Abstract
Quantification of inflammation in tissue samples can be a time-intensive bottleneck in therapeutic discovery and preclinical endeavors. We describe a versatile and rapid approach to quantitatively assay macrophage burden in intact tissue samples. Perfluorocarbon (PFC) emulsion is injected intravenously, and the emulsion droplets are effectively taken up by monocytes and macrophages. These ‘in situ’ labeled cells participate in inflammatory events in vivo resulting in PFC accumulation at inflammatory loci. Necropsied tissues or intact organs are subjected to conventional fluorine-19 (19F) NMR spectroscopy to quantify the total fluorine content per sample, proportional to the macrophage burden. We applied these methods to a rat model of experimental allergic encephalomyelitis (EAE) exhibiting extensive inflammation and demyelination in the central nervous system (CNS), particularly in the spinal cord. In a cohort of EAE rats, we used 19F NMR to derive an inflammation index (IFI) in intact CNS tissues. Immunohistochemistry was used to confirm intracellular colocalization of the PFC droplets within CNS CD68+ cells having macrophage morphology. The IFI linearly correlated to mRNA levels of CD68 via real-time PCR analysis. This 19F NMR approach can accelerate tissue analysis by at least an order of magnitude compared with histological approaches.
Keywords: inflammation, macrophage, EAE, perfluorocarbon, emulsion, 19F, NMR
Rapid, quantitative scoring of inflammation in tissue specimens is a common need in many facets of biomedical research. Discovery and preclinical studies often rely on histological processing and analysis of a panel of tissues obtained from animal models, and these procedures are often viewed as time-consuming and expensive, and a bottleneck in research and development endeavors. Here, we describe a platform to rapidly and quantitatively assay macrophage infiltration in intact tissue samples. In this approach, we utilize a perfluorocarbon (PFC) emulsion reagent that labels phagocytic monocytes and macrophages in vivo, and conventional fluorine-19 (19F) nuclear magnetic resonance (NMR) spectroscopy is used as a quantitative readout of inflammation in intact, excised tissue samples. The approach requires no tissue preparation other than an optional fixation step.
Assaying inflammation often involves tissue-destructive methods. Commonly, thin-sectioning (4–10 µm) of embedded tissues is used in combination with one of more stains, for example, using colorimetric hematoxylin and eosin (H&E), or using immunoreactive reagents that highlight cell-specific markers or molecular events. Organ analysis of inflammation often requires serial sectioning and the preparation of a large number of slides. Periodic sampling of serial sections for the sake of accelerating analysis may lead to bias or quantification inaccuracies, or may necessitate a large experimental group to achieve statistical significance. Thus, widespread biodistribution analysis of a large panel of tissues taken from a single animal—much less an experimental group—can be a laborious undertaking. Although great strides have been made to increase tissue histology throughput, section preparation and analysis can only be made semiautomated at this time.
Alternatively, ‘bulk’ cellular, biochemical, or molecular analyses of disrupted tissue samples have been developed to assay inflammation. For example, approaches include monitoring the expression levels of selected biomarkers in peripheral blood mononuclear cells using flow cytometry (1), mRNA profiling using real-time PCR, or ELISA to detect monocyte-specific markers or pro-inflammatory cytokines (2,3).
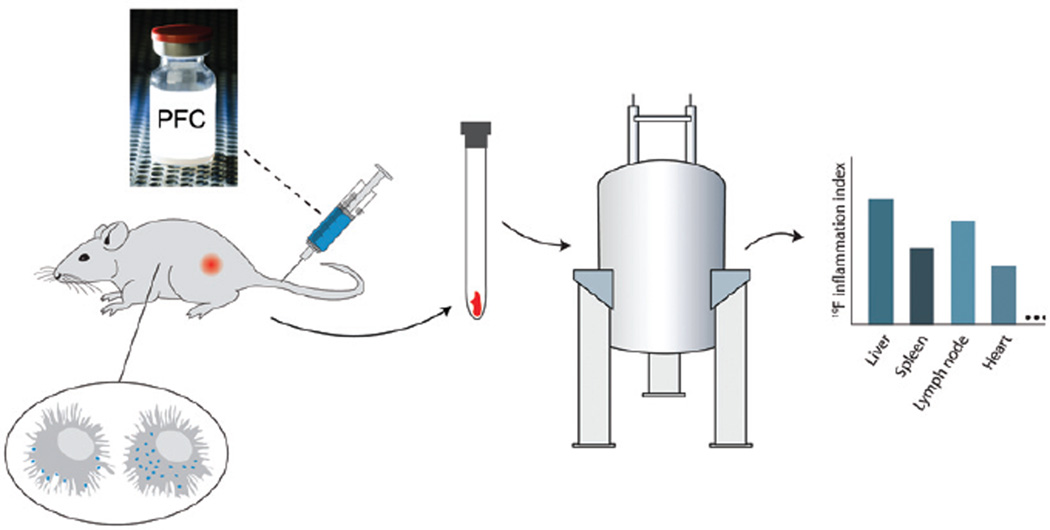
In this article, we describe an alternative approach to assay inflammation (Figure 1), without the need for tissue disruption, thereby potentially accelerating studies. The approach uses a PFC emulsion reagent comprised of a colloidal suspension of non-toxic PFC formulated into small (~150-nm diameter) emulsion droplets. Following i.v. injection, the droplets are taken up by phagocytic cells (4–7), predominately monocytes and macrophages, and to a small degree neutrophils. These ‘in situ’ labeled cells subsequently participate in inflammatory events in vivo, resulting in an accumulation of PFC at inflammatory loci. Necropsied tissues samples, or intact organs, are subjected to conventional 19F NMR analysis to quantitatively measure the total fluorine content per sample, which is proportional to the inflammatory burden. There is negligible natural abundance of 19F in soft tissues, thus this approach offers high specificity to inflammation.
