Abstract
MLO proteins are highly conserved proteins with seven trans-membrane domains. Specific MLO genes have been linked to plant disease susceptibility. Others are involved in plant reproduction and in root thigmomorphogenesis. Functions of the remaining MLOs are still unknown. Here we performed a genome-wide survey of the MLO family in eight legume species from different clades of the Papillionoideae sub-family. A total of 118 MLO sequences were identified and characterized. Their deduced protein sequences shared the characteristics of MLO proteins. The total number of MLO genes per legume species varied from 13 to 20 depending on the species. Legume MLOs were evenly distributed over their genomes and tended to localize within syntenic blocks conserved across legume genomes. Phylogenetic analysis indicated that these sequences clustered in seven well-defined clades. Comparison of MLO protein sequences revealed 34 clade-specific motifs in the variable regions of the proteins. Comparative analyses of the MLO family between legume species also uncovered several evolutionary differences between the tropical legume species from the Phaseoloid clades and the other legume species. Altogether, this study provides interesting new features on the evolution of the MLO family. It also provides valuable clues to identify additional MLO genes from non-sequenced species.
Grain and forage legumes are among the most important crops worldwide for both animal and human consumptions1. They are also important players of sustainable agriculture2. Their capacity to fix atmospheric nitrogen allows them to grow in poor soils without application of nitrogenous fertilizers. As a consequence, they contribute to reduce both fossil energy requirement and greenhouse gas emission3. Inclusion of legume crops in rotation impacts positively on subsequent crop production2. However, legume yield is constantly threatened by fungal diseases4.
Powdery mildew emerged as one of the most widespread and damaging legume diseases4. One of the most efficient and durable powdery mildew resistance mechanisms was originally found in barley5,6. Lines carrying homozygous recessive alleles at the Mlo locus showed an efficient penetration resistance to this pathogen5. mlo-based resistance is one of the few examples of monogenic traits that confer broad spectrum resistance in the field6,7,8. Recently, mlo-based resistance has been identified in other crops including pea (er1)9,10and tomato (ol-2)11. Penetration resistance was also detected in other legumes including, Medicago truncatula12 and Lathyrus belinensis13. Although the genetic base controlling the resistance in these species is not known, their phenotypes are reminiscent to that of mlo-based resistant accessions. It is thus possible that they arose from mutations in one MLO gene. Other species may, thus, contain natural mlo mutants that could be very useful to breed crops for resistance to powdery mildew.
The barley MLO is a seven trans-membrane domain protein that localizes at cell plasma membrane5. This gene belongs to a highly conserved family found in both monocots and eudicots14. To date, the total number of MLO genes identified varied from 11 to 31 according to the plant species15,16 (Table 1). The biological function of most MLO genes remains largely unknown. Phylogenetic analyses classified these genes in 6 to 8 clades14,17. All MLO genes with a function in powdery mildew susceptibility clustered in clades IV and V14,18. A clade V MLO gene, from pepper, CaMLO2, has also been associated with susceptibility to bacterial and oomycete pathogens, and to drought19,20. In addition, the expression of some Lathyrus sativus MLO transcripts were induced shortly after rust infection in resistant genotypes, which might suggest their involvement in rust resistance21. Although the exact role of clade V MLOs is still unclear, they might interfere with the plant immune response to stresses. This is similar to the Lr34 resistance gene that protects wheat by controlling the induction of multiple defense pathways7,22. Apart for the known functions of clade IV and VMLOs, two clade I MLO genes from A. thaliana (AtMLO4 and AtMLO11) were found to play an important role in root thigmomorphogenesis23,24. Two clade III genes, AtMLO7 and OsMLO12, were also shown to be required for normal pollen tube perception and pollen hydration, respectively25,26, which suggested a role of clade III MLOs in plant reproduction14. The MLO family may thus play a wider range of functions than initially thought. Isolating and characterizing new MLOs from other plant species is thus a promising approach to get new insights on this highly conserved family.
Table 1. MLO family members of legume and non-legume species and their phylogenetic classificationa.
| Plant species | Common name | Total | Clade |
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||
| Non-legume species | ||||||||||
| Arabidopsis thaliana | Thale cress | 15 | 3 | 3 | 5 | 0 | 3 | 1 | 0 | Chen et al.36 |
| Vitis vinifera | Grapevine | 14 | 3 | 3 | 2 | 1 | 4 | 2 | 2 | Feechan et al.37 |
| Cucumis sativus | Cucumber | 13 | 4 | 2 | 3 | 0 | 3 | 1 | 0 | Zhou et al.32 |
| Solanum lycopersicum | Tomato | 15 | 3 | 3 | 3 | 0 | 4 | 1 | 1 | Chen et al.31 |
| Hordeum vulgare | Barley | 11 | 2 | 7 | 1 | 1 | 0 | 0 | 0 | Kusch et al.16 |
| Capsicum annuum | Pepper | 2 | 2 | Kim and Huang19, Panstruga35, | ||||||
| Legume species | ||||||||||
| Medicago truncatula (Galegoid clade) | Barrel medic | 14 | 3 | 4 | 3 | 1 | 2 | 0 | 1 | This study |
| Cicer arietinum (Galegoid clade) | Chickpea | 13 | 3 | 3 | 3 | 1 | 2 | 0 | 1 | This study |
| Pisum sativum (Galegoid clade) | Pea | 4 | 1 | 1 | 1 | 1 | This study | |||
| Lupinus angustifolius (Genistoid clade) | Narrow-leaf lupin | 15 | 3 | 3 | 6 | 0 | 3 | 0 | 0 | This study |
| Arachis spp. (Dalbergioid clade) | Peanut | 14 | 4 | 4 | 2 | 1 | 2 | 0 | 1 | This study |
| Glycine max (Phaseoloid clade) | Soybean | 31b | 5 | 5 | 6 | 2 | 9 | 2 | 2 | Deshmukh et al.15 |
| Cajanus cajan (Phaseoloid clade) | Pigeonpea | 20 | 4 | 3 | 3 | 1 | 6 | 1 | 2 | This study |
| Phaseolus vulgaris (Phaseoloid clade) | Common bean | 19 | 4 | 3 | 3 | 1 | 6 | 1 | 1 | This study |
| Vigna radiata (Phaseoloid clade) | Mungbean | 18 | 3 | 3 | 3 | 1 | 6 | 1 | 1 | This study |
aMLO classification is based on Neighbor-Joining phylogenetic analysis and literature.
bActualized G. max MLO sequences after removing partial sequences and those classified by Genbank as obsolete sequences.
MLO genes have been intensively studied in some plant species. However, little is known about the MLO members of Fabaceae. Here, we performed the genome-wide characterization of the MLO gene family in eight legume species belonging to the major clades of the Papillionoideae sub-family (Genistoid, Dalbergioid, Phaseoloid and Galegoid). This included three species from the Galegoid clade (the temperate legumes, barrel medic, chickpea and pea), one from the Genistoid clade (narrow-leaf lupin), one from the Dalbergioid clade (peanut) and three from the Phaseoloid clade that regroups the tropical legumes (pigeon pea, common bean and mung bean) (Table 2). The newly identified sequences were then compared with previously identified MLOs to get insights about the evolution of the MLO family in legumes.
