Abstract
Intracellular vesicle trafficking is a fundamental process in eukaryotic cells. It enables cellular polarity and exchange of proteins between subcellular compartments such as the plasma membrane or the vacuole. Adaptor protein complexes participate in the vesicle formation by specific selection of the transported cargo. We investigated the role of the adaptor protein complex 3 (AP-3) and adaptor protein complex 4 (AP-4) in this selection process by screening for AP-3 and AP-4 dependent cargo proteins. Specific cargo proteins are expected to be mis-targeted in knock-out mutants of adaptor protein complex components. Thus, we screened for altered distribution profiles across a density gradient of membrane proteins in wild type versus ap-3β and ap-4β knock-out mutants. In ap-3β mutants, especially proteins with transport functions, such as aquaporins and plasma membrane ATPase, as well as vesicle trafficking proteins showed differential protein distribution profiles across the density gradient. In the ap-4β mutant aquaporins but also proteins from lipid metabolism were differentially distributed. These proteins also showed differential phosphorylation patterns in ap-3β and ap-4β compared with wild type. Other proteins, such as receptor kinases were depleted from the AP-3 mutant membrane system, possibly because of degradation after mis-targeting. In AP-4 mutants, membrane fractions were depleted for cytochrome P450 proteins, cell wall proteins and receptor kinases. Analysis of water transport capacity in wild type and mutant mesophyll cells confirmed aquaporins as cargo proteins of AP-3 and AP-4. The combination of organelle density gradients with proteome analysis turned out as a suitable experimental strategy for large-scale analyses of protein trafficking.
Individual cells within multicellular organisms need to interact with their neighbors to establish correct polarity during growth and development. Also the composition of the plasma membrane and vacuole is a dynamic and signal-dependent process requiring correct targeting and recycling of specific membrane components. These dynamic and well organized processes at membranes are based on a powerful targeting system of membrane proteins to their destination compartment.
All plasma membrane constituents undergo a mechanism called secretion by which proteins, phospholipids, and other membrane molecules are transported and delivered to their final destination membranes. The secretory pathway of eukaryotic cells consists of an interconnected series of intracellular membranes and the migration of proteins starts in the endoplasmic reticulum (ER) where integral membrane proteins are synthesized, followed by movement through the Golgi compartment where they obtain specific modifications (reviewed in (1)). Sorting of proteins and final targeting occurs in the trans-Golgi network (TGN). There are indications that the trafficking of proteins from the TGN to the plasma membrane involves the exocyst complex (2), exosomes and multivesicular bodies (3).
The adaptor protein complexes play a vital role in selection of protein cargo for different cellular compartments (4). The structure of adaptor protein complexes (AP) 1 is conserved in all eukaryotes. AP-complexes are hetero-tetramers consisting of four subunits called adaptins. Adaptins are involved in the formation of intracellular transport vesicles and in the selection of cargo for incorporation into the vesicles (5). Thereby, μ-adaptins and ρ-adaptins are smaller than the other subunits (α, β, δ, γ, ε). β-subunit of some adaptor protein complexes were found to have a major role in binding to clathrins. Clathrins form an outer layer to the coat and probably play a structural role in deforming the donor membrane (6). In general, clathrin-coated vesicles are required for receptor-mediated endocytosis at the plasma membrane. Specificity in this process is achieved through complex formation of the large subunits with the μ-adaptin.
Adaptor protein complex 1 (AP-1) was shown to play a role in vesicle trafficking between Golgi and endosomes. AP-1 was suggested to target proteins from the TGN to vacuole, particularly for proteins exposing a dileucine motif (7). Proteins could be efficiently re-routed from plasma membrane to vacuolar destination by expression of a dileucine motif. Adaptor protein complex 2 (AP-2) is the best characterized member of the adaptor complex family and is involved in endocytosis by binding to clathrin-coated vesicles (8). Recent work demonstrated a role of AP-2 in clathrin-mediated endocytosis (CME), which is the major process by which receptors and other integral membrane proteins and lipids are removed from the plasma membrane and delivered into the endosomal system (8–11). So far, the function of the adaptor protein complex 3 (AP-3) in plants has not been fully elucidated. AP-3 was suggested to be involved in transport of vesicles from the TGN to the vacuole without the involvement of endosomes or prevacuolar compartments (12). Mutants in the β- and δ-subunit of the AP-3 complex seemed to accumulate plasma membrane proteins in vacuolar compartments, and vacuole biogenesis was disturbed (13). However, storage proteins were correctly targeted to vacuoles in the ap-3β mutant. A complete set of adaptor protein complex 4 (AP-4) subunits has, besides in mammals, only been found in Arabidopsis thaliana (5) and recently, a role of the AP-4 complex in vacuolar protein sorting has been suggested (14).
