPM (fig. 2), individual transcripts (table 2), or toxin-like sequences with non-cnidarian counterparts (table 5). Among the most abundant transcripts in the tentacles, the type III KTx may be responsible for the reported pain felt upon envenomation (Ocana et al. 2004); however, these toxins are also found in high abundance in A. sulcata and H. crispa (fig. 2). Upon closer examination, our tissue-specific approach indicates that M. griffithsi apparently has no unique or functionally important venom composition (based on known or predicted toxins) that produces the symptoms observed in humans. Their potent envenomation capabilities may be linked to the extremely large and numerous basitrich nematocysts found in the balloon-like extensions of their branching tentacles, commonly called acrospheres (Ardelean and Fautin 2004). The large basitrichs (2–3 times larger than those of the other focal species) found within these structures may be capable of penetrating the epidermis, explaining the severe symptoms observed in humans (supplementary table S2, Supplementary Material online). A similar phenomenon has also been reported in two species of Cubozoa (Carybdea brevipedalia and Chironex yamaguchii): As nematocyst tubule length increased, so did the severity of acute pain (Kitatani et al. 2015).
Conclusion
Sea anemone toxins were initially characterized for their potential pharmaceutical and biomedical applications (Frazao et al. 2012). Like many venomous lineages, characterization of toxic components in sea anemones has been done mostly through an opportunistic approach, focusing on peptides and taxa that are easily accessible. Our tissue-specific approach to characterizing the toxin gene assemblage across multiple taxa produced some unexpected results: 1) Toxin assemblage varies depending on the tissue and species, warranting caution when making generalizations about species or toxin assemblages without broad sampling; 2) a high-throughput assay is needed for the toxin-like sequences that goes beyond simple transcriptomic approaches, combining proteomic data and functional screening of proteins to determine whether these candidate toxin genes are actually components of sea anemone venom; and 3) the large number of highly expressed transcripts with no known function indicates that genomic resources available for cnidarians are lacking. Our study shows that the venom assemblages within these species are quite complex. As transcriptomic approaches to studying sea anemone venom become more commonplace, it is necessary to understand the functional role of different toxins, the evolutionary history of these toxin genes, and potential ecological role toxins play.
Supplementary Material
Supplementary figures S1–S9 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by US National Science Foundation (DEB-1257796 to M.D.) and the OSUCCC CCSG grant (P30CA016058). The authors thank Dr Paulyn Cartwright, Sally Chang, Dr Joseph Ryan, and Dr Steven Sanders for advice about bioinformatic approach. They also thank Dr Mande Holford for her advice on figure 2. Finally, they thank Nicole Leier for assistance in acquiring the specimen of Megalactis griffithsi and Bernard Picton for his assistance in acquiring the specimen of Anemonia sulcata.
Literature Cited
- Ardelean A, Fautin DG. 2004. A new species of the anemone Megalactis (Cnidaria: Anthozoa: Actiniaria: Actinodendridae) from Taiwan and designation of a neotype for the type species of the genus. Proc Biol Soc Wash. 117:488–504. [Google Scholar]
- Assis R, Bachtrog D. 2015. Rapid divergence and diversification of mammalian duplicate gene functions. BMC Evol Biol. 15:138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach PS. 1997. Envenomations from jellyfish and related species. J Emerg Nurs. 23:555–568. [DOI] [PubMed] [Google Scholar]
- Basulto A, et al. 2006. Immunohistochemical targeting of sea anemone cytolysins on tentacles, mesenteric filaments and isolated nematocysts of Stichodactyla helianthus. J Exp Zool. 305A:253–258. [DOI] [PubMed] [Google Scholar]
- Beress L. 2004. Biologically active polypeptides of Anemonia sulcata—and of other sea anemones—tools in the study of excitable membranes. J Toxicol. 23:451–466. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillion M, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478:343–348. (16 co-authors). [DOI] [PubMed] [Google Scholar]