Table 2. Legume genomic databases used in this study.
| Species | Common name | Legume clade | Depository (Bioproject) | Version | Web address | Reference |
|---|---|---|---|---|---|---|
| Medicago truncatula | Barrel medic | Galegoid | JCVI | v.4 | http://jcvi.org/medicago/index.php | Young et al.54 |
| Cicer arietinum | Chickpea | Galegoid | NCBI (PRJNA190909) | v.1 | ftp://ftp.ncbi.nlm.nih.gov/genomes/Cicer_arietinum/ | Varshney et al.55 |
| GigaDB | http://gigadb.org/dataset/100076 | |||||
| Lupinus angustifolius | Narrow-leaf lupin | Genistoid | NCBI (PRJNA179231) | Draft | http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=AOCW01#contigs | Yang et al.56 |
| Arachis spp. | Peanut | Dalbergioid | PeanutBase | v.1 | http://www.peanutbase.org | Bertioli et al.57 |
| Cajanus cajan | Pigeonpea | Phaseoloid | GigaGB | v.5 | http://gigadb.org/dataset/100028 | Varshney et al.58 |
| Phaseolus vulgaris | Common bean | Phaseoloid | Phytozome | v.1 | http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Pvulgaris | Schmut et al.59 |
| Vigna radiata | Mungbean | Phaseoloid | NCBI (PRJNA243847) | v.1 | http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=JJMO01#contigs | Kang et al.60 |
Results
Identification of legume MLOs
Datamining of the different legume genomes (Table 2) identified from 14 to 23 sequences with homology to A. thaliana MLOs (Supplementary Table S1). In most genomes, several hits were predicted to encode for truncated proteins. This included the M. truncatula sequences MtMLO12 and MtMLO16 (Supplementary Table S1). Most of these truncated versions were located close to retro-transposon-like sequences. Thus, these shorter sequences were considered pseudogenes and they were not analysed further.
The remaining sequences were confirmed as putative full length MLOs. This led to the identification of 14 MLO genes in M. truncatula, 13 in Cicer arietinum, 15 in Lupinus angustifolius, 20 in Cajanus cajan, 19 in Phaseolus vulgaris, 18 in Vigna radiata and 13 in each Arachis genome (Table 1 and Supplementary Table S1). Interestingly, the sequences SSV2N, from A. duranensis, and MQE1N, from A. ipaensis, had no counterpart in the second Arachis genome (Supplementary Table S1). The peanut genome may, thus, contain 14 potential MLO members. They have been named ArMLO1 to ArMLO14 (Table 1 and Supplementary Table S1). To avoid redundancy, only one sequence for each Arachis MLO orthologue was used in the analysis.
Since the pea genome has not been sequenced yet, we used the large transcriptomic resources available to search for potential MLO genes in this species. We identified several pea transcripts showing homology with 11 MtMLO sequences. This suggested the presence of, at least, 11 potential MLOs in pea (data not shown). In addition to PsMLO1, three full length MLO genes could be reconstructed. These sequences, named PsMLO2, PsMLO3 and PsMLO4, showed high similarity to MtMLO9, MtMLO11 and MtMLO15, respectively (Supplementary Tables S1).
Organization and distribution of legume MLOs
The gene characteristics are summarized in Supplementary Table S1. Large variations in gene size were detected within and between legume species. The longest gene, ArMLO13, covered a genomic region of 28.05 kb, although this might be due to assembly errors. The mean genomic length of the other legume MLOs varied from 4.41 to 6.62 kb. The length of their coding regions varied from 1.63 to 1.65 kb on average distributed on 12 to 17 exons. Accordingly, the mean protein size varied from 539 to 547 amino acids (Supplementary Table S1).
One to four MLO genes were detected on almost all legume chromosomes indicating an even distribution over legume genomes. In addition, we observed that physically close MLO pairs, in any given species, had orthologous pairs in the corresponding chromosome of other legume species (Supplementary Table 1). For instance, the orthologous sequences of MtMLO5, MtMLO8 and MtMLO9, from M. truncatula chromosome 3, are located on the same chromosome in P. vulgaris, Cicer arietinum, V. radiata, Cajanus cajan and Arachis spp. (Supplementary Table S1). Similar situation was found for the orthologues of MtMLO2 and MtMLO6, located in M. truncatula chromosome 5, and those of MtMLO4 and MtMLO7, from chromosome 2, that were detected, in the same order, on the corresponding chromosomes of Cicer arietinum, P. vulgaris and V. radiata, respectively (Supplementary Table S1). This would suggest that at least some of the MLOs localized within syntenic blocks which are conserved across legume genomes.
Characterization of protein and domain organization
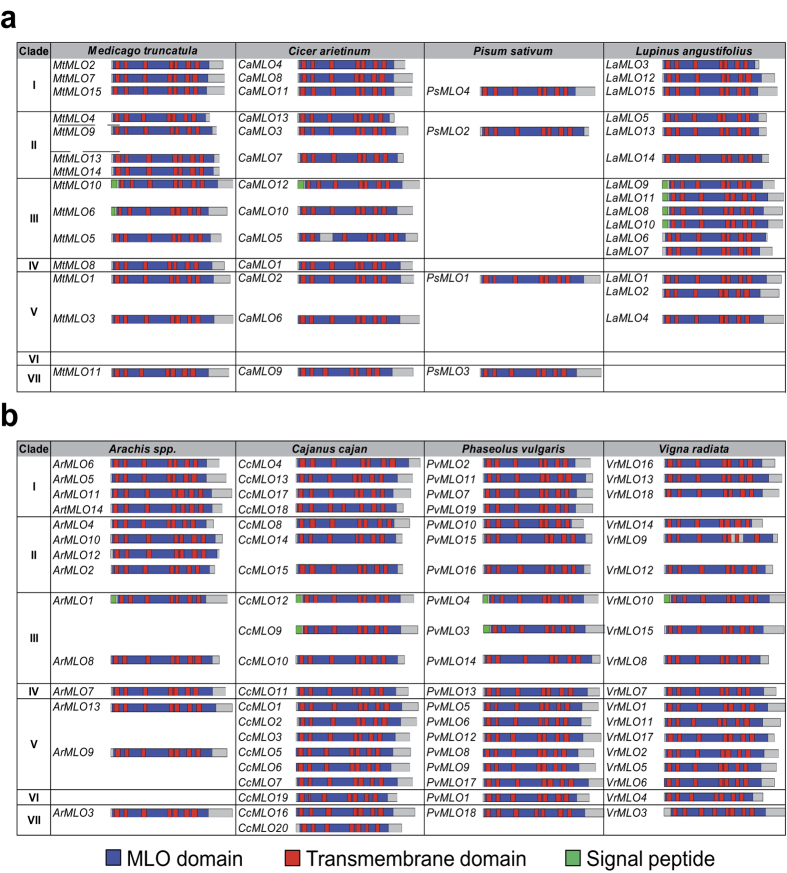
MLO proteins are characterized by the presence of seven trans-membrane (TM) domains and one MLO functional domain14. To determine whether the legume MLO genes shared these typical characteristics, their deduced amino acid sequences were subjected to different prediction servers (Supplementary Table S2). Almost all sequences were predicted to contain a single MLO domain covering most of the protein length. The sole exceptions were CaMLO5 and VrMLO9 for which two separated MLO domains were predicted (Supplementary Table S3 and Fig. 1). All potential MLOs were predicted to localise within cell membranes (Supplementary Table S3).
Figure 1. Domain organization of legume MLOs.
The figure represents the domain organization of all MLO protein sequences isolated from (a) Genistoid and Galegoid legumes and (b) Dalbergioid and Phaseoloid legumes. Sequences are drawn to scale with the IBS server51 following Interproscan 529, TMHMM27 and SignalP52 predictions (Supplementary Table S2).
