Abstract
Formins are widespread actin-polymerizing proteins that play pivotal roles in a number of processes, such as cell polarity, morphogenesis, cytokinesis, and cell migration. In agreement with their crucial function, formins are prone to a variety of regulatory mechanisms that include autoinhibition, post-translational modifications, and interaction with formin modulators. Furthermore, activation and function of formins is intimately linked to their ability to interact with membranes. In the budding yeast Saccharomyces cerevisiae, the two formins Bni1 and Bnr1 play both separate and overlapping functions in the organization of the actin cytoskeleton. In addition, they are controlled by both common and different regulatory mechanisms. Here we show that proper localization of both formins requires the redundant E3 ubiquitin ligases Dma1 and Dma2, which were previously involved in spindle positioning and septin organization. In dma1dma2 double mutants, formin distribution at polarity sites is impaired, thus causing defects in the organization of the actin cable network and hypersensitivity to the actin depolymerizer latrunculin B. Expression of a hyperactive variant of Bni1 (Bni1-V360D) rescues these defects and partially restores proper spindle positioning in the mutant, suggesting that the failure of dma1dma2 mutant cells to position the spindle is partly due to faulty formin activity. Strikingly, Dma1/2 interact physically with both formins, while their ubiquitin-ligase activity is required for formin function and polarized localization. Thus, ubiquitylation of formin or a formin interactor(s) could promote formin binding to membrane and its ability to nucleate actin. Altogether, our data highlight a novel level of formin regulation that further expands our knowledge of the complex and multilayered controls of these key cytoskeleton organizers.
Keywords: actin, formin, ubiquitylation, budding yeast
THE ability to polarize is a fundamental property of all types of cells, being crucial for numerous cellular processes such as proliferation, differentiation, and morphogenesis. Indeed, dysregulation of cell polarity can underlie developmental disorders and cancers (Wodarz and Nathke 2007). Cell polarization is strictly linked to the reorganization of the cytoskeleton and in particular of the actin network, whose dynamics must be tightly controlled for polarized processes to occur properly.
The unicellular budding yeast Saccharomyces cerevisiae divides asymmetrically and undergoes highly polarized cell growth throughout its life cycle. Most aspects of polarized growth in budding yeast arise from a precise arrangement of the cortical actin cytoskeleton during the cell cycle. Three main actin structures can be found in yeast cells: (i) actin patches, which are sites of active endocytosis, (ii) actin cables, which serve as tracks for polarized secretion and segregation of organelles, and (iii) the contractile acto-myosin ring, which is involved in cytokinesis (Adams and Pringle 1984; Kilmartin and Adams 1984; Bi et al. 1998; Lippincott and Li 1998). Once the future bud site has been selected in the G1 phase of the cell cycle, polarized growth is directed toward the growing bud by vesicle transport along actin cables (reviewed in Goode et al. 2015). Actin patches also cluster at the bud tip, to support efficient membrane trafficking and cooperate with secretion in the establishment of cell polarity (Jose et al. 2013). Later during the cell cycle, a switch from polarized to isotropic growth, where the bud expands in all directions, is triggered by mitotic cyclin B-CDK activity (Lew and Reed 1993). This implicates the spread-out redistribution of actin patches and cables within the bud and the extension of cables from the bud neck toward the mother cell. Finally, when cells exit mitosis upon inactivation of mitotic CDKs, actin repolarizes at the bud neck to support cytokinesis. This leads to assembly of the contractile actin ring, reorganization of the actin cables to direct secretion toward the division site for septum formation, and the convergence of actin patches at both sides of the bud neck, presumably for endocytic internalization and/or recycling of cytokinetic factors (Pruyne and Bretscher 2000a,b). Like in all eukaryotic cells, GTPases of the Rho family, i.e., Cdc42 and Rho1-5, are key regulators of actin organization and remodelling in yeast (reviewed in Perez and Rincon 2010). Among their effectors, the partially redundant PAK (p21-activated kinase) kinases Cla4 and Ste20 are required for actin polarization throughout the cell cycle downstream of Cdc42 (Benton et al. 1997; Holly and Blumer 1999; Lamson et al. 2002).
Formins are universal actin nucleators that can assemble actin filaments in vitro by virtue of their conserved formin homology 2 (FH2) domains, which dimerize into a donut-shaped catalytic core (Xu et al. 2004; Otomo et al. 2005b). The FH2 motif is always preceded by a proline-rich conserved region, referred to as formin homology 1 (FH1) domain, which accelerates actin polymerization by recruiting actin monomers bound to profilin (Sagot et al. 2002b; Romero et al. 2004). In addition to FH1 and FH2, formins contain several other regulatory domains (Higgs 2005; Goode and Eck 2007). Many formins, for instance, are in a close, autoinhibited conformation due to an intramolecular interaction between the diaphanous autoregulatory domain (DAD) located at the C terminus of the FH2 domain and a region called diaphanous inhibitory domain (DID) that resides at the N terminus of the protein (Alberts 2001; Li and Higgs 2005). Another region, called GTPase-binding domain (GBD), is located next to or partially overlapping with DID and binds to Rho GTPases, thereby relieving autoinhibition (Li and Higgs 2003; Lammers et al. 2005; Otomo et al. 2005a; Rose et al. 2005).
Budding yeast cells possess two formins named Bni1 and Bnr1 that assemble, respectively, two distinct arrays of actin cables, one polarized toward the bud cortex and the other polarized toward the bud neck (Kohno et al. 1996; Evangelista et al. 1997; Evangelista et al. 2002; Sagot et al. 2002a,b; Pruyne et al. 2004). Consistently, the localization pattern of these two formins differs, in that Bnr1 resides at the bud neck from bud emergence to mitotic exit (Kamei et al. 1998; Pruyne et al. 2004; Buttery et al. 2007), while Bni1 is found at the bud tip throughout most of the cell cycle until mitotic exit, when it replaces Bnr1 at the bud neck (Ozaki-Kuroda et al. 2001; Buttery et al. 2007). Although loss of either Bni1 or Bnr1 causes different phenotypes, in agreement with their different biochemical properties (Moseley and Goode 2005; Delgehyr et al. 2008; Wen and Rubenstein 2009), deletion of both formins is lethal, suggesting that they share at least one essential function. Conditional bni1bnr1 double mutants disrupt both polarized growth and the actin cable network in restrictive conditions (Imamura et al. 1997; Evangelista et al. 2002; Sagot et al. 2002a). Furthermore, they fail to assemble the contractile actin ring for cytokinesis (Tolliday et al. 2002).
Formin activity in budding yeast is controlled by the Rho GTPases Cdc42, Rho1, and Rho3–4 (Kohno et al. 1996; Evangelista et al. 1997; Imamura et al. 1997; Dong et al. 2003). Some of these GTPases might promote formin activation by direct binding to the GBD. Additionally, Rho1 is necessary for formin activity at high temperature (37°) through its effector protein kinase C (Pkc1) (Dong et al. 2003), while the Cdc42 effector Gic2 interacts with Bni1 and contributes to its polarized localization and/or activation (Jaquenoud and Peter 2000; Chen et al. 2012). At the bud tip, Bni1 interacts with several components of the polarisome, a protein complex involved in cell polarity that includes the proteins Spa2, Bud6, and Pea2 (Fujiwara et al. 1998; Sheu et al. 1998). Gic2 might be itself part of the polarisome, since it cofractionates and interacts with polarisome components (Jaquenoud and Peter 2000). Spa2 binds to a region of Bni1 (amino acids 826–987) referred to as Spa2-binding domain (SBD) and together with Pea2 recruits Bni1 to the bud tip (Fujiwara et al. 1998; Sagot et al. 2002a). The interaction of Bni1 with Bud6 through its Bud6-interacting domain (BBD, amino acids 1647–1953), which overlaps the DAD region, promotes Bni1-mediated actin polymerization through recruitment of actin monomers and contributes, to a lesser extent, to Bni1 localization (Sagot et al. 2002a; Moseley et al. 2004; Moseley and Goode 2005; Delgehyr et al. 2008; Graziano et al. 2011). Finally, Bni1 shares with other formins a tripartite formin homology region, referred to as FH3, that resides between the GBD and the FH1 domains of the protein, likely contributing to its cortical localization (Petersen et al. 1998).
Localization of Bnr1 at the bud neck requires septins (Pruyne et al. 2004; Gao et al. 2010), which at this site form a collar that acts as scaffold for most cytokinetic factors (reviewed in Oh and Bi 2010). Furthermore, the septin-associated kinases Gin4 and Elm1 promote efficient recruitment of Bnr1 to the bud neck, where this formin gets activated (Buttery et al. 2012). Several other factors have been involved in the specific regulation of Bnr1 vs. Bni1. The kinesin-like myosin-passenger protein Smy1 acts as a Bnr1 damper in vitro and in vivo by binding directly to Bnr1 (Chesarone-Cataldo et al. 2011; Eskin et al. 2016). A polarized complex made by Bud14 and the Kelch-domain proteins Kel1 and Kel2 displaces Bnr1 from actin filaments to support proper actin cable dynamics (Chesarone et al. 2009; Gould et al. 2014). Finally, The F-BAR protein Hof1, which controls septin organization and septum deposition (Kamei et al. 1998; Vallen et al. 2000; Oh et al. 2013), attenuates the actin-nucleating activity of Bnr1 in vitro and in vivo, thereby tuning the architecture of the actin cable network (Graziano et al. 2014).
The Dma1 and Dma2 proteins are paralogous E3 ubiquitin ligases that share 58% identity in their primary sequence. They carry a C-terminal RING finger domain that can catalyze both K48- and K63-linked ubiquitin chains (Loring et al. 2008) and a central FHA domain that is thought to bind Thr-phosphorylated proteins (Durocher et al. 2000). So far, Dma1 and Dma2 have been redundantly involved in spindle positioning, septin organization, and vacuole inheritance (Fraschini et al. 2004; Merlini et al. 2012; Chahwan et al. 2013; Yau et al. 2014). In this paper, we show that Dma1 and Dma2 also contribute to formin regulation. Indeed, they interact physically with Bni1 and Bnr1 and through their ubiquitin ligase activity contribute to the overall organization of the actin cable network as well as to proper formin distribution. Altogether our data indicate that a ubiquitination-dependent step modulates formin localization and activity.