- Calvete JJ. 2013. The continuing saga of snake venom disintegrins. Toxicon 62:40–49. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wuster W, Vonk FJ, Harrison RA, Fry BG. 2013. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 28:219–229. [DOI] [PubMed] [Google Scholar]
- Casteline M, et al. 2012. Macroevoltion of venom apparatus innovations in auger snails (Gastropoda: Conoidea; Terebridae). Mol Phylogenet Evol. 64:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi V, Pennington MW, et al. 2012. Development of a sea anemone toxin as an immunomodulator for therapy of autoimmune diseases. Toxicon 59:529–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization, and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Diochot S, et al. 2004. A new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive channel in sensory neurons. EMBO J. 23:1516–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Leys SP, Haddock SHD. 2015. The hidden biology of sponges and ctenophores. Trends Ecol Evol. 30:282–291. [DOI] [PubMed] [Google Scholar]
- Dunn DF. 1981. The clownfish sea anemones: Stichodactylidae (Coelenterata: Actiniaria) and other sea anemones symbiotic with pomacentrid fishes. Trans Am Philos Soc. 71:3–115. [Google Scholar]
- Dutertre S, et al. 2014. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 24:3521.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds M, Potts GW, Swinfen RC, Waters VL. 1976. Defensive behavior of sea anemones in response to predation by the opisthobranch mollusc Aeolidia papillosa. J Mar Biol Assoc UK. 56:65–83. [Google Scholar]
- Fautin DF. 1991. The anemonefish symbiosis: what is known and what is not. Symbiosis 10:23–46. [Google Scholar]
- Fautin DF. 2009. Structural diversity, systematics, and evolution of cnidae. Toxicon 54:1054–1064. [DOI] [PubMed] [Google Scholar]
- Fautin DF, Allen GR. 1997. Anemonefishes and their host sea anemones. Perth WA: Western Australian Museum; p. 160. [Google Scholar]
- Fautin DF, Mariscal RN. 1991. Microscopic anatomy of invertebrates. Vol. 2 New York: Wiley-Liss, Inc; p. 267–358. [Google Scholar]
- Frazao B, Vasconcelos V, Antunes A. 2012. Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: an overview. Mar Drugs. 10:1812–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry BG, et al. 2009. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 10:483–511. [DOI] [PubMed] [Google Scholar]
- Gacesa R, Chung R, Dunn SRet al. 2015. Gene duplications are extensive and contribute significantly to the toxic proteome of nematocysts isolated from Acropora digitifera (Cnidaria: Anthozoa: Scleractinia). BMC Genomics. 16:774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea C, Nguyen HM, Chandy GK, Smith BJ, Norton RS. 2014. Domain structure and function of matrix metalloprotease 23 (MMP23): role in potassium channel trafficking. Cell Mol Life Sci. 71:1191–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, MacCallum I, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Vaughn M, et al. 2011. The iPlant collaborative: cyberinfrasturcutre for plant biology. Front Plant Sci. 2:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves AD, Swain MT, Hegarty MJ, Logan DW, Mulley JF. 2014. Restriction and recruitment-gene duplication and the origin and evolution of snake venom toxins. Genome Biol Evol. 6:2088–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnett W, Harnett MM. 2008. Therapeutic immunomodulators from nematode parasites. Expert Rev Mol Med. 10:e18.. [DOI] [PubMed] [Google Scholar]
- Hessinger DA. 1988. Nematocyst venoms and toxins In: Hessinger DA, Lenhoff HM, editors. The biology of nematocysts. San Diego, CA: Academic Press; p. 333–368. [Google Scholar]
- Hessinger DA, Lenhoff HM. 1976. Mechanism of hemolysis induced by nematocyst venom: roles of phospholipase A and direct lytic factor. Arch Biochem Biophys. 173:603–613. [DOI] [PubMed] [Google Scholar]
- Honma T, et al. 2005. Novel peptide toxins from acrorhagi, aggressive organs of the sea anemone Actinia equina. Toxicon 46:768–774. [DOI] [PubMed] [Google Scholar]
- Honma T, et al. 2008. Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptide 29:536–544. [DOI] [PubMed] [Google Scholar]