The prediction servers used to estimate the number of TM domains (TMHMM27, Psort28 and InterProScan5)29 implemented different algorithms. This lead to some variations in the total number of TM domains predicted (Supplementary Table S3). Despite these small variations, all sequences, except VrMLO3, were predicted to contain seven TM domains. For 97 sequences, the prediction was supported by two or more servers (Supplementary Table S3 and Fig. 1). The TM domain distribution was largely similar between them and it fitted with the distribution of TM domains of typical MLO proteins (Fig. 1). Several putative MLOs were also predicted to contain a signal peptide at their N terminal region (Supplementary Table S3 and Fig. 1).
In parallel, the legume MLO sequences were subjected to the MEME suite server30 to identify conserved amino acid motifs and to uncover species-specific or legume-specific signatures. This identified 14 amino acid motifs common to most MLO sequences (Table 3). These motifs co-localized with the TM domains, the internal loops 2 and 3 and the calmodulin-binding region (CaMBD) (Supplementary Fig. S1). These motifs were also found in most MLO sequences from Glycine max and from non-legume species including Arabidopsis thaliana, Cucumis sativus, Solanum lycopersicum and Vitis vinifera. In addition, they were largely similar and overlapping with the motifs identified in previous studies15,16,31,32 (Supplementary Fig. S1).
Table 3. Conserved motifs common to all sequences as detected by MEME software.
| Motif | Sequence | e-value | N° sequence |
|---|---|---|---|
| 1 | RSL[EDA]ETPTWAVAVVC[TF]V[FIL][VLI][AL][IV]S | 6.4e−1500 | 208/212 |
| 2 | [LAI][ILV]E[RHK][SI]H[KR][LI]GKWLKKK[HN]KKAL[LYF] | 1.6e−997 | 205/212 |
| 3 | E[AS]LEK[IV]K[EA]ELMLLGFISLLLT[VF] | 1.6e−1654 | 209/212 |
| 4 | [SA][KR]IC[IV][PS][ES][KS]VA[DNS][ST][MW][LHF]PC | 1.4e−821 | 193/212 |
| 5 | P[LF][VLI]S[VY]EG[LI][HE]QLH[IR]FIF[VF]LA[VI][FT]H[VI]L[YF][SC][VIA][LI]T[MVL][LA]L[GA]R[AL]K[IM]R[RS] | 3.0e−2726 | 211/212 |
| 6 | WK[ARK]WE[EAD]ET[KS][TS][LH]EY[QE]F[ASY][NH]DP[ES]RFR[FL][AT][RH][EDQ]T[ST]F[GV]RRHLS | 2.5e−1389 | 127/212 |
| 7 | [CS]FFRQF[YF][GR]SVT[KR][VA]DYL[TA]LR[HL]GF | 1.2e−1793 | 212/212 |
| 8 | KF[DN]F[HQ]KY[IM]KRS[LM]E[DE]DFKV[VI]VG[IV]S[PW]PLW[FA][FS][VA]V | 2.9e−2348 | 210/212 |
| 9 | N[IVT][HN]GW[HYN][TS]YFW[LI][SPA]FIP[LV][IV][IL][ILV]L[LA]VGTKL[QE][HV][IV]I[TA] | 1.9e−2051 | 211/212 |
| 10 | M[AG]L[ERD]I[QAT][ED][RK][HG][AE]V[VI][KQ]GI[PL][LV]V[QE]P[SG]D | 1.6e−956 | 156/212 |
| 11 | FWF[NG][RK]P[RQ]L[VL]L[FH]LIH[FL][IV]LFQNAF | 1.7e−1699 | 212/212 |
| 12 | [AT][FY]F[FL]W[TIS]W[YW][EQT][FY]GFDSC[FI] | 3.8e−946 | 207/212 |
| 13 | R[LV][AI][LM]GV[FA][VI]Q[VF]LCSY[VSI]TLPLYA[LI]VTQMG[ST][TR]MK | 7.8e−2592 | 212/212 |
| 14 | K[ATS]IF[DN]E[QR][VT][ARS]KALK[KNG]WHK[TA][AV]KKKxKHKKxGSS | 2.2e−1128 | 189/212 |
A previous study by Elliot et al.33 identified 30 invariable amino acid residues within 38 MLO sequences. Twenty two of these residues were also invariable in legume MLOs. The other residues were also highly conserved since they only changed in one or two sequences per legume species (Table 4).
Table 4. Conservation of previously identified invariable amino acid residues33 in legume MLO sequences at species and clade level.
| Barley Residue | Legume species |
MLO clade |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mt | Ca | La | Ar | Cc | Pv | Vr | Gm | Ia | Ib | II | III | IV | V | VI | VII | |
| E35 | E/N | E/H | E/Q | E/H/D | E/Q/D | E/Q/D | E/Q | E/Q | Q/H/N | E | E | E | E | E/D | E | E |
| M65 | M | M | M | M/I | M | M | M | M | M | M | M | M | M | M/I | M | M |
| G68 | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
| S71 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| L74 | L/M | L/M | L/M | L/M | L/M | L/M | L/M | L/M | M | L | L/M | L | L/I | L | L | L |
| C86 | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| C98 | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| C114 | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| F135 | F/S | F/S | F | F/S | F/S | F/S | F/S | F/S | F | F | F | F | F | F | F | S |
| W158 | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W |
| E163 | E | E | E/Q | E | E/D | E/D | E/D | E/D | E | E | E | E/Q | E | E | D | E |
| F207 | F | F | F | F | F/L | F | F | F | F | F | F | F | F | F | F | F/L |
| Q210 | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q |
| Y220 | Y | Y | Y/F | Y | Y/F | Y | Y | Y | Y | Y | Y | Y/F | Y | Y | Y | Y |
| R224 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| F227 | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F |
| F240 | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F |
| Y243 | Y/F | Y/F | Y | Y/F | Y/F | Y/F | Y/F | Y/F | Y | Y | Y | Y | Y | Y | Y | F |
| W263 | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W |
| P287 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| F329 | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F |
| W330 | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W |
| P334 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| F346 | F/I | F/I | F | F/L | F/I | F/I | F/I | F/I | F | F | F | F | F | F | F | I/L |
| N348 | N | N | N/I | N/T | N | N | N | N | N | N | N/I | N | N | N | N | N/T |
| F350 | F | F | F/I | F | F | F | F | F | F | F | F/I | F | F | F | F | F |
| C367 | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| T393 | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T | T |
| P395 | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| W423 | W/L | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W |
Phylogenetic analysis of legume MLOs
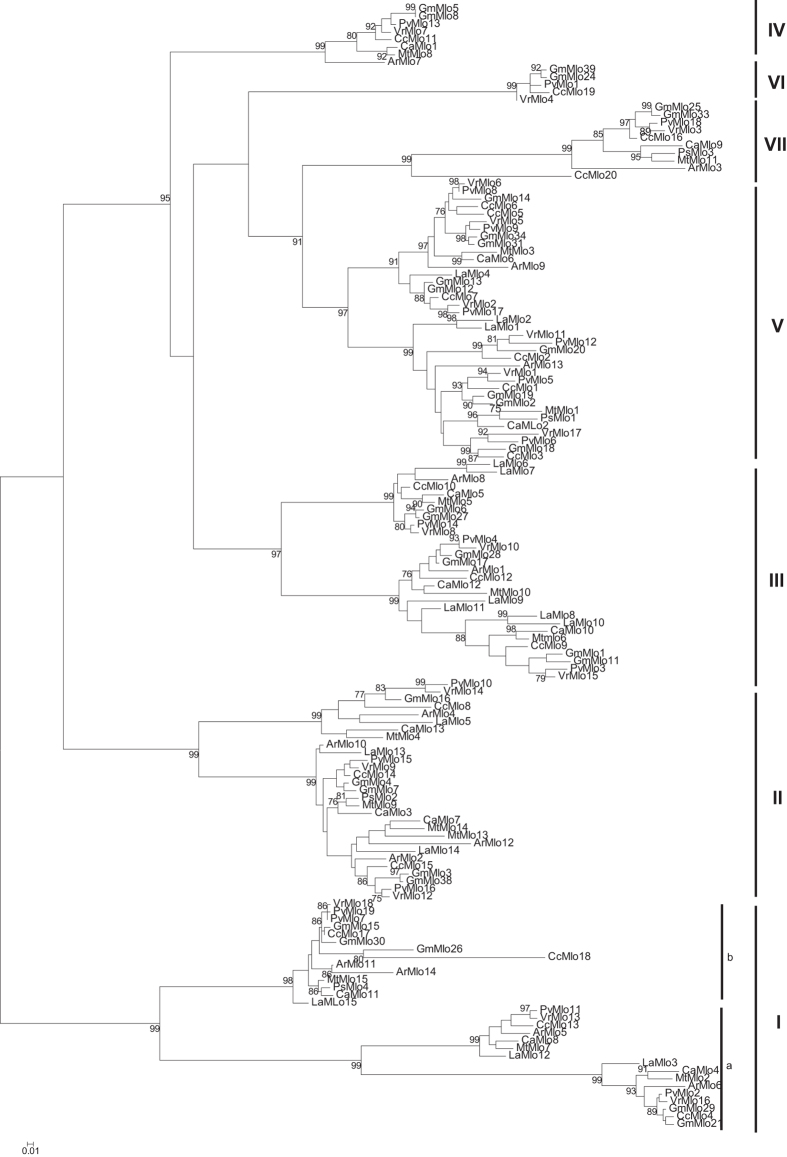
The MLO family was previously subdivided in six to eight clades14,17. To classify the legume MLOs, a Neighbor-Joining (NJ) phylogenetic analysis was performed. To this aim, their deduced protein sequences were aligned with already characterized MLO sequences (Table 1 and Supplementary Fig. S2).
