Abstract
Regulation of NF-kB nuclear translocation and stability is central to mounting an effective innate immune response. Here, we describe a novel molecular mechanism controlling NF-kB-dependent innate immune response. We show that a previously unknown protein, termed as Charon, functions as a regulator of antibacterial and antifungal immune defense in Drosophila. Charon is an ankyrin repeat-containing protein that mediates PARP-1-dependent transcriptional responses downstream of the innate immune pathway. Our results demonstrate that Charon interacts with the NF-kB orthologue Relish inside perinuclear particles and delivers active Relish to PARP-1-bearing promoters, thus triggering NF-kB/PARP-1-dependent transcription of antimicrobial peptides. Ablating the expression of Charon prevents Relish from targeting promoters of antimicrobial genes and effectively suppresses the innate immune transcriptional response. Together, these results implicate Charon as an essential mediator of PARP-1-dependent transcription in the innate immune pathway. Thus, our results are the first to describe the molecular mechanism regulating translocation of the NF-kB subunit from cytoplasm to chromatin.
Keywords: CG5118, IMD, Relish, NF-κB, PARP-1, Drosophila, Innate immunity
INTRODUCTION
Innate immune response is common throughout the animal kingdom, playing a central role in host defense against pathogenic diseases (1). A key function of innate immunity is recognition of microbial agents by host cell receptors, such as Toll-like receptors (TLRs). This elicits a cascade of signaling pathways to induce the rapid transcription of genes encoding antimicrobial peptides and/or proinflammatory cytokines (2, 3). In Drosophila, cleaved Spatzle fragments function as cytokine ligands for the Toll receptor. The fly sense system then processes the Toll-Spatzle complex, detecting the invading fungi and Gram-positive bacteria (4, 5). Once activated, the Toll receptor induces phosphorylation of Cactus, a homolog of mammalian inhibitors of NF-kB (IKB), further degrading it by polyubiquitination (6). Degradation of IKB then causes the release and translocation of NF-kB transcriptional factors (Dorsal and DIF) from the cytoplasm to the nucleus where DIF and/or Dorsal induce the transcription of antimicrobial peptide genes by binding to the promoters of these genes, such as Drosomycin (7, 8). The immune deficiency (IMD) pathway is integral to innate immune response against Gram-negative bacteria in Drosophila (9). In the IMD pathway, a receptor named peptidoglycan recognition protein (PGRP) recognizes peptidoglycan (PGN) present in the cell wall of most Gram-negative bacteria and recruits the adaptor protein IMD to initiate signaling (10–13). The IMD pathway culminates in the cleavage of Relish (Rel68; orthologous to mammalian NF-κB) and the translocation of the transactivating domain of Relish into the nucleus for the transcriptional induction of antimicrobial peptide genes, such as Diptericin (14, 15). The existence of crosstalk between Immune melanization protease (IMP) and Toll pathways has been reported, and Rel68 protein is suggested to control both pathways in Drosophila (16). The common theme of both Toll and IMP pathways is the rapid transcriptional induction of antimicrobial peptide genes through NF-κB transcription factors.
Poly(ADP-ribose) Polymerase 1 (PARP-1) has proven to be integral to NF-κB-mediated immune response (17). The failure of mouse PARP-1−/− macrophages to produce TNF-α in response to LPS stimulation suggests that PARP-1 is required for NF-κB-dependent transcriptional induction (18). In addition to its role in the transcriptional induction of mammalian proinflammatory cytokine genes, it has been shown that PARP-1 is required for the expression of antimicrobial peptides, such as Diptericin (Dpt) and Drosomycin (Drs), after bacterial infection in Drosophila (19). Since Drs expression is induced by the Toll pathway and Dpt by the IMD pathway, PARP-1 is involved in NF-κB-mediated transcription of antimicrobial peptide genes. However, it is not clear whether PARP-1 induces the transcription of antimicrobial peptide genes by direct binding to NF-κB or through the regulation of chromatin structure via its enzymatic activity.
Here we report the discovery of the CG5118 gene which encodes a previously unknown, evolutionarily conserved component of innate immune response. We named the CG5118 gene product Charon after the boatman in Greek mythology who delivered dead spirits across the River Styx to the underworld. Charon encodes an ankyrin domain-containing nuclear and perinuclear protein that interacts with Relish and delivers this transcription factor to PARP-1-occupied promoters to mediate transcription of antimicrobial genes, including Dpt and Drs. In Drosophila, Charon mediates PARP-1-dependent binding of Relish to promoters and transcription of Dpt and Drs genes encoding antimicrobial peptides. Previously, only two PARP-1-dependent mechanisms of gene expression regulation had been reported: 1) pADPr mediation of chromatin loosening (19–22) and 2) pADPr/hnRNPA1 regulation of RNA fate (23–25). Unlike those two pathways, PARP-1/Relish/Charon-dependent innate immune response does not require PARP-1 enzymatic activity. Our findings thus represent a third mechanism of PARP-1-dependent transcriptional regulation.
MATERIALS AND METHODS
Drosophila strains and genetics
Genetic markers are described in FlyBase 1999 (26), and stocks were obtained from the Bloomington Stock Center, except as indicated. pP{w1, UAST::PARP-1-DsRed}, called UAS::PARP-1-DsRed, was described in (19). The following GAL4 driver strains were used: 69B-GAL4 (27), arm::GAL4 (Bloomington stock no. 1560), da-Gal4 [a gift of A. Veraksa] and Act-Gal4 [Bloomington stock no. 4414]. Balancer chromosomes carrying Kr::GFP, i.e., TM3, P{w1, Kr-GFP} and FM7i, P{w1, Kr-GFP}, were used to identify heterozygous and homozygous parg27.1 and parpC03256 (28). siRNA transgenic Drosophila stocks #106292 (siRNA1) and #34937 (siRNA2) were obtained from VDRC (29). Transgenic stock expressing Diptericine-lacZ and Drosomycin-GFP reporter genes were obtained from Dr. D. Ferrandon (30,31).