For most of the adaptor protein complexes, individual cargo proteins are known, but systematic large scale analysis of protein cargo was not carried out. Therefore, the aim of this work was to use density gradient centrifugation to separate organellar membranes and their associated proteins and to systematically search for altered protein distributions in plants with mutations in β-subunits of the adaptor complexes AP-3 and AP-4 (12, 14). Density-based separation of organelle membranes is a well-established technique to identify protein complexes and protein distribution in cells (15–19) and has recently been extended to study the subcellular distribution of metabolite-protein complexes to organelles (20). This simultaneous separation and enrichment of endomembranes followed by the identification of their protein contents gives a comprehensive distribution profile of proteins and in addition, is sensitive to detect differences in protein distributions. Quantitation of the protein profiles can be achieved by label-free approaches (18) or pairwise isotope labeling (15). In previous work we already detected dynamic changes in the protein distribution across such density gradients to characterize targeting of membrane proteins at different time points during pollen germination using label-free quantification (21). Here, we use metabolic 15N-labeling to compare protein distributions in ap-3β and ap-4β mutants with wild type. Our results suggest a number of proteins, particularly aquaporins as cargo of adaptor protein complexes AP-3 or AP-4 in the cellular protein trafficking pathway.
EXPERIMENTAL PROCEDURES
Plant Material
Seeds of knock-out mutants in ap-3β (At3g55480) were obtained as SAIL_1258_G03 and knock-out mutant ap-4β (At5g11490) was used as SAIL_781_H01.
Metabolic Labeling of Seedling Cultures
Surface sterilized seeds of Arabidopsis thaliana Col-0, ap-3β, and ap-4β mutants were transferred into flasks with modified JPL medium (JPL-3: 1% w/v sucrose, 3 mm NH4NO3, 7 mm KNO3). The 15N-labeled media was prepared using 98.1atm% 15NH415NO3 and K15NO3 (Sigma Aldrich, Munich, Germany) replacing the natural nitrogen source (22). Metabolically 15N-labeled and unlabeled Arabidopsis cultures were grown under 8 h light/16 h dark at 22 °C with constant shaking at 80 rpm. After 3 weeks the seedling cultures were harvested, wrapped into aluminum foil and immediately frozen in liquid N2. Plant material was stored at −80 °C. To exclude any side-effects or artifacts of the labeling treatment and to minimize internal biological variations between the cultures all experiments were performed with reciprocal metabolic labeling of wild type and mutants (Fig. 1) (23).
Fig. 1.
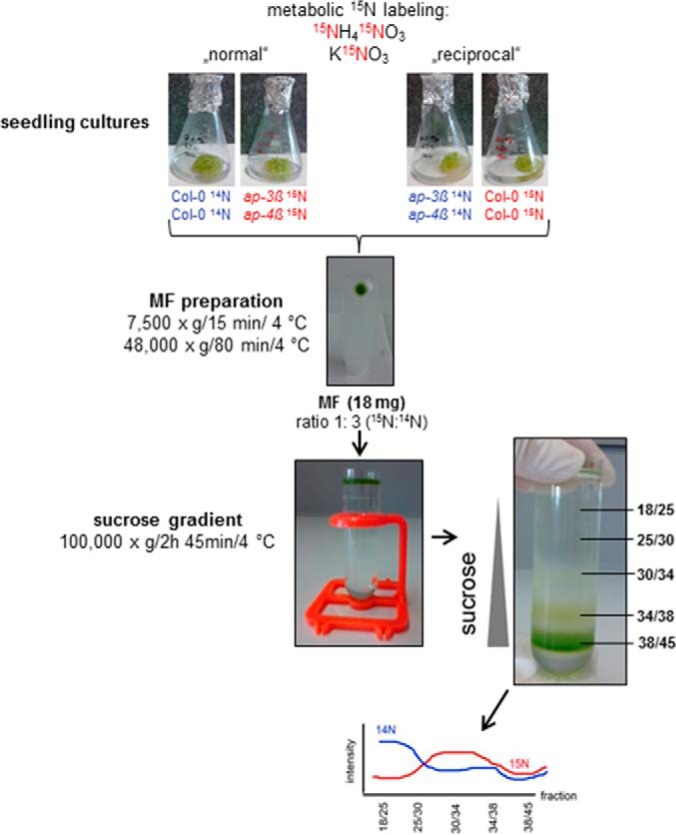
Experimental design of reciprocal metabolic labeling and workflow of protein preparation and analysis. See text for details.