Materials and Methods
Strains, media and reagents, and genetic manipulations
All yeast strains (Supplemental Material, Table S1) are derivatives of W303 (ade2-1, trp1-1, leu2-3,112, his3-11, 15 ura3, and ssd1), except for strains in Figure S2C that were derivatives of BY4741 (his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). W303 bears a frameshift mutation in the BUD4 gene, which encodes an anillin-related protein (Voth et al. 2005). Unless specified (Figure S2, A, B, and D), most strains were generated in the original bud4 W303 background.
Yeast cultures were grown at 25°–30°, unless otherwise specified, in either synthetic medium (SD) supplemented with the appropriate nutrients and 2% glucose or YEP (1% yeast extract, 2% bactopeptone, 50 mg/liter adenine) medium supplemented with 2% glucose (YEPD). α-Factor was used at 4 µg/ml at 25° and hydroxyurea at 0.2 M at 30°.
Standard techniques were used for genetic manipulations (Sherman 1991; Maniatis et al. 1992). Gene deletions were generated by one-step gene replacement (Wach et al. 1994). One-step tagging techniques (Janke et al. 2004; Sheff and Thorn 2004) were used to generate HA3-, Flag3-, eGFP-, or mCherry-tagged proteins. To generate Bni1-, Bnr1-, and Bud6-GFP, we integrated at the endogenous BNI1, BNR1, or BUD6 integrative plasmids carrying the 3′ end of each ORF fused to the coding sequence of GFP (Delgehyr et al. 2008). To generate a high copy number plasmid bearing GIC2 (pSP1260), the coding region of GIC2 including 500 bp of promoter region and 200 bp of 3′ UTR was amplified by PCR from the genome of W303 and subcloned in the polylinker of YEplac181 using artificial SalI and KpnI restriction sites.
Fluorescence microscopy
F-actin staining was performed on cells fixed with 3.7% formaldehyde for 30–60 min under shaking at 30°. F-actin was visualized with Alexa Fluor 546-labeled phalloidin (Molecular Probes) at 20 units/ml after a 30-min to 4-hr incubation at room temperature or overnight incubation at 4°.
Detection of Bni1-GFP, Bnr1-GFP, Myo1-mCherry, Spa2-eGFP, or GFP-Cdc12 was carried out on live cells growing in SD medium at 25°–30°. Detection of and Bud6-GFP was carried out on cells grown in SD medium at 25°, fixed in 70% ethanol at −20° overnight and washed with PBS.
Still digital images were taken with an oil immersion ×63 1.4 HCX Plan-Apochromat objective (Zeiss) with a Coolsnap HQ2 CDD camera (Photometrics) mounted on a Zeiss AxioimagerZ1 fluorescence microscope and controlled by the MetaMorph imaging system software. Z-stacks containing 11 planes were acquired with a step size of 0.3 μm and a binning of 1. Z-stacks were maximum-projected and calibrated using ImageJ.
Fluorescence intensities of Bni1-GFP, Bnr1-GFP, Spa2-eGFP, and Bud6-GFP were quantified with ImageJ on a single focal plane. Further details are indicated in the figure legends. Spindle distances from the bud neck were measured with ImageJ on max-projected stacks of 11 planes at a step size of 0.3 μm.
For time-lapse video microscopy, cells were mounted in SD medium on fluorodishes and filmed at room temperature with a DeltaVision OMX microscope using a ×63 1.4 N.A. oil immersion objective and the SoftWoRx software (Applied Precision). Z-stacks of 15–31 planes were acquired every 1–2 min with a step size of 0.2 μm and a binning of 1. Z-stacks were deconvolved with Huygens (Scientific Volume Imaging) and max-projected. Kymographs were generated with Metamorph (Molecular Devices) by creating a line from the bud tip to the bud neck of 5-pixel width.
Protein extracts and immunoprecipitation experiments
For immunoprecipitations, cell pellets from 50 ml of culture (107 cells/ml) were lysed at 4° with acid-washed glass beads in lysis buffer [50 mM Tris-Cl pH 7.5, NaCl 150 mM, 10% glycerol, 1 mM EDTA, 0.1% NP40, supplemented with protein inhibitors (Complete; Roche, Indianapolis, IN), 1 mM Na-orthovanadate, and 60 mM β-glycerophosphate]. Total extracts were cleared by spinning at 12,000 rpm for 10 min and quantified using NanoDrop (Thermo Scientific). Same amounts of protein extracts were subjected to immunoprecipitation with an anti-Flag antibody (M2; Sigma, St. Louis, MO) preadsorbed to protein A-sepharose. Immunocomplexes were washed three times in lysis buffer and twice in PBS before SDS page electrophoresis.
To measure stability of Bni1- and Bnr1-HA3 (Figure 8D), cells were grown to exponential phase at 25° and cycloheximide (250 μg/ml) was added to the cultures (time 0). Cell samples were collected at different time points and subjected to Western blot analysis.
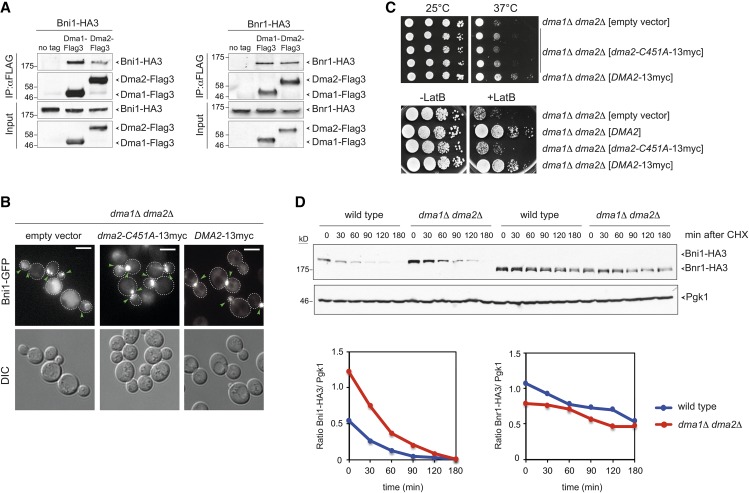
Figure 8.
Dma proteins interact physically with formins and their E3 ligase activity is required for proper Bni1 distribution. (A) Total lysates from logarithmically growing cells expressing at the same time Dma1-Flag3 or Dma2-Flag3 and Bni1-HA3 or Bnr1-HA3 were subjected to immunoprecipitation with anti-Flag antibody followed by Western blot analysis with anti-HA and anti-Flag antibodies. Inputs represent 1/50th of the lysate used for each pull-down. (B) dma1Δ dma2Δ cells expressing Bni1-GFP and carrying a centromeric plasmid to express the catalytically inactive myc-tagged Dma2-C451A (dma2-C451A-13myc) or wild-type Dma2 (DMA2-13myc) were imaged at 25°. Representative images from max-projected Z-stacks (11 planes at 0.3-μm spacing) are shown. Arrowheads indicate different localizations of Bni1. Bar, 5 μm. (C, upper panels) Serial dilutions of cells with the indicated genotypes were spotted on selective (SD −His) plates and incubated at the indicated temperatures. (Lower panels) Serial dilutions of cells with the indicated genotypes were spotted on YEPD either lacking or containing 5 μM LatB and incubated at 25° for 2 days. (D) Wild-type and dma1Δ dma2Δ cells expressing Bni1-HA3 or Bnr1-HA3 were treated with cycloheximide (CHX) at 25° (time 0) to monitor formin degradation by Western blot with anti-HA antibodies. Pgk1 was used as loading control. Ratios between Bni1-HA3 or Bnr1-HA3 levels and the Pgk1 over time were calculated with ImageJ.
Trichloroacetic acid protein extracts were prepared as previously described (Fraschini et al. 2006) for Western blot analysis. Proteins transferred to Protran membranes (Schleicher and Schuell) were probed with monoclonal anti-HA 16B12 (Babco), anti-Pgk1 (Molecular Probes, Eugene, OR), anti-Pkc1 (Santa Cruz) or anti-FLAG M2 (Sigma). Secondary antibodies were purchased from GE Healthcare and proteins were detected by a home-made enhanced chemiluminescence system.
Other techniques
Significance of the differences between fluorescence intensities was statistically tested by means of a two-tailed t-test, assuming unequal variances. Differences with P-values <0.05 were considered statistically significant (* P < 0.05; ** P < 0.01; *** P < 0.001).
Data and reagent availability
Strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
The ubiquitin ligases Dma1 and Dma2 are required for robust actin cables assembly
Although individually the two yeast formins Bni1 and Bnr1 are not essential for cell viability, lack of both is lethal (Imamura et al. 1997). Therefore, loss of factors that promote activation of one formin are expected to exacerbate the defects caused by deletion of the other one. This approach has proven successful to identify novel regulators of Bnr1 (Buttery et al. 2012).
Deletion of DMA1 and DMA2 together causes synthetic growth defects at high temperatures when combined with BNI1 deletion (Figure 1A, Figure S1A, and Fraschini et al. 2004), whereas it has no effect with BNR1 deletion (Figure 1B and Figure S1B). Thus, Dma ubiquitin ligases might cooperate with formins, and Bnr1 in particular, in the organization of the actin cytoskeleton.