Construction of transgenic Drosophila
To construct UAS::CG5118-EGFP, we generated full-length genomic fragment of CG5118 locus (Fig. 1A and Supplemental Fig. 1A) using PCR. We used wild-type Drosophila genomic DNA as a template for PCR. The resulting PCR products were cloned through The Drosophila Gateway™ Vector Cloning System (Carnegie Institution of Washington) into the corresponding vector for Drosophila transformation. Transformation was performed as described (32).
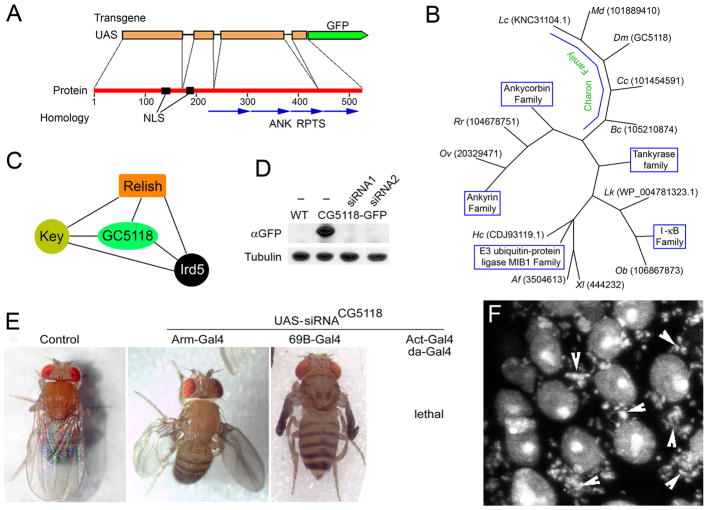
Figure 1. Charon (CG5118) gene encodes new component of IMD pathway.
A. Structure of Charon gene and protein and transgenic construct. ANK RPTS – Ankyrin repeats. NLS – nuclear localization signal. B. Evolutional tree of Charon homologs. Only representative sequences or known protein families are shown. Two first letters represent an organism, followed by sequence ID. Xl – Xenopus laevis; Af - Aspergillus fumigatus; Lc - Lucilia cuprina; Hc - Haemonchus contortus; Dm - Drosophila melanogaster; Rr - Rhinopithecus roxellana; Md - Musca domestica; Lk - Leptospira kirschneri; Cc - Ceratitis capitata; Bc - Bactrocera cucurbitae; Ob - Octopus bimaculoides; Ov - Opisthorchis viverrini. C. Charon protein interacts with components of IMD pathway [36,41]. D. The expression of Charon-GFP transgenic reporter and siRNA transgenes against Charon. Two different siRNA constructs against CG5118 were expressed using ubiquitous Act-Gal4 drivers in CG5118-GFP-expressing Drosophila. Total protein extracts from third instar larvae were subjected to Western blot analysis using anti-GFP antibody. Tubulin antibody was used as a loading control. E. Charon is required for Drosophila development. siRNA against CG5118 was expressed using tissue-specific (Arm-Gal4, 69B-Gal4) or ubiquitous (Act-Gal4, da-Gal4) drivers in wild-type flies. F. Charon is required for antifungal protection of Drosophila. Larval salivary gland cells expressing siRNA against Charon locus often become infected with bacteria and fungi (arrowheads). Nuclei and pathogens are stained with Draq5.
Western blot
The following antibodies were used for immunoblotting assays: anti-pADPr (Rabbit 1:4000, Calbiochem, #528815), anti-pADPr (Mouse monoclonal, 1:500, Tulip, #1020), anti-B-actin (Mouse monoclonal 1:5000, Sigma, #A5441), pAB anti-histone H4 (1:1000; Santa Cruz Biotech), mouse anti-Tubulin (1:1000,Sigma, #M1A), rabbit anti-GFP (Torrey Pines Biolabs, #TP401, 1:1000), anti-GFP (Mouse monoclonal, BD, #632380, 1:5000), mouse anti-Relish (C21F3,DSHB), rabbit anti-Relish (RayBiotech, #RB-14-0004), rabbit anti-Hrp38/hnRNP A1(1:10000, a gift from Dr. Dr. J. A. Steitz) (33), Rabbit anti-DsRed (1:500, Clontech), and mouse anti-LacZ antibody (1:1000, Promega #Z378A). Western blotting was done using the detection kit from Amersham/GE Healthcare (#RPN2106), according to manufacturer’s instructions.
Polytene chromosome immunostaining
Preparation and immunostaining of polytene chromosome squashes were performed exactly as described (19). The primary antibodies used were anti-GFP (Rabbit, Torrey Pines Biolabs, #TP401, 1:400), rabbit anti-Relish (RayBiotech, #RB-14-0004, 1:50). The secondary antibodies used were goat anti-rabbit Alexa-488, goat anti-rabbit Alexa-633, goat anti-mouse Alexa-488, and goat anti-mouse Alexa-633 (Molecular Probes (1:400). Slides were mounted in Vectashield (Vector Laboratories, Burlingame, CA) with propidium iodide at 0.05 mg/ml for DNA staining.