Preparation of Organelle Membrane Vesicles and Sucrose Density Gradient Centrifugation
Microsomal fractions (MF) of seedling cultures were prepared by differential centrifugation (24). Frozen Arabidopsis seedlings (∼20 g of fresh weight) from Col-0, ap-3β and ap-4β were smashed into small pieces and resuspended in ice-cold homogenisation buffer (330 mm sucrose, 100 mm KCl, 1 mm EDTA, 50 mm Tris (Tris(hydroxymethyl)aminomethane) adjusted with MES (2-(N-Morpholino)ethanesulfonic acid) to pH 7.5, 5 mm DTT), protease inhibitor mixture (Sigma-Aldrich), phosphatase inhibitor mixture 2 (Sigma-Aldrich) and phosphatase inhibitor mixture 3 (Sigma-Aldrich) were added from stock solutions (50 μl per 10 ml of the homogenization buffer just before use). Tissue was homogenized with a Teflon Potter-Elvehjem-type homogenizer on ice. The homogenate was filtered through a 21 μm nylon mesh, and centrifuged at 7500 × g for 15 min at 4 °C. Finally, the supernatant was centrifuged at 48,000 × g for 80 min at 4 °C. The resulting pellet was the microsomal fraction (MF) and stored at −80 °C.
MFs from labeled and unlabeled seedlings were mixed in a ratio of 1:3 (15N:14N) as “normal” pair with wild type as unlabeled partner (e.g. ap-3β 15N + Col-0 14N) or as “reciprocal” pair with the mutant as the unlabeled partner (e.g. Col-0 15N + ap-3β 14N). A total of 18 mg MF protein was loaded onto a discontinuous sucrose gradient (21). In brief, the sucrose step gradient was centrifuged at 100,000 × g for 2.75 h at 4 °C (Beckman UZ Optima XPN-80, swing-out rotor SW32Ti) and the resulting interphases (18/25, 25/30, 30/34, 34/38, and 38/45) representing organelle-enriched fractions were collected and their density was determined with a refractometer (Hanna Instruments, Vöhringen, Germany; HI96801). Finally, the membrane fractions were pelleted by centrifugation at 125,000 × g for 1.75 h at 4 °C. Pellets were resuspended in centrifugation buffer (1 mm MgSO4, 1 mm Tris, pH 7.2 adjusted with MES) and protein concentrations were determined using a Lowry-DC (BioRad, Munich, Germany) assay with BSA as protein standard. Samples were stored at −80 °C.
Sample Preparation for Mass Spectrometry
For in-solution digest the proteins (5 μg) of each organelle-enriched fraction were denatured using 6 m urea, 2 m thiourea, pH 8.0. After reduction in 0.5 m DTT and alkylation of cysteine residues by 2.5 mm iodoacetamide, proteins were digested for 3 h by LysC (Wako, Neuss, Germany) at room temperature. The solution was then diluted fourfold with 10 mm Tris-HCl, pH 8.0 followed by overnight digestion with trypsin (sequencing grade, Promega, Fitchburg, WI) at 37 °C under continuous shaking at 350 rpm. Finally, digested peptides were desalted over C18 STAGE-tips and vacuum-dried. For mass spectrometric analysis samples were resuspended in resuspension buffer (0.2% v/v TFA, 5% v/v acetonitrile). Sucrose gradient interphases were also used for phosphopeptide enrichments as described (25).