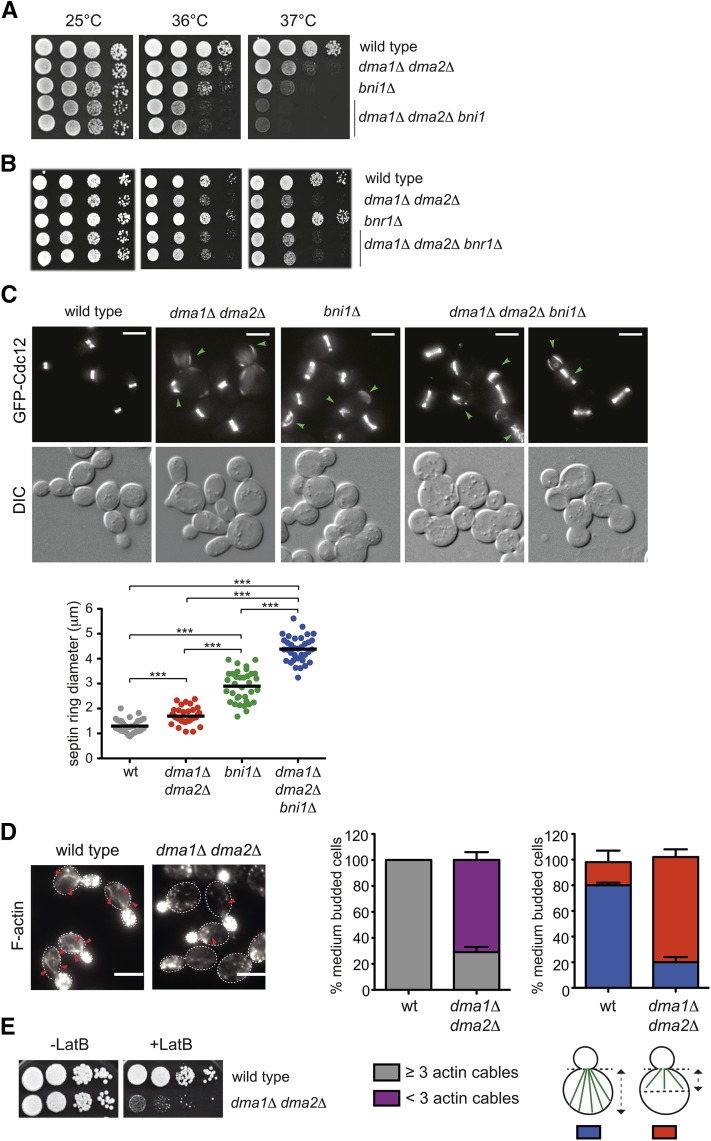
Figure 1.
Deletion of DMA1 and DMA2 causes synthetic defects in cells lacking the formin Bni1 and weakens the actin cable network. (A and B) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates and incubated at the indicated temperatures. (C) Cultures from the indicated strains all expressing GFP-Cdc12 were grown in SD medium at 25° and shifted to 37° for 2 hr. Representative GFP-Cdc12 images from Z-stack max-projection (11 planes at 0.3-μm spacing) and DIC from a single plane at 37° are shown. The dot plot indicates average diameters of septin rings in the different strains after 2 hr of incubation at 37° (n = 35). Green arrowheads indicate ectopic septin assemblies or abnormal rings. Bar, 5 μm. (D) F-actin was stained with Alexa-546 phalloidin on fixed asynchronous cells grown at 30°. Arrowheads indicate actin cables. Bar, 5 μm. Graphs indicate the percentage of medium-sized budded cells with ≥3 or <3 actin cables or with actin cables extending more or less than half the size of the mother cell (n ≥ 200; error bars: SD). (E) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates either lacking or containing 5 μM LatB and incubated at 25°.
The strain background that we use (W303) contains a frameshift mutation in the BUD4 gene (Voth et al. 2005), which encodes for a homolog of anillin that in animal cells localizes at the division site and links RhoA activity with actin, myosin, and septins (Piekny and Maddox 2010). We therefore tested if the bud4 allele present in W303 could affect the genetic interaction between BNI1 and DMA1/2 deletion that we observed. However, the slight temperature sensitivity of dma1Δ dma2Δ double mutant cells or that of the dma1Δ dma2Δ bni1Δ triple mutant was only mildly, if at all, influenced by the bud4 frameshift mutation of W303, as shown by similar degrees of growth defects in the presence or absence of wild-type BUD4 (Figure S2, A and B).
After 2 hr of incubation at 37° dma1Δ dma2Δ bni1Δ triple mutant cells displayed morphological defects, including chains of unseparated cells with broad bud necks, that are more severe than those observed in bni1Δ or dma1Δ dma2Δ cells (Figure 1C). Imaging of the septin Cdc12 fused to GFP showed that at 37° septin rings were large and often aberrant in bni1Δ single mutants and, to a higher extent, dma1Δ dma2Δ bni1Δ triple mutant cells. Furthermore, additional septin structures could be occasionally visualized (Figure 1C). Consistent with an additive defect of DMA1/2 and BNI1 deletion on bud neck organization, the diameter of septin rings was significantly higher in dma1Δ dma2Δ bni1Δ than in bni1Δ and dma1Δ dma2Δ cells (Figure 1C).
To investigate more directly if Dma proteins are involved in the organization of the actin cytoskeleton, we examined F-actin distribution in wild-type and dma1Δ dma2Δ cells by staining with Alexa-546 phalloidin. As expected, at 25°, wild-type cells exhibited the characteristic polarized actin patches concentrated in the bud and actin cables inside the mother cell (Figure 1D). While the overall distribution of actin patches was not affected, the number, length, and thickness of actin cables were strongly reduced in dma1Δ dma2Δ double mutant cells (Figure 1D). Accordingly, these cells, but not the single dma1Δ and dma2Δ mutant cells, were highly sensitive to latrunculin B (LatB), a drug that selectively depolymerizes actin cables (Figure 1E and Figure S2C). The LatB sensitivity of dma1Δ dma2Δ cells was unlinked to the presence or absence of a wild-type BUD4 allele in the strain (Figure S2D) and, accordingly, was also noticeable in the BY4741 background (Figure S2C).
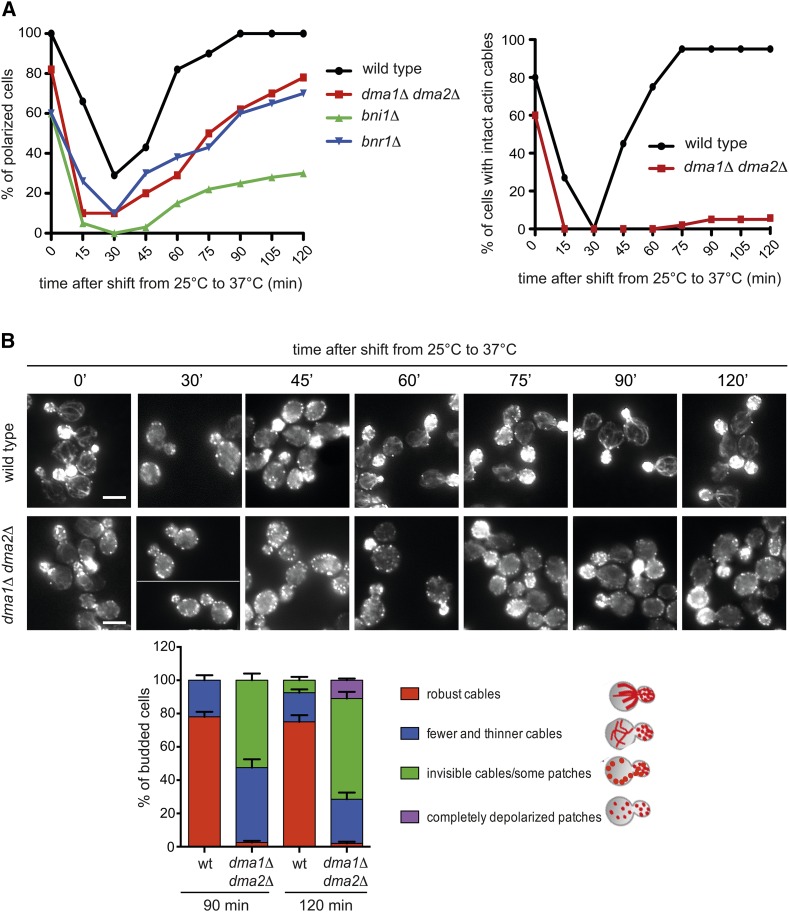
We then analyzed the impact of DMA1/2 deletion on actin cytoskeleton organization under thermal stress, which is known to cause a transient depolarization of actin (Delley and Hall 1999). Wild-type and dma1Δ dma2Δ cells, as well as bni1Δ or bnr1Δ cells as controls, were grown at 25° and then shifted to 37° to analyze the distribution of F-actin at different time points. As expected, in wild-type cells the actin cytoskeleton was rapidly depolarized (30 min after temperature shift) with disassembly of actin cables and redistribution of actin patches between the mother cell and the bud. Subsequently, actin gradually repolarized, showing a complete repolarization in 100% of the cells by 90 min after heat shock (Figure 2, A and B). In dma1Δ dma2Δ mutant cells, actin depolarized like in wild-type cells, but repolarization was slower and followed kinetics similar to bnr1Δ mutant cells. Furthermore, only actin patches succeeded in completely repolarizing, whereas robust actin cables failed to polymerize in most cells (Figure 2, A and B). Thus, Dma proteins are required for assembly of a robust actin cable network in unperturbed conditions and upon thermal stress. However, in spite of their apparent weakness, actin cables in dma1Δ dma2Δ cells were still able to drive polarized localization of Sec4-GFP, a marker for secretory vesicles (Figure S3; Schott et al. 2002).
Figure 2.
Dma proteins are required for proper actin cable organization after thermal stress. (A) Logarithmically growing cultures of cells with the indicated genotypes were grown in YEPD at 25° and then shifted to 37° at time 0. At different time points after release, cell samples were fixed and stained with Alexa-546 phalloidin to analyze F-actin structures. The graphs show the percentage of budded cells with polarized actin (>50% of actin patches inside the bud, left) and of budded cells with intact actin cables (right). Over 100 cells were scored for each time point. (B) Wild-type and dma1Δ dma2Δ cells were treated as in A. Bar, 5 μm. Organization of the actin cytoskeleton at 90 min and 120 min after release was further classified in different categories (bottom histograms). Over 100 cells were scored for each time point; error bars: SD.
