Abstract
Anemones of genus Exaiptasia are used as model organisms for the study of cnidarian-dinoflagellate (genus Symbiodinium) endosymbiosis. However, while most reef-building corals harbor Symbiodinium of clade C, Exaiptasia spp. anemones mainly harbor clade B Symbiodinium (ITS2 type B1) populations. In this study, we reveal for the first time that bleached Exaiptasia pallida anemones can establish a symbiotic relationship with a clade C Symbiodinium (ITS2 type C1). We further found that anemones can transmit the exogenously supplied clade C Symbiodinium cells to their offspring by asexual reproduction (pedal laceration). In order to corroborate the establishment of stable symbiosis, we used microscopic techniques and genetic analyses to examine several generations of anemones, and the results of these endeavors confirmed the sustainability of the system. These findings provide a framework for understanding the differences in infection dynamics between homologous and heterologous dinoflagellate types using a model anemone infection system.
Keywords: Endosymbiology, Marine biology, Microalgae
Introduction
The sea anemone Exaiptasia pallida is a widespread species that has been well-adopted as a model animal for the study of cnidarian endosymbiology, particularly with associations featuring the dinoflagellate algae Symbiodinium sp. (Weis et al., 2008; Grajales & Rodriguez, 2016). In the laboratory, bleached anemones can be prepared by cold shock treatment (Muscatine, Grossman & Doino, 1991) and then maintained for several years in laboratory culture. Recently, genetic examinations of field-collected specimens and laboratory infection demonstrate that E. pallida anemones primarily harbor Symbiodinium spp. of Symbiodinium minutum (ITS2 type B1) and Symbiodinium A4 (ITS2 type A4), and in rare cases, a mixed population of Symbiodinium B1 and C1 (Thornhill et al., 2013; Grajales, Rodriguez & Thornhill, 2016), which can be readily isolated from these anemones and cultured in vitro (Kinzie III et al., 2001; Wang et al., 2008; Peng et al., 2012; Xiang et al., 2013). By infecting bleached anemones with free-living Symbiodinium, the endosymbiotic relationship can then be re-established and tracked in order to understand the recognition processes that occur at the molecular level and culminate in successful mutualisms (Weis et al., 2008; Kinzie & Chee, 1979; Lin, Wang & Fang, 2000; Chen et al., 2004; Hong et al., 2009; Wang et al., 2013; Hambleton et al., 2014; Xiang et al., 2013).
In our previous research involving the infection of E. pallida anemones with various Symbiodinium sp., we discovered the uptake and consequent cellular proliferation of a cultured Symbiodinium of clade C, a lineage of dinoflagellates known to predominantly infect reef corals (Chen et al., 2005; LaJeunesse et al., 2003; LaJeunesse et al., 2004a; LaJeunesse et al., 2008; LaJeunesse et al., 2010; Lien, Fukami & Yamashita, 2012). This is a particularly interesting finding, and it needs to be confirmed if this Exaiptasia-clade C Symbiodinium association is a sustainable endosymbiotic relationship. Furthermore, if the nature of this association is proven to be similar to that of corals, this Exaiptasia-clade C Symbiodinium association deserves even more merit as a model system for understanding reef corals, which cannot be successfully bleached and re-infected due to the stress it imposes on the corals (i.e., they are obligately endosymbiotic).
Metabolic relationships between corals and Symbiodinium are functionally diverse, depending largely on the genetic identity of the latter (Baker et al., 2004; Abrego et al., 2008; Stat, Morris & Gates, 2008; Yuyama, Harii & Hidaka, 2012; Yuyama & Higuchi, 2014). For instance, corals associated with Symbiodinium of clade D have been shown to possess an enhanced degree of thermal tolerance (Baker et al., 2004). Understanding the physiological consequences of engaging in an endosymbiotic relationship with dinoflagellates of differing identity would then be useful in formulating predictions as to how anemones, or even reef corals, may respond to global climate change. To further corroborate our previously unpublished findings and gain greater insight into the ability to develop a heterologous anemone-Symbiodinium infection system, we co-cultured exogenously supplied Symbiodinium C1 with bleached anemones (infection trial). The Symbiodinium C1-infected anemones were then maintained in the laboratory for more than one year, and asexual reproduction (pedal laceration) of Symbiodinium C1-infected anemones was observed.