Immunohistochemistry
Immunochemistry was performed as described in (34).
Mononucleosome Chromatin Immunoprecipitation
Nuclei were isolated from ~200 adult flies as described in (35) with some modifications. In one 1.5ml Eppendorf tube, 50 larvae were homogenized with 300ul A1(60mM KCI,15mM NaCI, 5mM MgCI2, 15mM Tris-HCI (pH 7.5), 0.5% Triton X-100, 0.1mM EGTA, 0.5mM DTT and 25X complete protease inhibitors (Roche) EDTA-free/0.3M sucrose). The homogenates were passed over two layers of Miracloth tissue (Calbiochem) and centrifuged for 5 minutes at 8000g at 4°C. The pellet resuspended in 300ul A1 (0.5% Triton X-100)/0.3M sucrose) was loaded into a sucrose gradient (800ul A1 (0.5% Triton X-100)/0.8M sucrose + 150ul A1 (0.5% Triton X-100)/0.3M sucrose)) and further centrifuged for 6 minutes at 8000g at 4°C. The nuclei pellet resuspended in 100 ul micrococcal nuclease digestion buffer (60mM KCI, 15mM NaCI, 5mM MgCI2, 15mM Tris-HCI (pH 7.5), 1mM CaCI2, 0.5mM DTT, 0.34M sucrose and 25X complete protease inhibitors (Roche)) was pooled together, and the chromatin DNA concentration was measured in 0.1% SDS buffer. The nuclei were digested with micrococcal nuclease (NEB) with 2 gel units per 1 ug of chromatin for 6 minutes at 37°C in MNase digestion buffer (NEB). 5mM EDTA were added to stop the reaction. Immunoprecipitation was performed as described by Umlauf et al. (http://www.epigenomenoe.net/researchtools/protocol.php?protid=22).
After MNase digestion, the nuclei reactions were centrifuged for 10 minutes at 10K at 4°C. The supernatant fraction containing about 40 ug mononucleosome chromatin DNA was precleared with 60 ul protein A agarose (Invitrogen) for 1 hour at 4°C and then incubated with 10 ug anti-GFP rabbit polyclonal antibody (Torrey Pines Biolabs) or rabbit anti-Relish antibody (RayBiotech) overnight at 4°C and further incubated with 60 ul protein A agarose (Invitrogen) for 2 hours at 4°C. The IP complexes were washed with buffer A once, buffer B twice and buffer C twice, and then eluted with 500 ul elution buffer. The eluate and 10% input were digested with 5 ul of proteinase K (20 ug/ul) (Invitrogen) for 4 hours at 50°C, and DNA was extracted using the phenol:chloroform:iso-amylalcohol (25:24:1) method and precipitated by Ethanol with 2 ul Paintpellet (Novagen) for better visualization. The DNA pellet was resuspended in 40 ul of water and then treated with 1ul RNAse (2ug/ul) (Invitrogen) for 20 minutes at room temperature.
To detect Drosomycin or Diptericin DNA in ChIP experiments, we used 2 pairs of primers, as shown below, distributed along the promoter regions of Drosomycin and Diptericin genes, which have been demonstrated to bear NF-KB binding sites (36; 37). The primer sequences were as follows: 5′ TTTCGCTTACGCTTTTCGAT′ (forward) and 5′ ATAGTGCACCGATCCCTCAG 3′ (reverse) for Drosomycin promoter; 5′ GATCCCCTGGTGGTATTTG 3′ (forward) and 5′ CTTTCCAAAAGGAATCCCCG 3′ (reverse) for Diptericin promoter. Real-time PCR assays were performed using Power Sybr® Green PCR master mix and the StepOnePlus Real Time PCR System (Applied Biosystems). Cycling conditions were 95°C, 10 min, followed by 40 (3-step) cycles (95°C, 15 sec; 63°C, 30 sec; 72°C, 30 sec). Reactions were done in triplicate. All the experiments were repeated twice, and standard deviation was calculated.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Figures S1 – S4.
RESULTS
Charon encodes an evolutionarily conserved protein
Drosophila CG5118 gene (Fig. 1A and Supplemental Fig. 1A) encodes a putative protein containing 524 amino acids and four Ankyrin repeats (Supplemental Fig. 1B). It has close homologues broadly represented in the many genomes of the animal kingdom (Fig. 1B). The most conserved homologs are found in other insects (Supplemental Fig. 2). As shown in FlyBase (1999), the Charon protein is ubiquitously expressed in all tissues of D. melanogaster. Previous studies (38,39) reported that Charon interacts with components of the IMD pathway, including Kenny, Ird5 and Relish (Fig. 1C), suggesting that Charon is an as-yet uncharacterized component of this pathway. To study the functions of Drosophila Charon protein, we fused the genomic fragment, including four exons and three introns of Drosophila Charon gene, to a GFP tag under control of UASt promoter (Fig. 1A) and generated transgenic flies expressing Charon-GFP. We also used knockdown transgenic constructs to produce different nonoverlapping siRNAs directed against Charon mRNA. The expression of these siRNAs effectively eliminated Charon protein (Fig. 1D) and induced identical defects in Drosophila development (Fig. 1E). The ubiquitous expression of knocked down transgenes arrests the fly’s development at late third instar or pupal stages, while tissue-specific expression suggests organ-specific defects. These observations indicate that Charon is vital to Drosophila development. Besides developmental defects, Charon knockdowns demonstrate high sensitivity to bacterial and fungal invasion (Fig. 1F), hinting at the involvement of Charon in IMD signaling.