Dma1 and Dma2 are required for proper formin localization
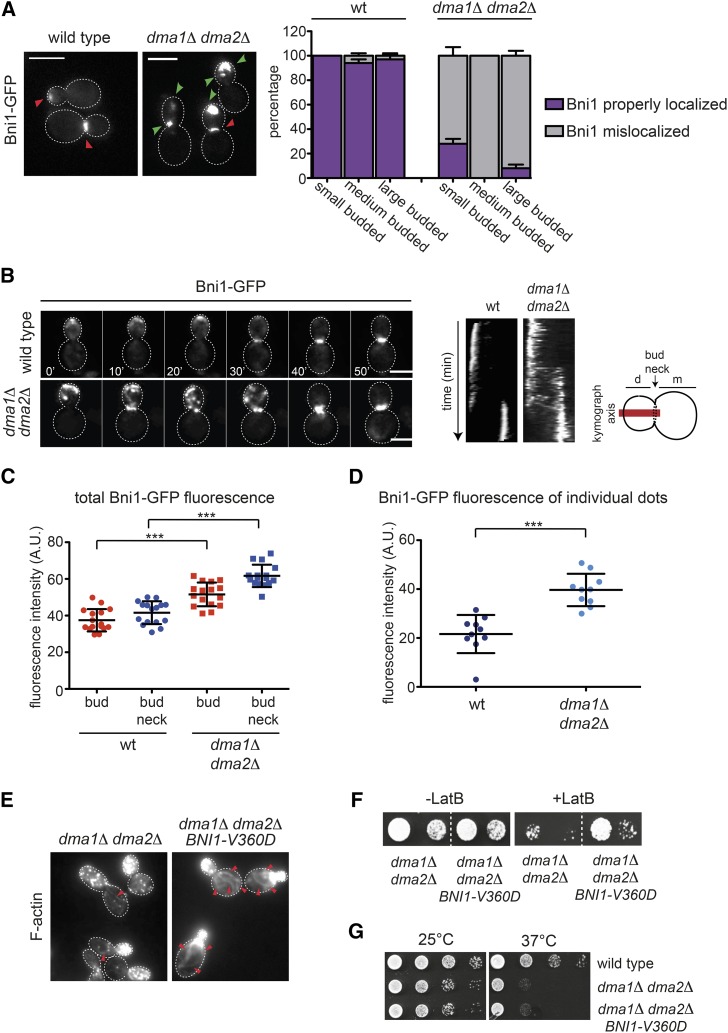
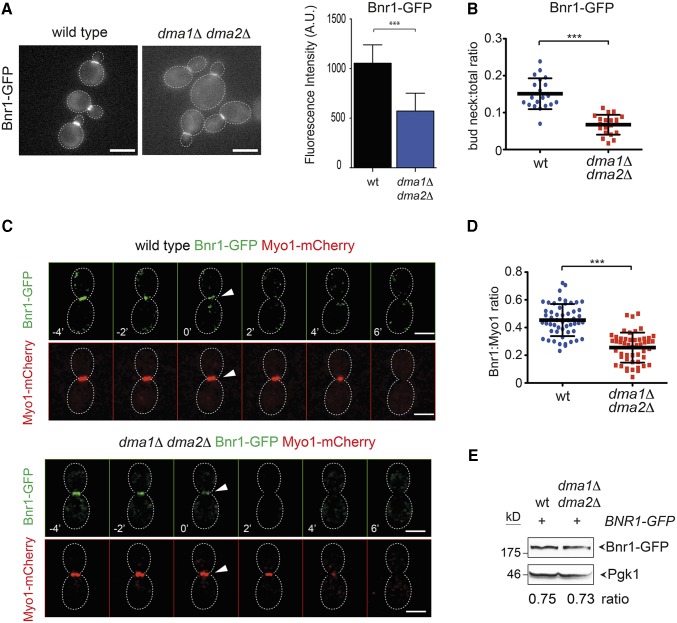
The results above raised the possibility that formins are misregulated in dma1Δ dma2Δ mutants. We therefore tagged with GFP Bni1 and Bnr1 in wild-type and dma1Δ dma2Δ cells to study their localization (Delgehyr et al. 2008). A preliminary analysis revealed that Bnr1 was at the bud neck of both strains, while Bni1-GFP was clearly mislocalized in the absence of Dma proteins. Indeed, while it was found at the bud tip of small/medium-budded cells and at the bud neck of large budded cells in the wild-type strain, it seemed to form big clusters spread around the bud that often coexisted with clusters of the protein at the bud neck in the dma1Δ dma2Δ mutant (Figure 3A). We then performed time-lapse video microscopy of wild-type and dma1Δ dma2Δ cells expressing Bni1-GFP or Bnr1-GFP. In agreement with previous data (Fujiwara et al. 1998; Ozaki-Kuroda et al. 2001; Pruyne et al. 2004; Buttery et al. 2007; Delgehyr et al. 2008), Bni1-GFP was detected at the site of bud emergence and at the tip of small buds early during the cell cycle and then it relocalized to the bud neck of large budded cells (n = 54, Figure 3B). In stark contrast, in dma1Δ dma2Δ mutant cells Bni1 accumulated in the bud as highly mobile aggregates that shuttled between the bud tip and the bud neck (n = 45), coexisting in both locations for at least 30 min (kymographs in Figure 3B). This behavior was never observed in wild-type cells and suggested that in the absence of Dma proteins, Bni1 might not be properly anchored to the cortex. Quantification of Bni1-GFP fluorescent signals in the bud and at the bud neck (Figure 3C), as well as in individual Bni1-GFP clusters (Figure 3D), revealed that higher levels of Bni1 were present at all these locations in dma1Δ dma2Δ relative to wild-type cells.
Figure 3.
Bni1 localization is impaired in dma1Δ dma2Δ mutant cells. (A) Logarithmically growing cultures of wild-type and dma1Δ dma2Δ expressing Bni1-GFP were imaged at 25°. Z-stacks max-projections (11 planes at 0.3-μm spacing) are shown. Red arrowheads indicate normal localization of Bni1, while green arrowheads indicate aberrant localization. Bar, 5 μm. The percentage of budded cells with properly localized or mislocalized Bni1 was scored in different categories of cells in relation to bud size (n ≥ 200; errors bars: SD). (B) Wild-type and dma1Δ dma2Δ expressing Bni1-GFP were filmed at room temperature (21°) with 1 min time lapse (n ≥ 45). Z-stacks (31 planes at 0.2-μm spacing) were deconvolved with Huygens and max-projected. Kymographs were created by drawing a 5-pixel-thick line across the daughter–mother axis, as indicated by the cartoon. Bar, 5 μm. (C) Fluorescence intensities of Bni1-GFP signals inside the bud or at the bud neck were measured with ImageJ in medium and large budded cells, respectively, within an oval region of 420 pixels in size after background subtraction (n = 16). A horizontal line in each dot plot indicates the mean ± SD. (D) Fluorescence intensities of Bni1-GFP signals were measured in individual dots within a region of 11 × 12 pixels after background subtraction (n = 10). A horizontal line in each dot plot indicates the mean ± SD. (E) dma1Δ dma2Δ cells lacking or carrying a centromeric plasmid to express the hyperactive BNI1-V360D allele were fixed and stained by Alexa-546 phalloidin to analyze F-actin structures. Arrowheads indicate visible actin cables. Bar, 5 μm. (F) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates either lacking or containing 5 μM LatB and incubated at 25°. (G) Serial dilutions of cells with the indicated genotypes were spotted on selective (SD −Trp) plates and incubated at 25° and 37°.
Since the actin and septin defects of dma1Δ dma2Δ mutant cells activate the morphogenesis checkpoint (Raspelli et al. 2011; Merlini et al. 2012), which delays activation of mitotic CDKs and mitotic entry through stabilization of the Wee1-like kinase Swe1 (Lew 2003), we asked if mislocalization of Bni1 in these cells was a secondary consequence of checkpoint activation. However, deletion of SWE1 did not restore proper Bni1 distribution (Figure S4, File S1, File S2, File S3, and File S4), suggesting that Dma1/2 might have a more direct role in this process.
The second formin Bnr1 was properly localized in both wild-type and dma1Δ dma2Δ cells at the bud neck from bud emergence to the onset of cytokinesis, when it disappeared concomitant with actomyosin ring contraction (AMR), which was monitored by tagging the myosin II Myo1 with mCherry (Figure 4, A and C; Kamei et al. 1998; Pruyne et al. 2004; Buttery et al. 2007). However, careful measurement of the fluorescence intensity of Bnr1-GFP at the bud neck showed that the amount of Bnr1-GFP was lower in dma1Δ dma2Δ cells relative to the wild-type control (Figure 4, A and B), while the steady-state levels of the protein were unchanged (Figure 4E). The decrease in Bnr1 levels at the bud neck of dma1Δ dma2Δ cells was further confirmed by measuring in individual cells arrested in S phase the intensity of the Bnr1-GFP signal relative to that of Myo1-mCherry (n ≥ 50, Figure 4D). Thus, localization of both formins is altered in the absence of Dma proteins, thereby causing defects in polymerization of actin cables. In agreement with this conclusion, expression of a hyperactive variant of Bni1 (BNI1-V360D) (kono et al. 2012) restored robust actin cables and rescued the sensitivity to LatB of dma1Δ dma2Δ cells (Figure 3, E and F), whereas it did not suppress their temperature sensitivity (Figure 3G). Furthermore, BNI1-V360D did not suppress the temperature sensitivity of dma1Δ dma2Δ cla4-75 cells (Figure S5) that we mainly ascribed to defects in septin organization (Merlini et al. 2012, 2015). Thus, formins likely act downstream of Dma proteins in the control of cell polarization.
Figure 4.
