Abstract
Mitochondrial intron patterns are highly divergent between the major land plant clades. An intron in the atp1 gene, atp1i361g2, is an example for a group II intron specific to monilophytes (ferns). Here, we report that atp1i361g2 is lost independently at least 4 times in the fern family Pteridaceae. Such plant organelle intron losses have previously been found to be accompanied by loss of RNA editing sites in the flanking exon regions as a consequence of genomic recombination of mature cDNA. Instead, we now observe that RNA editing events in both directions of pyrimidine exchange (C-to-U and U-to-C) are retained in atp1 exons after loss of the intron in Pteris argyraea/biaurita and in Actiniopteris and Onychium. We find that atp1i361g2 has significant similarity with intron rps3i249g2 present in lycophytes and gymnosperms, which we now also find highly conserved in ferns. We conclude that atp1i361g2 may have originated from the more ancestral rps3i249g2 paralogue by a reverse splicing copy event early in the evolution of monilophytes. Secondary structure elements of the two introns, most characteristically their domains III, show strikingly convergent evolution in the monilophytes. Moreover, the intron paralogue rps3i249g2 reveals relaxed evolution in taxa where the atp1i361g2 paralogue is lost. Our findings may reflect convergent evolution of the two related mitochondrial introns exerted by co-evolution with an intron-binding protein simultaneously acting on the two paralogues.
Keywords: group II introns, monilophytes, Pteridaceae, intron transfer, RNA editing, intron loss
Introduction
Group II introns can be found in mitochondrial and chloroplast genomes of plants, algae and fungi but also in the genomes of bacteria and archaea (Allet and Rochaix 1979; Dujon 1980; Michel and Dujon 1983; Bonen 2008; Simon et al. 2008; Lambowitz and Zimmerly 2011; McNeil et al. 2016). Group II introns may have originated in eubacteria and transmitted into eukaryotes via endosymbiosis from α-proteobacteria or cyanobacteria which developed into mitochondria or chloroplasts, respectively. Possibly, group II introns later migrated into the nucleus and gave rise to spliceosomal introns in eukaryotes (Cech 1986; Rogers 1990; Cavalier-Smith 1991; Doolittle 1991; Palmer and Logsdon 1991; Roger and Doolittle 1993; Logsdon 1998; Belshaw and Bensasson 2006).
The classification of group II introns is based on their characteristic secondary structures and the splicing mechanism. According to a widely accepted nomenclature proposal (Dombrovska and Qiu 2004), organelle introns are named after their host gene, the nucleotide position upstream of the insertion site (with the homologous liverwort Marchantia polymorpha sequence as reference) and the type of intron.
Group II introns feature six distinct domains which are positioned around a central wheel structure (Michel et al. 1989; Michel and Ferat 1995; Qin and Pyle 1998; Toor et al. 2001; Simon et al. 2008). The complex intron structure is based on highly conserved intra-intronic and intron-exon binding sites. Although some group II introns are known to be capable of self-splicing (Michel and Ferat 1995), plant organelle group II introns rely on nuclear co-factors or intron-encoded maturases for splicing (Matsuura et al. 2001; Brown et al. 2014). Maturases are generally encoded in domain IV and many of them possess reverse transcriptase domains (Michel and Lang 1985; Kennell et al. 1993; Matsuura et al. 2001). This allows group II introns to act as mobile genetic elements (Kennell et al. 1993; Lambowitz and Belfort 1993; Cousineau et al. 2000; Lambowitz and Zimmerly 2004, 2011; McNeil et al. 2016). Most plant organellar group II introns, however, do not contain maturase ORFs (Bonen and Vogel 2001).
The group II intron atp1i361g2 was discovered in an earlier study, which introduced the mitochondrial atp1 gene a new locus to investigate fern (monilophyte) phylogeny (Wikström and Pryer 2005). Intron atp1i361g2 was found in most ferns except Ophioglossales, Psilotales and Equisetales and Danaea elliptica (Wikström and Pryer 2005) and was later also found to be lacking in a subset of Pteridaceae (Rothfels and Schuettpelz 2014). Given its absence in Ophioglossales, Psilotales and Equisetales the most parsimonious explanation of the distribution of atp1i361g2 is a single gain in the joint clade of Marattiales and leptosporangiate ferns (Grewe et al. 2013; Knie et al. 2015).
The process of pyrimidine exchanges in RNA transcripts, termed RNA editing, is common in the organelles of land plants except the marchantiid liverworts as part of RNA maturation in chloroplasts and mitochondria (Chateigner-Boutin and Small 2011; Knoop 2011). In most cases, RNA editing restores conserved codons in mRNAs, but it has also been shown to be present in noncoding RNAs, including introns (Lippok et al. 1994; Carrillo and Bonen 1997; Carrillo et al. 2001; Castandet et al. 2010; Bégu et al. 2011; Farré et al. 2012; Oldenkott et al. 2014), rRNAs (Hecht et al. 2011) and tRNAs (Binder et al. 1994; Marchfelder et al. 1996; Maréchal-Drouard et al. 1996; Grewe et al. 2009, 2011). Intronic editing may restore critical nucleotides which are essential for the secondary structures and a prerequisite for splicing (Castandet et al. 2010; Bégu et al. 2011; Farré et al. 2012), although it should be noted that expected RNA editing to recreate highly conserved domain structures had not been observed in other cases (Carrillo and Bonen 1997)
The loss of RNA editing sites is a common phenomenon which can be explained by restoring point mutations or the recombination of matured cDNA into the organelle genomes. Intron losses are so far described as accompanied by a partial or complete loss of RNA editing sites in the vicinity of the insertion sites (Geiss et al. 1994; Itchoda et al. 2002; Lopez et al. 2007; Ran et al. 2010; Grewe et al. 2011; Oldenkott et al. 2014) and provide evidence for recombination events of in vivo retrotranscribed cDNA and genomic DNA.
In this study, we report on an extended investigation of the Pteridaceae finding at least four independent losses of atp1i361g2 within this family. Surprisingly, two of the atp1i361g2 losses did not cause a loss of the flanking RNA editing sites, hence in conflict with a model invoking a cDNA-mediated recombination event. We present a scenario postulating the more ancient intron rps3i249g2 as potential source of atp1i361g2 arising in the joint clade of Marattiales and leptosporangiate ferns. Finally, the detailed inspection of the two intron paralogues shows an intriguing co-evolution of secondary structure elements, notably their domains III, in the monilophyte clades in general and among Pteridaceae in particular.
Materials and Methods
Plant Material and Molecular Work
Plant material (table 1) was obtained from the Bonn Botanic Gardens, the Botanic Garden Marburg and the Botanic Garden München-Nymphenburg. Adiantum raddianum and Pteris biaurita were obtained from a commercial source (Exotic Plants, Mannheim: http://www.exotic-plants.de). Nucleic acids were extracted using the CTAB protocol (Doyle and Doyle 1990). Crude nucleic acid preparations were either treated with RNase A or DNase I (Thermo Scientific) to purify DNA or RNA, respectively. RNA was reverse transcribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) and random hexamer primers (Roth).
Table 1.