- Huebner LK, Dailey B, Titus BM, Khalaf M, Chadwick NE. 2012. Host preference and habitat segregation among Red Sea anemonefish: effects of sea anemone traits and fish life stages. Mar Ecol Prog Ser. 464:1–15. [Google Scholar]
- Jaques R, Schachter M. 1954. A sea anemone extract (thalassine) which liberates histamine and a slow contracting stubstance. Br J Pharmacol. 9:49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen SD, et al. 2010. Approaching marine bioprospecting in hexacorals by RNA deep sequencing. N Biotechnol. 27:267–275. [DOI] [PubMed] [Google Scholar]
- Jouiaei M, et al. 2015. Evolution of an ancient venom: recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemones. Mol Biol Evol. 32:1598–1610. [DOI] [PubMed] [Google Scholar]
- Junqueira-de-Azevedo IL, et al. 2015. Venom-related transcripts from Bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol Biol Evol. 32:754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass-Simon G, Scappaticci Jr AA. 2002. The behavioral and developmental physiology of nematocysts. Can J Zool. 80:1772–1794. [Google Scholar]
- Katoh S. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitatani R, Yamada M, Kamio M, Nagai H. 2015. Length is associated with pain: jellyfish with painful sting have longer nematocyst tubules than harmless jellyfish. PLoS One 10:e0135015.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov SA, et al. 2009. New polypeptide components from the Heteractis crispa sea anemone with analgesic activity. J Biol Chem. 35:711–719. [DOI] [PubMed] [Google Scholar]
- Kozlov S, Grishin E. 2011. The mining of toxin-like polypeptides from EST database by single residue distribution analysis. BMC Genomics 12:88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov S, Grishin E. 2012. Convenient nomenclature of cysteine-rich polypeptide toxins from sea anemones. Peptides 33:240–244. [DOI] [PubMed] [Google Scholar]
- Kuhn M, Beyer A. 2014. Conservation of expression regulation throughout the animal kingdom. bioRxiv. doi: 10.1101/007252. [Google Scholar]
- Lang J. 1973. Coral reef project—papers in memory of Dr. Thomas F. Goreau. 11. Interspecific aggression by scleractinian corals. 2. Why the race is not only to the swift. Bull Mar Sci. 23:260–279. [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacManes MD, Eisen MB. 2013. Improving transcriptome assembly through error correction of high-throughput sequence reads. Peer J. 1:e113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrander J, Broe M, Daly M. 2015. Multi-copy hidden de novo transcriptome assemblies, a cautionary tale with the snakelocks sea anemone Anemonia sulcata (Pennant, 1977). Toxicon 108:184–188. [DOI] [PubMed] [Google Scholar]
- Macrander J, Brugler MR, Daly M. 2015. A RNA-seq approach to identify putative toxins from acrorhagi in aggressive and non-aggressive Anthopleura elegantissima polyps. BMC Genomics 16:221.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin IN, Britayev TA, Anker A. 2004. Pontoniine shrimps associated with cnidarians: new records and list of species from coastal waters of Viet Nam. Arthropoda Sel. 13:199–218. [Google Scholar]
- Mariscal RN. 1974. Nematocysts In: Muscatine L, Lenhoff HM. editors. Coelenterate biology: reviews and new perspectives. San Diego (CA: ): Academic Press; pp. 129–178. [Google Scholar]
- Mathias AP, Ross DM, Schachter M. 1960. The distribution of 5-hydrotryptamine, etramethylammonium, homarine, and other substances in sea anemones. J Physiol. 151:296–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, et al. 2011. Neurotoxin localization to ectodermal gland cells uncovers an alternative mechanism of venom delivery in sea anemones. Proc R Soc B. 279:1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, et al. 2013. Analysis of soluble protein contents from the nematocysts of model sea anemone sheds light on venom evolution. Mar Biotechnol. 15:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenga A, Azad AF. 2005. The molecular and biological analysis of ixodid ticks histamine release factors. Exp Appl Acarol. 37:215–229. [DOI] [PubMed] [Google Scholar]
- Nagai H, et al. 2002. A new polypeptide from the nematocyst venom of an Okinawan sea anemone Phyllodiscus semoni (Japanese name “unbachi-isoginchaku”). Biosci Biotechnol Biochem. 66: 2621–2625. [DOI] [PubMed] [Google Scholar]