Tryptic peptide mixtures were analyzed by LC-MS/MS using nanoflow HPLC (Easy nLC 1000, Thermo Scientific, Waltham, MA) and a hybrid quadrupole-orbitrap mass spectrometer (Q Exactive Plus, Thermo Scientific) as a mass analyzer. Peptides were eluted from a 75 μm x 50 cm C18 analytical column (PepMap® RSLC C18, Thermo Scientific) on a gradient using 0.5% acetic acid as aqueous phase and 0.5% acetic acid in 80% acetonitrile as organic phase. The flow rate was set to 250 nL per minute. Peptides were eluted on a linear gradient running from 4 to 64% acetonitrile in 240 min. Spectra were using information-dependent acquisition of fragmentation spectra of multiple charged peptides within the m/z range of 300–1600. Up to twelve data-dependent MS/MS spectra were acquired for each full-scan spectrum acquired at 70,000 full-width half-maximum resolution. Fragment spectra were acquired at a resolution of 35,000.
Protein identification and ion intensity quantitation was carried out by MaxQuant version 1.4.1.2 (26). Spectra were matched against the Arabidopsis proteome (TAIR10, 35386 entries) using Andromeda (27). In the search process, carbamidomethylation of cysteine was set as a fixed modification; oxidation of methionine and N-terminal protein acetylation was set as variable modifications. Phosphorylation of serine, threonine and tyrosine was additionally used as a variable modification in data analysis of the phosphopeptide enrichment samples. Spectra were matched using trypsin as enzyme specificity allowing up to two missed cleavages. Mass tolerance for the database search was set to 10 ppm on full scans and 0.5 Da for fragment ions. Multiplicity was set to 1. Retention time matching between runs was chosen within a time window of 2 mins, and “match unidentified features” option was selected. Peptide false discovery rate (FDR) and protein FDR were set to 0.01. The FDR was calculated based on the posterior error probability derived from hit distributions in forward and reverse database searches. Hits to known contaminants (e.g. keratins) and reverse hits identified by MaxQuant were excluded from further analysis. Spectra of identified phosphopeptides were submitted to the PhosPhAt database (28), and spectra are presented in supplemental Fig. S5. Localization of phosphorylation sites was accepted based on the PTM-scores calculated by MaxQuant. Sites were accepted with a PTM-score probability greater than 0.75. In all cases, for further data mining of subcellular locations and functions, we used the “leading razor protein” as a single protein inferred from identified peptides. Identified protein groups are listed in supplemental Table S5 including the number of peptides and sequence coverage. Raw files and search results have been deposited to the ProteomeXchange Consortium via the PRIDE (29) partner repository with the data set identifier PXD003905 for the AP-3 and PXD003894 for the AP-4 experiments.
Quantitative Analysis of 15N Labeled Peptides
Peak ion intensities were used for quantitation. An in-house script was developed in C# matching identified unlabeled peptides (14N) with their 15N counterparts from quantified, but unidentified features in “matched features.txt”. Thereby, requirements for retention time match within 1 min were made, and matching masses were required to fall within a 97.5atm% to 100atm% window of the expected 15N masses. Performance and coverage of this script is presented in the results section (Fig. 2). For further data analysis, ratios of 15N and 14N ions were calculated. For clarity of data presentation, in some cases, profiles of labeled and unlabeled peptides were not plotted as ratios, but as normalized extracted ion intensity profiles.
Fig. 2.
15N quantitation using MaxQuant based on evidence. A, Boxplot of calculated log2 (15N/14N) for each peptide in three replica raw files of protein with different mixing ratios. B, Number of quantified peptides in three replica raw files of different mixing ratios.
Experimental Design and Statistical Rationale
The experiments followed a typical pairwise metabolic labeling setup (Fig. 1) including a label-swap replication. Thus, each pair of wild type and mutant was analyzed in two biological replicates and because wild type was used together with both mutants, there were four biological replicates of wild type data and two replicates for each mutant. Ion intensity ratios of label-swap replicates were expressed as mutant versus wild type ratios and averaged. The ratios obtained from different gradient fractions followed a normal distribution and were statistically analyzed using t-tests and one-way ANOVA.