Taxa and Loci
| Species | Botanic garden accession numbers | matK | rbcL | atpA | atpB | rps3 | atp1 | atp1 cDNA |
|---|---|---|---|---|---|---|---|---|
| Cheilanthes fraseri | PE-0-BONN-29933 | – | KU744807 | – | KU744792 | – | KU744752 | KU744740 |
| Astrolepis sinuata | xx-0-BONN-34363 | KF289577 | EF452141 | EF452079 | EF452021 | KU744816 | KU744753 | KU744739 |
| Myriopteris microphylla | xx-0-BONN-17356 | KU744798 | KF961807 | – | KU744791 | – | KU744754 | – |
| Doryopteris pedata | xx-0-BONN-34193 | KU744799 | U27206 | KU744781 | KU744797 | – | KU744755 | – |
| Pellaea falcata | xx-0-BONN-17727 | – | GU136794 | – | GU136775 | – | KU744756 | – |
| Adiantum capillus-veneris | xx-0-BONN-22292 | AY178864 | AY178864 | AY178864 | AY178864 | KU744819 | KU744760 | KU744738 |
| Adiantum raddianum | Commercial source | KJ605550 | KU744803 | EF452071 | GU136769 | KU744820 | KU744764 | – |
| Adiantum peruvianum | xx-0-B-0436988 | KJ605530 | EF452133 | EF452070 | EF452013 | KU744822 | KU744759 | KU744742 |
| Adiantum reniforme | ES-0-BONN-1097 | KJ605551 | KU744804 | KJ742797 | JF935364 | KU744821 | KU744763 | – |
| Adiantum trapeziforme | MX-0-BONN-17375 | KU744800 | KU744805 | – | KU744789 | KU744823 | KU744761 | KU744743 |
| Adiantum pedatum | US-0-BONN-33986 | KJ605529 | KU744802 | EF452069 | EF452012 | KU744824 | KU744762 | – |
| Vittaria lineata | xx-0-BONN-17295 | – | KU744813 | KU744782 | KJ716384 | – | KU744757 | – |
| Antrophyum mannianum | RW-0-BONN-31914 | – | KU744814 | KU744785 | KU744790 | KU744818 | KU744758 | KU744741 |
| Pteris argyraea | xx-0-BONN-3977 | KF289554 | EF452169 | EF452117 | EF452054 | KU744826 | KU744770 | KU744745 |
| Pteris quadriaurita | xx-0-BONN-17387 | KF289541 | EF452173 | EF452121 | KU744795 | – | KU744768 | – |
| Pteris cretica | xx-0-BONN-17520 | KF289524 | KU744810 | EF452118 | EF452055 | KU744825 | KU744765 | KU744747 |
| Pteris biaurita | Commercial source | KF289546 | KF289546 | KU744783 | – | – | KU744771 | – |
| Pteris ryukyuensis | MB-1992/478 | KF289492 | AB574842 | KU744780 | HM582601 | – | KU744767 | KU744744 |
| Pteris vittata | MB-1975/82 | KF289512 | KU744812 | EF452123 | KU744796 | KU744827 | KU744769 | KU744748 |
| Pteris umbrosa | MB-1979/1674 | KF289518 | KU744811 | – | – | – | KU744766 | KU744746 |
| Pityrogramma sulphurea | AR-0-MB-2011/0060 | – | KU744809 | KU744786 | KU744794 | KU744828 | KU744772 | KU744749 |
| Actiniopteris dimorpha | xx-0-M-1996/3147 | KF289571 | EF452130 | EF452066 | KU744788 | – | KU744773 | – |
| Onychium japonicum | xx-0-BONN-26126 | KF289514 | KU744808 | EF452107 | EF452045 | KU744817 | KU744774 | KU744750 |
| Onychium siliculosum | BG München | KF289580 | KU744815 | KU744784 | – | – | KU744775 | – |
| Ceratopteris thalictroides | xx-0-BONN-1069 | KJ772645 | KU744806 | – | – | KU744830 | KU744776 | – |
| Acrostichum aureum | xx-0-BONN-1096 | KF848284 | KU744801 | JF303991 | KU744787 | KU744829 | KU744777 | KU744751 |
| Coniogramme japonica | xx-0-M-S/1230 | JF303920 | KC700111 | JF303990 | KU744793 | – | KU744778 | – |
| Outgroups | ||||||||
| Polypodium cambricum | TR-0-BONN-15737 | HE970727 | FJ825703 | JF832137 P. vulgare | EF463510 P. vulgare | KU744831 | KJ944565 | – |
| Woodwardia radicans | xx-0-BONN-3522 | JF303937 W. japonica | AY137667 | EF463623 W. virginica | EF463359 W. virginica | KU744832 | KJ944567 | – |
| Dicksonia antarctica | xx-0-BONN-20037 | HM021802 | U05919 | AM176442 | U93829 | KU744834 | AJ548853 | – |
| Azolla filiculoides | xx-0-BONN-16921 | – | U24185 A. caroliniana | DQ390547 A. pinnata | AY612689 | KU744835 | KJ944561 | – |
| Vandenboschia radicans | PT-0-BONN-17818 | JF303901 | AF275650 | DQ390581 | AY612715 | KU744836 | KJ944574 | – |
| Todea barbara | xx-0-BONN-20306 | KM925082 | AY612686 | DQ390580 | AY612714 | KU744833 | KJ944575 | – |
| Angiopteris madagascariensis | SC-0-BONN-17551 | DQ821119 A. evecta | EF463239 A. evecta | DQ390544 A. evecta | EF463485 A. evecta | KU744837 | KJ944577 | – |
| Ophioglossum petiolatum | xx-0-BONN-167 | HF585134 | AF313582 O. reticulatum | DQ390571 O. reticulatum | U93825 O. reticulatum | KU744838 | KJ944580 | – |
| Equisetum arvense | xx-0-BONN-3965 | JN968380 | JN968380 | JN968380 | JN968380 | KU756282 E. giganteum | KJ944581 E. giganteum | – |
Note.—Database accessions are given for all sequences investigated in this study. Accession numbers in bold indicate new sequences obtained in this study, “–” indicates that no data are available. Plant material was obtained from the Botanic Garden Bonn or from commercial sources. Accession numbers of plant material from the botanic garden are also shown. The respective epithet is indicated below the accession number when originating from a different species of the same genus.
Mitochondrial DNA and cDNA amplicons were amplified by Touchdown (RT)-PCR with the Go-Taq polymerase (Promega). A list of primers for PCR amplification is provided as supplementary table S1, Supplementary Material online. An initial annealing temperature of 55 °C was lowered to 45 °C in one degree centigrade steps per cycle. The elongation times were between 2 min and 2 min 30 s. After gel electrophoresis in 0.8% agarose gels the PCR products were cut out and purified with the NucleoSpin Extract II Kit (Macherey Nagel). The recovered PCR products were cloned into the pGEM-T Easy Vector (Promega) and transformed into Escherichia coli XLI blue cells. Plasmids were isolated using the BibDo method (Birnboim and Doly 1979). Plasmid insert sequencing was done at Macrogen Europe (Amsterdam, The Netherlands).
RNA Editing Analysis
Exon RNA editing sites were determined from at least two independent cDNA clones per taxon. Incongruencies in editing patterns, e. g. owing to partial editing, were evaluated by parallel direct sequencing of the respective RT-PCR products. Editing sites in introns were determined with two overlapping RT-PCRs using one primer binding within the intron and one to an edited exon sequences followed by merging of the separate sequences. All new sequences are deposited in GenBank (table 1). Editing analysis was aided by using PREPACT available under www.prepact.de (Lenz et al. 2010; Lenz and Knoop 2013). RNA editing sites are labelled following a standardizing nomenclature (Rüdinger et al. 2009; Lenz et al. 2010). Lacking cDNA sequences were supplemented by predictions with the alignment prediction tool of PREPACT and the sequence of Pteris umbrosa as reference. Using diverse alternative references, including those of liverworts, mosses or angiosperms, for editing prediction in the highly conserved atp1 gene had no significant impact. All detected exon editing sites are shown in supplementary table S2, Supplementary Material online. Intron RNA editing sites are labeled with the name of the intron, the nucleotide resulting from editing (eU or eC) and the intron nucleotide position. All detected intronic RNA editing sites are shown in supplementary table S3, Supplementary Material online.
Phylogenetic Analysis
Sequence handling and alignment was performed with MEGA 5.21 (Tamura et al. 2011) using the implemented ClustalW algorithm (Thompson et al. 1994) with subsequent manual corrections. Phylogenetic tree inference was done with IQ-Tree (Nguyen et al. 2015), model selection was set auto and bootstrapping (1,000 replicates) was done with the ultrafast bootstrapping algorithm (Minh et al. 2013). The best fitting model according to the Bayesian Information Criterion (Schwarz 1978) and the corrected Akaike Information Criterion (Burnham and Anderson 2004) was the GTR + Γ+I substitution model (Rodríguez et al. 1990) with four discrete gamma categories. Phylogenetic tree construction was independently done with PhyML using the SPR branch-swapping option and evaluating node support with 150 bootstrap replicates. The resulting tree was fully congruent with the one shown in figures 2 and 3. Significant bootstrap supports of at least 70% deviating by >7% only affected nodes placing Saccoloma inaequale as sister of Pteridaceae, Dennstaedtiaceae and Eupolypodiales (98–83), Davallia solida as sister of Nephrolepis cordifolia (100–91), Cryptogrammoids as sister of all other Pteridaceae (96–72), the monophyletic clade of Anetium citrifolium, the genera Vittaria and Antrophyum (100–78), uniting all taxa of the genus Pteris (100–78) and the node placing Botrychium lunaria sister to Helminthostachys zeylanica and Ophioglossum californicum (89–76).