Charon is a nuclear, chromatin-associated protein that is regulated by PARP-1
The sequence analysis of Charon and its close homologues predicted conserved nuclear localization signals (NLS) located at 155–161aa and 191–197aa positions (Fig. 1A and Supplemental Fig. 1AB). To monitor the subcellular localization of Charon protein, we expressed UAS-Charon-GFP reporter using ubiquitous GAL4 driver. An immunoblot analysis using the antibody to a GFP tag (αGFP) showed that the transgene produces a single 79 kDa protein (Fig. 1C and Supplemental Fig. 3). This expression is well tolerated by flies and has no effects on development, viability or health. Confocal microscopy of dissected Drosophila tissue identified Charon as a nuclear, chromatin-associated protein (Fig. 2A). Quantification revealed that Charon occupies approximately 400 loci in the euchromatic fractions of polytene chromosomes in Drosophila salivary gland tissue (Supplemental Fig. 3A). Additionally, significant amounts of Charon protein are located in soluble nucleoplasm (Fig. 2A, arrow) and in perinuclear bodies (Fig. 2A, arrowheads).
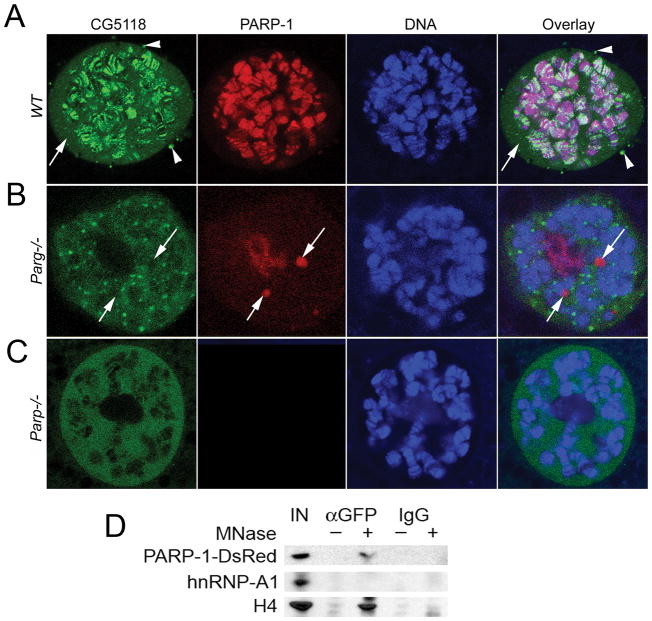
Figure 2. Charon encodes a PARP-1-dependent chromatin-associated protein.
A–C. Charon protein is a nuclear protein. In wild-type Drosophila tissues (A), it bound to ~ 400 sites in chromatin, presenting in soluble nucleoplasm (arrow) and accumulating in cytoplasmic perinuclear bodies (arrowheads). It accumulates in nucleoplasmic bodies in parg27.1 mutants (B) and in soluble nucleoplasm in parp-1C03265 mutants (C). D. Charon protein interacts with PARP-1 protein in chromatin. Co-immunoprecipitation experiments: nuclei were purified from animals expressing CG5118-GFP and PARP-1-DsRed proteins; samples were treated with MNase (+) to digest chromatin to mononucleosomal state; untreated nuclei were used as a control (−); chromatin was extracted, and protein complexes were precipitated using anti GFP antibody or IgG; samples were subjected to Western blot analysis using anti-DsRed, anti hnRNP A1 and anti-histone H4 antibodies.
We found that mutating the components of poly(ADP-ribosyl)ating pathways completely disrupts the localization of Charon. Drosophila that lack the Poly(ADP-ribose) glycohydrolase (PARG) gene (parg27.1) accumulate Charon in dispersed nucleoplasmic bodies (Fig. 2B), while mutants in the PARP-1 encoding gene (parpC03256) retain Charon only in soluble nucleoplasm (Fig. 2C). These data establish that the distribution of cellular Charon is dependent on PARP-1. Unlike other PARP-1-dependent proteins, Charon does not localize to Cajal bodies in parg27.1 mutants (Fig. 2B, arrows) and is not covalently bound with pADPr (Supplemental Fig. 3B). Charon does not colocalize to components of the previously described PARP-1-dependent hnRNP pathway (Supplemental Fig. 4A) and does not demonstrate any effect on PARP-1-dependent heat shock transcriptional response (Supplemental Fig. 4B). Co-immunoprecipitation experiments revealed that Charon interacts with PARP-1 in chromatin, but not in soluble nucleoplasm (Fig. 2D). Taken together, these data strongly support the hypothesis that Charon is an integral part of a novel PARP-1-dependent pathway based on the vitality it affords Drosophila development and antimicrobial response. The Charon/PARP-1 axis also demonstrates the dependency of PARP-1 protein interactions in chromatin.