Materials and Methods
Preparation of clade C Symbiodinium-infected anemones
The sea anemones, Exaiptasia pallida, were collected from the tanks in the Husbandry Centre of the National Museum of Marine Biology and Aquarium. The origin of the anemones comes from the location beneath the native habitat (N22 03 00.08 E120 41 42.88) of Exaiptasia pallida, which harbor Symbiodinium minutum (ITS2 type B1) (Grajales & Rodriguez, 2014; Grajales & Rodriguez, 2016; Grajales, Rodriguez & Thornhill, 2016). Clade C Symbiodinium (CCMP 2466) were purchased from the National Center for Marine Algae and Microbiota (NCMA), which were originally isolated from the corallimorph Discosoma sanctithomae in the Caribbean Sea (https://ncma.bigelow.org/ccmp2466). The Symbiodinium were cultured in the laboratory according to a previously published protocol (Peng et al., 2012) for several years, and its genetic identity was confirmed to be ITS2 type C1 (Fig. S1; Krueger et al., 2015). Briefly, Symbiodinium cells were cultured in Guillard’s (f/2) media (without silica, Cat. G0154, Sigma-Aldrich, USA) containing antibiotics (10 mg ml−1 streptomycin and 10 units ml−1 penicillin; Cat. 15140-122, Gibco, USA) and maintained at 25 °C with a photoperiod of 12 h light (∼40 µmol m−2 s−1): 12 h dark (12L/12D). The Symbiodinium cultures were changed every week. The infection trials were performed by collecting f/2 media-cultured clade C Symbiodinium in the early stationary phase via centrifugation (800 xg for 5 min) and then re-suspending them in filtered seawater (FSW, 0.22 µm). Bleached, juvenile sea anemones (from a clonal Exaiptasia pallida line; ∼2–3 mm in height) were prepared via repeated cold shock treatment until they were completely bleached (Muscatine, Grossman & Doino, 1991). Then, the bleached anemones were maintained in the dark. In order to confirm there was no contamination of Symbiodinium cells within the bleached anemone before the infection trial, the anemones were cultured under a photoperiod of 12 h light (∼40 µmol m−2 s−1): 12 h dark (12L/12D) for one week. This period allowed Symbiodinium cells, if any, to replicate within the anemone. The presence of multiple Symbiodinium cells would more easily be observed. After the incubation period, the anemones were checked by the fluorescent stereomicroscope (AxioCam SteREO Discovery.V8; Zeiss, Germany) to ensure the absence of Symbiodinium cells. The bleached anemones were incubated with the clade C Symbiodinium (3 × 105 cells/ml) in a sterile 6-well cell culture plate (10∼15 anemones per well with 1.5 mL of infection solution). After 1 h of co-culturing, most of the anemones were infected with the clade C Symbiodinium. The infected sea anemones were washed twice with FSW to remove any Symbiodinium cells that were not taken up before being moved to a new tank.
Culturing of infected anemones and their offspring
Juvenile anemones infected with the clade C Symbiodinium were cultured in a tank (GEX GENOAH100, Japan; 10.8 × 10.8 × 12.9 cm; volume: 800 ml, with a transparent cover) under a photoperiod of 12 h light (∼34 µmol m−2 s−1): 12 h dark (12L/12D) at a temperature of 25 °C ± 1 °C and fed brine shrimp larvae once a week. The FSW in the tanks was changed twice a week. The clade C-infected anemones, which make up the first generation (G1), were maintained in the laboratory for more than one year, during which they matured and underwent asexual reproduction (pedal laceration). The production of lacerates (offspring) from the clade C-infected anemones was checked daily. The newly produced lacerates (the second generation, G2) were moved to a new tank for further development. When the G2 anemones grew up and began to produce lacerates, the lacerates (the third generation, G3) were moved to a new tank or a 6-well plate for further development. Lacerate development of the same specimen was observed in a 6-well plate and recorded by a fluorescent stereomicroscope until they became juveniles (∼9 days). The juvenile anemones were maintained in the same conditions as described above.
Restriction fragment length polymorphism (RFLP) analysis
For RFLP analysis, three individual clade-C Symbiodinium infected anemones or specimens, as well as their offspring, were collected. Several tentacles from both first generation (G1), the clade C Symbiodinium-infected anemones and second generation (G2, the offspring of G1) anemones were collected. Since the Symbiodinium number in a single tiny anemone of the third generation (G3) was not enough for efficient DNA extraction, 5–10 juvenile anemones were collected, pooled, and processed as a single specimen for analysis. To demonstrate that the anemone which originally harbors clade B Symbiodinium can now harbor clade C Symbiodinium, DNA from one representative anemone, which is from the same clonal line as the clade C-infected anemone and cultured under the same conditions but not subject to cold shock bleaching, was also analyzed. Genomic DNA of all specimens was extracted using the Plant Genomic DNA Extraction Miniprep System (Cat. GPG1001; Viogene, Taiwan) according to the manufacturer’s recommendations. Genomic fragments from small-subunit ribosomal RNA genes (18S rDNA) were amplified from nucleic acid samples using the Symbiodinium-specific PCR primers ss5z and ss3z as previously described (Rowan & Powers, 1991a). Briefly, the DNA amplification was performed in a DNA Thermal Cycler (GeneAmp PCR System 2700; Applied Biosystems) with the following thermal cycles: 1 cycle of 94 °C for 4 min, 55 °C for 30 s, 72 °C for 2 min; and then 29 cycles of 94 °C for 45 s, 55 °C for 30 s, 72 °C for 2 min; plus a final 5 min extension at 72 °C after the 29 cycles. PCR products were purified by High Pure PCR Product Purification Kit (Roche, Germany) and then digested with either the restriction enzyme Taq I or Sau3A I (New England Biolabs, USA) according to the manufacturer’s recommendations. The digested DNA fragments of each sample were separated by electrophoresis (110 V, ∼20 min) on 1.5% 0.5x TAE buffer (Amresco, USA) agarose gels, to generate the RFLP pattern. Comparing the obtained RFLP patterns with known patterns of Symbiodinium B1 and C1 (Rowan & Powers, 1991a; Rowan & Powers, 1991b; Rowan & Knowlton, 1995) enabled the confirmation of the genetic identities of the dinoflagellates.