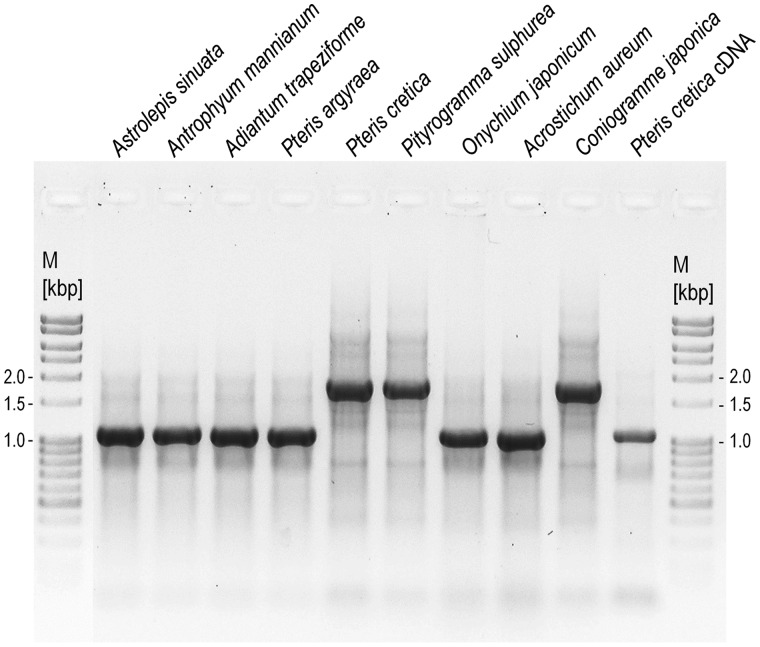
Fig. 1.—
PCR amplification of the mitochondrial atp1 gene, here exemplarily shown for a DNA sampling of nine Pteridaceae species and the Pteris cretica cDNA. The different product sizes indicate variable presence of intron atp1i361g2, subsequently confirmed by cloning and sequencing.
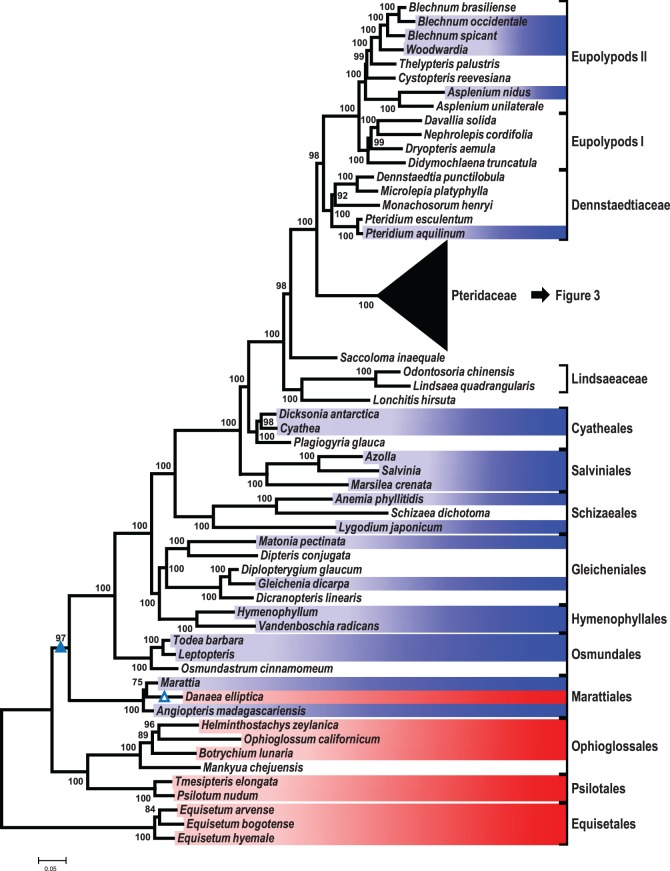
Fig. 2.—
Phylogeny of monilophytes infered with IQ-Tree (Nguyen et al. 2015), based on an alignment of chloroplast loci atpA, atpB, rbcL and matK. Bootstrapping was performed with the ultrafast bootstrap algorithm (Minh et al. 2013) with 1,000 replications. Only bootstrap node supports of at least 70 are shown. Shading highlights species investigated for the atp1 locus, indicating the presence (blue) or absence (red) of the atp1i361g2 intron. The likely gain of intron atp1i361g2 in the common ancestor of Marattiales and leptosporangiate ferns is shown with a filled triangle, its loss in Danaea elliptica is shown with an open triangle. The phylogeny of Pteridaceae is shown in detail in figure 3.
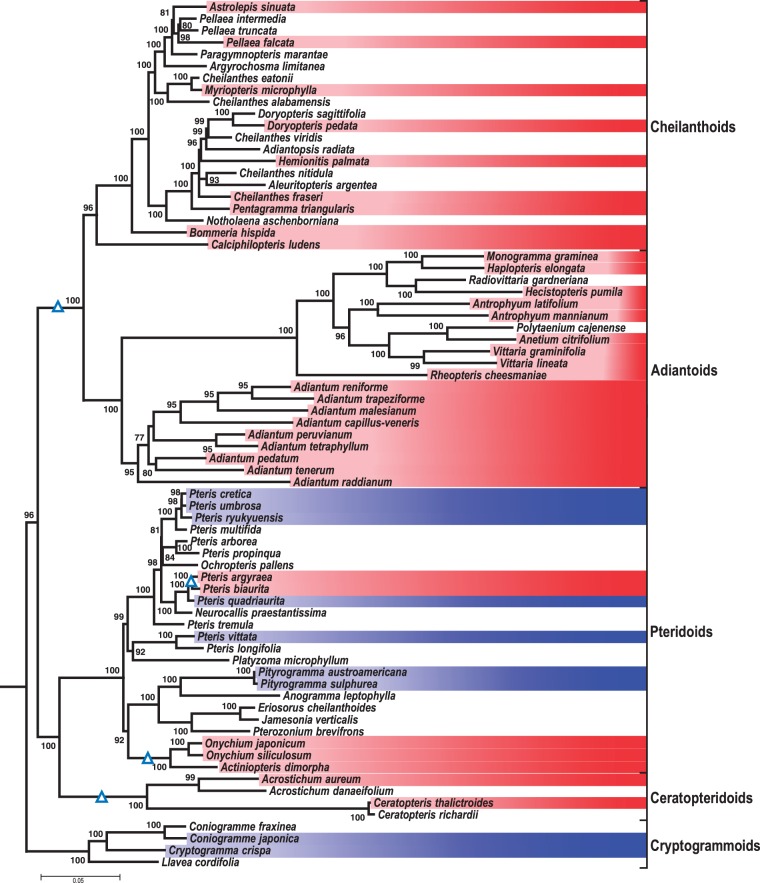
Fig. 3.—
Phylogeny of the Pteridaceae and losses of the atp1i361g2 intron. The phylogram shown is an expanded view of the clade collapsed in figure 2. Shading highlights species investigated for the atp1 locus, indicating the presence (blue) or absence (red) of the atp1i361g2 intron. We could show that atp1i361g2 is absent in the AD/CH clade, the Ceraptopteridoids, Actiniopteris dimorpha, Onychium and Pteris argyraea and P. biaurita. According to this phylogeny, atp1i361g2 was lost at least 4 times in the Pteridaceae.
Results
Phylogeny of Monilophytes Focusing on Pteridaceae: Five Independent Losses of atp1i361g2
Extending the taxon sampling for the mitochondrial atp1 gene among monilophytes (table 1), we observed a variability of PCR product sizes suggesting a variance in the presence of intron atp1i361g2 among Pteridaceae as exemplarily shown in figure 2. In order to elucidate the evolutionary history of the monilophyte-specific intron atp1i361g2 in monilophytes and particularly in Pteridaceae, we extended the original data set of Schuettpelz et al. (2007) with an expanded taxon sampling (table 1 and supplementary table S4, Supplementary Material online) and included matK for phylogenetic analysis, which has previously been used successfully in different studies of monilophyte and Pteridaceae phylogeny (Kuo et al. 2011; Rothfels et al. 2012; Chao et al. 2014; Knie et al. 2015). The resulting phylogenetic trees are shown in figures 2 and 3.
Recent insights on monilophyte backbone phylogeny placing horsetails (Equisetales) as sister to all remaining monilophytes (Knie et al. 2015; Rothfels et al. 2015) are used to root the tree in figure 2. The phylogenetic tree is fully congruent with a modern understanding of monilophyte phylogeny. Noteworthy are high bootstrap values supporting sister group relationships of Gleicheniales and Hymenophyllales and Cyatheales and Polypodiales, respectively. In our study, Pteridaceae are identified as sister group to a clade comprising Dennstaedtiaceae and Eupolypod clades I and II (fig. 2). Lindsaeaceae and Lonchitis are identified as a monophylum, sister to all other Polypodiales. The isolated taxon Saccoloma inequale alone is sister to the large clade comprising Dennstaedtiaceae, Pteridaceae and Eupolypods, the “core polypods”. Except for the Pteridaceae, which we will consider separately (fig. 3), the evolutionary history of the atp1i361g2 intron is most parsimoniously explained with a unique gain in the common ancestor of Marattiales and the leptosporangiate lineage, followed by an early loss in Danaea elliptica (fig. 2; Wikström and Pryer 2005; Grewe et al. 2013; Knie et al. 2015).