This analysis separated the MLO proteins into seven well-supported clades (Supplementary Fig. S2). The MLO members from clade I further clustered in two well-separated sub-clades (Ia and Ib). At least one MLO protein, from each legume species, was found in clade IV that contains the barley MLO. Several members from each legume species also clustered in clade V with the powdery mildew susceptibility genes of Arabidopsis thaliana. Surprisingly, only sequences from tropical legumes clustered in clade VI with AtMLO3. By contrast, the last group (clade VII) was nearly exclusively composed of legume sequences except for the tomato protein SlMLO2 (Supplementary Fig. S2).
To confirm this classification and to analyse further the evolution of the MLO family in legumes, a more detailed phylogenetic analysis was performed using the maximum likelihood (ML) or maximum parsimony (MP) algorithms. The two approaches (ML and MP) retrieved very similar tree topologies, thus only the ML tree is shown (Fig. 2). This approach also grouped the legume MLOs in seven clades with clade I further divided in two well-supported branches (cluster Ia and Ib) (Fig. 2). Clades I, II and III were represented by three to four MLOs per legume species. By contrast, clades IV, VI and VII were only represented by one sequence per species (Fig. 2 and Table 1). As already observed after the NJ phylogenetic analysis, clade VI only contained MLO sequences from tropical legumes including G. max, Cajanus cajan, P. vulgaris and V. radiata (Fig. 2 and Supplementary Fig. S2). The ML phylogenetic tree also showed the expansion of clade V MLOs in tropical legumes. In these Phaseoloid species, six clade V genes were detected while the other legume species had only two genes (Fig. 2 and Table 1).
Figure 2. Phylogenetic relationship of legume MLOs.
The phylogenetic relationship of legume MLO protein sequences was estimated with the Maximum likelihood (ML) method with MEGA648 software with 1,000 bootstrap independent replicates. The tree was drawn to scale, with branch lengths measured as the number of substitutions per site. Number on a node indicates the percentage of bootstrap when higher than 75%.
Conservation of MLO members within clades and identification of clade-specific motifs
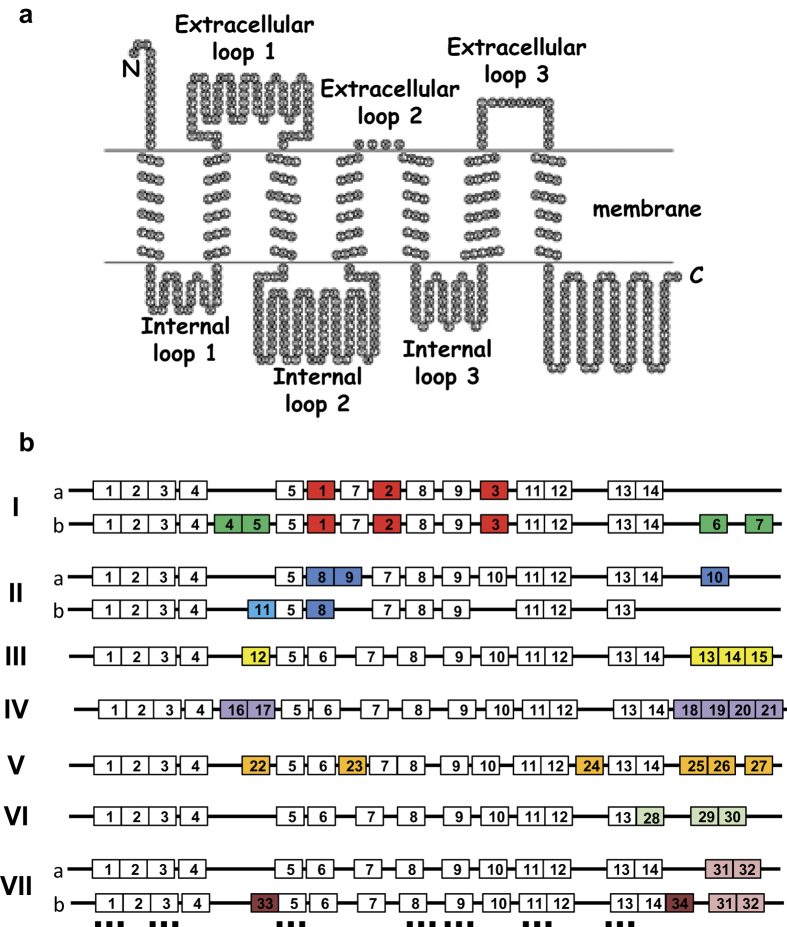
To determine the presence of clade-specific motifs within legume MLOs, they were classified according to the phylogenetic tree and subjected to MEME (Fig. 3). In parallel, all MLO orthologues were aligned with Clustal W34 to visually assess their overall conservation and to locate the conserved motifs (Supplementary Figs S3 to S9). The MEME analysis revealed 34 clade-specific motifs (Fig. 3B and Table 5). According to this analysis, each clade can be recognized by the presence of two to six motifs. These motifs mostly localised within the first extracellular loop, the second intracellular loop and the C-terminal region (Fig. 3 and Supplementary Figs S3 to S9). Among the clade-specific motifs, six were specific to clade V. Three of these motifs localised at the C terminal end of the proteins. Among them, domain 25 and 27 covered the three distinctive regions identified in the MLOs associated with powdery mildew susceptibility16,35 (Supplementary Fig. S7). On the other hand, four motifs (motifs 4 to 7) could distinguish between the two sub-types of clade I MLOs. Other motifs distinguished between two sub-types within the MLO members of clades II and VII (Fig. 3B). Interestingly, one of the clade VII sub-types was characterized by the presence of two motifs (motifs 33 and 34) that were only detected in Phaseoloid species (Fig. 3 and Supplementary Fig. S9). Many of these motifs were also found in non-legume species including Vitis vinifera, Cucumis sativus and Solanum lycopersicum. This includes all clade V-specific motifs. Nevertheless, 12 motifs (motifs 5, 10, 11, 14, 15, 16, 19, 20, 21, 29, 30 and 32) were only found in legume sequences (Table 5).