Swelling Assay of Mesophyll Cells
Arabidopsis seedlings of wild type and mutants were grown on modified JPL medium (30) with 1% w/v agar for 10 days at 22 °C under 8 h light/16 h dark. Separated leaves of 30 seedlings were collected in 3 ml JPL with 300 mm mannitol and then transferred to 300 m solution (300 mm mannitol, 10 mm MES/KOH pH5.8, 10 mm KCl, 10 mm CaCl2) with 1% w/v cellulase (Onozuka R10) and 1% w/v macerozyme, both from Duchefa (RV Haarlem, The Netherlands). The leaves were further cut into small pieces and incubated at room temperature for 1.5 to 2 h under gentle shaking. Protoplasts were separated from tissue debris by filtration through a 50 μm nylon net, centrifuged at 80 × g, for 10 min at 4 °C and washed three times by centrifugation with 500 μl ice cold 300 m solution. Aliquots of protoplasts were transferred into 200 μl 300 m solution in a perfusion chamber mounted on a video microscope (Zeiss Primovert with DinoLite camera) and were let settled down onto the glass bottom of the chamber. The chamber was then slowly perfused with 300 m solution, a protoplasts sticking to the glass slide was selected, the time lapsed video was started and the chamber was perfused with 150 m solution (same as 300 m solution but with only 150 mm mannitol). Images were taken every 3 s for a duration of 5 min. The diameter of protoplasts was manually measured using imageJ software and the protoplast volumes as well as the volume increase rates (ΔVΔt−1) were calculated using Sigma Plot software v.11.
Cloning of DNA Constructs
For generation of fusion proteins between PIP2A and GFP, the full cDNA of PIP2A was amplified by PCR from Arabidopsis thaliana cDNA using the primers AtPIP2A-GW-f(5′-CACCATGGCAAAGGATGTGGAAG-3′) and AtPIP2A-r 5′-GACGTTGGCAGCACTTCTGAATGA-3′. The amplified fragment was cloned into pENTR/d-TOPO (Invitrogen, Karlsruhe, Germany), verified by sequencing, and recombined into destination vector pMDC43 (31) for GFP fusion to the N terminus.
Protoplast Isolation, Polyethylene Glycol-mediated Transformation of Protoplasts
For transient transformation, Arabidopsis mesophyll protoplasts were generated (32) and transformed (33) as described. Transformed Arabidopsis protoplasts were incubated in the dark at 22 °C prior to confocal analysis.
Confocal Microscopy
Images of protoplasts were taken 2 days after transformation on a confocal laser scanning microscope (Leica TCS SP5; Leica Microsystems) using 488 nm laser light for excitation, and processed with LAS AF Version 2.7.29586. The detection window for GFP ranged from 495 nm to 553 nm. Maximum projections were generated from z-stacks with at least 30 steps and a step size of ∼0.5 μm–1 μm. On these images, each showing several transformed protoplasts, every cell with visible GFP fluorescence was scored as either localization of PIP2A to the plasma membrane (evenly distributed fluorescence) or “other” (fluorescence mostly concentrated in punctae, patches, or network-like structures within the cell). To exclude bias, all pictures were numbered (randomly varying the order of the genotypes) prior to scoring by a third person (single-blind).
RESULTS
The chosen experimental strategy aimed for an unbiased and systematic identification of proteins with altered distribution profiles between wild type and the adaptor protein complex mutants ap-3β and ap-4β.
Experimental design and 15N ratio quantitation
To compare differences in protein distribution across different membrane compartments in ap-3β and ap-4β mutants with wild type plants we used a reciprocal metabolic labeling experimental design (34). We used pairwise experimental setups of wild type compared with ap-3β and wild type compared with ap-4β, each repeated twice in label swap experimental design (Fig. 1). To minimize putative artificial effects of differential centrifugation and protein extraction efficiencies, wild type material was mixed with labeled mutant material at the level of purified microsomal protein. The mixing ratio of labeled to unlabeled material was 1:3 leading to enhanced protein identification rates with more unlabeled protein being present in the combined complex sample (34).
Although 14N forms of the peptides can readily be quantified using MaxQuant, the software is unable to directly assign pairs of 15N-labeled peptides. However, using the “matched features” file containing unassigned peaks with their retention times and ion intensities, the respective 15N-peptide can be assigned with the identified corresponding 14N-peptide by matching ion features based on retention time alignment and the expected mass of the 15N-labeled peptide. This procedure was validated using existing raw files of different protein mixtures ranging from 1:5 to 5:1 of 15N protein mixed with 14N protein that were previously used in an extensive evaluation of quantitation in 15N metabolic labeling (34). For the matching of unidentified features to identified peptide sequences, a retention time alignment window of 1 min was used and a mass tolerance window of expected 15N-labeled peptide masses ranging from 97.5atm% to 100atm% 15N (supplemental Fig. S1). Using these parameters, in the existing test files the average calculated 15N ratios represented the expected mixing ratios (Fig. 2A). In mixtures with high content of unlabeled protein (1:5 mixture) more peptides could be quantified than in the mixtures containing high proportion of 15N peptides (Fig. 2B). Our quantitation results are in line with previously published features of these raw files (34) indicating that the assignment of unidentified ion features to identified peptide sequences was reasonable with the parameters chosen.