Microscopic examination of Symbiodinium cells
Three juvenile G3 anemone specimens harboring clade C Symbiodinium and three juvenile anemone specimens harboring clade B Symbiodinium were anaesthetized with 7% MgCl2 in FSW for 5 min. The specimens were then placed on slides and covered with a 22 × 22-mm coverslip for observation with an upright microscope (Axioskop 2 Plus, Zeiss, Germany), and three differential interference contrast (DIC) images of each specimen’s tentacles were obtained. Using the open source, Java-based software package ImageJ (National Institutes of Health, Bethesda, MD, USA), the diameters of all Symbiodinium cells in each image were measured, and the average Symbiodinium cell size was calculated. A student’s t-test was used to identify differences between the two groups, which resulted in a p value < 0.01 indicating significance. All results have been expressed as mean ± SD. For reference, free-living cultured clade B and C Symbiodinium were centrifuged to concentrate the cells. Aliquots of each Symbiodinium solution was dropped on slides for imaging and cell size measurement as above.
Results and Discussion
To determine whether the clade C Symbiodinium (CCMP2466; ITS2 type C1) could be taken up by Exaiptasia anemones and then be transmitted to offspring after proliferation through the adult tissues, anemones infected with the clade C Symbiodinium were cultured for more than one year. During this period, juvenile anemones infected with Symbiodinium C1 grew from 2–3 mm in height to 2–3 cm in height, and Symbiodinium cells proliferated throughout the bodies of the specimens (Fig. 1B). Asexual reproduction (pedal laceration) was also observed and recorded (Fig. 1B). An aboral view of a representative anemone clearly illustrates the process of asexual reproduction, pedal laceration, in which the newly budded lacerates surround the pedal disk of the anemone (Fig. 1B). Furthermore, Symbiodinium cells were found to aggregate within the lacerates (Fig. 1B).
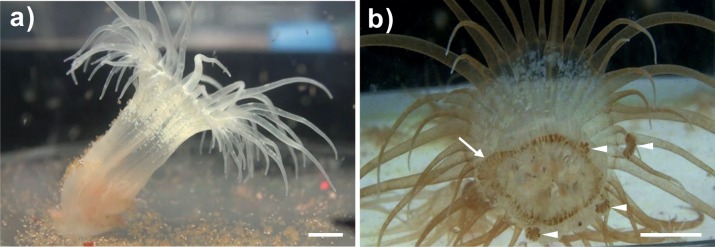
Figure 1. A representative bleached and a representative clade C Symbiodinium-infected anemone and its lacerates.
(A) Representative image of a bleached anemone that had lost its brownish coloration following the expulsion of Symbiodinium cells during cold shock-induced bleaching. (B) Aboral view of a representative clade C Symbiodinium-infected anemone showing brownish Symbiodinium cells distributed throughout the body, with a notable degree of dinoflagellate aggregation in the margins of the pedal disk (arrow), as well as within the newly budded lacerates (triangles). Scale bars: 0.5 cm.
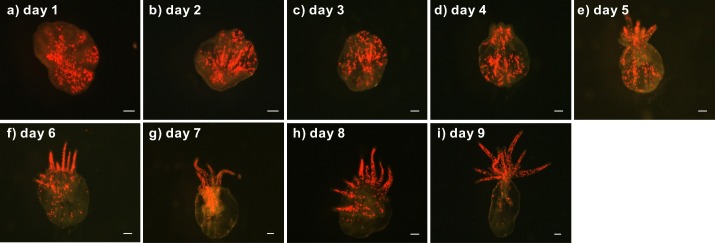
The lacerates of Symbiodinium C1-infected anemones developed into juvenile anemones within 9 days (Fig. 2). On day 1, red autofluorescence of the Symbiodinium cells was already readily observed with a fluorescence stereomicroscope (Fig. 2A). On days 3 and 4, tentacle tissue began forming and extended out from the top of the lacerates, as shown in Figs. 2C and 2D. Between days 5 and 9, the tentacle tissue extended significantly to form the shape of a juvenile anemone (Figs. 2E–2I). During lacerate development, Symbiodinium cells were re-distributed to the tentacles, where they are predominantly localized in healthy, adult anemones. Such localization suggests that these clade C Symbiodinium cells had successfully established a symbiotic relationship with the juvenile anemones.