Bnr1 recruitment to the bud neck is affected by DMA1 and DMA2 deletion. (A) Logarithmically growing wild-type and dma1Δ dma2Δ cells expressing Bnr1-GFP were imaged at 25°. Acquired Z-stacks (11 planes at 0.3-μm spacing) were max-projected. Bar, 5 μm. Fluorescence intensities of Bnr1-GFP at the bud neck were quantified on one single in-focus plane in medium-budded cells after drawing a line across the bud neck along the mother–bud axis and measuring the integrated density of the resulting histogram after background correction (n ≥ 14; error bar: SD; *** P < 0.001). (B) Ratios between bud neck and total Bnr1-GFP were calculated after measuring fluorescence intensities in medium-budded cells by ImageJ using the “analyse particles” function applied to a single in-focus plane after background subtraction (n = 20; *** P < 0.001). (C) Wild-type and dma1Δ dma2Δ cells expressing Bnr1-GFP and Myo1-mCherry were imaged every minute at 21°. Z-stacks (31 planes at 0.2-μm spacing) were deconvolved with Huygens and max-projected. Arrowheads indicate the start of AMR contraction that coincides with complete disappearance of Bnr1 at the bud neck. Bar, 3 μm. (D) Wild-type and dma1Δ dma2Δ cells were arrested in S phase by hydroxyurea. Fluorescence intensities of Bnr1-GFP and Myo1-mCherry were measured with ImageJ as in B. A horizontal line in each dot plot indicates the mean ± SD (n ≥ 50; *** P < 0.001). (E) Steady-state levels of Bnr1-GFP were quantified by Western blot analysis in wild-type and dma1Δ dma2Δ cells. Ratios between Bnr1-GFP and Pgk1 (loading control) levels are averaged from three independent blots.
The polarisome components Spa2 and Bud6 are not affected by loss of Dma proteins
Bni1 is part of the polarisome, together with the Spa2, Bud6, and Pea2 proteins (Fujiwara et al. 1998; Sheu et al. 1998). Furthermore, polarisome components promote Bni1 recruitment to the bud tip (Fujiwara et al. 1998; Sagot et al. 2002a; Moseley et al. 2004; Moseley and Goode 2005; Delgehyr et al. 2008; Graziano et al. 2011). Since Bni1 is mislocalized in the absence of Dma1/2, we wondered if other polarisome components were similarly affected. Distribution of Spa2-eGFP (Figure S6A) and Bud6-GFP (Figure S6D) showed a similar pattern in wild-type and dma1Δ dma2Δ mutant cells, accumulating at the bud cortex of small and medium-budded cells and being relocalized to the bud neck of large budded cells. Quantification of fluorescent signals of either protein in the bud or at the bud neck showed no major differences in the presence or absence of Dma proteins (Figure S6, B, E, and G), although a small but significant increase in the amount of Bud6-GFP in small budded cells could be observed in dma1Δ dma2Δ relative to wild-type cells (Figure S6G). Finally, the levels of the two proteins in total extracts of wild-type and dma1Δ dma2Δ mutant cells were comparable, as assessed by Western blot analysis (Figure S6, C and F). Thus, loss of Dma proteins does not seem to affect the whole polarisome.
Dma proteins control actin cable assembly and Bni1 localization independently of the Elm1 kinase
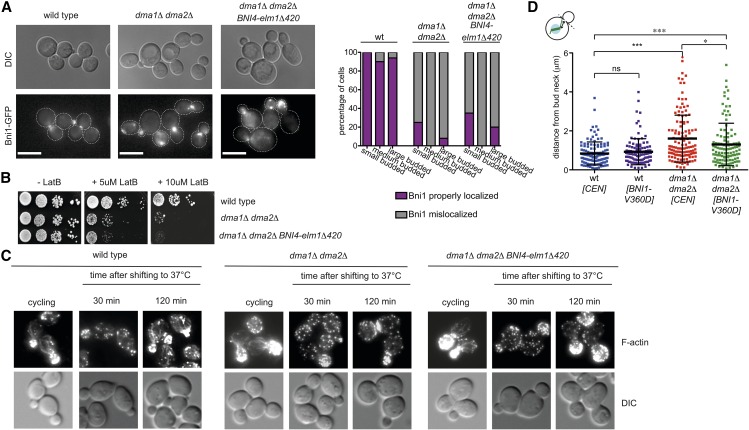
We previously showed that dma1Δ dma2Δ mutant cells are defective in the recruitment to the bud neck of the Elm1 kinase (Merlini et al. 2012), which in turn is involved in septin organization, timely mitotic entry, and the checkpoint responding to spindle mispositioning (Sreenivasan and Kellogg 1999; Bouquin et al. 2000; Thomas et al. 2003; Gladfelter et al. 2004; Caydasi et al. 2010; Moore et al. 2010). Artificial tethering of Elm1 to the bud neck by the Bni4-Elm1Δ420 chimeric protein (Moore et al. 2010) significantly rescued the septin and spindle position defects of dma1Δ dma2Δ cells (Merlini et al. 2012). Since Elm1 also contributes to bud neck localization of Bnr1, especially at high temperatures (Buttery et al. 2012), we wondered if inefficient Elm1 recruitment to the neck could be responsible for the formin mislocalization and actin cable defects of dma1Δ dma2Δ mutant cells. This does not seem to be the case. Indeed, expression of the Bni4-Elm1Δ420 chimeric protein did not restore proper Bni1 localization (Figure 5A) and rescued neither the LatB sensitivity nor the actin cable defects of dma1Δ dma2Δ mutant cells (Figure 5, B and C), suggesting that Dma proteins control formin activity through proteins other than Elm1.
Figure 5.
Artificial tethering of Elm1 to the bud neck does not rescue the actin defects of cells lacking Dma proteins, while Bni1 hyperactivation partially restores proper spindle positioning in dma1Δ dma2Δ cells. (A) Wild-type and dma1Δ dma2Δ cells expressing Bni1-GFP and either lacking or carrying the ELM1-BNI4-elm1Δ420 construct were grown at 30° and imaged. The graph shows the percentage of cells with properly localized or mislocalized Bni1 classified according to bud size. n ≥ 80. (B) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates either lacking or containing the indicated concentrations of LatB and incubated at 25°. (C) Wild-type and dma1Δ dma2Δ cells either lacking or carrying the ELM1-BNI4-elm1Δ420 construct were grown at 25° and shifted to 37°. At the indicated times, cells were fixed and stained by Alexa-546 phalloidin to analyze F-actin structures. (D) Wild-type and dma1Δ dma2Δ cells either carrying an empty vector (CEN) or a centromeric plasmid bearing the hyperactive BNI1-V360D allele were grown in selective (SD −Trp) medium at 30° and then shifted to YEPD containing 0.2 M hydroxyurea to arrest cells in S phase. Cells were washed with PBS and imaged to measure the distance between the proximal spindle pole and the bud neck (n ≥ 108). A horizontal line in each dot plots indicates the mean ± SD (* P ≤ 0.05; *** P ≤ 0.001; ns, not significant).
Since Bni1 has been implicated in spindle positioning (Lee et al. 1999), we wondered if Bni1 mislocalization could partly explain the spindle position defects of dma1Δ dma2Δ cells. Interestingly, expression of the hyperactive Bni1-V360V protein partially, but significantly, decreased the average spindle distance from the bud neck in dma1Δ dma2Δ mutant cells (1.3 ± 1.1 μm, n = 108 in dma1Δ dma2Δ BNI1-V360D cells vs. 1.6 ± 1.2 μm, n = 124 in dma1Δ dma2Δ cells, Figure 5D). Thus, inefficient spindle positioning in dma1Δ dma2Δ cells is likely due to both Elm1 and formin localization defects.
Hyperactivation of Rho1, Cdc42, or Pkc1, as well as GIC2 overexpression, do not restore proper Bni1 localization and actin cable network in dma1Δ dma2Δ cells
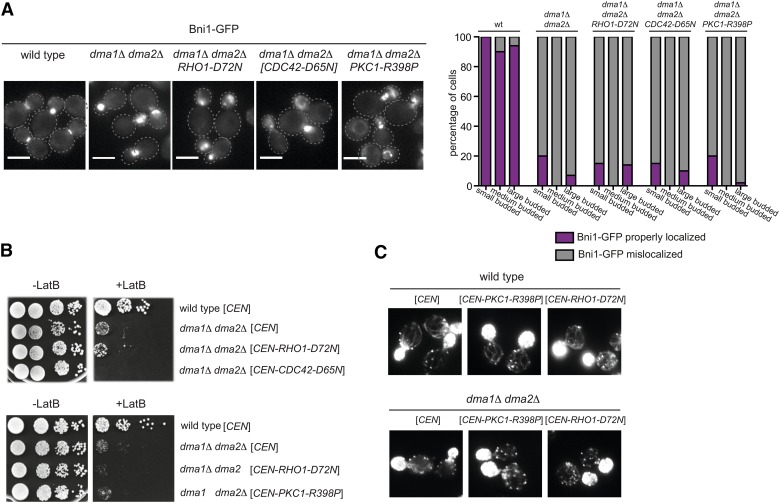
We recently showed that hyperactivation of the Rho1 GTPase and its effector protein kinase C (Pkc1) recue the temperature sensitivity and septin defects of dma1Δ dma2Δ cla4-75 triple mutant cells (Merlini et al. 2015). Since Rho1 and Pkc1 are known formin regulators together with Cdc42 (Kohno et al. 1996; Evangelista et al. 1997; Imamura et al. 1997; Dong et al. 2003), we asked if hyperactivation of these proteins could rescue the mislocalization of Bni1 and impaired actin cable organization of dma1Δ dma2Δ double mutant cells. To address this question, we used the dominant hyperactive alleles RHO1-D72N (Merlini et al. 2015), PKC1-R398P (Nonaka et al. 1995), and CDC42-D65N (Mosch et al. 2001). None of these mutant alleles restored normal Bni1 localization (Figure 6A), sensitivity to LatB (Figure 6B), or robust actin cable organization (Figure 6C) in dma1Δ dma2Δ cells, suggesting that the polarity defects of this mutant are probably unlinked to a reduced activity of the above factors.
Figure 6.