- Nakasugi K, et al. 2013. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One 8:e59534.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesher N, Zlotkin E, Hochner B. 2014. The sea anemone toxin AdE-1 modifies both the sodium and potassium currents of rat cardiomyocytes. Biochem J. 461:51–59. [DOI] [PubMed] [Google Scholar]
- Nevalainen TJ, et al. 2004. Phospholipase A2 in Cnidaria. Comp Biochem Phys. 139:731–735. [DOI] [PubMed] [Google Scholar]
- Nicosia A, Maggio T, Mazzola S, Cuttitta A. 2013. Evidence of accelerated evolution and ectodermal-specific expression of presumptive BDS toxin cDNAs from Anemonia viridis. Mar Drugs. 11:4213–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocana M, Cendan CM, Cobos EJ, Entrena JM, Baeyens JM. 2004. Potassium channels and pain: present realities and future opportunities. Eur J Pharmacol. 500:203–219. [DOI] [PubMed] [Google Scholar]
- Oliveira JS, Fuentes D, King GF. 2012. Development of a rational nomenclature for naming peptide and protein toxins from sea anemones. Toxicon 60:539–550. [DOI] [PubMed] [Google Scholar]
- Orts DJ, et al. 2013. Biochemical and electrophysiological characterization of two sea anemone type 1 potassium toxins from a geographically distant population of Bunodosoma caissarum. Mar Drugs. 11:655–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro N, et al. 2004. A new membrane-attack complex/perforin (MACPF) domain lethal toxin from the nematocyst venom of the Okinawan sea anemone Actineria villosa. Toxicon 43:225–228. [DOI] [PubMed] [Google Scholar]
- Osmakov DI, Kozlov SA, et al. 2013. Sea anemone peptide with uncommon β-hairpin structure inhibits acid-sensing ion channel 3 (ASIC3) and reveals analgesic activity. J Biol Chem. 288:23116–23127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T-L, Groger H, Schmid V, Spring J. 1998. A toxin homology domain in an astacin-like metalloproteinase of the jellyfish Podocoryne carnea with a duel role in digestion and development. Dev Genes Evol. 208:259–266. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Peigneur S, et al. 2012. A natural point mutation changes both target selectivity and mechanism of action of sea anemone toxins. FASEB J. 26:5141–5151. [DOI] [PubMed] [Google Scholar]
- Pennington MW, et al. 1996. Identification of three separate binding sites on SHK toxin, a potent inhibitor of voltage-dependent potassium channels in human T-lymphocytes and rat brain. Biochem Biophys Res Commun. 219:696–701. [DOI] [PubMed] [Google Scholar]
- Pennington MW, et al. 2012. A C-terminally amidated analogue of ShK is a potent and selective blocker of the voltage-gated potassium channel Kv1.3. FEBS Lett. 586:3996–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachamim T, et al. 2014. The dynamically evolving nematocyst content of an anthozoan, a scyphozoan, and a hydrozoan. Mol Biol Evol. 32:740–753. [DOI] [PubMed] [Google Scholar]
- Rangaraju S, Khoo KK, et al. 2010. Potassium channel modulation by a toxin domain in matrix metalloprotease 23. J Biol Chem. 285:9124–9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattmann YD, et al. 2008. Vascular permeability and vasodilation induced by the Loxosceles intermedia venom in rats: involvement of mast cell degranulation, histamine and 5-HT receptors. Toxicon 51:363–372. [DOI] [PubMed] [Google Scholar]
- Razpotnik A, et al. 2010. A new phospholipase A2 isolated from the sea anemone Urticina crassicornis-its primary structure and phylogenetic classification. FEBS J. 277:2641–2653. [DOI] [PubMed] [Google Scholar]
- Reft AJ, Daly M. 2012. Morphology, distribution, and evolution of apical structure of nematocysts in Hexacorallia. J Morphol. 273:121–136. [DOI] [PubMed] [Google Scholar]
- Reyes-Velasco J, et al. 2015. Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol Biol Evol. 32:173–183. [DOI] [PubMed] [Google Scholar]
- Richet C. 1903a. Des poisons contenus dans les tentacules des Actinies (congestine et thalassine). CR Soc Biol Paris. 55:246–248. [Google Scholar]
- Richet C. 1903b. De la thalassine toxine cristalliseé et pruritogène. CR Soc Biol Paris. 55:707–710. [Google Scholar]