Separation of Microsomal Membranes on a Sucrose Step Gradient
Generally, and irrespective of the labeling scheme, more spectra were sampled for wild type than for adaptor protein complex mutants. Lowest numbers of proteins were identified for the ap-4β mutant. The reproducibility of membrane separation and the interphase collection was very high with only small variations in sucrose density and total protein content of the interphases (Table I). Protein abundances in each interphase of the step gradient (21) were quantified and compared between wild type and respective mutants. In total, for each interphase over thousands of proteins were identified ranging from 1500 in interphase 18/25 up to over 4000 proteins identified in interphases 30/34, 34/38 and 38/45 (Table II, supplemental Table S1).
Table I. Densities of the collected interphases and their protein concentrations of 8 sucrose density gradients (n = 8).
| Interphase | % Sucrose (w/w)±S.D. | Protein conc. μg μl−1 |
|---|---|---|
| 18/25 | 20.2 ± 1.4 | 0.28 ± 0.17 |
| 25/30 | 27.9 ± 0.4 | 0.24 ± 0.15 |
| 30/34 | 31.9 ± 0.5 | 0.31 ± 0.25 |
| 34/38 | 36.6 ± 0.2 | 1.08 ± 0.65 |
| 38/45 | 40.4 ± 0.5 | 9.48 ± 3.13 |
Table II. Overview of identified proteins. From each interphase 5 μg protein was digested and analyzed by mass spectrometry.
| No. of identified proteins (mean) ±S.D. |
|||
|---|---|---|---|
| col-0 (n = 4) | ap-3ß (n = 2) | ap-4ß (n = 2) | |
| 18/25 | 1658 ± 309 | 1384 ± 172 | 1874 ± 23 |
| 25/30 | 2497 ± 1088 | 2250 ± 158 | 2793 ± 14 |
| 30/34 | 3965 ± 491 | 2783 ± 72 | 4042 ± 116 |
| 34/38 | 4358 ± 446 | 2657 ± 180 | 4444 ± 214 |
| 38/45 | 4492 ± 373 | 2963 ± 334 | 4517 ± 165 |
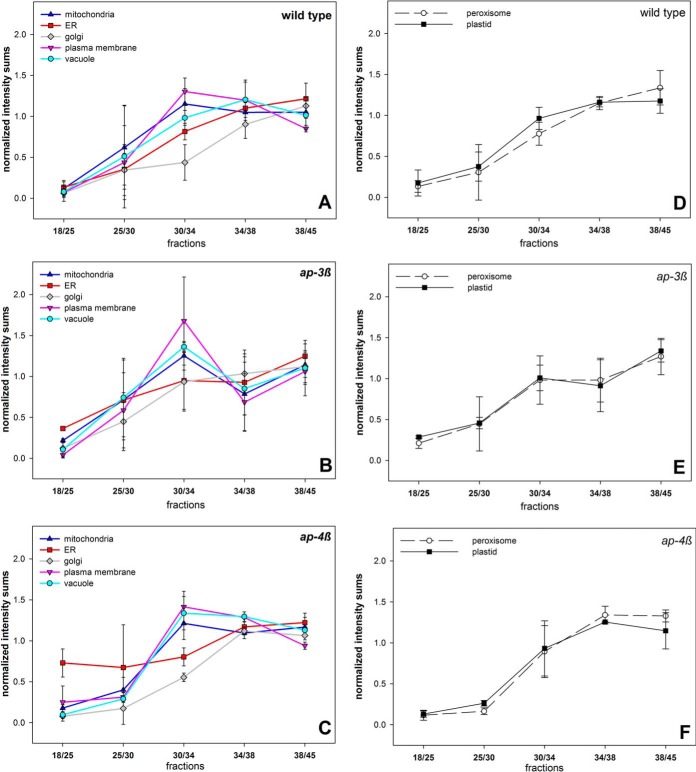
To assign the interphases of the step gradient to organelle membrane-enriched fractions, the distribution of typical organelle marker proteins across the gradient was analyzed. Therefore, well characterized membrane proteins with known location to different subcellular compartments, such as mitochondria, ER, Golgi apparatus, plasma membrane and vacuole were chosen. For example, known residents of the plasma membrane were members of the PM H+ ATPase family (AHA1 AT2G18960, AHA3 AT5G57350, AHA4 AT3G47950, AHA11 AT5G62670) and aquaporins (PIP1B AT2G45960, PIP1C AT1G01620, PIP3 AT4G35100). Markers for the endoplasmic reticulum were ER-localized Ca2+-ATPases (ECA1 AT1G07810, ECA2 AT4G00900, ECA4 AT1G07670), calreticulins (CRT1 AT1G56340, CRT2 AT1G09210, CRT3 AT1G08450) and ER-localized members of the HSP70 family (BIP1 AT5G28540, BIP2 AT5G42020) (35, 36). The full lists of marker proteins used for the corresponding subcellular compartment are listed