Figure 1. Overview of inflammation quantification of intact tissue samples using 19F NMR.
PFC emulsion is injected i.v. and is taken up by monocytes and macrophages. These labeled cells participate in inflammatory events in vivo resulting in an accumulation of 19F at inflammatory loci. Conventional 19F NMR spectroscopy of panels of intact tissue samples is used to measure histogram profiles of the inflammatory index (IFI), proportional to tissue macrophage burden.
PFC-based reagents are biologically safe, even at very high doses. PFC emulsions have been studied clinically for many years as potential artificial blood substitutes in humans (8,9), although the PFC molecule used in the present study is different than those used for blood substitutes and has been optimized for 19F NMR-MRI applications. Numerous in vitro studies have shown that intracellular labeling with PFC does not affect cell phenotype and function (10–14).
As an example of the applicability of these methods, we deliver PFC emulsion reagent intravenously to the dark agouti (DA) rat model of experimental allergic encephalomyelitis (EAE) (15), a widely used model for human multiple sclerosis (MS). EAE has many clinical and histopathological similarities to MS (16). EAE is most commonly induced by immunizing animals with myelin proteins or their disease-inducing peptides (16). Clinical disease develops when primed CD4+ T cells enter the central nervous system (CNS) and recognize their cognate self-antigen presented in the context of MHC class II molecules. The resulting perivascular and parenchymal infiltrations in the CNS, particularly in the spinal cord, include a large number of macrophages and eventually lead to demyelination and clinical paralysis. In the EAE model, the ability to rapidly assay the inflammatory biodistribution in the CNS would be tremendously beneficial in attempts to understand the pathogenesis of autoimmunity and to design therapeutic interventions. Overall, the methods described herein can dramatically reduce the time to evaluate macrophage involvement in a wide variety of acute and chronic inflammatory models.
Materials and methods
EAE model and PFC administration
Experiments were carried out in accordance with the guidelines provided by the Carnegie Mellon Institutional Animal Care and Use Committee (IACUC) and the National Institute of Health Guide for the Care and Use of Laboratory Animals. The EAE rat model was generated using previously reported methods (15). Adult female DA rats (Harlan, Indianapolis, IN, USA), 11 weeks old, were inoculated with emulsified syngeneic spinal cord. The inoculant was prepared by homogenizing frozen DA spinal cord (50 mg/rat) with incomplete Freud’s adjuvant (IFA; cat. no. DF0639–60–6; Difco, Detroit, MI, USA) at 200 µL/rat and Mycobacterium tuberculosis (H37Ra; cat. no. DF3114–33–8; Difco) at 2 mg/mL. The rat’s tail base was shaved, and a single injection of the homogenate was delivered subcutaneously in a total of n = 11 rats. A control group of rats (n = 3) were also prepared by inoculating with adjuvant in the same manner, except spinal cord homogenate and H37Ra were excluded. After induction, rats were monitored daily for body weight and clinical symptoms of EAE. Symptoms were scored according to the convention: 0 = normal; 1 = limp tail; 2 = paraparesis with a clumsy gait; 3 = hindlimb paralysis; 4 = hind- and forelimb paralysis; 5 = moribund.