Figure 2. Development of a representative lacerate collected from a clade C Symbiodinium-infected anemone.
First, the lacerate was transferred to a new dish immediately following laceration from the pedal disk of a clade C Symbiodinium-infected anemone. The development of the lacerate and spread of Symbiodinium was recorded daily using a fluorescent stereomicroscope (A–I). The red spots in the images indicate chlorophyll autofluorescence of the Symbiodinium cells. Scale bars: 100 µm.
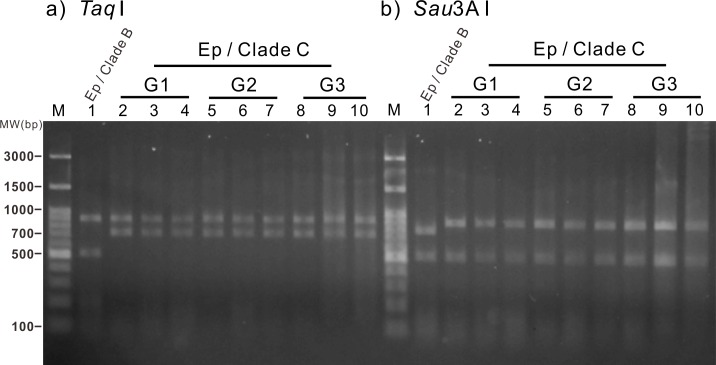
To confirm the sustainability and genetic identity of the clade C Symbiodinium within the infected anemones and their offspring, specimens of three generations of anemones (Fig. 3) were subjected to RFLP analysis of partially digested Symbiodinium 18S rDNA. Our results reveal that all three generations of the clade C Symbiodinium-infected anemones had the same RFLP pattern (Fig. 4). The major fragments were ∼900 and ∼750 bp when digested with TaqI and ∼900 and ∼500 bp when digested with Sau3AI (Fig. 4 lanes 2–10); such patterns are diagnostic of Symbiodinium C1 (Rowan & Powers, 1991a; Rowan & Powers, 1991b; Rowan & Knowlton, 1995). Furthermore, RFLP patterns of the clade C Symbiodinium-infected anemones were well-differentiated from the representative clade B Symbiodinium-infected anemone (Fig. 4, Lane 1), which came from the same clonal line as the clade C-infected anemones but was not subject to bleaching and re-infection. The RFLP patterns were ∼900 and ∼500 bp upon digestion with TaqI and ∼800 and 500 bp upon digestion with Sau3AI (Fig. 4, lane 1). These results clearly demonstrate that the anemones which originally harbored clade B Symbiodinium now harbor clade C and transmitted the symbiont for many generations.
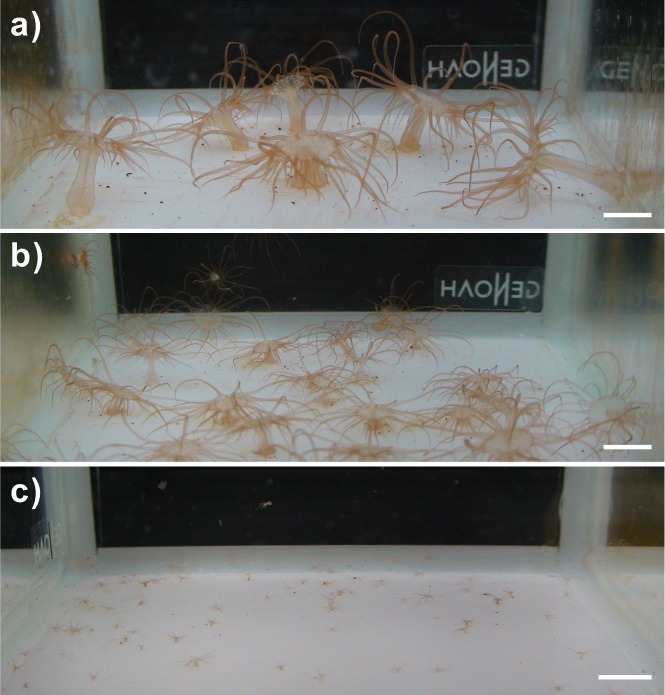
Figure 3. Images of the clade C Symbiodinium-infected anemones and their offspring.
(A) Generation 1 (G1) anemones maintained in the laboratory for more than one year; (B) Generation 2 (G2) anemones cultured for more than three months; (C) Generation 3 (G3) of anemone cultured for 15 days following laceration. Scale bar: 1 cm.
Figure 4. Restriction fragment length polymorphism (RFLP) analysis of three generations of the clade C Symbiodinium-infected anemones.
Amplified genomic fragments of small subunit ribosomal RNA genes (18S rDNA) from Symbiodinium were digested using the restriction enzymes TaqI and Sau3A I (A and B, respectively). Lane 1 (control): Normal anemone harboring clade B Symbiodinium. Lanes 2–4: Generation 1 (G1) anemones. Lanes 5–7: Generation 2 (G2) anemones. Lanes 8–10: Generation 3 (G3) anemones. Ep, Exaiptasia pallida; MW, molecular weight.