Within Pteridaceae, the clades consisting of Adiantoids and Cheilanthoids and the Pteridoids and Ceratopteridoids, respectively, are sister to each other (fig. 3). Cryptogrammoids are the sister group to all other Pteridaceae. The support for the positions of Cryptogrammoids as the earliest-branching taxon in Pteridaceae and Ceratopteridoids sister to Pteridoids in our analysis is significantly higher compared with the IQ-Tree derived supports using the Schuettpelz data set, which in our analysis receives support values of 93 and 81 (not shown). The genus Pteris is paraphyletic with Neurocallis, Ochropteris and Platyzoma considered as separate genera (Schuettpelz et al. 2007). The extended genus Pteris sensu lato (Zhang et al. 2015) is well supported in our analysis (fig. 3). The genus Onychium is identified to be monophyletic in contrast to an earlier study on Pteridaceae phylogeny (Chao et al. 2014).
Group II intron atp1i361g2 has previously been assumed to be absent in Adiantoids and Cheilanthoids (AD/CH) but present in Cryptogrammoids and Pteridoids (Rothfels and Schuettpelz 2014). We now find that atp1i361g2 is also absent in all investigated Ceratopteridoids, in the genus Onychium, in Actiniopteris dimorpha and in Pteris argyraea and P. biaurita. In particular, the absence of atp1i361g2 in P. biaurita and P. argyraea and in Actiniopteris dimorpha and Onychium but its retention in their respective sister species is striking and indicates (at least) two evolutionary recent losses on top of two deeper losses in the Ceratopteridoids and in the AD/CH stem lineage.
Searches for coexisting atp1 alleles using specific primers across exon–intron borders in species which lack the atp1i361g2 intron revealed an additional, intron-containing but pseudogenized atp1 gene copy only in Onychium japonicum, which is frameshifted and, as cDNA studies suggested, not transcribed. No evidence for such additional intron-containing alleles of atp1 was found for the other intron-lacking species.
RNA Editing in Pteridaceae atp1 and Its Correlation to Intron Losses
Plant organelle intron losses are often accompanied by loss of RNA editing events in the flanking exons, assumed to result from recombination of cDNA derived from matured transcripts. We expected that comparing RNA editing patterns could be particularly interesting for atp1 of the Pteridaceae, given that we postulate two deep and two shallow losses of atp1i361g2. Our RNA editing analyses of atp1 show that the numbers of RNA editing sites vary widely between the monophyletic subgroups of the Pteridaceae, most notably comparing species Antrophyum mannianum and Acrostichum aureum to the basal Cryptogrammoids (CR). Many RNA editing sites are conserved among the species of the different subclades, in particular among the early-branching Cryptogrammoids and Pteridoids (fig. 4).
Fig. 4.—
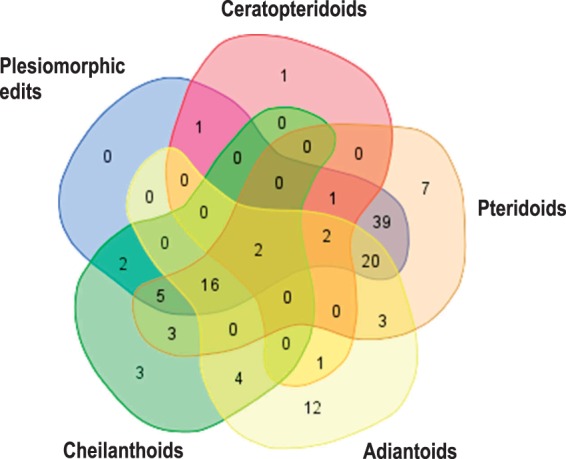
Venn diagram showing the occurrence of RNA editing sites in the five subclades of Pteridaceae. Editing sites which are shared between the early-branching Cryptogrammoids and at least one of the other four subclades is considered to be an ancient plesiomorphic editing state among Pteridaceae. Most editing sites observed in the Pteridaceae are therefore plesiomorphic. The Adiantoid and Pteridoid clades feature 14 and 8 unique editing sites, respectively, adding to the more ancient editing sites shared with the other clades.
We consider edits shared between the CR subclade and at least one of the other subclades to represent a plesiomorphic editing state among Pteridaceae. In total, we count 124 different sites of RNA editing in the atp1 gene of Pteridaceae. Of these, we assume 88 edits to be ancient plesiomorphic editing events and consider 13 as clade-specific edits (which occur in at least two genera) and 23 as “unique” genus-specific sites.
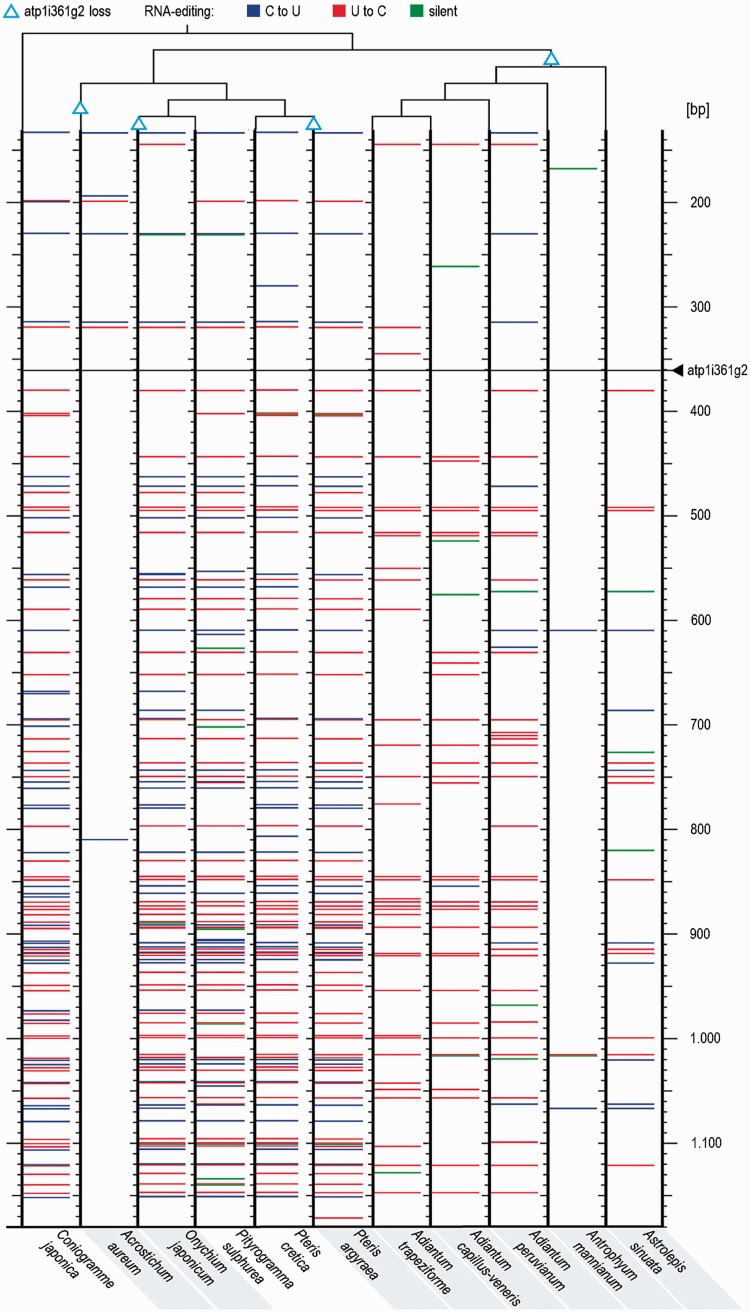
Overall, it is evident that the AD and CH clades show particularly diverging editing patterns. A detailed investigation of the RNA editing sites in selected taxa of the different Pteridaceae clades makes this more obvious. In species, which have lost atp1i361g2, numbers of RNA editing sites are overall drastically reduced (fig. 5). Most notably, Acrostichum aureum lacking atp1i361g2 is nearly completely devoid of editing sites downstream of the previous intron insertion site as could be expected for a cDNA-mediated recombination extending far into the downstream exon. Surprisingly, however, such a correlation in the absence of intron and editing sites is not found for Pteris argyraea/biaurita and Actiniopteris dimorpha and Onychium (fig. 5). In these taxa, nearly all editing sites in comparison to the respective closest relatives, including the edits in close vicinity to the intron insertion site, are retained. This is particularly intriguing given the close relation of Pteris argyraea and P. biaurita and its intron-containing sister taxon P. quadriaurita.