Hyperactivation of Rho GTPases and Pkc1 does not rescues the mislocalization of Bni1 and the actin defects of dma1Δ dma2Δ cells. (A) Cells with the indicated genotypes and expressing Bni1-GFP were grown at 25° and imaged. The graph shows the percentage of cells with properly localized or mislocalized Bni1 classified according to bud size (n ≥ 100). (B) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates either lacking or containing 5 μM LatB and incubated at 25°. (C) F-actin was stained with Alexa-546 phalloidin on fixed asynchronous cells with the indicated genotype.
The polarity protein Gic2, which together with its paralogue Gic1 is an effector of Cdc42 (Brown et al. 1997; Chen et al. 1997), has been implicated in Bni1 localization and activity by direct binding to a region of the protein (referred to as ND2) that is unrelated to the other known regulatory domains (GBD, SBD, BBD, FH1, and FH2; (Jaquenoud and Peter 2000; Chen et al. 2012). Furthermore, deletion of GIC2, or GIC1 and GIC2 together, displays synthetic interactions with BNI1 deletion (Bi et al. 2000; Jaquenoud and Peter 2000), similar to deletion of DMA1 and DMA2. Upon crossing a gic1Δ gic2Δ deletion strain with a bni1Δ deletion strain, followed by tetrad analysis, we confirmed that the gic1Δ gic2Δ bni1Δ triple mutant is inviable also in our strain background (Figure S7A, Bi et al. 2000), while a gic1Δ gic2Δ bni1-ts mutant (alias bni1-FH2#1, Sagot et al. 2002a) is temperature sensitive already at 30° (Figure S7B). We then tested if deletion of DMA1 and DMA2 displays synthetic growth defects in gic1Δ gic2Δ cells. After tetrad dissection of a diploid strain heterozygous for all four gene knockouts, we found that gic1Δ gic2Δ dma1Δ dma2Δ quadruple mutants were mostly inviable (Figure S7, C and D), while the only quadruple mutant that we obtained out of 38 tetrads was slow growing and temperature sensitive (Figure S7E). Altogether, these genetic interactions suggest that Dma1/2 and Gic1/2 might be part of parallel pathways in the control of Bni1 localization/activity. We therefore decided to test if high levels of GIC2 on a 2μ high copy number plasmid could restore proper Bni1 localization and sensitivity to LatB of dma1Δ dma2Δ mutant cells. Although our 2μ-GIC2 plasmid could efficiently rescue the temperature sensitivity of gic1Δ gic2Δ mutant cells, it did not rescue the slight temperature sensitivity of dma1Δ dma2Δ cells (Figure S8A), their Bni1 mislocalization (Figure S8B), and LatB sensitivity (Figure S8C), suggesting that Gic2, even at presumably higher levels, might not compensate for loss of Dma1/2 in cell polarity.
The catalytic domain of Bni1 is insufficient to provide essential formin functions in the absence of Dma1/2
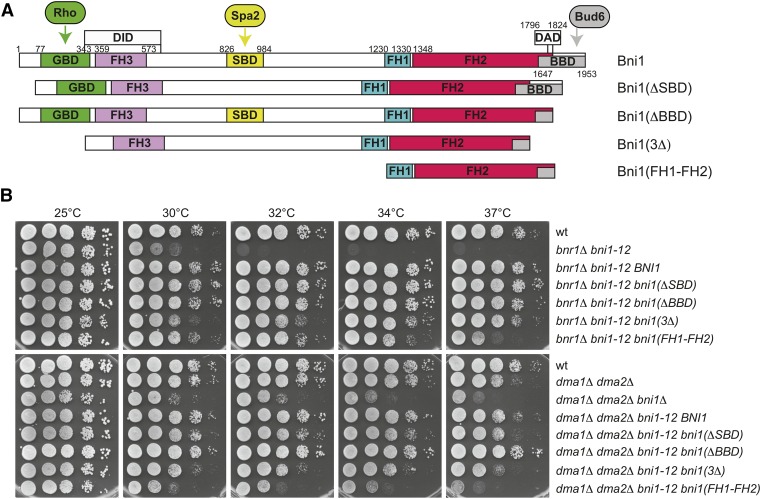
To gain further insights into the mechanism by which Dma proteins could control formin function, we integrated at the bni1-12 locus of a bnr1Δ bni1-12 double mutant or of a dma1Δ dma2Δ bni1-12 triple mutant various BNI1 alleles expressing different portions of Bni1 (Figure 7A, Chen et al. 2012). The alleles lacking the Spa2-binding domain [bni1(ΔSBD)], the Bud6-binding domain [bni1(ΔBBD)], or the Rho-, Spa2- and Bud6-binding domains together [bni1(3Δ)], which are dispensable for Bni1 essential functions (Chen et al. 2012), could efficiently rescue the temperature sensitivity of both a bnr1Δ bni1-12 and dma1Δ dma2Δ bni1-12 cells (Figure 7B). In striking contrast, a construct expressing only the catalytic FH1 and FH2 domains [bni1(FH1-FH2)], which can normally provide the essential formin functions in yeast (Gao and Bretscher 2009; Chen et al. 2012), could efficiently rescue the temperature sensitivity of a bnr1Δ bni1-12 but not of dma1Δ dma2Δ bni1-12 cells, which displayed at all temperatures the same growth defects of a dma1Δ dma2Δ bni1Δ mutant (Figure 7B). Thus, in the absence of Dma1/2 the FH1-FH2 fragment of Bni1 is no longer able to suffice for formin’s crucial functions, suggesting that Dma proteins might impact on the actin-polymerizing activity of formin rather than on other regulatory inputs.
Figure 7.
The catalytic domain of Bni1 is not sufficient to provide essential formin functions in the absence of Dma1/2. (A) Schematic representation of Bni1 indicating its relevant regulatory domains and the constructs used in B (Chen et al. 2012). All constructs were tagged at the N terminus with three HA and one GFP. GBD, GTPase-binding domain; SBD, Spa2-binding domain; BBD, Bud6-binding domain; DID, diaphanous inhibitory domain; DAD, diaphanous autoregulatory domain; FH1–FH3, formin homology 1–3. (B) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates and incubated at the indicated temperatures.
Dma proteins interact physically with formins to control their localization and/or activity likely through ubiquitylation
Dma proteins may control formin dynamics by interacting physically with them. To test this possibility, we immunoprecipitated Dma1 or Dma2 tagged at the C terminus with Flag3 epitopes (Dma1-Flag3 and Dma2-Flag3) from cells coexpressing endogenous Bni1 or Bnr1 tagged at the C terminus with a triple HA epitope (Bni1-HA3 and Bnr1-HA3). Remarkably, both Dma1 and Dma2 co-immunoprecipitated Bni1 and Bnr1 (Figure 8A), suggesting that these proteins interact in yeast cells.
The formin Bni1 was identified as an ubiquitylated protein in a large-scale proteomic study (Kolawa et al. 2013), although the E3 ubiquitin-ligase(s) involved is unknown. Additionally, an N-terminal fragment of Bni1 was shown to be ubiquitylated in vitro by Rsp5 and its protein levels critical for actin reorganization during wound healing (Kono et al. 2012). Finally, the mammalian formin mDia2 is targeted to ubiquitin-mediated degradation (DeWard and Alberts 2009). It is therefore possible that Dma proteins control formin activity and/or localization through ubiquitylation. Despite our efforts, we could not detect Bni1 ubiquitylation in yeast cells. We therefore asked if the ubiquitin-ligase activity of Dma1/2 was required for proper formin distribution by expressing wild-type DMA2 or the catalytically inactive dma2-C451A allele, both tagged with 13 myc epitopes at the C terminus (Loring et al. 2008; Chahwan et al. 2013) in dma1Δ dma2Δ mutant cells. Consistent with a role for ubiquitylation in Bni1 distribution, dma1Δ dma2Δ cells expressing dma2-C451A showed the same mislocalization of Bni1-GFP and sensitivity to high temperature and LatB observed in the same cells transformed with the empty vector, while expression of wild-type DMA2 restored normal Bni1 distribution, as well as resistance to high temperature and LatB (Figure 8, B and C). Therefore, proper formin distribution and robust actin cable assembly depends on the ubiquitin-ligase activity of Dma proteins. In contrast, Dma proteins were not required for Bni1 proteolysis (Figure 8D), which instead involves Rsp5 (Kono et al. 2012).
Discussion
A novel role for Dma proteins in the regulation of the actin cytoskeleton
The redundant Dma1 and Dma2 proteins are E3 ubiquitin ligases of the RING finger family (Joazeiro and Weissman 2000) that had been previously implicated in a number of polarized processes, such as septin organization, spindle positioning, and vacuole inheritance (Fraschini et al. 2004; Merlini et al. 2012; Chahwan et al. 2013; Yau et al. 2014). Here we show that they are additionally involved in the architecture of the actin cable network, thus emphasizing their involvement in the fine regulation of cell polarity. Indeed, dma1dma2 double mutants show reduced number and thickness of actin cables in unperturbed conditions and are hypersensitive to LatB, which depolymerizes actin cables more efficiently than actin patches (Irazoqui et al. 2005). Upon heat shock, which triggers a transient actin depolarization (Lillie and Brown 1994), the actin defects of dma1dma2 cells get even more pronounced. In spite of their apparent scarcity and thinness, actin cables are still proficient at delivering Sec4 to polarity sites in dma1Δ dma2Δ cells, suggesting that they can promote polarized exocytosis. At the moment, however, we cannot rule out that other cable-mediated directional movements, such as retrograde transport or organelle inheritance, are impaired in the absence of Dma1/2.
We further show that in dma1Δ dma2Δ mutant cells the distribution of both formins, which are responsible for actin cable polymerization, is severely perturbed, thus accounting for their actin defects. In contrast, localization of the polarisome components Spa2 and Bud6 is not affected in the absence of Dma proteins. While in dma1dma2 cells Bnr1 is present at the bud neck with normal kinetics during the cell cycle, albeit at reduced levels, Bni1 localization is strongly affected, with the protein forming mobile clusters that shuttle between bud tip and bud neck, suggesting that Bni1 might not be stably retained at membranes. Although this behavior is never observed in wild-type cells, Bni1 must be still at least partly functional, as underscored by the viability of dma1dma2bnr1 triple mutant cells that contrasts with the lethality of bni1bnr1 double mutants. On the contrary, simultaneous deletion of DMA1 and DMA2 causes synthetic growth defects in bni1Δ cells, as well as wider bud necks and septin rings, suggesting that reduced levels of Bnr1 at the bud neck in the absence of Dma proteins do have a physiological impact.