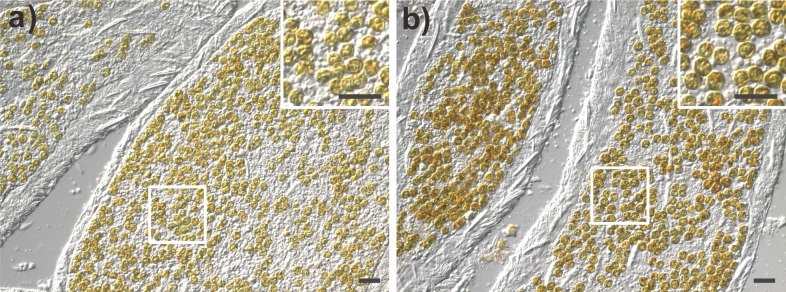
In addition to the RFLP analysis, the bright field images (Fig. 5) and diameter measurement of Symbiodinium further support the presence of clade C Symbiodinium cells in the anemones in contrast with the presence of clade B Symbiodinium. As shown in Fig. 5, the clade C Symbiodinium cells appear more darkly brownish than clade B cells. Moreover, the clade C Symbiodinium cells averaged 8.30 ± 0.09 µm (n = 855) in diameter in hospite, significantly larger (p < 0.01) than in hospite clade B cells (7.72 ± 0.13 µm, n = 992). Since the cell size of the clade C Symbiodinium (6.70 ± 0.84 µm, n = 1,000) is significantly larger (p < 0.01) than clade B Symbiodinium (6.00 ± 0.68 µm, n = 1,000) in free-living cultures, the larger Symbiodinium cells in the anemones demonstrate it harbors the clade C Symbiodinium instead of the clade B. It also demonstrates that like the clade B Symbiodinium (Pasaribu et al., 2015), the cell size of the clade C Symbiodinium increase in endosymbiotic condition. This experimental evidence reveals that the exogenously supplied clade C Symbiodinium can be transmitted to multiple generations of progeny via pedal laceration under laboratory conditions. We thus conclude that the Exaiptasia anemone species used in this study can establish a symbiotic relationship with the clade C Symbiodinium (CCMP2466; ITS2 type C1). In addition, our study and previous work that has infected Exaiptasia with heterologous Symbiodinium types originally from other host species (Schoenberg & Trench, 1980; Hawkins et al., 2016) also demonstrate that Exaiptasia pallida could potentially establish symbiotic relationships with other clades of Symbiodinium.
Figure 5. The morphology of clade B and clade C Symbiodinium cells within their host anemone tissue.
(A) Tentacular tissues harboring clade B Symbiodinium; (B) Tentacular tissues harboring clade C Symbiodinium. Scale bar: 20 µm.
Recently, the scientific name of several anemones used widely as a model system for the study of cnidarian-dinoflagellate endosymbiosis has been revised to Exaiptasia pallida (Grajales & Rodriguez, 2014; Grajales & Rodriguez, 2016; Grajales, Rodriguez & Thornhill, 2016). This change was based on the updated morphological and genetic population study of the newly collected specimen from around the world. According to these global comparative investigations, the anemones used in our study, which originate from the same wild population as the studies cited above (N22 03 00.08 E120 41 42.88), belong to a widespread lineage that hosts Symbiodinium minutum (ITS2 type B1). These updated studies also found that, in addition to the previously known Florida lineage of Exaiptasia pallida which hosts Symbiodinium A4, B1, and a mixed population of B1 and C1 (Thornhill et al., 2013), Exaiptasia hosts a mixed population of Symbiodinium B1 and C1 in Bermuda and Symbiodinium type A4 in Mexico and the Bahamas (Grajales & Rodriguez, 2016; Grajales, Rodriguez & Thornhill, 2016). Therefore, although the present study is not the first to document Exaiptasia spp. hosting clade C Symbiodinium, it does show for the first time that the Exaiptasia pallida anemones predominantly known to host the clade B Symbiodinium (LaJeunesse, Parkinson & Reimer, 2012; Thornhill et al., 2013; Kinzie et al., 2001) can be bleached and re-infected with exogenously supplied dinoflagellate algae of the clade C Symbiodinium.
The clade C Symbiodinium (CCMP2466) used in this study was originally isolated from a corallimorph (Discosoma sanctithomae), not a reef-building coral, but the genetic data shows that it belongs to a line (ITS2 type C1; Krueger et al., 2015) of diverse clade C Symbiodinium that are mainly harbored by reef-building corals (Van Oppen et al., 2001; LaJeunesse et al., 2003; LaJeunesse et al., 2004b; Chen et al., 2005; LaJeunesse et al., 2010) and are sensitive to thermal stress (Baker et al., 2004; Litmann, Bourne & Willis, 2010). Since CCMP2466 could infect aposymbiotic larvae of a reef-building coral, Acropora tenuis, and then establish a monoclonal Symbiodinium C1-infected coral association (Yuyama & Higuchi, 2014), CCMP2466 has been successfully applied to the study of thermal tolerance of corals when it harbors the thermal sensitive clade C (CCMP2466) or thermal tolerant clade D Symbiodinium (Yuyama et al., 2016). These updated reports and this present study imply that the Exaiptasia-Symbiodinium C1 association is an interesting and useful system for studying the functional diversity between cnidarian hosts and their symbionts.