Upon the first signs that an animal reached clinical stage 2—typically 16–20 days after induction in 93% of the subjects—rats received a single i.v. injection (0.5 mL) of PFC emulsion (Cat. no. VS-580; Celsense, Inc., Pittsburgh, PA, USA) via the jugular vein. This sterile reagent contains emulsified perfluoro-15-crown-5 ether at a volume fraction of 20% (v/v), and a mean droplet size of 145 nm. Two days after injection with PFC rats were sacrificed.
In a subset of animals (n = 4) the PFC emulsion was rendered fluorescent prior to injection to aid in histological analysis. This was achieved via adding a simple premix step of the PFC emulsion with a lipophilic dialkylcarbocyanine (DiI; cat. no. V22885; Molecular Probes, Inc., Eugene, OR, USA) fluorophore. The fluorescent inoculant was prepared by mixing 5 mL emulsion with a 2-µL/mL DiI stock solution prepared according to the manufacturer’s instructions. After incubating the emulsion with DiI for 20 min at room temperature under gentle agitation, the DiI molecules became associated with the PFC emulsion droplets (14). The fluorescent emulsion was then injected into the stage 2 EAE rat.
Tissue preparation for NMR
Two days after PFC injection, rats (n = 8) were anesthetized and sacrificed by transcardial perfusion with PBS followed by 4% paraformaldehyde (PFA). The intact dorsal columns and brain were excised and stored in 4% PFA for >24 h. Using a razor blade, the entire dorsal column, except the sacral region, was partitioned at intervertebral discs into a total of 15 segments, where each segment contained 1–3 vertebrae. The spinal cord was dissected from each segment, discarding the vertebral bone and spinal nerves, and each segment was weighed. Using a scalpel, the intact brain stem and cerebellum was also dissected from the excised brain and weighed. Other organs were also necropsied and fixed in control rats for PFC biodistribution analysis.
19F NMR
Fixed CNS samples and other tissues from EAE and control rats were subjected to 19F NMR analysis. The weighed tissue samples were placed at the bottom of quartz 5-mm NMR tubes (Cat. no. WG-1000; Wilman-Labglass, Vineland, NJ, USA). Importantly, the specimen size was small enough to fit entirely within the homogenous receptive field of the NMR probe so that all 19F was detected. (The spinal cord segments of 1–3 vertebrae in length, brain stem, and cerebellum easily met this criterion.) A 10-µL, 2% (v/v) aliquot of trifluoroacetic acid (TFA) was placed in a flame-sealed 1 mm–diameter glass capillary tube (Cat. no. 34507-99; Kimble Kontes, Vineland, NJ, USA) and positioned inside the NMR tube next to the tissue sample; this TFA reference contained a total of NF = 1.2 × 1019, where NF is the number of fluorine atoms. A one-dimensional 19F NMR spectrum was acquired for each sample at 470 MHz using a Bruker spectrometer (Bruker Biospin, Billerica, MA, USA) with a delay time of 10 s and 32 averages. Two distinct peaks were observed at approximately −76 ppm for TFA and −92 ppm for the PFC. The total fluorine content (CF) of the sample was calculated from the integrated peak areas for the PFC (IPFC) and TFA (ITFA) reference using the formula CF = IPFCNF/ITFA. Results were normalized to the tissue weight (w), yielding an inflammation index (IFI) of IFI = CF/w, expressed as the number of 19F atoms per gram of tissue.
Histology and immunohistochemistry
After NMR analysis, selected spinal cords were paraffin-embedded and sectioned at 10-µm thickness and stained with H&E using standard procedures. Immumohistochemistry was also performed on selected spinal cords that were flash-frozen in OCT compound (Qiagen, Valencia, CA, USA) and cryosectioned at 6-µm thickness. Sections were stained for CD68 using ED1 as the primary antibody (MCA341R; AbD Serotec, Inc., Raleigh, NC, USA), followed by a secondary horse anti-mouse antibody conjugated to FITC dye (Cat. no. FL-2000; Vector Laboratories, Burlingame, CA, USA). We also stained for astrocytes using rabbit anti-GFAP (Cat. no. ab7260; Abcam, Inc., Cambridge, MA, USA) follow by a secondary Alexa Fluor488–conjugated, donkey anti-rabbit antibody (Cat. no. A21206; Invitrogen, Carlsbad, CA, USA). Stained sections were imaged using a Zeiss 510 LSM UV Duoscan microscope (Carl Zeiss MicroImaging, Thornwood, NJ, USA).