Fig. 5.—
Intron losses and RNA editing patterns in atp1 of selected taxa of the Pteridaceae. Phylogenetic relationships as determined (fig. 3) and the events of intron losses (open triangles) are shown with the simplified cladogram on top. Species without atp1i361g2 are shaded in grey. Acrostichum aureum has only one editing site downstream of the intron insertion site. In the intron-less species Pteris argyraea and Actiniopteris dimorpha no reduction of RNA editing sites is observed. Editing sites of Coniogramme japonica are predicted.
Although the majority of the editing sites in the intron-less species of the Cheilanthoids and Adiantoids can be considered to be plesiomorphic edits also found in the intron-containing species and intron-lacking species that are not affected by RNA editing loss, we also found at least 21 new editing sites. These unique edits, mostly found in the genus Adiantum, may have evolved after the loss of atp1i361g2 in the common ancestor of Cheilanthoids and Adiantoids. A particularly interesting case is the editing pattern of Adiantum trapeziforme given that exclusively U-to-C editing could be observed within the atp1 amplicon. To our knowledge, this is the first example of a plant organelle transcript region featuring exclusively U-to-C editing. However, other mitochondrial cDNAs of Adiantum trapeziforme, including the here investigated rps3 gene, show the usual mix of C-to-U and U-to-C editing typically identified in monilophytes.
Structural Analysis of atp1i361g2 and Its Likely Source Paralogue, rps3i249g2
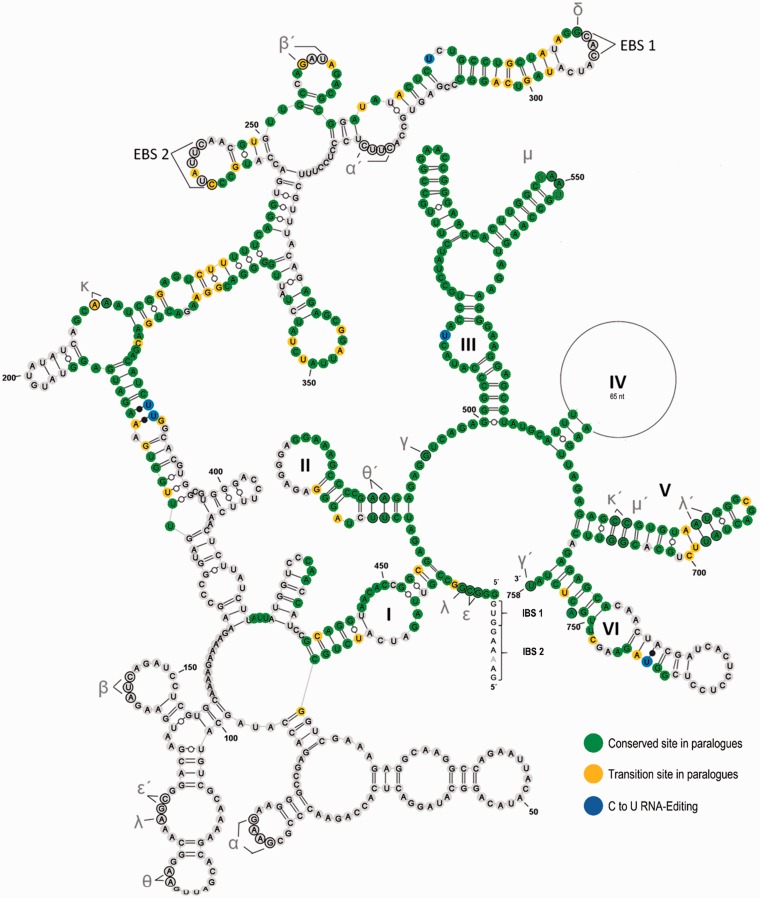
Its unique presence in the monilophytes and its independent losses connected to, or disconnected from, the loss of flanking editing sites prompted us to investigate atp1i361g2 more closely. Where present, atp1i361g2 features conserved canonical group II intron structures, with sequence or structural differences pronounced mainly in domain IV among Marattiales and Polypodiales. The atp1i361g2 secondary structure is exemplarily shown for Pteris cretica (fig. 6).
Fig. 6.—
Secondary structure model of atp1i361g2 in Pteris cretica and similarities with its paralogue rps3i249g2. The intron features all typically conserved structures in group II introns (Michel et al. 1989; Michel and Ferat 1995; Qin and Pyle 1998; Toor et al. 2001; Simon et al. 2008). The six intron domains are labelled with Roman numerals (I–VI) and the tertiary interaction sites with Greek letters. Exon binding sites (EBS) and corresponding intron binding sites (IBS) in the 5′-exon are indicated. Nucleotides which are identical to the rps3i249g2 intron are shown in green. Differences in nucleotide sequences that can be explained by transitions (A/G and C/U) are shown in yellow. Four experimentally verified C-to-U editing sites are shaded in blue and the three newly created A–U bindings in stem regions of domain I and domain VI are marked with black dots. Nucleotides in black circles highlight tertiary base-pairing interactions. Introns were manually folded and the drawing was made using the VARNA software (Darty et al. 2009).
We noted that atp1i361g2 shows significant primary sequence similarities to rps3i249g2, a mitochondrial group II intron conserved in Adiantum capillus-veneris, the lycophyte Phlegmariurus squarrosus and gymnosperms (Ran et al. 2010; Regina and Quagliariello 2010; Liu et al. 2012; Bonavita and Regina 2016). We targeted the rps3i249g2 insertion site for a phylogenetic wide sampling of monilophytes, finding it highly conserved and without evidence for intron losses (table 1). For closer comparison to atp1i361g2 we cloned and sequenced several monilophyte rps3 amplicons with a focus on the Pteridaceae. A secondary structure model of rps3i249g2 in Pteris cretica is given in supplementary figure S1, Supplementary Material online, for direct comparison to its atp1i361g2 counterpart in the same species (fig. 6). The most significant similarities between the two intron paralogues are found in the region encompassing the 3′-part of domain I up to the beginning of domain IV and in their near-identical domains V (fig. 6 and supplementary fig. S1, Supplementary Material online). Given the now observed wide conservation of rps3i249g2 among monilophytes and its presence also in lycophytes and gymnosperms, we conclude that an intron-copying event via reverse splicing of rps3i249g2 into atp1 in the joint stem lineage of Marattiales and leptosporangiate ferns may be a likely evolutionary scenario to explain the existence of atp1i361g2 in monilophytes. However, it should be noted that the present dissimilar exon binding sites of the two intron paralogues do not immediately support such a reverse splicing scenario. Possibly, an alternative intron folding within an ancestral rps3i249g2 domain I may have created novel EBS sites for interacting at the new location in atp1 (supplementary fig. S2, Supplementary Material online). An alternative scenario of large-scale genomic recombination appears less likely given that similarities are entirely restricted to the intron sequences and no conversion of flanking exon sequences is discernible.