In conclusion, the present study opens up a window for future studies to determine whether the molecular pathways and the type of symbiosis underlying the establishment of the Exaiptasia–Symbiodinium B1 association are similar to the Exaiptasia–Symbiodinium C1 endosymbiotic association. If this relationship is later shown to be mutualistic, as is Exaiptasia-clade B Symbiodinium association, then this could serve as a potentially valuable model for the study of cnidarian-Symbiodinium endosymbiosis.
Supplemental Information
Amplified genomic fragments of small subunit ribosomal RNA genes (18S rDNA) from Symbiodinium were digested using the restriction enzymes TaqI. Lane 1: The free-living cultured clade C Symbiodinium (CCMP 2466). Lane 2: Free-living cultured clade B Symbiodinium (originally from Exaiptasia pallida). M: marker. MW: molecular weight.
Acknowledgments
We express appreciation to Mr. Yu-Bao Wang for technical support, editors and reviewers for constructive comments, and Ms. Cherilyn Gan for edits on this manuscript.
Funding Statement
This work was supported by grants from Taiwan’s Ministry of Science and Technology (NSC 102-2311-B-291-002-MY3 to WNUC; NSC 102-2311-B-291-001 & MOST 103-2311-B-291-001-MY3 to SEP), as well as intramural funding from the National Museum of Marine Biology and Aquarium. ABM was supported by a postdoctoral research fellowship from the Khaled bin Sultan Living Oceans Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Wan-Nan U. Chen conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Ya-Ju Hsiao performed the experiments.
Anderson B. Mayfield wrote the paper, reviewed drafts of the paper.
Ryan Young performed the experiments, wrote the paper, prepared figures and/or tables.
Ling-Lan Hsu performed the experiments, prepared figures and/or tables.
Shao-En Peng conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Data Availability
The following information was supplied regarding data availability:
All of the raw data are presented in the manuscript. A supplementary figure has been included as Fig. S1.
References
- Abrego et al. (2008).Abrego D, Ulstrup KE, Willis BL, Van Oppen MJH. Species–specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proceedings of the Royal Society B: Biological Sciences. 2008;275:2273–2282. doi: 10.1098/rspb.2008.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker et al. (2004).Baker AC, Starger C, McClanahan T, Glynn PW. Corals’ adaptive response to climate change. Nature. 2004;430:741. doi: 10.1038/430741a. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2004).Chen MC, Cheng YM, Hong MC, Fang LS. Molecular cloning of Rab5 (ApRab5) in Aiptasia pulchella and its retention in phagosomes harboring live zooxanthellae. Biochemical and Biophysical Research Communications. 2004;324:1024–1033. doi: 10.1007/s00338-004-0389-7. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2005).Chen CA, Yang YW, Wei NV, Tsai WS, Fang LS. Symbiont diversity in scleractinian corals from tropical reefs and subtropical non-reef communities in Taiwan. Coral Reefs. 2005;24:11–22. doi: 10.1016/j.bbrc.2004.09.151. [DOI] [Google Scholar]
- Grajales & Rodriguez (2014).Grajales A, Rodriguez E. Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actinaria, Metridioidea) Zootaxa. 2014;3826:55–100. doi: 10.11646/zootaxa.3826.1.2. [DOI] [PubMed] [Google Scholar]
- Grajales & Rodriguez (2016).Grajales A, Rodriguez E. Elucidating the evolutionary relationships of the Aiptasiidae, widespread cnidarian-dinoflagellate model system. Molecular Phylogenetics and Evolution. 2016;94:252–263. doi: 10.1016/j.ympev.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Grajales, Rodriguez & Thornhill (2016).Grajales A, Rodriguez E, Thornhill DJ. Patterns of Symbiodinium spp. associations within the family Aiptasiidae, a monophyletic lineage of symbiotic of sea anemones (Cnidaria, Actiniaria) Coral Reefs. 2016;35:345–355. doi: 10.1007/s00338-015-1352-5. [DOI] [Google Scholar]