in supplemental Table S2. Thereby, for wild type plants, typical distributions of plasma membrane proteins peaked at interphases 30/34 and 34/38 whereas vacuolar proteins peaked in interphase 34/38. Proteins from ER and Golgi showed highest abundance in interphase 38/45, but with different abundance profiles in lighter interphases (Fig. 3A). These general distribution profiles of proteins were altered in the ap-3β mutant plants (Fig. 3B). Particularly proteins with known plasma membrane and vacuolar location showed similar distribution profiles in the ap-3β mutant with highest abundances in interphase 30/34. In ap-3β mutants, proteins with expected ER and Golgi location showed highest abundance in interphase 38/45, similar to wild type, but with enhanced abundance of Golgi proteins also in interphase 30/34. In the ap-4β mutant, protein distribution again differed from wild type profiles. Also in this mutant, plasma membrane proteins and vacuolar proteins showed very similar distribution patterns, with the distribution peak of vacuolar proteins being shifted from interphase 34/38 to 30/34 (Fig. 3C). ER proteins of ap-4β showed unusual high abundance in interphases 18/25 and 25/30. The distribution of mitochondrial, plastidial and peroxisomal proteins was less or almost not affected in the mutants compared with wild type (Fig. 3D, 3E, 3F). Thus, correct targeting of proteins with vacuolar or plasma membrane location as well as location in the secretory pathway from ER to Golgi is disturbed in the ap-3β and ap-4β mutants.
Fig. 3.
Distribution across the gradient of proteins localized to different subcellular compartments. Distribution of proteins localized to mitochondria, ER, Golgi apparatus, plasma membrane and vacuole for (A) wild type, (B) ap-3β mutant, and (C) ap-4β mutant. Distribution of proteins localized to peroxisomes and plastid for (D) wild type, (E) ap-3β mutant, and (F) ap-4β mutant. Averages for all proteins of a given subcellular location with standard deviation are shown.
The identification of proteins with altered distribution profiles was further confirmed by an unsupervised k-means clustering of protein abundance profiles. Six clusters were generated based on a figure of merit analysis (supplemental Fig. S2A). Especially many of the proteins isolated from the ap-3β mutant were found in different clusters compared with the respective wild type protein (supplemental Fig. S2B), and also proteins from ap-4β mutant were found in different clusters compared with wild type proteins. In general, pairwise analysis of profile differences and the clustering analysis gave highly overlapping results (supplemental Fig. S2C).