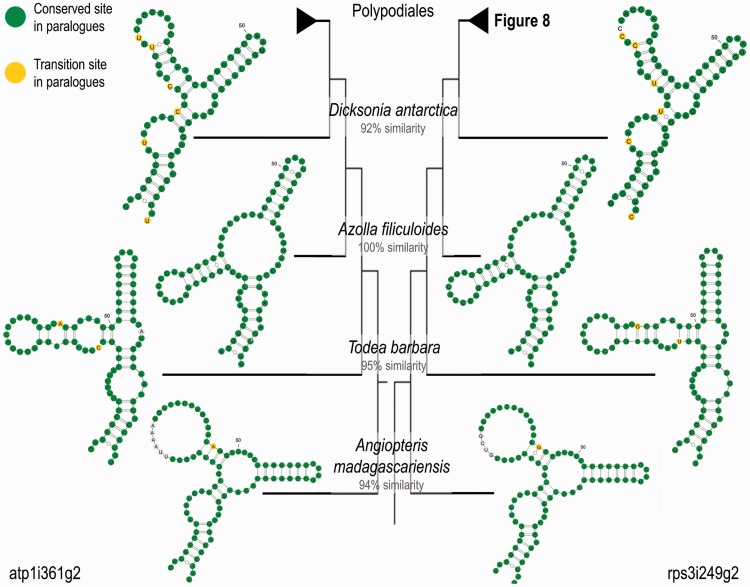
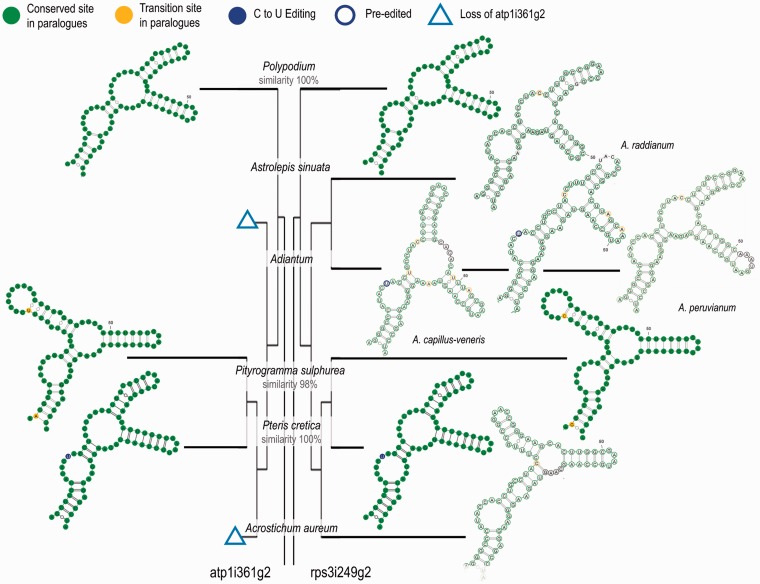
Convergent Evolution of Domains III of atp1i361g2 and rps3i249g2
As our preliminary data suggested that the two paralogues would not decrease but rather even increase in similarity over evolutionary time, we extended the monilophyte sampling for atp1i361g2 and rps3i249g2 also outside of the Pteridaceae (table 1). The comparative analysis revealed a striking convergent evolution of the region most similar between the two intron paralogues, most notably with respect to the structure of their domains III. In fact, the domains III of the two intron paralogues within one species are extremely similar, even nearly or completely identical in Polypodiales, whereas the orthologous regions differ even between closely related taxa (figs. 7 and 8; supplementary fig. S3, Supplementary Material online). The independent unique losses of atp1i361g2 are interesting cases to investigate the evolutionary fate of the rps3i249g2 paralogue. The losses of atp1i361g2 indeed seem to allow domain III of rps3i249g2 to evolve more freely as reflected by higher sequence divergence among related species. The only exception in our sampling are rps3i249g2 domains III of Pteris agyraea and Pteris quadriaurita, two species which have split only very recently in evolution.
Fig. 7.—
Convergent evolution of domains III of atp1i361g2 and rps3i249g2. Domains III of the two group II introns are compared in four selected fern species. Examples within the Polypodiales are displayed in detail in figure 8. Identical nucleotides in paralogues are shown in green and transitions between both domains are shown in yellow, analogous to figure 6. The paralogous domains III are nearly identical whereas orthologous domains are highly different in the analysed monilophyte taxa.
Fig. 8.—
Convergent evolution of domains III of atp1i361g2 and rps3i249g2 in Polypodiales and Pteridaceae. Graphic display is as in figure 7. Convergent evolution of the structure of domain III is observed in species with both introns. When atp1i361g2 is lost (open triangles), domain III of rps3i249g2 undergoes structural changes. A conserved C-to-U editing site in a bulge of the stem region in Pteris cretica is highlighted in blue. Black circles indicate inserted nucleotides.
Intron RNA Editing
High numbers of RNA editing sites in plant organelle coding regions generally indicate that intron sequences might be affected by RNA editing, too. Given the high abundance of RNA editing identified in the atp1 coding sequences, we exemplarily checked for RNA editing in partially matured, unspliced pre-mRNAs. We identified five sites of C-to-U and two sites of U-to-C editing in Pteris cretica and P. vittata (see supplementary table S3, Supplementary Material online). One of these sites is of particular interest as it affects a bulged region close to the conserved proximal stem of domain III, which shows convergent evolution with its rps3i249g2 counterpart (fig. 8). The position of C-to-U editing identified in the two Pteris species is “pre-edited” with a T present on DNA level in this position in the atp1i361g2 orthologues of other taxa including most distantly related genera Angiopteris, Marattia and Todea. Intriguingly, this position is also pre-edited in the paralogue domain III of rps3i249g2, including the adiantoids Adiantum capillus-veneris and A. trapeziforme taxa where atp1i361g2 is lost.
Discussion
Here, we have described the evolutionary history of monilophyte-specific intron atp1i361g2 and its likely source paralogue rps3i249g2. Our data are discussed on the basis of an extended monilophyte phylogeny based on four chloroplast loci (atpA, atpB, rbcL and matK). The resulting phylogenetic trees (figs. 2 and 3) are fully congruent with recent insights on monilophyte phylogeny and even further contribute to open issues of fern phylogeny such as a likely sister grouping of Gleicheniales and Hymenophyllales, of Cyatheales and Polypodiales, and, among the latter, in supporting the serial sister group relationships of Dennstaedtiaceae, Pteridaceae, Saccoloma and Lindsaeaceae (and Lonchitis) to the Eupolypods. We have used this encompassing monilophyte phylogeny and the detailed phylogeny of the Pteridaceae as the framework to investigate the evolutionary fates of the two mitochondrial introns, which are of rather restricted occurrence among land plants. Importantly, critical nodes such as the position of the Ceratopteridoids are now perfectly supported. The well-supported phylogeny enables to identify an evolutionary scenario, in which intron atp1i361g2 has been lost 4 times independently in the Pteridaceae (fig. 3).
Introns in plant mitochondrial DNAs are strikingly different between the major land plant clades and have contributed to elucidate the embryophyte backbone phylogeny. Particularly intriguing, for example, are the intron patterns supporting the sister group relationships of liverworts and all other land plants (Qiu et al. 1998) and of hornworts and vascular plants (Groth-Malonek et al. 2005), respectively. Notably, not one single mitochondrial intron of altogether 100 recognized among bryophytes is shared between all of their three distinct clades, the liverworts, mosses and hornworts (Knoop 2010, 2013). In contrast to the intron gain and loss dynamic along the backbone of land plant phylogeny, mitochondrial introns are much more stable once a clade is established. For example, most of 27 introns in the mitochondrial DNA of the gymnosperm Cycas taitungensis (with the exception of rps3i249g2 and the chloroplast-derived trnVi36g2) are conserved among flowering plants. Exceptions of introns lost more frequently and independently among angiosperms are cox2i373g2, cox2i691g2 and nad4i976g2 (Unseld et al. 1997; Kubo et al. 2000; Itchoda et al. 2002; Kudla et al. 2002; Sloan et al. 2010; Hepburn et al. 2012). A recently identified unique exception to the overall stability of mitochondrial introns within a clade is the gymnosperm Welwitschia mirabilis, which has lost more than half of the ancient seed plant mitochondrial intron set (Guo et al. 2016).
The here investigated atp1i361g2 intron is unique to monilophytes. We now suggest that it originated in the joint stem lineage of Marattiales and leptosporangiate ferns from its more ancient counterpart rps3i249g2, which likely emerged much earlier with the origin of tracheophytes. Alternative scenarios, for example, invoking events of horizontal gene transfer (HGT) are less likely given that the phylogenies of both introns are fully congruent with the organismic phylogeny (supplementary fig. S4, Supplementary Material online). The rps3i249g2-to-atp1i361g2 intron copying event likely is a further molecular synapomorphy for the joint clade of Marattiales and leptosporangiate ferns, which is well supported from independent recent phylogenetic studies (Knie et al. 2015; Rothfels et al. 2015). Whereas rps3i249g2 has never been found in flowering plants and was likely lost in the angiosperm stem lineage, we here report that it is highly conserved among monilophytes.
The here identified independent losses of atp1i361g2 in a moderate sampling of 42 monilophyte taxa, four of which occur in the Pteridaceae alone, are unusual for a mitochondrial group II intron. Intriguingly though, nad5i1242g2 has also been found lost at least 3 times independently in a sampling of 27 monilophytes (Vangerow et al. 1999; Knie et al. 2015). Possibly, monilophytes may ultimately reveal more dynamic intron loss scenarios than seed plants.
Losses of organelle introns are usually accompanied by a loss of RNA editing sites in the closer or wider vicinity of the intron insertion site (Geiss et al. 1994; Itchoda et al. 2002; Lopez et al. 2007; Ran et al. 2010; Grewe et al. 2011; Oldenkott et al. 2014). The comparative studies suggest that the losses of RNA editing sites in the environment of the lost introns are within a range of 210 bp to >1,600 bp (Geiss et al. 1994; Itchoda et al. 2002; Ran et al. 2010; Grewe et al. 2011; Oldenkott et al. 2014). Integration of matured and reverse transcribed RNA into the genomic DNA is assumed to cause such a simultaneous loss of editing sites together with the intron. This scenario is also plausible for two of the observed deep losses of atp1i361g2 in the CE and in the joint AD/CH clade. In the AD/CH clade, we see a sporadic re-occurrence of what we classified as plesiomorphic, ancient editing sites. We assume that the common ancestor of the AD/CH clade has lost atp1i361g2 and most flanking RNA editing sites in a single, ancient cDNA recombination event. The nuclear-encoded RNA editing factors responsible for the individual RNA editing sites, however, initially remain present and will turn into pseudogenes and ultimately disappear only if not needed over a longer period of time. A re-appearing pyrimidine transition at an ancient editing site can immediately be addressed, when the respective editing factor is still present to correct the mutation and evolutionary pressure on its maintenance is regained. Notably, we recently found that editing factors are more strongly maintained in the course of angiosperm evolution when addressing multiple sites on different transcripts in parallel (Hein et al. 2016), a scenario that is very likely for the abundant editing observed in monilophytes.