- Hambleton et al. (2014).Hambleton EA, Guse A, Pringle JR. Similar specificities of symbiont uptake by adults and larvae in an anemone model system for coral biology. The Journal of Experimental Biology. 2014;217:1613–1619. doi: 10.1242/jeb.095679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins et al. (2016).Hawkins TD, Hagemeyer JCG, Hoadley DK, Marsh AG, Warner ME. Partitioning of respiration in an animal-algal symbiosis: implications for different aerobic capacity between Symbiodinium spp. Frontiers in Physiology. 2016;7 doi: 10.3389/fphys.2016.00128. Article 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong et al. (2009).Hong MC, Huang YS, Lin WW, Fang LS, Chen MC. ApRab3, a biosynthetic Rab protein, accumulates on the maturing phagosomes and symbiosomes in the tropical sea anemone, Aiptasia pulchella. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2009;152:249–259. doi: 10.1016/j.cbpb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Kinzie & Chee (1979).Kinzie RA, Chee GS. The effect of different zooxanthellae on the growth of experimentally reinfected hosts. The Biological Bulletin. 1979;156:315–327. doi: 10.2307/1540920. [DOI] [PubMed] [Google Scholar]
- Kinzie et al. (2001).Kinzie III RA, Takayama M, Santos SR, Coffroth MA. The adaptive bleaching hypothesis: experimental tests of critical assumptions. The Biological Bulletin. 2001;200:51–58. doi: 10.2307/1543084. [DOI] [PubMed] [Google Scholar]
- Krueger et al. (2015).Krueger T, Fisher PL, Becker S, Pontasch S, Dove S, Hoegh-Guldberg O, Leggat W, Davy SK. Transcriptomic characterization of the enzymatic antioxidants FeSOD, MnSOD, APX, and KatG in the dinoflagellate genus Symbiodinium. BMC Evolutionary Biology. 2015;15:48. doi: 10.1186/s12862-015-0326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse et al. (2004a).LaJeunesse TC, Bhagooli R, Hidaka M, DeVantier L, Done T, Schmidt GW, Fitt WK, Hoegh-Guldberg O. Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Marine Ecology Progress Series. 2004a;284:147–161. doi: 10.3354/meps284147. [DOI] [Google Scholar]
- LaJeunesse et al. (2008).LaJeunesse TC, Bonilla HR, Warner ME, Wills M, Schmidt GW, Fitt WK. Specificity and stability in high latitude eastern Pacific coral–algal symbioses. Association for the Sciences of Limnology and Oceanography. 2008;53:719–727. doi: 10.4319/lo.2008.53.2.0719. [DOI] [Google Scholar]
- LaJeunesse et al. (2003).LaJeunesse TC, Loh WKW, Van Woesik R, Hoegh-Guldberg O, Schmidt GW, Fitt WK. Low symbiont diversity in southern Great Barrier Reef corals relative to those of the Caribbean. Association for the Sciences of Limnology and Oceanography. 2003;48:2046–2054. doi: 10.4319/lo.2003.48.5.2046. [DOI] [Google Scholar]
- LaJeunesse, Parkinson & Reimer (2012).LaJeunesse TC, Parkinson JE, Reimer JD. A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with Cnidaria. Journal of Phycology. 2012;48:1380–1391. doi: 10.1111/j.1529-8817.2012.01217.x. [DOI] [PubMed] [Google Scholar]
- LaJeunesse et al. (2010).LaJeunesse TC, Pettay DT, Sampayo EM, Phongsuwan N, Brown B, Obura DO, Hoegh-Guldberg O, Fitt WK. Long-standing environmental conditions, geographic isolation and host-symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. Journal of Biogeography. 2010;37:785–800. doi: 10.1111/j.1365-2699.2010.02273.x. [DOI] [Google Scholar]
- LaJeunesse et al. (2004b).LaJeunesse TC, Thornhill DJ, Cox EF, Stanton FG, Fitt WK, Schmidt GW. High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral Reefs. 2004b;23:596–603. doi: 10.1007/s00338-004-0428-4. [DOI] [Google Scholar]
- Lien, Fukami & Yamashita (2012).Lien YT, Fukami H, Yamashita Y. Symbiodinium clade C dominates zooxanthellate corals (Scleractinia) in the temperate region of Japan. Zoological Science. 2012;29:173–180. doi: 10.2108/zsj.29.173. [DOI] [PubMed] [Google Scholar]
- Lin, Wang & Fang (2000).Lin KL, Wang JT, Fang LS. Participation of glycoproteins on zooxanthellal cell walls in the establishment of a symbiotic relationship with the sea anemone, Aiptasia pulchella. Zoological Studies. 2000;39:172–178. [Google Scholar]
- Litmann, Bourne & Willis (2010).Litmann RA, Bourne DG, Willis BL. Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Molecular Ecology. 2010;19:1978–1990. doi: 10.1111/j.1365-294X.2010.04620.x. [DOI] [PubMed] [Google Scholar]
- Muscatine, Grossman & Doino (1991).Muscatine L, Grossman D, Doino J. Release of symbiotic algae by tropical sea anemones and corals after cold shock. Marine Ecology Progress Series. 1991;77:233–243. doi: 10.3354/meps077233. [DOI] [Google Scholar]