Charon protein is required for Relish-dependent transcriptional response
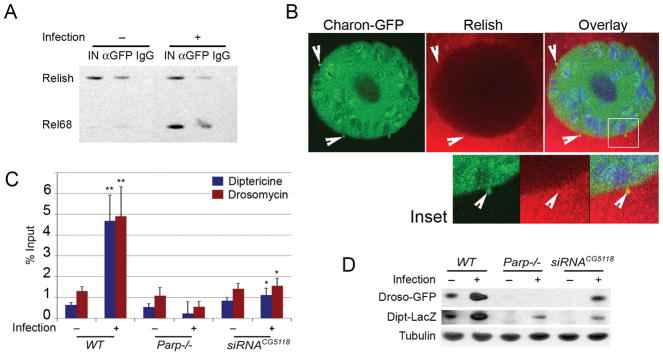
To study the roles of Charon in antimicrobial response, we first needed to establish its interaction with Relish protein. To accomplish this, we compared Charon/Relish interaction in unchallenged animals and Drosophila larvae exposed to E. coli. Co-immunoprecipitation experiments show that Charon interacts with full-length Relish, as well as with C-terminal transactivation domain (Fig. 3A). Immunostaining of dissected Drosophila tissues established that the interaction with full-length Relish protein only occurred in the cytoplasm of perinuclear bodies (Fig. 3B, Inset) in a manner that precedes the activation of signaling pathways and cleaving of Relish (14, 15) into its Rel68 fragment, which appears only after signal pathway activation (14, 15). These observations suggest that the interaction of Charon and Relish occurs in perinuclear bodies prior to pathway activation and that subsequent to such activation, Charon accompanies Rel68 into nucleoplasm and chromatin. Relish protein controls the transcription of antimicrobial peptides by binding promoters and inducing transcription (40, 41). We then investigated the effect of parp-1 or Charon mutation on Relish protein binding. As expected, chromatin immunoprecipitation (ChIP) assay, using anti-Relish antibody, shows that Relish protein binds to the promoters of Dpt and Drs genes following bacterial infection in wild-type animals (Fig. 3C). Mutation of parp-1 or Charon removes Relish from the Dpt and Drs promoter (Fig. 3C) and, hence, impairs the expression of antibacterial and fungal peptides following infection (Fig. 3D and Supplemental Fig.S4C).
Figure 3. Charon controls Relish protein delivery from cytoplasm to chromatin.
A. Charon protein interacts with full-length Relish prior to pathway activation, as well with Rel68 after infection. Co-immunoprecipitation assay using anti-GFP and IgG antibody is shown. B. Charon and full- length Relish are colocalized in perinuclear bodies (arrows). Immunostaining of Drosophila salivary glands using monoclonal anti-GFP (green); polyclonal anti-Relish (red) antibodies: a single cell is shown; DNA is blue. Arrowheads show perinuclear bodies. Inset: Magnification of the perinuclear body outlined with white rectangle. C. Mutating PARP-1 or Charon abolishes Rel68 binding in promoter of Dpt and Drs locus. ChIP assay using polyclonal anti-Relish antibody. *: p≤0.05; **: p≤0.01. D. Charon and PARP-1 are required to express innate immunity genes. Levels of NF-κB-dependent innate immunity gene reporters Dipt-LacZ and Droso-GFP were quantitated using epitope-specific antibodies; Drosophila larvae expressing reporters were cultured 60 hours with a sublethal dose of E. coli bacteria (+). Western blot reveals that both immunity genes are strongly induced by infection, relative to Tubulin control, in wild-type (wt), but not in Parp-1 or Charon mutant larvae.
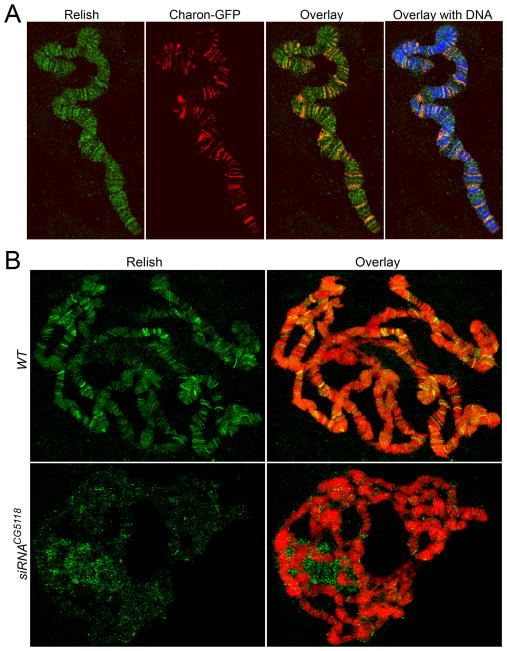
We next examined the co-occupancy of Charon and Relish in Drosophila chromatin. For this, we performed immunostaining of polytene chromosomes in Drosophila salivary gland tissue for Relish and Charon-GFP. Notably, about 60% of Charon-positive sites are occupied by Relish (Fig. 4A). In wild-type salivary glands, Relish bound approximately 100 sites following infection (Fig. 4B). However, when we disrupted the expression of Charon protein, we found that the binding of Relish was disrupted on a genome-wide scale (Fig. 4C). This is compelling evidence that Charon controls Relish protein targeting to promoters.
Figure 4. Charon is required for Rel68 protein binding in salivary gland polytene chromosomes.
A. Charon and Relish proteins are co-localized in chromatin: Drosophila larvae expressing Charon-GFP were cultured for 60hrs with E.coli bacteria; salivary glands were dissected from third instar larvae, squashed and stained with monoclonal anti-GFP (Red) and polyclonal anti-Relish (green) antibodies; DNA was detected using propidium iodide (blue). B–C. Charon is required for Rel68 protein binding in chromatin: wild type (WT) or Charon knockdown (siRNACG5118) larvae were treated as described above; the localization of Relish in polytene chromosomes was detected using the polyclonal anti-Relish (green) antibodies; DNA was detected using propidium iodide (red).
DISCUSSION
Here we demonstrated that Charon is an essential component of IMD signaling and necessary for Relish translocation to nucleoplasm and to PARP-1-bound promoters following cleavage of Relish protein. Our data reveal that Charon is required for Rel68 binding to promoters and the expression of Relish-dependent antimicrobial peptides after bacterial infection.