Pteris argyraea, its close relative Pteris biaurita, as well as the genus Onychium and Actiniopteris dimorpha are intriguing cases, because these taxa lost the atp1i361g2 intron without an apparent effect on editing patterns in the vicinity of the former insertion site. A scenario as described earlier with a complete regain of all editing sites by pyrimidine transitions appears very unlikely. A possible explanation for the observations could be efficient splicing of unedited transcripts giving rise to cDNA substrates for recombination in the ancestors of the taxa lacking the intron but retaining RNA editing. However, for partially matured atp1 transcripts in intron-containing species, we rather observed partially or even completely edited but unspliced variants in our cDNA analyses. It appears more likely that the Pteris argyraea/biaurita and Onychium/Actiniopteris dimorpha cases represent examples of recombination mediated through very short stretches of homology between the most proximal editing sites flanking the intron insertion site, here 18-bp downstream and 42-bp upstream. Such a concept of recombination across very short nucleotide stretches was previously also proposed to explain intron and editing site losses in Silene noctiflora (Sloan et al. 2010).
Monilophyte-specific intron atp1i361g2 shows significant similarity to rps3i249g2, its likely source paralogue. Particularly intriguing are the structural similarities in domains III, which do not decrease but rather increase over evolutionary time after creation of the atp1i361g2 paralogue (figs. 7 and 8; supplementary fig. S3, Supplementary Material online). Additional support for a scenario of convergent evolution comes from the relaxed evolution of rps3i249g2 domain III once its paralogue atp1i361g2 is lost and from an editing event further increasing similarity in domain III. Domain III plays a key role in positioning domain I and V during splicing (Fedorova and Pyle 2005). Micro-recombinational events of cDNA across short sequence stretches, as assumed here for the loss of introns without the loss of flanking editing sites and also proposed previously (Sloan et al. 2010), could be the cause for the extraordinary sequence similarities of the intron domain III paralogues. However, if mediated by cDNA, this would require a significant amount of unspliced immature RNA as a source for cDNA synthesis. Moreover, the lack of extensive sequence similarities downstream of domain III that could serve as an anchoring guide for homology-mediated recombination makes this mechanism less likely in our eyes. The frequent recombination in plant mtDNA on genomic level generally relies on much larger and perfect repeat sequences that create reciprocal exchanges of the flanking unique sequences. We rather assume a splicing-factor or an otherwise RNA-binding protein affecting both intron paralogues simultaneously to be the underlying cause for our observations. Numerous such splicing factors affecting individual or multiple organelle introns simultaneously have already been identified in model taxa like Arabidopsis thaliana, Physcomitrella patens, rice or maize (Ostersetzer et al. 2005; Keren et al. 2009, 2012; Brown et al. 2014; Cohen et al. 2014). If such a factor pre-existed for the more ancient rps3i249g2 to assist splicing, it likely also serves the new atp1i361g2 intron paralogue. Unfortunately, the restricted occurrence of rps3i249g2 only in the lycophyte Phlegmariurus, in gymnosperms and, as here shown, also in ferns, all of which are clades not represented by prominent model organisms, does hitherto not provide hints to a relevant splicing factor.
Supplementary Material
Supplementary tables S1–S4 and Supplementary figures S1–4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We are highly grateful to our colleagues at the Botanic gardens in Bonn, Marburg (Dr Andreas Titze and Ms Marion Reich) and Munich (Dr Andreas Gröger) for providing fern plant material. In particular, we wish to thank the curator of the Bonn Botanic Garden, Dr Wolfram Lobin, and his team, especially Mr Bernd Reinken, for support and expert consultation. We gratefully acknowledge the expert technical assistance of Ms Monika Polsakiewicz. The study relied exclusively on basic funding through the University of Bonn.
Literature Cited
- Allet B, Rochaix JD. 1979. Structure analysis at the ends of the intervening DNA sequences in the chloroplast 23S ribosomal genes of C. reinhardii. Cell 18(1):55–60. [DOI] [PubMed] [Google Scholar]
- Bégu D, Castandet B, Araya A. 2011. RNA editing restores critical domains of a group I intron in fern mitochondria. Curr Genet. 57(5):317–325. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Bensasson D. 2006. The rise and falls of introns. Heredity 96(3):208–213. [DOI] [PubMed] [Google Scholar]
- Binder S, Marchfelder A, Brennicke A. 1994. RNA editing of tRNA(Phe) and tRNA(Cys) in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol Gen Genet. 244(1):67–74. [DOI] [PubMed] [Google Scholar]
- Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7(6): 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonavita S, Regina TMR. 2016. The evolutionary conservation of rps3 introns and rps19-rps3-rpl16 gene cluster in Adiantum capillus-veneris mitochondria. Curr Genet. 62(1):173–184 [DOI] [PubMed] [Google Scholar]
- Bonen L. 2008. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8(1):26–34. [DOI] [PubMed] [Google Scholar]
- Bonen L, Vogel J. 2001. The ins and outs of group II introns. Trends Genet. 17(6):322–331. [DOI] [PubMed] [Google Scholar]
- Brown GG, Des Colas Francs-Small C, Ostersetzer-Biran O. 2014. Group II intron splicing factors in plant mitochondria. Front Plant Sci. 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. 2004. Model selection and multimodel inference. New York (NY: ): Springer New York. [Google Scholar]
- Carrillo C, Bonen L. 1997. RNA editing status of nad7 intron domains in wheat mitochondria. Nucleic Acids Res. 25(2):403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo C, Chapdelaine Y, Bonen L. 2001. Variation in sequence and RNA editing within core domains of mitochondrial group II introns among plants. Mol Gen Genet. 264(5):595–603. [DOI] [PubMed] [Google Scholar]
- Castandet B, Choury D, Bégu D, Jordana X, Araya A. 2010. Intron RNA editing is essential for splicing in plant mitochondria. Nucleic Acids Res. 38(20):7112–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1991. Intron phylogeny: a new hypothesis. Trends Genet. 7(5):145–148. [PubMed] [Google Scholar]
- Cech TR. 1986. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 44(2):207–210. [DOI] [PubMed] [Google Scholar]
- Chao Y-S, Rouhan G, Amoroso VB, Chiou W-L. 2014. Molecular phylogeny and biogeography of the fern genus Pteris (Pteridaceae). Ann Bot. 114(1):109–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin A-L, Small I. 2011. Organellar RNA editing. Wiley Interdiscip Rev. 2(4):493–506. [DOI] [PubMed] [Google Scholar]
- Cohen S, et al. 2014. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78(2):253–268. [DOI] [PubMed] [Google Scholar]
- Cousineau B, Lawrence S, Smith D, Belfort M. 2000. Retrotransposition of a bacterial group II intron. Nature 404(6781):1018–1021. [DOI] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. 2009. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics 25(15):1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrovska O, Qiu Y-L. 2004. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol Phylogenet Evol. 32(1):246–263. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. 1991. The origins of introns. Curr Biol. 1(3):145–146. [DOI] [PubMed] [Google Scholar]
- Doyle LL, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus 12:13–15. [Google Scholar]
- Dujon B. 1980. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell 20(1):185–197. [DOI] [PubMed] [Google Scholar]
- Farré J-C, Aknin C, Araya A, Castandet B. 2012. RNA editing in mitochondrial trans-introns is required for splicing. PloS One 7(12):e52644.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Pyle AM. 2005. Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3. The EMBO journal. 24(22):3906–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss KT, Abbas GM, Makaroff CA. 1994. Intron loss from the NADH dehydrogenase subunit 4 gene of lettuce mitochondrial DNA: evidence for homologous recombination of a cDNA intermediate. Mol Gen Genet. 243(1):97–105. [DOI] [PubMed] [Google Scholar]
- Grewe F, et al. 2011. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 39(7):2890–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Guo W, Gubbels EA, Hansen AK, Mower JP. 2013. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol Biol. 13:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe F, Viehoever P, Weisshaar B, Knoop V. 2009. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 37(15): 5093–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth-Malonek M, Pruchner D, Grewe F, Knoop V. 2005. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol Biol Evol. 22(1):117–125. [DOI] [PubMed] [Google Scholar]