Figure 3. Motif organization of legume MLOs.
The figure shows the predicted topology of a typical MLO protein (a) and the schematic organization of the common and specific motifs for each MLO clade (b). Common and clade-specific motifs are represented by white and colored boxes, respectively. These motifs were identified by scanning the MLO sequences with the MEME suite software30 (Supplemental Table S1). Common and clade-specific amino acid motifs are listed in Tables 3 and 4 respectively. Localization of transmembrane domains is shown as dashed horizontal lines.
Table 5. Clade-specific motifs in MLO sequences as detected by MEME software.
| Clade | Motif | Sequence | e-value | N° Sequence |
|---|---|---|---|---|
| I | 1 | [DN][SG][LN]S[QE][IS][TK][RKS][ES][LIK][TR][ML]RR[QL][ST]TF[VI][FK][HS]H[TA]S[HN]P[WL]S[RHK][NH][KSP] | 1.2e−405 | 42/212 |
| I | 2 | I[TM][NE]HNL[PS]L[KTS] | 1.1e−100 | 29/212 |
| I | 3 | [TK]LA[LV]E[NI]A[GE][IQR][TC][GP][FP][FM][SKP][EPR][AHT][KQ][LFV][RKN][PL]RDELFWF[KNG]KP[ERD] | 8.5e−565 | 40/212 |
| I | 4 | TRS[EQ]ID[EK][EQ][MI]E[ED]NGSE[EG]RKLL[MT]A | 4.1e−151 | 18/212 |
| I | 5 | [YA][PY][HR][LV][IF][RG]RML[ND]G[IM]NR[SN][ST] | 3.9e−082 | 13/212 |
| I | 6 | TIHTDTSTVLSIEEDDQLID[DAT]PE | 1.2e−161 | 16/212 |
| I | 7 | [AT]VT[SA]TPSPIANETSSRA[VA]TPLLRPSASISS[SV][HQV][PCS][SF]S | 1.0e−190 | 14/212 |
| II | 8 | G[AL][RK]IR[QE]WKHWEDSIAK[QE]NYETx | 2.1e−334 | 50/212 |
| II | 9 | [RP]VL[KE]P[KT]VT[HN]V[HQ]QH[ADE]FI | 8.3e−146 | 26/212 |
| II | 10 | GIQLGS[VI]F[RKQ][KR][AR][SA][AS][PA][EP][DE] | 3.7e−126 | 22/212 |
| II | 11 | [AKN][KR][KR][KR][GL]L[KRS][AG]D[SGN][NQ][SHP][SGQ][HS][GC]S | 8.1e−111 | 28/212 |
| III | 12 | [EG][EG]EH[RH]R[KR]LLSYERR[YF]L[AS][AG][DG][GTA][TG][SG] | 8.1e−314 | 33/212 |
| III | 13 | [DGS]ST[VI]HSSGPTLHR[FY]KTTGHSTR | 7.1e−258 | 26/212 |
| III | 14 | Y[DE]D[QD]D[DE]Y[HEQ]SDIE | 4.5e−079 | 18/212 |
| III | 15 | [PQ]T[AT][SNT][LI][IV][VT]RVD[HN][GD][ED]Q[QE][AQ]EE[EN]E[HE]H | 3.0e−144 | 26/212 |
| IV | 16 | [FD][DE][DE]N[MLV][EV]WRRVLA | 7.3e−076 | 11/212 |
| IV | 17 | A[AS][SG]G[GD]DYCS[QN]KGKV[PS]LISQSGV | 9.5e−091 | 9/212 |
| IV | 18 | SGE[TA]TPSQGTSP[LI]HLL[HQ]K[YF]KPS | 1.2e−099 | 12/212 |
| IV | 19 | [HQ]TDTDSVLYSPRSYQSD | 1.1e−080 | 8/212 |
| IV | 20 | TD[LF]S[DE]TEGS[ST]HQLN[EL]I[TQ][QI][TM][HS]Q[PA] | 3.4e−066 | 8/212 |
| IV | 21 | P[RN]N[GQ][EL]THNI[DE]FSF[VD][KS]P | 2.5e−044 | 8/212 |
| V | 22 | LA[TAG][GK]GYDKC | 8.5e−169 | 28/212 |
| V | 23 | FW[TS][QK][SN][PT][IV][LS][LV]WIV | 2.7e−203 | 50/212 |
| V | 24 | FHSTT[EA]D[VI]VIR[IL] | 2.4e−198 | 51/212 |
| V | 25 | STTPFSSRP[ST]TPTHGMSP[VA]HLL | 4.4e−360 | 30/212 |
| V | 26 | [APR][GRS][RHE]SDS[AFP][QP]TSPR[TAR]SNY[ED]NEQWD | 3.0e−275 | 50/212 |
| V | 27 | P[ITV][SR][SHT][QE][HIL]EI[NR][IV][SA][SL][SK][ED]FSF[EDG][KR][RG][HP][THI] | 9.7e−142 | 24/212 |
| VI | 28 | [SC]KALAK[IM]L[KR]QWH[VL]EVRERR | 1.4e−131 | 5/212 |
| VI | 29 | [QN][RELQ][KE][LQ][VL]KSFSF[RS][HR] | 1.3e−009 | 5/212 |
| VI | 30 | MSSEWSQGNKSAP[ED]FSSTL[CR]E[SN][IANT]RSSDEGEIVEELEH[MPRS][VDEM]KTKA[SCNT]SSSDPP | 2.3e−103 | 5/212 |
| VII | 31 | N[PG]KIITRG[TI]YDGEISFGS[SY][WV][KG][NS] | 9.5e−100 | 10/212 |
| VII | 32 | SSRGI[GR]EI[GV]SI[TAI]EE[DE]D | 2.7e−046 | 9/212 |
| VII | 33 | AT[RH]TSTS[EGQ][LF]D[VI]A[PH]ATN[EQ]S[TAEN][IV]E[VF] | 8.2e−038 | 5/212 |
| VII | 34 | NNSTSSKHSDSLHSK[EG][GC]DNS[AV]RG[ST][VM]DSVH[TN]PDNV[VA][LV]T[SN][NP]P[SF][PH] | 1.8e−071 | 5/212 |
On the other hand, the level of conservation of specific amino acid residues was also compared between clades (Table 4 and Supplementary Table S4). This comparison indicated that five of the 30 residues, previously described as invariable33, were modified on a clade specific basis (Table 4). For instance, the barley E135 residue was systematically changed to a D in clade VI sequences. Similarly, the Y243 and F135 were changed, in clade VII sequences, to F and S, respectively. Most of these punctual changes (three out of five punctual changes) were detected in clade VII. In the other clades, these amino acid positions remained invariable, as previously described33 (Table 4).
