Abstract
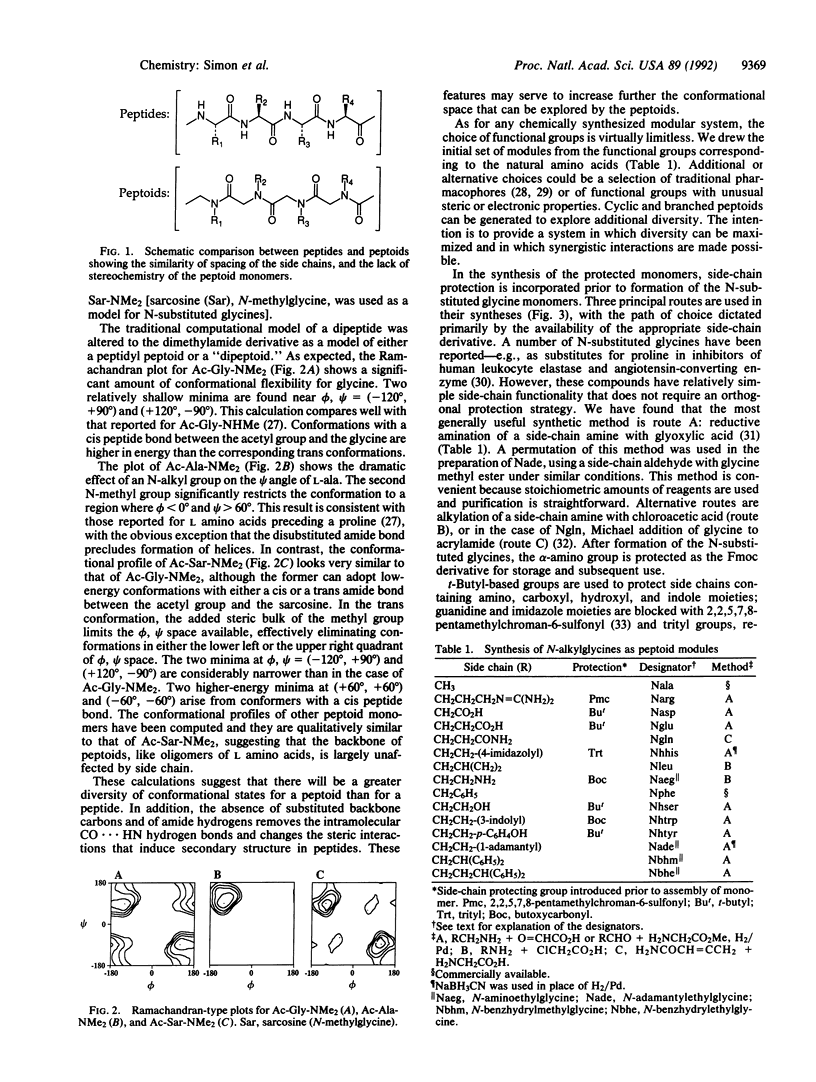
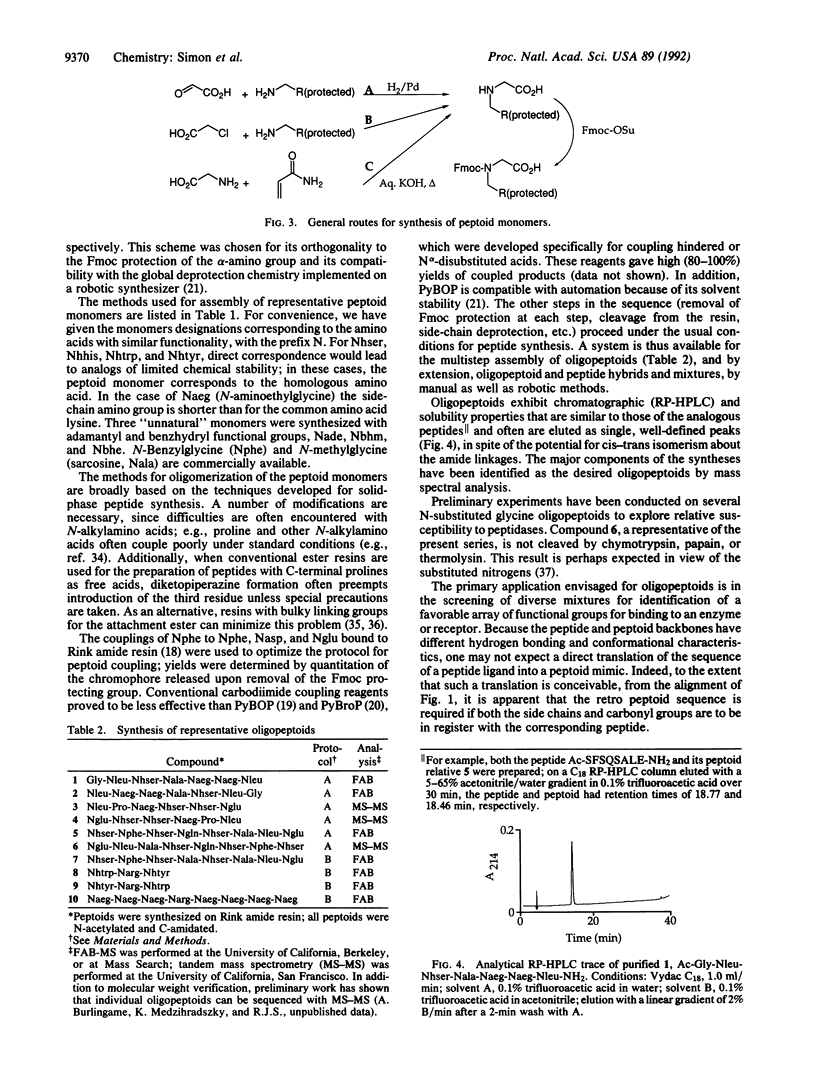
Peptoids, oligomers of N-substituted glycines, are described as a motif for the generation of chemically diverse libraries of novel molecules. Ramachandran-type plots were calculated and indicate a greater diversity of conformational states available for peptoids than for peptides. The monomers incorporate t-butyl-based side-chain and 9-fluorenylmethoxy-carbonyl alpha-amine protection. The controlled oligomerization of the peptoid monomers was performed manually and robotically with in situ activation by either benzotriazol-1-yloxytris(pyrrolidino)phosphonium hexafluorophosphate or bromotris(pyrrolidino)phosphonium hexaflurophosphate. Other steps were identical to peptide synthesis using alpha-(9-fluorenylmethoxycarbonyl)amino acids. A total of 15 monomers and 10 oligomers (peptoids) are described. Preliminary data are presented on the stability of a representative oligopeptoid to enzymatic hydrolysis. Peptoid versions of peptide ligands of three biological systems (bovine pancreatic alpha-amylase, hepatitis A virus 3C proteinase, and human immunodeficiency virus transactivator-responsive element RNA) were found with affinities comparable to those of the corresponding peptides. The potential use of libraries of these compounds in receptor- or enzyme-based assays is discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barlos K., Chatzi O., Gatos D., Stavropoulos G. 2-Chlorotrityl chloride resin. Studies on anchoring of Fmoc-amino acids and peptide cleavage. Int J Pept Protein Res. 1991 Jun;37(6):513–520. [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Cwirla S. E., Peters E. A., Barrett R. W., Dower W. J. Peptides on phage: a vast library of peptides for identifying ligands. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber-Osguthorpe P., Roberts V. A., Osguthorpe D. J., Wolff J., Genest M., Hagler A. T. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins. 1988;4(1):31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- Devlin J. J., Panganiban L. C., Devlin P. E. Random peptide libraries: a source of specific protein binding molecules. Science. 1990 Jul 27;249(4967):404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990 Aug 30;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fodor S. P., Read J. L., Pirrung M. C., Stryer L., Lu A. T., Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991 Feb 15;251(4995):767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Geysen H. M., Meloen R. H., Barteling S. J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghten R. A., Pinilla C., Blondelle S. E., Appel J. R., Dooley C. T., Cuervo J. H. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991 Nov 7;354(6348):84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- Joyce G. F. Amplification, mutation and selection of catalytic RNA. Gene. 1989 Oct 15;82(1):83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Lam K. S., Salmon S. E., Hersh E. M., Hruby V. J., Kazmierski W. M., Knapp R. J. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991 Nov 7;354(6348):82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- Malcolm B. A., Chin S. M., Jewell D. A., Stratton-Thomas J. R., Thudium K. B., Ralston R., Rosenberg S. Expression and characterization of recombinant hepatitis A virus 3C proteinase. Biochemistry. 1992 Apr 7;31(13):3358–3363. doi: 10.1021/bi00128a008. [DOI] [PubMed] [Google Scholar]
- Pei D. H., Ulrich H. D., Schultz P. G. A combinatorial approach toward DNA recognition. Science. 1991 Sep 20;253(5026):1408–1411. doi: 10.1126/science.1716784. [DOI] [PubMed] [Google Scholar]
- Petithory J. R., Masiarz F. R., Kirsch J. F., Santi D. V., Malcolm B. A. A rapid method for determination of endoproteinase substrate specificity: specificity of the 3C proteinase from hepatitis A virus. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11510–11514. doi: 10.1073/pnas.88.24.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G. N., Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Scott J. K., Smith G. P. Searching for peptide ligands with an epitope library. Science. 1990 Jul 27;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Seigner C., Prodanov E., Marchis-Mouren G. The determination of subsite binding energies of porcine pancreatic alpha-amylase by comparing hydrolytic activity towards substrates. Biochim Biophys Acta. 1987 Jun 17;913(2):200–209. doi: 10.1016/0167-4838(87)90331-1. [DOI] [PubMed] [Google Scholar]
- Skiles J. W., Fuchs V., Miao C., Sorcek R., Grozinger K. G., Mauldin S. C., Vitous J., Mui P. W., Jacober S., Chow G. Inhibition of human leukocyte elastase (HLE) by N-substituted peptidyl trifluoromethyl ketones. J Med Chem. 1992 Feb 21;35(4):641–662. doi: 10.1021/jm00082a005. [DOI] [PubMed] [Google Scholar]
- Topliss J. G. A manual method for applying the Hansch approach to drug design. J Med Chem. 1977 Apr;20(4):463–469. doi: 10.1021/jm00214a001. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Zuckermann R. N., Kerr J. M., Siani M. A., Banville S. C., Santi D. V. Identification of highest-affinity ligands by affinity selection from equimolar peptide mixtures generated by robotic synthesis. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4505–4509. doi: 10.1073/pnas.89.10.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]