Synthetic lethality/sickness combining given mutations with deletion of either formin has been extensively used to assign the function of the corresponding proteins in the actin polymerization pathway of Bni1 or Bnr1 (Kamei et al. 1998; Jaquenoud and Peter 2000; Chesarone et al. 2009; Buttery et al. 2012). However, the existence of redundant controls over formin function precludes drawing definitive conclusions. For instance, Spa2 and Gic2 are involved in Bni1 recruitment to sites of cell polarity and/or activation, yet their deletion shows genetic interactions with BNI1 deletion (Fujiwara et al. 1998; Jaquenoud and Peter 2000; Sagot et al. 2002a; Chen et al. 2012). In this context it is worth considering that effectors of the Cdc42 GTPase, which include many formin regulators and formins themselves, are part of positive feedback loops that enhance Cdc42 activation at the polarity site, thereby contributing to the establishment of a robust polarity axis (Wedlich-Soldner et al. 2003; Wedlich-Soldner et al. 2004; Kozubowski et al. 2008; Freisinger et al. 2013). Thus, some of the genetic interactions reported in the literature could be ascribed to polarity defects, rather than to problems in actin organization. In this context, it is quite remarkable that deletion of DMA1 and DMA2 shows synthetic interactions with several effectors of Cdc42 (i.e., Cla4, Bni1, Gic1, and Gic2), suggesting that Dma1/2 might contribute to such feedback mechanisms. Thus, the interpretation of genetic interactions is not always straightforward and we speculate that Dma proteins likely regulate recruitment, and possibly activity, of both formins at membranes. This conclusion is in line with our finding that Dma1/2 interact physically with both formins (see below).
Formin and actin defects of dma1dma2 cells are unlinked to the previously discovered function of Dma1/2 in the localization of the Elm1 kinase to the bud neck (Merlini et al. 2012), as they are not rescued by a chimeric protein that drives constitutive recruitment of the Elm1 kinase domain to the bud neck (Bni4-Elm1Δ420, Moore et al. 2010). In contrast, expression of a dominant hyperactive variant of Bni1 (Bni1-V360D, Kono et al. 2012) in dma1dma2 cells restores apparently normal actin cables and wild-type sensitivity to LatB, indicating that the actin defects in these mutant cells are due to dysfunctional formins. Remarkably, we find that the spindle positioning defects of dma1dma2 mutant cells are also partially suppressed by expression of the hyperactive Bni1-V360D variant, suggesting that they are partly caused by a sluggish actin cable network. In contrast, Bni1-V360D does not rescue the lethality of dma1dma2cla4 triple mutant cells that was primarily attributed to septin defects (Merlini et al. 2012, 2015).
How could Dma proteins control formins?
We show that the ubiquitin-ligase activity of Dma proteins is required for proper Bni1 distribution and resistance to LatB, suggesting that specific targets are likely ubiquitylated to modulate formin function. Given the physical interaction between Dma1/2 and both formins, we speculate that Dma1/2 promote ubiquitylation of formins themselves or a formin interactor, thereby impacting on their ability to interact with membranes. Indeed, Bni1 was found to be ubiquitylated in a large-scale proteomic study (Kolawa et al. 2013), although the E3 ubiquitin-ligase(s) involved is unknown. Furthermore, a truncated variant of Bni1 is ubiquitylated in vitro by the Rsp5 E3 ligase and Bni1 is degraded in an Rsp5-dependent manner to bring about actin reorganization under stress conditions and wound healing (Kono et al. 2012). Finally, the mammalian formin mDia2 is targeted to ubiquitin-mediated degradation (DeWard and Alberts 2009), suggesting that formin ubiquitylation and degradation might be a conserved mechanism to remodel the actin cytoskeleton. Interestingly, we find that Dma proteins are dispensable for Bni1 proteolysis, indicating that Dma1/2 might act differently from Rsp5 for what concerns formin regulation. Importantly, Dma proteins were shown to promote both K48- and K63-linked polyubiquitylation in vitro (Loring et al. 2008), which have been classically associated with proteasome-mediated degradation and nonproteolytic modulation of protein function, respectively (Komander and Rape 2012). Thus, Dma1/2 could impact on formin activity by regulating its interaction with the cortex and/or its conformation. For instance, Dma-dependent ubiquitylation could regulate the dimerization properties of formin that in turn depend on the FH2 domain of the protein. Despite our efforts, we could not detect Bni1 ubiquitylation in yeast cells. Thus, further work will be required to assess the possible involvement of Dma proteins in ubiquitylation of formins or formin regulators.
Formins exist in an autoinhibited, close conformation held by intramolecular interactions between the N- and the C-terminal part of the protein. Autoinhibition is thought to be relieved by formin binding to Rho GTPases (reviewed in Chesarone et al. 2010). We find that hyperactivation of Cdc42, Rho1, or Pkc1 does not rescue Bni1 mislocalization, hypersensitivity to LatB, and actin cable defects of dma1Δ dma2Δ mutant cells, suggesting that Dma-dependent regulation of formins exploits a distinct mechanism from that used by Rho GTPases. Additional mechanisms, besides binding to Rho GTPases, are necessary for complete formin activation in vitro (Li and Higgs 2003; Martin et al. 2007). For instance, autoinhibition of mammalian formins prevents their membrane localization and actin assembly activity. Membrane interaction can be restored either by binding to Cdc42 or through a Rho-independent mechanism that implicates the interaction with a membrane-associated factor (Seth et al. 2006). An attractive hypothesis is that such a mechanism involves Dma1/2 in budding yeast.
So far, we found no evidence indicating the possible involvement of other known formin regulators (i.e., Gic2, Spa2, Bud6, or Elm1) in the Dma-dependent control of formins. Interestingly, the catalytic FH1–FH2 portion of Bni1, which normally suffices for the essential functions of formin, requires Dma1/2 for its activity, suggesting that Dma proteins might impact on these protein domains rather than on other regulatory regions, such as GBD, SBD, and BBD. Thus, we speculate that Dma-dependent ubiquitylation might impact a novel regulatory input to formin regulation that is unlinked to the ones described to date.
In summary, our data uncover a potentially novel level of regulation of formin activation that adds up to its overwhelming complexity. Considering the critical role of formins in a variety of diverse cellular processes, such as cell polarity, cell migration, cytokinesis, spindle positioning, and cytokinesis, it not surprising that formins are subject to elaborate and multilayered controls.
Acknowledgments
We are grateful to Y. Barral, G. Braus, J. Cooper, S. Gravel, M. Hall, D. Lew, M. P. Longhese, M. Muzi-Falconi, D. Pellman, E. Schwob, and M. Segal for strains and plasmids; to G. Rancati for insightful discussions; and to V. Georget and J. Mateos Langerak for invaluable help with video microscopy and image processing at the Montpellier RIO Imaging platform. This work has been supported by Fondation ARC (Cancer Research Association) (grant PJA 20141201926 to S.P.) and the Ligue Régionale contre le Cancer (Comité de l’Hérault). M.A.J. was supported by a postdoctoral fellowship from the Fondation pour la Recherche Médicale and the Merlion Programme.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.189258/-/DC1.