Real-time PCR
In selected EAE rats (n = 3), fresh spinal cord segments were retrieved and partitioned, as above. The tissue segments were then further cut in half, where one part was fixed and subjected to 19F NMR measurement as described in the “19F NMR” section, and the other half was preserved in RNAlater (Ambion, Austin, TX, USA) for real-time PCR analysis. Total RNA was extracted for the PCR samples using TRIzol (Invitrogen) following the manufacturer’s instructions. From each sample, 2 µg total RNA were reverse-transcribed into first strand cDNA with SuperScript III (Invitrogen) and poly-T primers. Multiplexed, real-time PCR was performed on a Stratagene MX3000 (Stratagene, La Jolla, CA, USA) to evaluate the expression of CD68, a biomarker for monocytes and tissue macrophages, as well as astrocyte-specific glial fibrillary acidic protein (GFAP). Expression levels of CD68 and GFAP in each sample were normalized to expression of the ‘housekeeping gene’ glyceraldehyde 3-phosphate dehydrogenase (gapdh). The primer sequences are listed below (where F is a forward primer and R is a reverse primer): CD68 F: 5′-GCATAGTTCTTTCTCCAGCAATTCACCT-3′, CD68 R: 5′-GAGAGGCAGCAAGAGAGATTGGTCA-3′, and CD68 probe: 5′-FAM-AGGGACACTTCGGGCCATGCTTCTCTT-Iowa FQ; GFAP F: 5′-GCCTTGACCTGCGACCTTGAGT-3′, GFAP R: 5′-GTGCCTCCTGGTAACTCGCCG-3′, and GFAP probe: 5′-Cy5-CGGCACGAACGAGTCCTTGGAGAGGC-IAbRQSp-3′; GAPDH F: 5′- TGAACGGGAAGCTCACTGGCAT-3′, GAPDH R: 5′-CGCCTGCTTCACCACCTTCTTG-3′, and GAPDH probe: 5′-HEX-CCGCCTGGAGAAACCTGCCAAGTATGATGAC-Iowa FQ.
Results and discussion
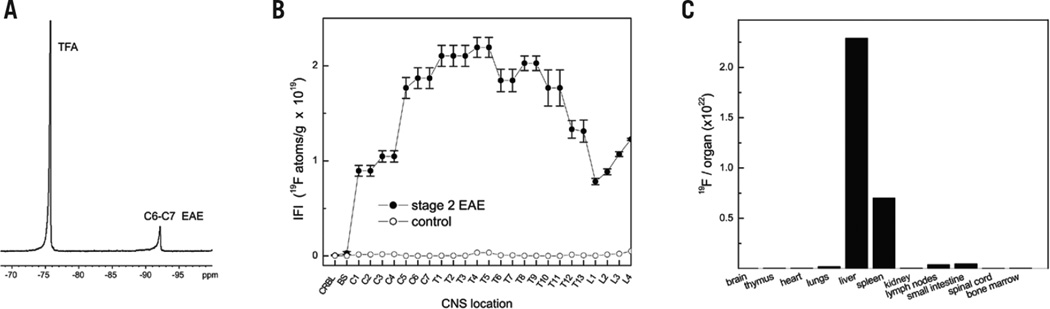
We used NMR measurements on the tissue samples to quantify macrophage burden. Figure 2A displays a typical 19F spectrum of a spinal cord segment; it shows a single PFC peak at −92 ppm and a TFA peak at −76 ppm from the sealed reference capillary placed alongside the sample. From such data, we calculated the IFI along the length of the spinal cord (Figure 2B), where the IFI values represent the apparent number of 19F nuclei per tissue weight. These data represent the mean of n = 8 spinal cords, where the vertebrae are enumerated along the abscissa. The 19F content of the tissue—and thus IFI—is linearly proportional to the amount of macrophage present in the tissue. The IFI results (closed circles, Figure 2B) are consistent with the known topology of the inflammation distribution in the CNS in this EAE rat model, which is particularly prevalent in the spinal cord (15). The EAE CNS, especially the spinal cord, is generally the focus of study when evaluating therapeutic candidates for MS.
Figure 2. NMR results in the EAE (stage 2) and control rats.
(A) 19F NMR spectrum of a fixed, intact cervical spinal cord segment from EAE rat. The PFC displays a single peak at −92 ppm and the TFA reference peak is at −76 ppm. (B) Average inflammation index (IFI) along spinal cord axis in EAE (n = 8) and control (n = 3) rats (closed and open circles, respectively). CRBL, cerebellum; BS, brainstem. The IFI is expressed as the number of 19F atoms per gram of tissue. (C) 19F NMR measurements of organ and tissue biodistribution of the PFC emulsion in a control rat shows RES uptake.