We also challenged the specificity of the 73 amino acids that discriminated between the powdery mildew susceptibility MLO of monocots and eudicots, according to Appiano et al.18 (Supplementary Table S4). As indicated previously, these sequences belonged to two different clades (clade IV and V). Thus we aimed not only to validate these specific changes but also to determine whether they could discriminate between monocot and eudicot sequences or between clades. For this, we aligned all clade IV and V legume sequences with the sequences included in Appiano et al.18 (Supplementary Table S4). Thirty six of these amino acid positions differed between clade IV and V sequences. For instance, the monocot valine and serine residues at position 32 and 145, were also found in clade IV legume sequences (Supplementary Table 4). However, they were systematically changed to isoleucine and glycine, in clade V sequences (Supplementary Table 4). These 36 amino acid positions could be instrumental to discriminate between clades IV and V (Supplementary Table S4). In addition, 17 amino acid positions discriminated between monocots and eudicot sequences. These residues were found unchanged in all eudicot sequences from clades IV and V, but, differed, within clade IV, between monocot and eudicot sequences. This is the case for the proline and leucine residues found at position 234 and 271. While these residues were found unchanged in all clade IV legume sequences, they were replaced by a glutamine and a phenylalanine residue in the monocot sequences (Supplementary Table 4). Interestingly, four additional residues varied not only between clade IV and V legume sequences, but also, between monocot and eudicot sequences within clade IV (Supplementary Table 4). For instance the S325 residue found in monocot clade IV sequences was modified to asparagine in clade IV legume sequences whereas it was changed to glycine in clade V sequences (Supplementary Table 4). The remaining 16 variable positions did not follow any distinctive pattern.
Discussion
MLO is a large protein family highly conserved across plant kingdom. Apart from the well-documented role of some MLOs in powdery mildew susceptibility, the biological functions of MLOs remain largely unknown14. Besides providing hints on their potential functions, studying the diversification and multiplication of MLOs in a given species may give clues on its genome evolution. Thus we performed a genome-wide characterisation of the MLO family in eight legume species belonging to different clades and ecological habitats.
Mining legume genomic databases allowed the identification and characterization of 118 MLO sequences. The total number of MLO sequences varied from 13 in chickpea to 20 in pigeon pea (Table 1 and Supplementary Table S1). This is broadly similar to the situation found in other eudicot species that demonstrated the presence of 15 MLO genes in A. thaliana36, 17 in grapevine37 and tomato31, 14 in cucumber32, 18 in strawberry17 or 19 in peach17. The highest number of MLOs was identified in soybean with 31 full length MLOs15 (Table 1). The phylogenetic analysis showed, in most cases, a pair of soybean MLO genes clustering together, for any given MLO orthologue (Fig. 2). Thus the large MLO expansion in soybean is likely consequence of its recent genome duplication15. We also detected the presence of shorter truncated sequences with homology to MLO genes. Since most of them were close to retrotransposon-like sequences, we concluded that they were inactive pseudogenes. However, shorter MLO-like sequences have been described in many plant species including tomato31, cucumber32, soybean15, strawberry17 and apple17. Thus, these sequences might represent a new family of membrane-proteins not considered before.
The MLO genes were widely distributed over the legume genomes. They were found on almost all chromosomes of any given species (Supplementary Table S1). In addition, most legume MLO orthologues were located, within conserved syntenic blocks, in related chromosomes in the different legumes (Supplementary Table S1). The MLO distribution supported the high level of micro- and macro-synteny that exist between legume genomes38,39. It also further illustrated the assumption that most legume genes are likely located in syntenic regions40 as previously demonstrated for most phenylpropanoid genes of soybean and common bean41. This distribution also suggested that they mainly arose from segmental duplication as it was already assumed for rice and several Rosaceae species17,42. Tandem duplication may have also played a minor role in MLO evolution since we detected evidence of a few tandem duplication events such as the gene pairs PvMLO5/PvMLO6 and VrMLO5/VrMLO6 in P. vulgaris and V. radiata, respectively (Supplementary Table S1).
The phylogenetic analysis classified the legume MLOs in seven clades (Figs 2 and 3), which is in accordance with previous studies14,17,37. The largest clades were clades I, II III and V that contained two to six MLO genes, in each legume species (Fig. 2 and Supplementary Fig. S2). In our analysis, the clade I was further divided in two well-supported sub-clades. These sub-clades can also be distinguished by the examination of their sequences (Fig. 2 and Supplementary Figs S2 and S3). One MLO per legume species was found in clade IV that was originally thought to be restricted to monocots14. Clade VI, characterized by the presence of AtMLO3, contained only a small number of legume sequences (Figs 2 and 3). This supports its recent addition to the MLO family14. The legume sequences in this clade were only from the Phaseoloid legumes including common bean, mung bean, pigeon pea and soybean (Fig. 2 and Supplementary Fig. S2). AtMLO3 orthologues have been found in all eudicot species studied so far17,31,32,37. Lack of AtMLO3 orthologues in the other legume species is thus surprising. This could be explained by either loss of these orthologues in lupin, barrel medic, chickpea and peanut or by their specific incorporation in the genome of the Phaseoloid species. Lupin and peanut belongs to the early-diverging clades, Genistoid and Dalbergioid, respectively43. Their separation from the other Papillionidoids clades has been estimated some 55–56 million years ago44. The evolution of the tropical (Phaseoloids) and temperate legumes (Galegoids) is more recent43,44. It has been estimated to have taken place approximately 52.8 and 50 million years ago, respectively44. According to this, it appears more likely that the Phaseoloid species (common bean, pigeon pea, mung bean and soybean) have incorporated this MLO clade during speciation. The phylogenetic study also revealed a seventh clade that was mainly represented by legume MLO. This is in accordance with recent studies that also identified a seventh clade in cucumber32 and tomato31. Another recent study on Rosaceae MLO identified two new clades apparently restricted to Rosaceae species (clades VII and VIII)17. However, MLO sequences from soybean, cucumber or tomato were not included in their analysis17. Thus, a more global analysis of MLO sequences, over plant kingdom, would be necessary to determine whether evolution of MLO sequences led to the apparition of genera-specific clades.
Previous studies identified several conserved motifs15,16,31,32,35 that we also detected in the legume MLO protein sequences (Table 3 and Supplementary Fig. S1). One of these common motifs, located at the C-terminal region, was previously shown to bind to the calcium-sensing protein calmodulin35. Here, we confirmed that the calmodulin binding site was conserved in all MLO clades (Supplementary Figs S3 to S9), since it was found within the common conserved motif 14 in all legume sequences (Fig. 3). In addition to these common motifs, our study identified 34 clade-specific motifs and several clade-specific amino acid residues. These motifs located in the extracellular loops 1 and 3, the intracellular loop 2 and the C terminal region (Table 4, Fig. 3 and Supplementary Table S4). For instance, six clade V-specific motifs were detected (Table 4, Fig. 3 and Supplementary Fig. S7). Two of them, motifs 25 and 27, contained the previously identified consensus clade V sequences16,35. This confirmed the efficacy of the method used. Interestingly, the conserved tetrad [E/D]FSF35 was also detected in clade IV and VI MLO sequences (Supplementary Figs S6, S7 and S8). The presence of this motif in clade IV sequences may have been expected since it contains the powdery mildew susceptibility genes of monocots. By contrast, its presence in the more divergent clade VI is surprising. It might indicate a common mechanism of action of these three clades. The identification of clade-specific motifs is very useful to isolate MLO orthologues in plant species not yet sequenced.
Beyond finding interesting new features about the MLO gene family, our study also showed diverging features between the tropical legumes (Phaseoloids) and the other legume species. One of the most striking differences was the total number of MLOs found in each type of legumes. Legumes from the Genistoid, Dalbergioid and Galegoid clades were characterized by 13 to 15 genes while tropical legumes contained from 18 to 31 genes (Table 1 and Supplementary Table S1). Almost all additional genes from tropical legumes clustered in clade V (Fig. 2 and Table 1). Given the importance of this clade in disease susceptibility, the specific multiplication of clade V MLOs in tropical legumes may reflect a greater pathogenic variability and pressure in tropical regions. Another phylogenetic difference was the lack of clade VI MLO genes in legumes from the Genistoid, Dalbergioid and Galegoid clades. The significance of the absence of this particular clade is not known. Clades V and VI correspond to the most recent diversification of the MLO family14. At this respect, our data suggested that the MLO family evolved differently depending on the legume clade considered. The tropical legume diverged after the separation from lupin and peanut ancestors but before that of the temperate legumes44. They are the only ones to have incorporated the clade VI MLO in its genome and to have followed a significant expansion of this family (Fig. 2 and Supplementary Fig. 2). Our data also indicate that the Genistoid, Dalbergoid and Galegoids, have evolved in parallel. These assumptions were also supported by the detection of two specific motifs (motifs 33 and 34) only found in clade VII MLOs of tropical legumes.