- Guo W, et al. 2016. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol Biol Evol. 33(6):1448–1460. [DOI] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V. 2011. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 3(0):344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein A, Polsakiewicz M, Knoop V. 2016. Frequent chloroplast RNA editing in early-branching flowering plants: pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors. BMC Evol Biol. 16(1):23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn NJ, Schmidt DW, Mower JP. 2012. Loss of two introns from the Magnolia tripetala mitochondrial cox2 gene implicates horizontal gene transfer and gene conversion as a novel mechanism of intron loss. Mol Biol Evol. 29(10):3111–3120. [DOI] [PubMed] [Google Scholar]
- Itchoda N, Nishizawa S, Nagano H, Kubo T, Mikami T. 2002. The sugar beet mitochondrial nad4 gene. An intron loss and its phylogenetic implication in the Caryophyllales. Theor Appl Genet. 104(2-3): 209–213. [DOI] [PubMed] [Google Scholar]
- Kennell JC, Moran JV, Perlman PS, Butow RA, Lambowitz AM. 1993. Reverse transcriptase activity associated with maturase-encoding group II introns in yeast mitochondria. Cell 73(1):133–146. [DOI] [PubMed] [Google Scholar]
- Keren I, et al. 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 71(3):413–426. [DOI] [PubMed] [Google Scholar]
- Keren I, et al. 2009. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15(12): 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knie N, Fischer S, Grewe F, Polsakiewicz M, Knoop V. 2015. Horsetails are the sister group to all other monilophytes and Marattiales are sister to leptosporangiate ferns. Mol Phylogenet Evol. 90:140–149. [DOI] [PubMed] [Google Scholar]
- Knoop V. 2010. Looking for sense in the nonsense: a short review of non-coding organellar DNA elucidating the phylogeny of bryophytes. Bryophyte Divers Evol. 31(1):51. [Google Scholar]
- Knoop V. 2011. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci. 68(4):567–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V. 2013. Plant mitochondrial genome peculiarities evolving in the earliest vascular plant lineages. J Syst Evol. 51(1):1–12. [Google Scholar]
- Kubo T, et al. 2000. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 28(13): 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Albertazzi FJ, Blazevic D, Hermann M, Bock R. 2002. Loss of the mitochondrial cox2 intron 1 in a family of monocotyledonous plants and utilization of mitochondrial intron sequences for the construction of a nuclear intron. Mol Genet Genomics 267(2):223–230. [DOI] [PubMed] [Google Scholar]
- Kuo L-Y, Li F-W, Chiou W-L, Wang C-N. 2011. First insights into fern matK phylogeny. Mol Phylogenet Evol. 59(3):556–566. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. 1993. Introns as mobile genetic elements. Annu Rev Biochem. 62:587–622. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. 2004. Mobile group II introns. Annu Rev Genet. 38:1–35. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. 2011. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harbor Perspect Biol. 3(8):a003616.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz H, et al. 2010. Introducing the plant RNA editing prediction and analysis computer tool PREPACT and an update on RNA editing site nomenclature. Curr Genet. 56(2):189–201. [DOI] [PubMed] [Google Scholar]
- Lenz H, Knoop V. 2013. PREPACT 2.0: predicting C-to-U and U-to-C RNA editing in organelle genome sequences with multiple references and curated RNA editing annotation. Bioinform Biol Insights 7:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippok B, Brennicke A, Wissinger B. 1994. Differential RNA editing in closely related introns in Oenothera mitochondria. Mol Gen Genet. 243(1):39–46. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. 2012. The mitochondrial genome of the lycophyte Huperzia squarrosa: the most archaic form in vascular plants. PloS One 7(4):e35168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon JM. 1998. The recent origins of spliceosomal introns revisited. Curr Opin Genet Dev. 8(6):637–648. [DOI] [PubMed] [Google Scholar]
- Lopez L, Picardi E, Quagliariello C. 2007. RNA editing has been lost in the mitochondrial cox3 and rps13 mRNAs in Asparagales. Biochimie. 89(1):159–167. [DOI] [PubMed] [Google Scholar]
- Marchfelder A, Brennicke A, Binder S. 1996. RNA editing is required for efficient excision of tRNA(Phe) from precursors in plant mitochondria. J Biol Chem. 271(4):1898–1903. [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard L, Kumar R, Remacle C, Small I. 1996. RNA editing of larch mitochondrial tRNA(His) precursors is a prerequisite for processing. Nucleic Acids Res. 24(16):3229–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM. 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J. 20(24):7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil BA, Semper C, Zimmerly S. 2016. Group II introns. Versatile ribozymes and retroelements. WIREs RNA 7(3): 341–355. [DOI] [PubMed] [Google Scholar]
- Michel F, Dujon B. 1983. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 2(1):33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Ferat JL. 1995. Structure and activities of group II introns. Annu Rev Biochem. 64:435–461. [DOI] [PubMed] [Google Scholar]
- Michel F, Lang BF. 1985. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature 316(6029): 641–643. [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. 1989. Comparative and functional anatomy of group II catalytic introns – a review. Gene 82(1):5–30. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, Haeseler A, von Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, Knie N, Knoop V. 2014. Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20(10):1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A. 2005. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell 17(1):241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Logsdon JM. 1991. The recent origins of introns. Curr Opin Genet Dev. 1(4):470–477. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. 1998. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol. 8(3):301–308. [DOI] [PubMed] [Google Scholar]
- Qiu YL, Cho Y, Cox JC, Palmer JD. 1998. The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394(6694):671–674. [DOI] [PubMed] [Google Scholar]
- Ran J-H, Gao H, Wang X-Q. 2010. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol Phylogenet Evol. 54(1):136–149. [DOI] [PubMed] [Google Scholar]
- Regina TMR, Quagliariello C. 2010. Lineage-specific group II intron gains and losses of the mitochondrial rps3 gene in gymnosperms. Plant Physiol Biochem. 48(8):646–654. [DOI] [PubMed] [Google Scholar]
- Rodríguez F, Oliver JL, Marín A, Medina JR. 1990. The general stochastic model of nucleotide substitution. J Theor Biol. 142(4):485–501. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Doolittle WF. 1993. Molecular evolution. Why introns-in-pieces? Nature 364(6435):289–290. [DOI] [PubMed] [Google Scholar]
- Rogers JH. 1990. The role of introns in evolution. FEBS Lett. 268(2): 339–343. [DOI] [PubMed] [Google Scholar]
- Rothfels CJ, et al. 2012. Overcoming deep roots, fast rates, and short internodes to resolve the ancient rapid radiation of eupolypod II ferns. Syst Biol. 61(3):490–509. [DOI] [PubMed] [Google Scholar]
- Rothfels CJ, et al. 2015. The evolutionary history of ferns inferred from 25 low-copy nuclear genes. Am J Bot. 102(7):1089–1107. [DOI] [PubMed] [Google Scholar]
- Rothfels CJ, Schuettpelz E. 2014. Accelerated rate of molecular evolution for vittarioid ferns is strong and not driven by selection. Syst Biol. 63(1):31–54. [DOI] [PubMed] [Google Scholar]
- Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V. 2009. RNA editing: only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol Genet Genomics 281(5):473–481. [DOI] [PubMed] [Google Scholar]
- Schuettpelz E, Schneider H, Huiet L, Windham MD, Pryer KM. 2007. A molecular phylogeny of the fern family Pteridaceae: assessing overall relationships and the affinities of previously unsampled genera. Mol Phylogenet Evol. 44(3):1172–1185. [DOI] [PubMed] [Google Scholar]
- Schwarz G. 1978. Estimating the dimension of a model. Ann Stat. 6(2):461–464. [Google Scholar]
- Simon DM, et al. 2008. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA 14(9):1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. 2010. Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics 185(4):1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S. 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7(8):1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. 1997. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 15(1):57–61. [DOI] [PubMed] [Google Scholar]
- Vangerow S, Teerkorn T, Knoop V. 1999. Phylogenetic information in the mitochondrial nad5 gene of pteridophytes: RNA editing and intron sequences. Plant Biol. 1(2):235–243. [Google Scholar]
- Wikström N, Pryer KM. 2005. Incongruence between primary sequence data and the distribution of a mitochondrial atp1 group II intron among ferns and horsetails. Mol Phylogenet Evol. 36(3):484–493. [DOI] [PubMed] [Google Scholar]
- Zhang L, et al. 2015. A global plastid phylogeny of the brake fern genus Pteris (Pteridaceae) and related genera in the Pteridoideae. Cladistics 31(4):406–423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.