The control group (open circles, Figure 2B) receiving a CFA injection, but no spinal cord homogenate, showed minimal 19F uptake in the CNS, indicating that the PFC emulsion does not cross the intact blood-brain barrier or enter into the CNS in the absence of autoimmune disease.
Overall, in these experiments the total preparation and analysis time per rat CNS was ~6 h, which is vastly accelerated compared with most conventional histological analyses. Moreover, utilization of robotic or automated NMR sample-changing devices, commonplace in many NMR laboratories, can enable rapid data acquisition of tissue panels.
The PFC emulsion droplets are generally taken up by the reticuloendothelial system (RES) (4–7). This behavior was confirmed by 19F NMR using excised tissues and organs from a control DA rat (Figure 2C).
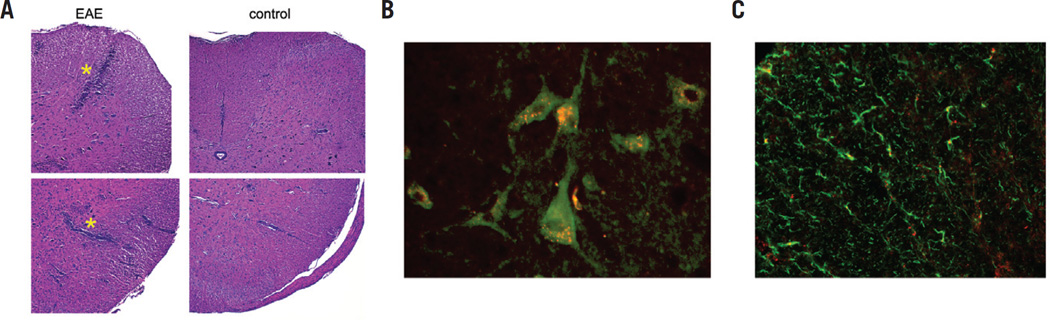
To validate the cell type incorporating the PFC emulsion droplets, the same tissues were examined histologically following NMR. Figure 3A shows H&E staining in the EAE (left panel) and control (right panel) cervical spinal cord white matter; the panels show the characteristic EAE pathology with a large number of perivascular mononuclear infiltrates compared with control.
Figure 3. Histological sections of cervical EAE (stage 2) and control spinal cords.
(A) H&E stained section (10×) in EAE (left panel) and control (right panel) spinal cords, where the top and bottom panels are dorsal and ventral white matter, respectively. EAE pathology (asterisks) shows significant perivascular infiltrates (blue) compared with control. (B) Fluorescent images (40×) of immunostained white matter regions showing punctate deposits of PFC-DiI (red) within CD68+ cells (green). Data show that PFC droplets colocalized within CD68+ cells that have morphologies consistent with monocytes and macrophages. (C) Fluorescent micrograph (40×) showing an absence of DiI-PFC (red) colocalization in GFAP-stained cells (green).
Using immunohistochemistry in the EAE spinal cord, we also confirmed that the PFC emulsion droplets colocalized within CD68+ cells that have morphologies consistent with monocytes and macrophages. For these experiments, we used stage 2 EAE rats that were injected with pretreated PFC emulsion such that the droplets had surface-bound fluorescent DiI. The fluorescent microscopy images of white matter regions of the spinal cord display punctate deposits of DiI (red, Figure 3B) within immunostained CD68+ cells (green, Figure 3B) consistent with perinuclear vesicular localization of the PFC. The intracellular localization of PFC is reminiscent of what has been previously observed in dendritic cells (14,17). In contrast, DiI-PFC was not observed in glial cells that were immunostained for GFAP (Figure 3C). This indicates that no apparent PFC was taken up by astrocytes.