- Pasaribu et al. (2015).Pasaribu B, Weng LC, Lin IP, Camargo E, Tzen JTC, Tsai CH, Ho SL, Lin MR, Wang LH, Chen CS, Jiang PL. Morphological variability and distinct protein profiles of cultured and endosymbiotic Symbiodinium cells isolated from Exaiptasia pulchella. Scientific Reports. 2015;5 doi: 10.1038/srep15353. Article 15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng et al. (2012).Peng SE, Chen CS, Song YF, Huang HT, Jiang PL, Chen WNU, Fang LS, Lee YC. Assessment of metabolic modulation in free-living versus endosymbiotic Symbiodinium using synchrotron radiation-based infrared microspectroscopy. Biology Letters. 2012;23:434–437. doi: 10.1098/rsbl.2011.0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan & Knowlton (1995).Rowan R, Knowlton N. Intraspecific diversity and ecological zonation in coral-algal symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2850–2853. doi: 10.1073/pnas.92.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan & Powers (1991a).Rowan R, Powers DA. Molecular genetic identification of symbiotic dinoflagellates (zooxanthellae) Marine Ecology Progress Series. 1991a;71:65–73. doi: 10.3354/meps071065. [DOI] [Google Scholar]
- Rowan & Powers (1991b).Rowan R, Powers DA. A molecular genetic classification of zooxanthellae and the evolution of animal-algal symbioses. Science. 1991b;15:1348–1351. doi: 10.1126/science.251.4999.1348. [DOI] [PubMed] [Google Scholar]
- Schoenberg & Trench (1980).Schoenberg DA, Trench RK. Genetic variation in Symbiodinium (=Gymnodinium) microadriaticum Freudenthal, and specificity in its symbiosis with marine invertebrates. III. Specificity and infectivity of Symbiodinium microadriaticum. Proceedings of the Royal Society B—Biological Sciences. 1980;207:445–460. doi: 10.1098/rspb.1980.0033. [DOI] [Google Scholar]
- Stat, Morris & Gates (2008).Stat M, Morris E, Gates RD. Functional diversity in coral-dinoflagellate symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9256–926. doi: 10.1073/pnas.0801328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill et al. (2013).Thornhill DJ, Xiang Y, Pettay DT, Zhong M, Santos SR. Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Molecular Ecology. 2013;22:4499–4515. doi: 10.1111/mec.12416. [DOI] [PubMed] [Google Scholar]
- Van Oppen et al. (2001).Van Oppen MJH, Palstra FP, Piquet AMT, Miller DJ. Patterns of coral-dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host-symbiont selectivity. Proceedings of the Royal Society B: Biological Sciences. 2001;268:1759–1767. doi: 10.1098/rspb.2001.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2013).Wang LH, Lee HH, Fang LS, Mayfield AB, Chen CS. Fatty acid and phospholipid syntheses are prerequisites for the cell cycle of Symbiodinium and their endosymbiosis within sea anemones. PLoS ONE. 2013;8(8):e2358. doi: 10.1371/journal.pone.0072486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2008).Wang LH, Liu YH, Ju YM, Hsiao YY, Fang LS, Chen CS. Cell cycle propagation is driven by light-dark stimulation in a cultured symbiotic dinoflagellate isolated from corals. Coral Reefs. 2008;27:823–835. doi: 10.1007/s00338-008-0434-z. [DOI] [Google Scholar]
- Weis et al. (2008).Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. Cell biology in model systems as the key to understanding corals. Trends in Ecology and Evolution. 2008;23:369–376. doi: 10.1016/j.tree.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Xiang et al. (2013).Xiang T, Hambleton EA, DeNofrio JC, Pringle JR, Grossman AR. Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. Phycological Society of America. 2013;49:447–458. doi: 10.1111/jpy.12055. [DOI] [PubMed] [Google Scholar]
- Yuyama, Harii & Hidaka (2012).Yuyama I, Harii S, Hidaka M. Algal symbiont type affects gene expression in juveniles of the coral Acropora tenuis exposed to thermal stress. Marine Environmental Research. 2012;76:41–47. doi: 10.1016/j.marenvres.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Yuyama & Higuchi (2014).Yuyama I, Higuchi T. Comparing the effects of symbiotic algae (Symbiodinium) clades C1 and D on early growth stages of Acropora tenuis. PLoS ONE. 2014;9:e2358. doi: 10.1371/journal.pone.0098999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuyama et al. (2016).Yuyama I, Nakamura T, Higuchi T, Hidaka M. Different stress tolerances of juveniles of the coral Acropora tenuis associated with clades C1 and D Symbiodinium. Zoological Studies. 2016;55 doi: 10.6620/ZS.2016.55-19. Article 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amplified genomic fragments of small subunit ribosomal RNA genes (18S rDNA) from Symbiodinium were digested using the restriction enzymes TaqI. Lane 1: The free-living cultured clade C Symbiodinium (CCMP 2466). Lane 2: Free-living cultured clade B Symbiodinium (originally from Exaiptasia pallida). M: marker. MW: molecular weight.
Data Availability Statement
The following information was supplied regarding data availability:
All of the raw data are presented in the manuscript. A supplementary figure has been included as Fig. S1.