Altogether, our study characterized 118 MLO sequences from eight different legume species with different habitats and agronomic characteristics. This comparative analysis revealed interesting new phylogenetic features that may be the base to further determine the function of this gene family. We also detected several differences between tropical and the other legume species that might reflect different evolutionary pressures. In addition, we identified from three to seven genes in clades IV and V that contains the genes associated with powdery mildew susceptibility. These new sequences are very valuable to identify new gene variants to confer powdery mildew resistance in these species and to identify new susceptibility genes in additional legume species.
Methods
Identification, annotation and validation of legume MLOs
M. truncatula MLO sequences were identified by mining the JCVI M. truncatula genomic project v4.0 database through BLAST searches with the 15 Arabidopsis thaliana MLO sequences as templates. All potential MLO sequences from the other legume species (Table 2) were retrieved by BLAST45 using the M. truncatula MLO CDS and protein sequences. In all cases, the lowest limit of significance (e-value) for any potential hits was set at 1e−20. All potential MLO sequences were systematically validated by reciprocal BLAST on the M. truncatula JCVI Mt4.0 (http://jcvi.org/medicago/index.php) and A. thaliana TAIRv10 databases (http://www.arabidopsis.org).
Upon validation, the genomic sequence of each potential MLO was examined to reconstruct the full length CDS and correct potential annotation errors. Each genomic sequence was then aligned to its corresponding transcripts by BLAST against its respective transcriptomic (TSA) and EST databases that are stored at the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). In parallel, validated MLO sequences, from unannotated legume genomes (Lupinus angustifolius and Vigna radiata), were analyzed with Fgenesh46 using the “Medicago legume gene” model (Supplementary Table S2). Manual correction of the annotation was also performed, if necessary, to improve sequence quality. SeqBuilder v12.0 (DNASTAR, Madison, WI) was used to draw and correct the resulting exon-intron organization of each sequence. Supplementary Figs S10 and S11 show the CDS and deduced protein sequences of the legume MLOs, respectively.
Conservation and phylogenetic analyses
Global pair-wise analysis was performed on the deduced protein sequences to determine their level of conservation to their closest homologue in M. truncatula and A. thaliana with Geneious R8 (http://www.geneious.com)47. Multiple protein sequence alignments were performed with ClustalW34. The alignments were manually corrected before phylogenetic reconstruction.
To assign each potential MLO to its clade, all identified MLO protein sequences were aligned with the MLO sequences from soybean and several non-legume species (Table 1). This alignment was used to calculate a p-distance matrix after pair-wise deletion of gaps using the MEGA6 software48. Then, a phylogenetic tree was reconstructed based on the p-distance matrix with the NJ algorithm. This analysis was performed with 1,000 bootstrap replicates with the MEGA6 software48. The phylogenetic relationship of legume MLO was also established using the MP and ML methods implemented in the MEGA 6 software. The search for the most parsimonious tree (MP method) was performed on 10 initial trees with the subtree-pruning-re-grafting method and 1,000 bootstrap replicates. Prior to ML analysis, all gaps and divergent regions were removed from the protein alignment with Gblocks version 0.91b49. The resulting alignment was then used to estimate the optimum substitution model with ProtTest 3.450. Subsequently, the ML tree was obtained on 1,000 bootstrap replicates using the JTT substitution model with gamma distribution of 8 categories and α = 1.05 following the ProtTest estimation.
Protein characterization and motif prediction
The deduced amino acid sequences of the potential MLO genes were subjected to several prediction programs to determine their sub-cellular localizations28,51, protein topologies27,28,29,52 and to identify functional domains29. The prediction servers used in this study are listed in Supplemental Table S2. Except otherwise stated, the prediction server were run with default settings. The result of these predictions was then used to draw the protein organization of each potential MLO protein on the IBS server53 (Supplementary Table S2).
Conserved motifs were determined with the MEME algorithm30 (Supplementary Table S2). The MEME parameters were set to search for a maximum of 15 motifs with a motif width comprised between five and 50 residues. Presence or absence of the conserved motifs in each MLO sequence was then determined using FIMO and MAST algorithms also available from the MEME suite web server30 (Supplementary Table S2).
Additional Information
How to cite this article: Rispail, N. and Rubiales, D. Genome-wide identification and comparison of legume MLO gene family. Sci. Rep. 6, 32673; doi: 10.1038/srep32673 (2016).
Supplementary Material
Acknowledgments
This work was supported by the European project LEGATO (FP7-KBBE2013.1.2-02-613551), and the Spanish project AGL2014-52871-R from the Spanish Ministry of Economy and Competitiveness (MINECO) and co-financed by European fund for regional development (FEDER). NR is holder of a Ramón y Cajal postdoctoral position from the Spanish Ministry of Economy and Competitiveness.
Footnotes
Author Contributions N.R. designed and performed the experiments, analysed and interpreted the data and wrote the manuscript; D.R. participated in the experimental designs, supervised the experiments and contributed in drafting and revising the manuscript. All authors read and approved the final manuscript.