- Richet C. 1905. De l’action de la congestine (virus des Actinies) sur les lapins et de ses effects anaphylactiques. CR Soc Biol Paris. 58:109–112. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AA, Cassoli JS, et al. 2012. Peptide fingerprinting of the neurotoxic fractions isolated from secretions of sea anemones Stichodactyla helianthus and Bunodosoma granulifera. New members of the APetx-like family identified by a 454 pyrosequencing approach. Peptides 34:26–38. [DOI] [PubMed] [Google Scholar]
- Rodriguez AA, Salceda E, et al. 2014. A novel sea anemone peptide that inhibits acid-sensing ion channels. Peptides 53:3–12. [DOI] [PubMed] [Google Scholar]
- Romero L, Marcussi S, et al. 2010. Enzymatic and structural characterization of basic phospholipase A2 from the sea anemone Condylactis gigantea. Biochimie. 92:1063–1071. [DOI] [PubMed] [Google Scholar]
- Sade YB, et al. 2012. Molecular cloning, heterologous expression and functional characterization of a novel translationally-controlled tumor protein (TCTP) family member from Loxosceles intermedia (brown spider) venom. Int J Biochem Cell Biol. 44:170–177. [DOI] [PubMed] [Google Scholar]
- Sanders S, Shcheglovitova M, Cartwright P. 2014. Differential gene expression between functionally specialized polyps of the colonial hydrozoan Hydractinia symbiolongicarpus (Phylum Cnidaria). BMC Genomics 15:406.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. 1972. Die Nesselkapseln der Anthozoen und ihre Bedeutung für die phylogenetische Systematik. Helgol Wiss Meeres. 23:422–458. [Google Scholar]
- Schweitz H, et al. 1995. Kalicludines and Kaliseptine. J Biol Chem. 42:25121–25126. [DOI] [PubMed] [Google Scholar]
- Shiomi K, et al. 1997. Novel polypeptide toxins with crab lethality from the sea anemone Anemonia erythraea. Biochim Biophys Acta. 1335:191–198. [DOI] [PubMed] [Google Scholar]
- Shiomi K, et al. 2003. An epidermal growth factor-like toxin and two sodium channel toxins from the sea anemone Stichodactyla gigantea. Toxicon 41:229–236. [DOI] [PubMed] [Google Scholar]
- Siebert S, et al. 2011. Differential gene expression in the siphonophore Nanomia bijuga (Cnidaria) assessed with multiple next-generation sequencing workflows. PLoS One 6:e22953.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WL, Wheeler WC. 2006. Venom evolution widespread in fishes: a phylogenetic road map for the bioprospecting of piscine venoms. J Hered. 97:206–217. [DOI] [PubMed] [Google Scholar]
- Sunagar K, Morgenstern D, Reitzel AM, Moran Y. 2016. Ecological venomics: how genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J Proteomics. 135:62–72. [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talvinen KA, Nevalainen TJ. 2002. Cloning of novel phospholipase A2 from the cnidarian Adamsia carciniopados. Comp Biochem Phys B. 132:571–578. [DOI] [PubMed] [Google Scholar]
- Turk T, Kem WR. 2009. The phylum Cnidaria and investigations of its toxins and venoms until 1990. Toxicon 54:1031–1037. [DOI] [PubMed] [Google Scholar]
- Urbaravo I, et al. 2012. Digital marine bioprospecting: mining new neurotoxin drug candidates from the transcriptomes of cold-water sea anemones. Mar Drugs. 10:2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay N, Poelstra JW, Kunstner A, Wolf JBW. 2013. Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol Ecol. 22:620–634. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131:281–285. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yap LL, Chua KL, Khoo HE. 2008. A multigene family of Heteractis magnificalysins (HMGS). Toxicon 51:1374–1382. [DOI] [PubMed] [Google Scholar]
- Yan L, Fei K, Bridge D, Sarras MP. 2000. A cnidarian homologue of translationally controlled tumor protein (P23/TCTP). Dev Genes Evol. 210:507–511. [DOI] [PubMed] [Google Scholar]
- Yang Y, Smith SA. 2013. Optimizing de novo assembly of short-read RNA-seq for phylogenomics. BMC Genomics 14:328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Hasegawa Y, Honma T, Nagashima Y, Shiomi K. 2010. Screening and cDNA cloning of Kv1 potassium channel toxins in sea anemones. Mar Drugs. 8:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharenko AJ, et al. 2008. Proteomics of the neurotoxic fraction from the sea anemone Bunodosoma cangicum venom: novel peptides belonging to new classes of toxins. Comp Biochem Physiol Part D Genomics Proteomics. 3:219–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.