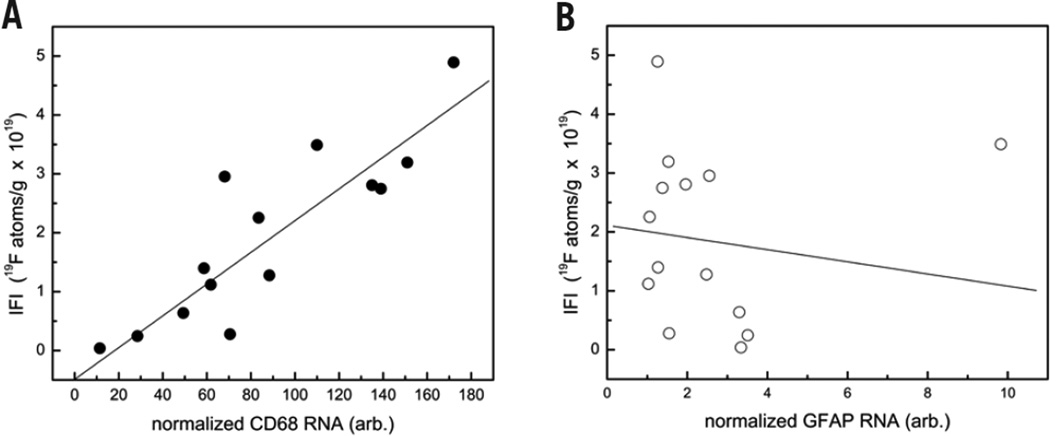
To confirm that the IFI linearly correlates to the density of monocyte/macrophage, we conducted quantitative PCR studies in EAE CNS tissue. Figure 4A shows a representative correlation analysis from a single stage 2 EAE rat, where the ordinate displays IFI obtained from NMR, and the abscissa displays the CD68 mRNA levels normalized to the GAPDH housekeeping gene. A linear regression analysis yields an R correlation coefficient of 0.89, showing a high degree of linearity between IFI and CD68 levels (Figure 4A). Overall, the average R correlation coefficient calculated for all animals studied (n = 3) was 0.68.
Figure 4. Quantitative analysis of macrophage burden in EAE spinal cord tissue using real-time PCR in a single rat.
(A) Correlation between IFI, obtained from 19F NMR, and CD68 mRNA levels. Data shows a linear correlation (R = 0.89), consistent with the PFC internalization into macrophage. The mRNA levels are normalized to gapdh. (B) Plot of IFI versus GFAP mRNA shows no correlation (R = −0.13), suggesting that PFC is not taken up by astrocytes.
Since astrocytes can also express low levels of CD68, to rule out their involvement in PFC uptake we also performed real-time PCR with the astrocyte-specific marker GFAP in a multiplexed, real-time PCR reaction along with CD68 and GAPDH. We did not observe any correlation between GFAP and 19F uptake (R = −0.13; Figure 4B).
The elevation of the CD68 mRNA level in EAE spinal cord was ≥100-fold higher compared with spinal cord samples from control rats (data not shown), where only minimal CD68 mRNA levels were detected. Thus, CD68 expressed in astrocytes and other glial cells is relatively low, and the CD68 detected in EAE spinal cord was mainly contributed by EAE-associated monocyte or macrophage infiltrates. The mRNA profiling data, in combination with the histological observation of PFC localization only within CD68+ cells with monocyte-macrophage morphology, suggest that PFC uptake and the measured IFI reflects the inflammation burden.
The PFC molecules used are not degraded in vivo by any known enzyme or at low pH (5,6). The 19F isotope has 100% natural abundance and is stable. In contrast, 18F-based positron emission tomography (PET) probes used in EAE studies have a limited half-life (18). The blood half-life of the PFC emulsion formulation used is ~9.4 ± 2.6 h in a DA rat (19), but may depend on species/strain and inflammation status of the subject. As shown in the control rats, the emulsion does not appear to cross the intact blood-brain barrier. The PFC emulsion is taken up by macrophages and monocytes, possibly in blood, and then carried into inflammatory loci. Generally, clearance of the PFC emulsion is via the reticuloendothelial system and exhalation through the lungs (4–6).
Prior work has reported the use of in vivo labeling and imaging of macrophage using PFC emulsion in various animal models (7,20–22), including early work in an EAE model (7); these studies mainly focus on the use of in vivo 19F MRI techniques. However, small animal MRI instruments are much less commonplace compared with ubiquitous NMR spectrometers. Moreover, the cell detection sensitivity of high-field NMR instruments greatly exceeds that of MRI instrumentation for detection of sparse cell numbers. We estimate that one can detect on the order of 103 cells per sample using high-field NMR instrumentation (e.g., 11.7 T), whereas high-field MRI detection limits are closer to 103–104 cells per voxel (13).
This study used tissue samples that were small enough to fit inside a standard 5-mm NMR tube and remain in the homogenous region of NMR coil. In mouse, intact organ tissues (spleen, lymph nodes, thymus, etc.) meet these criteria. For larger tissue samples (midbrain, heart, liver, etc.), several options are available: (i) the total organ or tissue can be manually segmented into discrete samples, each measured separately [this is the approach that we used for our study of CNS tissues (Figure 2B)]; (ii) one can dissect and weigh a piece of the whole tissue or organ of interest that fits into the NMR tube and use this for the IFI measurement [this approach assumes that the inflammation (i.e., 19F) distribution is roughly homogenous throughout the whole tissue or organ]; (iii) a tissue homogenate can be made by mechanical disruption, and a weighed aliquot is placed in the NMR tube to assay IFI of the sample; (iv) one can use larger-diameter NMR hardware (i.e., NMR tube and probe); for example, a standard 10-mm NMR tube can readily accommodate an intact mouse brain.