Relish is a key transcription factor in the induction of antimicrobial peptides, and our data suggest that Charon is a critical partner of Rel68 in the nucleus for transcription activation of antimicrobial peptide (AMP) genes. Full-length Relish undergoes endoproteolytic cleavage by the caspase-8 Dredd to generate two isoforms (Rel68 and Rel49) (14, 15). Once Rel68 translocates into the nucleus, it serves as a transcription activator by binding to promoters of many antimicrobial genes, such as Drosomycin (36, 41). In addition, phosphorylation of Rel68 at Ser-528 and -529 by the IKK complex comprised of IRD5, a catalytic kinase subunit, and Kenny, a regulatory subunit, is also critical for Rel68 transcription activation (42). Our findings suggest that Charon interacts and colocalizes with Rel68 in the promoters of AMP genes. Previous studies showed that a nuclear protein, Akirin, is required for Rel68-dependent immune response (43). However, it appeared that Akirin did not interact directly with Relish in the nucleus (43). Recent studies showed that Akirin regulates chromatin structure by recruiting the Osa-containing SWI/SNF-like Brahma complex (BAP) to AMP genes (44). In future studies, we will investigate how Charon works with Akirin in a concerted manner to regulate the expression of AMP genes.
Our study further reveals the role of PARP-1 in innate immune response. Previous studies showed that PARP-1 is essential for NF-kB-dependent immune response genes in Drosophila (19) and in mammals (17). However, no consensus has been reached on the mechanism underlying the control of PARP-1 over immune response, especially from the perspective of its enzymatic activity. The earlier study suggested that PARP-1 may directly bind to both p50 and p65 subunits of NF-κB for transcription activation, even though its enzymatic activity and DNA-binding domain had no effect on this process (45). A recent study showed that the cleavage of PARP-1 by Caspase-7 releases it from the promoters of some NF-κB-dependent genes, permitting transcription upon LPS stimulation (46). However, several studies have also demonstrated that PARP-1 enzymatic activity is very important for transcription of NF-κB-dependent genes (46–48). It has been demonstrated that LPS can activate PARP-1 to modify histones through poly(ADP-ribosyl)ation, destabilizing chromatin structure and inducing NF-κB-dependent promotion of proinflammatory cytokines such as IL-1 (47, 48). Data presented herein show that PARP-1 is required for proper Charon localization in chromatin and Relish binding to the promoter region of Drosomycin gene after infection. However, it appears that PARP-1 enzymatic activity is not required for Charon localization in chromatin. Therefore, our results support the model in which PARP-1 directly interacts with Relish and Charon to recruit these two proteins to the promoter region of the AMP gene (Fig. 5). The IKK kinase complex, comprised of Kenny and Ird5, is a key regulator of NF-κB. Proteomics studies have revealed that it forms a complex with Relish and Charon in the unchallenged state (38; 39). Interestingly, structural studies have shown that the IKK complex binds to Relish through six Ankyrin repeats of its C-terminus (49). It turns out that Charon also contains four Ankyrin repeats. Therefore, it is possible that the IKK complex also binds to Charon through its Ankyin repeats. These findings allow us to propose that Charon is a component of the IKK complex with inactive Relish in the cytoplasm in the unchallenged state (Fig. 5A). However, once challenged by Gram-negative bacteria, the IKK complex helps to cleave Relish to produce the Rel68 fragment. Then, as a partner of Rel68, Charon binds with PARP-1 to recruit Rel68 to the promoter region of AMP genes for transcription activation (Fig. 5B).
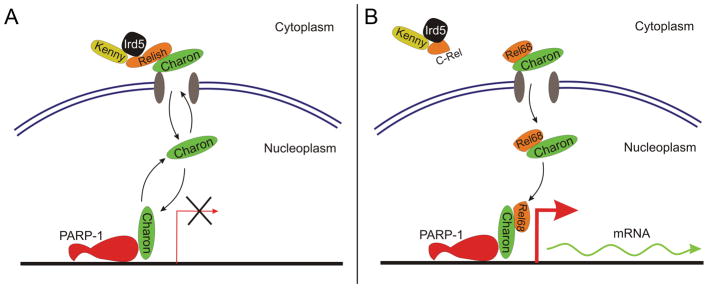
Figure 5. Charon protein delivers Rel68 to PARP-1-regulated promoters.
A. Charon interacts with Relish-Kenny-Imd5 complex in perinuclear bodies and shuttles between them and PARP-1-bearing promoters. B. Upon IMD pathways activation Relish protein cleaved into C-terminal domain, which remains bound with inhibitor-complex and Rel68 N-terminal domain which remains bound with Charon but looses interaction with Kenny/Imd5. Under control of Charon NLS Rel68 is transported to chromatin and by mediation of Charon localized into PARP-1 occupied promoter. Binding of Relish in promoters triggers transcription on innate immune genes.