References
- Rubiales D. & Mikic A. Introduction: legumes in sustainable agriculture. Crit. Rev. Plant. Sci. 34, 2–3, 10.1080/07352689.2014.897896 (2015). [DOI] [Google Scholar]
- Jensen E. S. et al. Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. A review. Agron. Sustain. Dev. 32, 329–364, 10.1007/s13593-011-0056-7 (2012). [DOI] [Google Scholar]
- Peix A., Ramirez-Bahena M. H., Velazquez E. & Bedmar E. J. Bacterial associations with legumes. Crit. Rev. Plant. Sci. 34, 17–42, 10.1080/07352689.2014.897899 (2015). [DOI] [Google Scholar]
- Rubiales D. et al. Achievements and challenges in legume breeding for pest and disease resistance. Crit. Rev. Plant. Sci. 34, 195–236, 10.1080/07352689.2014.898445 (2015). [DOI] [Google Scholar]
- Hueckelhoven R. & Panstruga R. Cell biology of the plant-powdery mildew interaction. Curr. Opin. Plant Biol. 14, 738–746, 10.1016/j.pbi.2011.08.002 (2011). [DOI] [PubMed] [Google Scholar]
- Lyngkjaer M. F., Newton A. C., Atzema J. L. & Baker S. J. The barley mlo-gene: an important powdery mildew resistance source. Agronomie 20, 745–756, 10.1051/agro:2000173 (2000). [DOI] [Google Scholar]
- Chauhan H. et al. The wheat resistance gene Lr34 results in the constitutive induction of multiple defense pathways in transgenic barley. Plant J. 84, 202–215, 10.1111/tpj.13001 (2015). [DOI] [PubMed] [Google Scholar]
- Pavan S., Jacobsen E., Visser R. G. F. & Bai Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breeding 25, 1–12, 10.1007/s11032-009-9323-6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphry M., Reinstaedler A., Ivanov S., Bisseling T. & Panstruga R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant Pathol. 12, 866–878, 10.1111/j.1364-3703.2011.00718.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan S. et al. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Genet. 123, 1425–1431, 10.1007/s00122-011-1677-6 (2011). [DOI] [PubMed] [Google Scholar]
- Bai Y. et al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of Mlo function. Mol. Plant Microbe In. 21, 30–39, 10.1094/mpmi-21-1-0030 (2008). [DOI] [PubMed] [Google Scholar]
- Prats E., Llamas M. J. & Rubiales D. Characterization of resistance mechanisms to Erysiphe pisi In Medicago truncatula. Phytopathology 97, 1049–1053, 10.1094/phyto-97-9-1049 (2007). [DOI] [PubMed] [Google Scholar]
- Poulter R., Harvey L. & Burritt D. J. Qualitative resistance to powdery mildew in hybrid sweet peas. Euphytica 133, 349–358, 10.1023/a:1025734428660 (2003). [DOI] [Google Scholar]
- Acevedo-Garcia J., Kusch S. & Panstruga R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 204, 273–281, 10.1111/nph.12889 (2014). [DOI] [PubMed] [Google Scholar]
- Deshmukh R., Singh V. K. & Singh B. D. Comparative phylogenetic analysis of genome-wide Mlo gene family members from Glycine max and Arabidopsis thaliana. Mol. Genet. Genomics 289, 345–359, 10.1007/s00438-014-0811-y (2014). [DOI] [PubMed] [Google Scholar]
- Kusch S., Pesch L. & Panstruga R. Comprehensive phylogenetic analysis sheds light on the diversity and origin of the MLO family of integral membrane proteins. Genome Biol. Evol. 8, 878–895, 10.1093/gbe/evw036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessina S. et al. Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics 15, 618, 10.1186/1471-2164-15-618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appiano M. et al. Monocot and dicot MLO powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant Biol. 15, 257, 10.1186/s12870-015-0639-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. S. & Hwang B. K. The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J. 72, 843–855, 10.1111/tpj.12003 (2012). [DOI] [PubMed] [Google Scholar]
- Lim C. W. & Lee S. C. Functional roles of the pepper MLO protein gene, CaMLO2, in abscisic acid signaling and drought sensitivity. Plant Mol. Biol. 85, 1–10, 10.1007/s11103-013-0155-8 (2014). [DOI] [PubMed] [Google Scholar]
- Almeida N. F. et al. Allelic diversity in the transcriptomes of contrasting rust-infected genotypes of Lathyrus sativus, a lasting resource for smart breeding. BMC Plant Biol. 14, 376, 10.1186/s12870-014-0376-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risk J. M. et al. The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol. J. 11, 847–854, 10.1111/pbi.12077 (2013). [DOI] [PubMed] [Google Scholar]
- Bidzinski P. et al. Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana Mildew Locus O genes. Plant Cell Environ 37, 2738–2753, 10.1111/pce.12353 (2014). [DOI] [PubMed] [Google Scholar]
- Chen Z. et al. Two seven-transmembrane domain Mildew Resistance Locus O proteins cofunction in arabidopsis root thigmomorphogenesis. Plant Cell 21, 1972–1991, 10.1105/tpc.108.062653 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S. A. et al. Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968–971, 10.1126/science.1195211 (2010). [DOI] [PubMed] [Google Scholar]
- Yi J., An S. & An G. OsMLO12, encoding seven transmembrane proteins, is involved with pollen hydration in rice. Plant Reprod. 27, 169–180, 10.1007/s00497-014-0249-8 (2014). [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G. & Sonnhammer E. L. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580, 10.1006/jmbi.2000.4315 (2001). [DOI] [PubMed] [Google Scholar]
- Nakai K. & Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–35, 10.1016/s0968-0004(98)01336-x (1999). [DOI] [PubMed] [Google Scholar]
- Jones P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240, 10.1093/bioinformatics/btu031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Johnson J., Grant C. E. & Noble W. S. The MEME suite. Nucleic Acids Res. 43, W39–49, 10.1093/nar/gkv416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang Y. & Zhang H. Genome-wide analysis of the Mildew Resistance Locus O (MLO) gene family in tomato (Solanum lycopersicum L.). Plant Omics 7, 87–93 (2014). [Google Scholar]
- Zhou S. J., Jing Z. & Shi J. L. Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet. Mol. Res. 12, 6565–6578, 10.4238/2013.December.11.8 (2013). [DOI] [PubMed] [Google Scholar]
- Elliott C. et al. Conserved extracellular cysteine residues and cytoplasmic loop-loop interplay are required for functionality of the heptahelical MLO protein. Biochem. J. 385, 243–254, 10.1042/BJ20040993 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680, 10.1093/nar/22.22.4673 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R. Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol. Biol. 59, 485–500, 10.1007/s11103-005-0353-0 (2005). [DOI] [PubMed] [Google Scholar]
- Chen Z. Y. et al. Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol. Biol. 60, 583–597, 10.1007/s11103-005-5082-x (2006). [DOI] [PubMed] [Google Scholar]
- Feechan A., Jermakow A. M., Torregrosa L., Panstruga R. & Dry I. B. Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct. Plant Biol. 35, 1255–1266, 10.1071/fp08173 (2008). [DOI] [PubMed] [Google Scholar]
- Bertioli D. J. et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics 10, 45, 10.1186/1471-2164-10-45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazkiewicz M. et al. Remnants of the legume ancestral genome preserved in gene-rich regions: insights from Lupinus angustifolius physical, genetic, and comparative mapping. Plant Mol. Biol. Rep. 33, 84–101, 10.1007/s11105-014-0730-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S. B., May G. D. & Jackson S. A. Three sequenced legume genomes and many crop species: rich opportunities for translational genomics. Plant Physiol. 151, 970–977, 10.1104/pp.109.144659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinprecht Y. et al. In silico comparison of genomic regions containing genes coding for enzymes and transcription factors for the phenylpropanoid pathway in Phaseolus vulgaris L. and Glycine max L. Merr. Front. Plant Sci. 4, 317, 10.3389/fpls.2013.00317 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. & Zhu H. Molecular evolution of the MLO gene family in Oryza sativa and their functional divergence. Gene 409, 1–10, 10.1016/j.gene.2007.10.031 (2008). [DOI] [PubMed] [Google Scholar]
- Smykal P. et al. Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant. Sci. 34, 43–104, 10.1080/07352689.2014.897904 (2015). [DOI] [Google Scholar]
- Lavin M., Herendeen P. S. & Wojciechowski M. F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Systematic Biol. 54, 575–594, 10.1080/10635150590947131 (2005). [DOI] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402, 10.1093/nar/25.17.3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V., Kosarev P., Seledsov I. & Vorobyev D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7 Suppl 1, S10, 10.1186/gb-2006-7-s1-s10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649, 10.1093/bioinformatics/bts199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729, 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165, 10.1093/bioinformatics/btr088 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K.-C. & Shen H.-B. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. Plos One 5, e11335, 10.1371/journal.pone.0011335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786, 10.1038/nmeth.1701 (2011). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361, 10.1093/bioinformatics/btv362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D. et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524, 10.1038/nature10625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K. et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246, 10.1038/nbt.2491 (2013). [DOI] [PubMed] [Google Scholar]
- Yang H. et al. Draft genome sequence, and a sequence-defined genetic linkage map of the legume crop species Lupinus angustifolius L. Plos One 8, e64799, 10.1371/journal.pone.0064799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli D. J. et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446, 10.1038/ng.3517 (2016). [DOI] [PubMed] [Google Scholar]
- Varshney R. K. et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 30, 83–89, 10.1038/nbt.2022 (2012). [DOI] [PubMed] [Google Scholar]
- Schmutz J. et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46, 707–713, 10.1038/ng.3008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. J. et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5, 5443, 10.1038/ncomms6443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.