Communicating editor: D. J. Lew
Literature Cited
- Adams A. E., Pringle J. R., 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98: 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts A. S., 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 276: 2824–2830. [DOI] [PubMed] [Google Scholar]
- Benton B. K., Tinkelenberg A., Gonzalez I., Cross F. R., 1997. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 17: 5067–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D. J., Salmon E. D., McMillan J. N., et al. , 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142: 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Chiavetta J. B., Chen H., Chen G. C., Chan C. S., et al. , 2000. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell 11: 773–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin N., Barral Y., Courbeyrette R., Blondel M., Snyder M., et al. , 2000. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 113(Pt 8): 1435–1445. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Jaquenoud M., Gulli M. P., Chant J., Peter M., 1997. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11: 2972–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery S. M., Yoshida S., Pellman D., 2007. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell 18: 1826–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery S. M., Kono K., Stokasimov E., Pellman D., 2012. Regulation of the formin Bnr1 by septins anda MARK/Par1-family septin-associated kinase. Mol. Biol. Cell 23: 4041–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi A. K., Kurtulmus B., Orrico M. I., Hofmann A., Ibrahim B., et al. , 2010. Elm1 kinase activates the spindle position checkpoint kinase Kin4. J. Cell Biol. 190: 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahwan R., Gravel S., Matsusaka T., Jackson S. P., 2013. Dma/RNF8 proteins are evolutionarily conserved E3 ubiquitin ligases that target septins. Cell Cycle 12: 1000–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. C., Kim Y. J., Chan C. S., 1997. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 11: 2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Kuo C. C., Kang H., Howell A. S., Zyla T. R., et al. , 2012. Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p. Mol. Biol. Cell 23: 3814–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M., Gould C. J., Moseley J. B., Goode B. L., 2009. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev. Cell 16: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M. A., DuPage A. G., Goode B. L., 2010. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11: 62–74. [DOI] [PubMed] [Google Scholar]
- Chesarone-Cataldo M., Guerin C., Yu J. H., Wedlich-Soldner R., Blanchoin L., et al. , 2011. The myosin passenger protein Smy1 controls actin cable structure and dynamics by acting as a formin damper. Dev. Cell 21: 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Lopes C. S., Moir C. A., Huisman S. M., Segal M., 2008. Dissecting the involvement of formins in Bud6p-mediated cortical capture of microtubules in S. cerevisiae. J. Cell Sci. 121: 3803–3814. [DOI] [PubMed] [Google Scholar]
- Delley P. A., Hall M. N., 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWard A. D., Alberts A. S., 2009. Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J. Biol. Chem. 284: 20061–20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Pruyne D., Bretscher A., 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161: 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Taylor I. A., Sarbassova D., Haire L. F., Westcott S. L., et al. , 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6: 1169–1182. [DOI] [PubMed] [Google Scholar]
- Eskin J. A., Rankova A., Johnston A. B., Alioto S. L., Goode B. L., 2016. Common formin-regulating sequences in Smy1 and Bud14 are required for the control of actin cable assembly in vivo. Mol. Biol. Cell 27: 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Blundell K., Longtine M. S., Chow C. J., Adames N., et al. , 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276: 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A., 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4: 260–269. [DOI] [PubMed] [Google Scholar]
- Fraschini R., Bilotta D., Lucchini G., Piatti S., 2004. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol. Biol. Cell 15: 3796–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., D’Ambrosio C., Venturetti M., Lucchini G., Piatti S., 2006. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 172: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisinger T., Klunder B., Johnson J., Muller N., Pichler G., et al. , 2013. Establishment of a robust single axis of cell polarity by coupling multiple positive feedback loops. Nat. Commun. 4: 1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Tanaka K., Mino A., Kikyo M., Takahashi K., et al. , 1998. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9: 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Bretscher A., 2009. Polarized growth in budding yeast in the absence of a localized formin. Mol. Biol. Cell 20: 2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Liu W., Bretscher A., 2010. The yeast formin Bnr1p has two localization regions that show spatially and temporally distinct association with septin structures. Mol. Biol. Cell 21: 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Zyla T. R., Lew D. J., 2004. Genetic interactions among regulators of septin organization. Eukaryot. Cell 3: 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B. L., Eck M. J., 2007. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76: 593–627. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Eskin J. A., Wendland B., 2015. Actin and endocytosis in budding yeast. Genetics 199: 315–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould C. J., Chesarone-Cataldo M., Alioto S. L., Salin B., Sagot I., et al. , 2014. Saccharomyces cerevisiae Kelch proteins and Bud14 protein form a stable 520-kDa formin regulatory complex that controls actin cable assembly and cell morphogenesis. J. Biol. Chem. 289: 18290–18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano B. R., DuPage A. G., Michelot A., Breitsprecher D., Moseley J. B., et al. , 2011. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol. Biol. Cell 22: 4016–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano B. R., Yu H. Y., Alioto S. L., Eskin J. A., Ydenberg C. A., et al. , 2014. The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol. Biol. Cell 25: 1730–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs H. N., 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30: 342–353. [DOI] [PubMed] [Google Scholar]
- Holly S. P., Blumer K. J., 1999. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 147: 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H., Tanaka K., Hihara T., Umikawa M., Kamei T., et al. , 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16: 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J. E., Howell A. S., Theesfeld C. L., Lew D. J., 2005. Opposing roles for actin in Cdc42p polarization. Mol. Biol. Cell 16: 1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., et al. , 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. [DOI] [PubMed] [Google Scholar]
- Jaquenoud M., Peter M., 2000. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p). Mol. Cell. Biol. 20: 6244–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C. A., Weissman A. M., 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102: 549–552. [DOI] [PubMed] [Google Scholar]
- Jose M., Tollis S., Nair D., Sibarita J. B., McCusker D., 2013. Robust polarity establishment occurs via an endocytosis-based cortical corralling mechanism. J. Cell Biol. 200: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T., Tanaka K., Hihara T., Umikawa M., Imamura H., et al. , 1998. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273: 28341–28345. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E., 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98: 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H., Tanaka K., Mino A., Umikawa M., Imamura H., et al. , 1996. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15: 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kolawa N., Sweredoski M. J., Graham R. L., Oania R., Hess S., et al. , 2013. Perturbations to the ubiquitin conjugate proteome in yeast deltaubx mutants identify Ubx2 as a regulator of membrane lipid composition. Mol. Cell. Proteomics 12: 2791–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M., 2012. The ubiquitin code. Annu. Rev. Biochem. 81: 203–229. [DOI] [PubMed] [Google Scholar]
- Kono K., Saeki Y., Yoshida S., Tanaka K., Pellman D., 2012. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell 150: 151–164. [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Saito K., Johnson J. M., Howell A. S., Zyla T. R., et al. , 2008. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 18: 1719–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M., Rose R., Scrima A., Wittinghofer A., 2005. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 24: 4176–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson R. E., Winters M. J., Pryciak P. M., 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 22: 2939–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Klee S. K., Evangelista M., Boone C., Pellman D., 1999. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J. Cell Biol. 144: 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15: 648–653. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I., 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Higgs H. N., 2003. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13: 1335–1340. [DOI] [PubMed] [Google Scholar]
- Li F., Higgs H. N., 2005. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 280: 6986–6992. [DOI] [PubMed] [Google Scholar]
- Lillie S. H., Brown S. S., 1994. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J. Cell Biol. 125: 825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R., 1998. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140: 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring G. L., Christensen K. C., Gerber S. A., Brenner C., 2008. Yeast Chfr homologs retard cell cycle at G1 and G2/M via Ubc4 and Ubc13/Mms2-dependent ubiquitination. Cell Cycle 7: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E. F., Sambrook J., 1992. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY. [Google Scholar]
- Martin S. G., Rincon S. A., Basu R., Perez P., Chang F., 2007. Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell 18: 4155–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Fraschini R., Boettcher B., Barral Y., Lucchini G., et al. , 2012. Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. PLoS Genet. 8: e1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Bolognesi A., Juanes M. A., Vandermoere F., Courtellemont T., et al. , 2015. Rho1- and Pkc1-dependent phosphorylation of the F-BAR protein Syp1 contributes to septin ring assembly. Mol. Biol. Cell 26: 3245–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. K., Chudalayandi P., Heil-Chapdelaine R. A., Cooper J. A., 2010. The spindle position checkpoint is coordinated by the Elm1 kinase. J. Cell Biol. 191: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch H. U., Kohler T., Braus G. H., 2001. Different domains of the essential GTPase Cdc42p required for growth and development of Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Goode B. L., 2005. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 280: 28023–28033. [DOI] [PubMed] [Google Scholar]
- Moseley J. B., Sagot I., Manning A. L., Xu Y., Eck M. J., et al. , 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15: 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka H., Tanaka K., Hirano H., Fujiwara T., Kohno H., et al. , 1995. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14: 5931–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Bi E., 2010. Septin structure and function in yeast and beyond. Trends Cell Biol. 21: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Schreiter J., Nishihama R., Wloka C., Bi E., 2013. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol. Biol. Cell 24: 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T., Otomo C., Tomchick D. R., Machius M., Rosen M. K., 2005a Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell 18: 273–281. [DOI] [PubMed] [Google Scholar]
- Otomo T., Tomchick D. R., Otomo C., Panchal S. C., Machius M., et al. , 2005b Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433: 488–494. [DOI] [PubMed] [Google Scholar]
- Ozaki-Kuroda K., Yamamoto Y., Nohara H., Kinoshita M., Fujiwara T., et al. , 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez P., Rincon S. A., 2010. Rho GTPases: regulation of cell polarity and growth in yeasts. Biochem. J. 426: 243–253. [DOI] [PubMed] [Google Scholar]
- Petersen J., Nielsen O., Egel R., Hagan I. M., 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A. J., Maddox A. S., 2010. The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 21: 881–891. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A., 2000a Polarization of cell growth in yeast. J. Cell Sci. 113(Pt 4): 571–585. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A., 2000b Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113(Pt 3): 365–375. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A., 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15: 4971–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspelli E., Cassani C., Lucchini G., Fraschini R., 2011. Budding yeast Dma1 and Dma2 participate in regulation of Swe1 levels and localization. Mol. Biol. Cell 22: 2185–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., et al. , 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119: 419–429. [DOI] [PubMed] [Google Scholar]
- Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M. R., et al. , 2005. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 435: 513–518. [DOI] [PubMed] [Google Scholar]
- Sagot I., Klee S. K., Pellman D., 2002a Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4: 42–50. [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D., 2002b An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4: 626–631. [DOI] [PubMed] [Google Scholar]
- Schott D. H., Collins R. N., Bretscher A., 2002. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J. Cell Biol. 156: 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A., Otomo C., Rosen M. K., 2006. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J. Cell Biol. 174: 701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff M. A., Thorn K. S., 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21: 661–670. [DOI] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Sheu Y. J., Santos B., Fortin N., Costigan C., Snyder M., 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18: 4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan A., Kellogg D., 1999. The elm1 kinase functions in a mitotic signaling network in budding yeast. Mol. Cell. Biol. 19: 7983–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. L., Blacketer M. J., Edgington N. P., Myers A. M., 2003. Assembly interdependence among the S. cerevisiae bud neck ring proteins Elm1p, Hsl1p and Cdc12p. Yeast 20: 813–826. [DOI] [PubMed] [Google Scholar]
- Tolliday N., VerPlank L., Li R., 2002. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr. Biol. 12: 1864–1870. [DOI] [PubMed] [Google Scholar]
- Vallen E. A., Caviston J., Bi E., 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth W. P., Olsen A. E., Sbia M., Freedman K. H., Stillman D. J., 2005. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot. Cell 4: 1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Altschuler S., Wu L., Li R., 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299: 1231–1235. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Wai S. C., Schmidt T., Li R., 2004. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 166: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen K. K., Rubenstein P. A., 2009. Differential regulation of actin polymerization and structure by yeast formin isoforms. J. Biol. Chem. 284: 16776–16783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Nathke I., 2007. Cell polarity in development and cancer. Nat. Cell Biol. 9: 1016–1024. [DOI] [PubMed] [Google Scholar]
- Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., et al. , 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116: 711–723. [DOI] [PubMed] [Google Scholar]
- Yau R. G., Peng Y., Valiathan R. R., Birkeland S. R., Wilson T. E., et al. , 2014. Release from myosin V via regulated recruitment of an E3 ubiquitin ligase controls organelle localization. Dev. Cell 28: 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]