PARP-1 has been linked to several human inflammatory diseases, such as Rheumatoid arthritis (RA), inflammatory bowel disease and asthma (50). PARP-1−/− mice had lower expression level of IL-1β and the chemokine MCP-1 in a model of arthritis induced by anti-collagen antibodies (CAIA) (51). Indeed, PARP-1 inhibition significantly reduced the production of TNF-induced cytokine and chemokine expression in fibroblast-like synoviocytes (FLS) derived from patients with RA (52). A recent study has demonstrated that PARP-1 knockout mice were resistant to dextran-sulfate sodium (DSS)-induced colitis, which is associated with changing transcriptional profiles, colonic microbiota and the frequency of mucosal CD4(+)CD25(+) Foxp3(+) regulatory T cells in the colon (53). It has also been reported that PARP-1−/− mice and PARP-1 inhibition by Olaparib obstruct development of asthma-like traits in mice in response to house dust mite (HDM) as a result of reduced production of T helper 2 (Th2) cytokine (54). Our finding that Charon controls the production of PARP-1-mediated AMP implies that the contribution of PARP-1 to these inflammatory diseases should be examined to determine if decreased expression of AMP from PARP-1 knockout could affect the pathogenesis of inflammatory diseases. Moreover, in human cells, our previous study has shown that PARP-1 binding sites in chromatin often coincide with NFATC2 transcriptional factor binding sites on a genome-wide scale (55). NFATC2 function is linked to the NF-κB pathway and controls the expression of IL2 in T cells (56). These data suggest that PARP-1/NF-κB-dependent transcriptional control is broadly conserved across evolution.
Besides its role in the induction of the innate immunity response, Charon is also required for development in Drosophila because RNAi knockdown of Charon causes developmental defects or total lethality, depending on the use of GAL4 drivers. Even in the unchallenged condition, Charon is localized in many transcriptional activation puffs (Supplemental Fig. 3), suggesting a developmental role in transcription control. Besides ankyrin repeats, Charon is lack of the conserved function domains although it was identified as a component in the transcription network (24). Therefore, future work should investigate how Charon behaves as a transcription factor.
Supplementary Material
Acknowledgments
We thank Dr. D. Ferrandon for the Drosomycin-GFP- and Diptericin-LacZ-expressing Drosophila line. Drs. David Wiest and Siddharth Balachandran contributed valuable comments on the manuscript.
The research was supported by grants from the National Institutes of Health (R01 GM077452 and R01 DK082623) to A.V.T.
Footnotes
DISCLOSURES
The authors have no financial conflicts of interest.
References
- 1.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clinical Microbiology Reviews. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 4.Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nature Reviews Immunology. 2007;7:862–874. doi: 10.1038/nri2194. [DOI] [PubMed] [Google Scholar]
- 5.Lindsay SA, Wasserman SA. Conventional and non-conventional Drosophila Toll signaling. Developmental & Comparative Immunology. 2014;42:16–24. doi: 10.1016/j.dci.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belvin MP, Jin Y, Anderson KV. Cactus protein degradation mediates Drosophila dorsal-ventral signaling. Genes & development. 1995;9:783–793. doi: 10.1101/gad.9.7.783. [DOI] [PubMed] [Google Scholar]
- 7.Ip YT, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 8.Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-κB factor. Genes & Development. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleino A, Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Developmental & Comparative Immunology. 2014;42:25–35. doi: 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe KM, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 11.Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–448. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 12.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 13.Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Reports. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stöven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engström Y, Maniatis T, Hultmark D. Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proceedings of the National Academy of Sciences. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Molecular and Cellular Biology. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassa PO, Hottiger MO. The functional role of poly (ADP-ribose) polymerase 1 as novel coactivator of NF-κB in inflammatory disorders. Cellular and Molecular Life Sciences. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver FJ, Ménissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly (ADP-ribose) polymerase-1 deficient mice. The EMBO Journal. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tulin A, Spradling A. Chromatin loosening by poly (ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 20.Kim MY, Mauro S, Gévry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Ji Y, Tulin AV. The roles of PARP-1 in gene control and cell differentiation. Current Opinion in Genetics & Development. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas CJ, Kotova E, Andrake M, Adolf-Bryfogle J, Glaser R, Regnard C, Tulin AV. Kinase-mediated changes in nucleosome conformation trigger chromatin decondensation via poly (ADP-ribosyl) ation. Molecular Cell. 2014;53:831–842. doi: 10.1016/j.molcel.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji Y, Tulin AV. Poly (ADP-ribosyl) ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Research. 2009;37:3501–3513. doi: 10.1093/nar/gkp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji Y, Tulin AV. Poly (ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nature communications. 2012;3:760. doi: 10.1038/ncomms1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji Y, Jarnik M, Tulin AV. Poly (ADP-ribose) glycohydrolase and poly (ADP-ribose)-interacting protein Hrp38 regulate pattern formation during Drosophila eye development. Gene. 2013;526:187–194. doi: 10.1016/j.gene.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.FlyBase. The FlyBase database of the Drosophila genome projects and community literature. Nucl Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp AV, Yang M, Glover D, Kaiser K, Palter K. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Developmental Dynamics. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 28.Casso D, Ramirez-Weber FA, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mechanisms of Development. 1999;88:229–232. doi: 10.1016/s0925-4773(99)00174-4. [DOI] [PubMed] [Google Scholar]