These same whole-tissue NMR methods can also be useful for ‘counting’ transplanted therapeutic or diagnostic cells that have been labeled with PFC emulsion ex vivo prior to transfer in vivo (13,23). A multitude of regenerative medicine and immunotherapeutic cell therapy studies could benefit from a tool that does not rely on laborious histology or flow cytometry to accurately assay cell biodistribution in tissue panels. Moreover, ex vivo labeled, phenotypically defined, immune cells can also be used for inflammation diagnostic purposes when infused into a subject. Using ex vivo labeling approaches, the mean PFC uptake, expressed as 19F/cell, is often measured as part of the cell labeling protocol development (13), and this parameter can be used to calculate the apparent number of cells present in tissue samples directly from the 19F NMR data (13,23).
In summary, we show that it is feasible to rapidly assay monocyte/macrophage burden in intact tissues using PFC emulsion administered in vivo and a conventional NMR scanner. These methods can be used to accelerate routine analysis of inflammation in discovery and preclinical studies.
Acknowledgments
We thank Kevin Hitchens and Virgil Simplaceanu for helpful discussion. This work was supported by the National Institutes of Health (NIH; grant nos. R01-CA134633, R01-EB003453, P01-HD047675, P41-EB001977). This paper is subject to the NIH Public Access Policy.
Footnotes
Competing interests
E.T.A. is a consultant for Celsense, Inc. All other authors declare no competing interests.
Author Reprints
Make the most of your hard work by ordering reprints of your article published in BioTechniques. Reprints are an inexpensive and easy way to distribute your findings to students and colleagues alike.
Corporate Reprints
Leverage BioTechniques, the most powerful brand in the market. Reprints help support your sales effort by utilizing articles that spotlight your brand/products to educate customers at meetings and industry trade events.
More info:
References
- 1.Afanasyeva M, Georgakopoulos D, Belardi DF, Ramsundar AC, Barin JG, Kass DA, Rose NR. Quantitative analysis of myocardial inflammation by flow cytometry in murine autoimmune myocarditis - correlation with cardiac function. Am. J. Pathol. 2004;164:807–815. doi: 10.1016/S0002-9440(10)63169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berti R, Williams AJ, Moffett JR, Hale SL, Velarde LC, Elliott PJ, Yao CP, Dave JR, Tortella FC. Quantitative real-time RT-PCR analysis of inflammatory gene expression associated with ischemia-reperfusion brain injury. J. Cereb. Blood Flow Metab. 2002;22:1068–1079. doi: 10.1097/00004647-200209000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Osuchowski MF, Siddiqui J, Copeland S, Remick DG. Sequential ELISA to profile multiple cytokines from small volumes. J. Immunol. Methods. 2005;302:172–181. doi: 10.1016/j.jim.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Lowe KC. Engineering blood: Synthetic substitutes from fluorinated compounds. Tissue Eng. 2003;9:389–399. doi: 10.1089/107632703322066570. [DOI] [PubMed] [Google Scholar]
- 5.Riess JG. Understanding the fundamentals of perfluorocarbons and perfluorocarbon emulsions relevant to in vivo oxygen delivery. Artif. Cells Blood Substit. Immobil. Biotechnol. 2005;33:47–63. doi: 10.1081/bio-200046659. [DOI] [PubMed] [Google Scholar]
- 6.Srinivas M, Turner MS, Janjic JM, Morel PA, Laidlaw DH, Ahrens ET. In vivo cytometry of antigen-specific T cells using F-19 MRI. Magn. Reson. Med. 2009;62:747–753. doi: 10.1002/mrm.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helfer BM, Balducci A, Nelson AD, Janjic JM, Gil RR, Kalinski P, De Vries IJM, Ahrens ET, Mailliard RB. Functional assessment of human dendritic cells labeled for in vivo F-19 magnetic resonance imaging cell tracking. Cytotherapy. 2010;12:238–250. doi: 10.3109/14653240903446902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janjic JM, Ahrens ET. Fluorine-containing nanoemulsions for MRI cell tracking. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2009;1:492–501. doi: 10.1002/wnan.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn. Reson. Med. 2007;58:725–734. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- 10.Ahrens ET, Flores R, Xu HY, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat. Biotechnol. 2005;23:983–987. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- 11.Lorentzen JC, Issazadeh S, Storch M, Mustafa MI, Lassman H, Linington C, Klareskog L, Olsson T. Protracted, relapsing and demyelinating experimental autoimmune encephalomyelitis in DA rats immunized with syngeneic spinal cord and incomplete Freund’s adjuvant. J. Neuroimmunol. 1995;63:193–205. doi: 10.1016/0165-5728(95)00153-0. [DOI] [PubMed] [Google Scholar]
- 12.Martin R, McFarland HF. Immunological aspects of experimental allergic encephalomyelitis and multiple sclerosis. Crit. Rev. Clin. Lab. Sci. 1995;32:121–182. doi: 10.3109/10408369509084683. [DOI] [PubMed] [Google Scholar]
- 13.Janjic JM, Srinivas M, Kadayakkara DKK, Ahrens ET. Self-delivering nanoemulsions for dual fluorine-19 MRI and fluorescence detection. J. Am. Chem. Soc. 2008;130:2832–2841. doi: 10.1021/ja077388j. [DOI] [PubMed] [Google Scholar]
- 14.Krafft MP. Fluorocarbons and fluorinated amphiphiles in drug delivery and biomedical research. Adv. Drug Deliv. Rev. 2001;47:209–228. doi: 10.1016/s0169-409x(01)00107-7. [DOI] [PubMed] [Google Scholar]
- 15.Krafft MP, Riess JG. Perfluorocarbons: Life sciences and biomedical uses: dedicated to the memory of Professor Guy Ourisson, a true RENAISSANCE man. J. Polym. Sci. Part A: Polym. Chem. 2007;45:1185–1198. [Google Scholar]
- 16.Radu CG, Shu CJ, Shelly SM, Phelps ME, Witte ON. Positron emission tomography with computed tomography imaging of neuroinflammation in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA. 2007;104:1937–1942. doi: 10.1073/pnas.0610544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitchens TK, Ye Q, Eytan DF, Janjic JM, Ahrens ET, Ho C. 19F MRI detection of acute allograft rejection with in vivo perfluorocarbon labeling of immune cells. Magn. Reson. Med. 2011 doi: 10.1002/mrm.22702. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGoron AJ, Pratt R, Zhang J, Shiferaw Y, Thomas S, Millard R. Perfluorocarbon distribution to liver, lung and spleen of emulsions of perfluorotributylamine (FTBA) in pigs and rats and perfluorooctyl bromide (PFOB) in rats and dogs by F-19 NMR spectroscopy. Artif. Cells Blood Substit. Immobil. Biotechnol. 1994;22:1243–1250. doi: 10.3109/10731199409138822. [DOI] [PubMed] [Google Scholar]
- 19.Noth U, Morrissey SP, Deichmann R, Jung S, Adolf H, Haase A, Lutz J. Perfluoro-15-crown-5-ether labelled macrophages in adoptive transfer experimental allergic encephalomyelitis. Artif. Cells Substit. Immobil. Biotechnol. 1997;25:243–254. doi: 10.3109/10731199709118914. [DOI] [PubMed] [Google Scholar]
- 20.Lanza GM, Winter PM, Neubauer AM, Caruthers SD, Hockett FD, Wickline SA. 1H/19F magnetic resonance molecular imaging with perfluorocarbon nanoparticles, pp. 58–78. In: Ahrens ET, editor. In Vivo Cellular and Molecular Imaging. San Diego: Elsevier; 2005. [DOI] [PubMed] [Google Scholar]
- 21.Flogel U, Ding Z, Hardung H, Jander S, Reichmann G, Jacoby C, Schubert R, Schrader J. In vivo monitoring of inflammation after cardiac and cerebral ischemia by fluorine magnetic resonance imaging. Circulation. 2008;118:140–148. doi: 10.1161/CIRCULATIONAHA.107.737890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebner B, Behm P, Jacoby C, Burghoff S, French BA, Schrader J, Flögel U. Early assessment of pulmonary inflammation by 19F MRI in vivo. Circ. Cardiovasc. Imaging. 2010;3:202–210. doi: 10.1161/CIRCIMAGING.109.902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadayakkara DK, Beatty PL, Turner MS, Janjic JM, Ahrens ET, Finn OJ. Inflammation driven by overexpression of the hypoglycosylated abnormal mucin 1 (MUC1) links inflammatory bowel disease and pancreatitis. Pancreas. 2010;39:510–515. doi: 10.1097/MPA.0b013e3181bd6501. [DOI] [PMC free article] [PubMed] [Google Scholar]