- 29.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 30.Ferrandon D, Jung AC, Criqui MC, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart JM, Hoffmann JA. A drosomycin–GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. The EMBO Journal. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun A, Hoffmann JA, Meister M. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc Natl Acad Sci USA. 1998;95:14337–14342. doi: 10.1073/pnas.95.24.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 33.Borah S, Wong AC, Steitz JA. Drosophila hnRNP A1 homologs Hrp36/Hrp38 enhance U2-type versus U12-type splicing to regulate alternative splicing of the prospero twintron. PNAS. 2009;106:2577–2582. doi: 10.1073/pnas.0812826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotova E, Lodhi N, Jarnik M, Pinnola AD, Ji Y, Tulin AV. Drosophila histone H2A variant (H2Av) controls poly (ADP-ribose) polymerase 1 (PARP-1) activation in chromatin. Proceedings of the National Academy of Sciences. 2011;108:6205–6210. doi: 10.1073/pnas.1019644108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinnola A, Naumova N, Shah M, Tulin AV. Nucleosomal core histones mediate dynamic regulation of PARP-1 protein binding to chromatin and induction of PARP-1 enzymatic activity. J Biol Chem. 2007;282:32511–32519. doi: 10.1074/jbc.M705989200. [DOI] [PubMed] [Google Scholar]
- 36.Tanji T, Yun EY, Ip YT. Heterodimers of NF-κB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proceedings of the National Academy of Sciences. 2010;107:14715–14720. doi: 10.1073/pnas.1009473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kappler C, Meister M, Lagueux M, Gateff E, Hoffmann JA, Reichhart JM. Insect immunity. Two 17 bp repeats nesting a kappa B-related sequence confer inducibility to the Diptericin gene and bind a polypeptide in bacteria-challenged Drosophila. EMBO J. 1993;12:1561–1568. doi: 10.1002/j.1460-2075.1993.tb05800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee DY, Cho DY, Zhai B, Slattery M, Ma L, Mintseris J, Wong CY, White KP, Celniker SE, Przytycka TM, Gygi SP. Transcription factor networks in Drosophila melanogaster. Cell Reports. 2014;8:2031–2043. doi: 10.1016/j.celrep.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busse MS, Arnold CP, Towb P, Katrivesis J, Wasserman SA. A κB sequence code for pathway-specific innate immune responses. The EMBO Journal. 2007;26:3826–3835. doi: 10.1038/sj.emboj.7601798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiklund ML, Steinert S, Junell A, Hultmark D, Stöven S. The N-terminal half of the Drosophila Rel/NF-κB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Developmental & Comparative Immunology. 2009;33:690–696. doi: 10.1016/j.dci.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Ertürk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stöven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proceedings of the National Academy of Sciences. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goto A, Matsushita K, Gesellchen V, El Chamy L, Kuttenkeuler D, Takeuchi O, Hoffmann JA, Akira S, Boutros M, Reichhart JM. Akirins are highly conserved nuclear proteins required for NF-κB-dependent gene expression in drosophila and mice. Nature Immunology. 2008;9:97–104. doi: 10.1038/ni1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonnay F, Nguyen XH, Cohen-Berros E, Troxler L, Batsche E, Camonis J, Takeuchi O, Reichhart JM, Matt N. Akirin specifies NF-κB selectivity of Drosophila innate immune response via chromatin remodeling. The EMBO Journal. 2014;33:2349–2362. doi: 10.15252/embj.201488456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. Journal of Biological Chemistry. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 46.Erener S, Pétrilli V, Kassner I, Minotti R, Castillo R, Santoro R, Hassa PO, Tschopp J, Hottiger MO. Inflammasome-activated caspase 7 cleaves PARP-1 to enhance the expression of a subset of NF-κB target genes. Molecular Cell. 2012;46:200–211. doi: 10.1016/j.molcel.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Zamudio R, Ha HC. Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Molecular and Cellular Biology. 2012;32:2490–2502. doi: 10.1128/MCB.06667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martínez-Zamudio R, Ha HC. PARP-1 enhances inflammatory cytokine expression by alteration of promoter chromatin structure in microglia. Brain and Behavior. 2014;4:552–565. doi: 10.1002/brb3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs MD, Harrison SC. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 50.Ba X, Garg NJ. Signaling Mechanism of Poly(ADP-Ribose) Polymerase-1 (PARP-1) in Inflammatory Diseases. The American Journal of Pathology. 2010;178:946–955. doi: 10.1016/j.ajpath.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García S, Bodaño A, González A, Forteza J, Gómez-Reino JJ, Conde C. Partial protection against collagen antibody-induced arthritis in PARP-1 deficient mice. Arthritis research & therapy. 2006;8:R14–22. doi: 10.1186/ar1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García S, Bodaño A, Pablos JL, Gómez-Reino JJ, Conde C. Poly (ADP-ribose) polymerase inhibition reduces tumor necrosis factor-induced inflammatory response in rheumatoid synovial fibroblasts. Annals of the rheumatic diseases. 2008;67:631–637. doi: 10.1136/ard.2007.077040. [DOI] [PubMed] [Google Scholar]
- 53.Larmonier CB, Shehab KW, Laubitz D, Jamwal DR, Ghishan FK, Kiela PR. Transcriptional reprogramming and resistance to colonic mucosal injury in Poly (ADP-ribose) polymerase 1 (PARP1)-deficient mice. Journal of Biological Chemistry. 2016;291:8918–30. doi: 10.1074/jbc.M116.714386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghonim MA, Pyakurel K, Ibba SV, Wang J, Rodriguez P, Al-Khami AA, Lammi MR, Kim H, Zea AH, Davis C, Okpechi S. PARP is activated in human asthma and its inhibition by olaparib blocks house dust mite-induced disease in mice. Clinical Science. 2015;129:951–62. doi: 10.1042/CS20150122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lodhi N, Kossenkov AV, Tulin AV. Bookmarking promoters in mitotic chromatin: poly (ADP-ribose) polymerase-1 as an epigenetic mark. Nucleic acids research. 2014;42:7028–38. doi: 10.1093/nar/gku415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walters RD, Drullinger LF, Kugel JF, Goodrich JA. NFATc2 recruits cJun homodimers to an NFAT site to synergistically activate interleukin-2 transcription. Molecular immunology. 2013;56:48–56. doi: 10.1016/j.molimm.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.