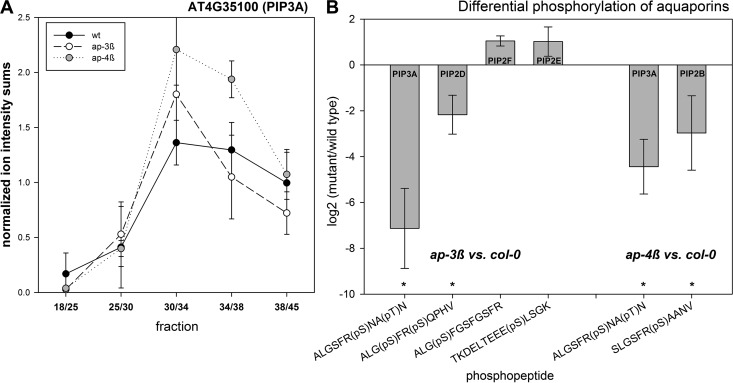
In a pairwise comparison, proteins with altered distribution in ap-3β were over-represented (Fisher-exact test) for proteins with ATPase function, e.g. vacuolar and plasma membrane H+-ATPases, p < 3.9E−10), aquaporins (p < 7.5E−9), but also for storage proteins (p < 1.2E−5), and proteins in secondary metabolism (p < 6.8E−5). For instance, although in wild type the aquaporin PIP3A peptides showed highest abundances in interphases 30/34 and 34/38, this distribution in both mutants was shifted to peak in interphase 30/34 (Fig. 4A). Receptor kinases, cell wall proteins and proteins involved in vesicle trafficking (e.g. SYPs) were completely depleted in ap-3β compared with wild type (Table III, supplemental Table S3). In the ap-4β mutant, proteins with ATPase function (p < 7.8E−8), aquaporins (p < 3.8E−4), proteins of lipid metabolism (p < 1.3E−4), vesicle trafficking proteins (p < 3.8E−4) and peroxidases (p < 3.9E−4) were found with altered abundance profiles compared with wild type. Receptor kinases, cell wall proteins and proteins of the secretory pathway were also depleted in ap-4β compared with wild type (Table III).
Fig. 4.
Aquaporins as putative targets of the AP-complex. A, Distribution and localization of aquaporin PIP3A (AT4G35100) in wild type (black), ap-3β (white) and ap-4β (gray). Values describe the mean with standard deviation of four independent gradient preparations for col-0 and two gradients for ap-3β and ap-4β mutants. B, Alterations in phosphorylation of C- and N-terminal residues of different aquaporin proteins. Values express the average log2 ratios of ap-3β or ap-4β mutant versus wild type. Error bars indicate standard deviation of two to four independent gradient preparations.
Table III. Over-representation analysis (Fisher exact test) of protein functions with altered or depleted protein distribution profile in ap-3ß or ap-4ß compared to wild type.
| Mutant | BIN | Binname | p value | changed vs wild type |
|---|---|---|---|---|
| 34.1 | Transport.p- and v-ATPases | 3.597E-10 | shifted distribution | |
| 34.19.1 | Transport.Major Intrinsic Proteins.PIP | 7.56E-09 | shifted distribution | |
| 34.1.1 | Transport.p- and v-ATPases.H+-transporting two-sector ATPase | 3.575E-06 | shifted distribution | |
| ap-3ß | 33.1 | Development.storage proteins | 1.196E-05 | shifted distribution |
| 16.5.1.3.1 | Secondary metabolism.sulfur-containing.glucosinolates.degradation.myrosinase | 6.858E-05 | shifted distribution | |
| 30.2 | Signalling.receptor kinases | 1.567E-08 | depleted | |
| 10 | Cell wall | 2.473E-05 | depleted | |
| 31.4 | Cell.vesicle transport | 0.0001364 | depleted | |
| 34.1 | Transport.p- and v-ATPases | 7.866E-08 | shifted distribution | |
| 11.9.3 | Lipid metabolism.lipid degradation.lysophospholipases | 0.0001395 | shifted distribution | |
| 34.19.1 | Transport.Major Intrinsic Proteins.PIP | 0.000375 | shifted distribution | |
| ap-4ß | 31.4 | Cell.vesicle transport | 0.0003806 | shifted distribution |
| 26.12 | Misc.peroxidases | 0.0003969 | shifted distribution | |
| 26.1 | Misc.cytochrome P450 | 0.0019244 | depleted | |
| 30.2.14 | Signalling.receptor kinases.leucine rich repeat XIV | 0.0030147 | depleted | |
| 10.4 | Cell wall.pectin synthesis | 0.0070206 | depleted | |
| 29.3.4.1 | protein.targeting.secretory pathway.ER | 0.0149856 | depleted |
Altered Phosphorylation Profiles
Besides alterations in protein abundance profiles, differences in phosphorylation of putative cargo proteins were analyzed (Table IV). Mainly proteins with plasma membrane or vacuolar location not only showed altered abundance profiles in mutants compared with wild type, but also were differentially phosphorylated at specific sites (supplemental Table S4). In the ap-3β mutant, four aquaporins (PIP3A AT4G35100, PIP2D AT3G54820, PIP2E AT2G39010, PIP2F AT5G60660) were found with differential phosphorylation compared with wild type. In the ap-4β mutant, two aquaporins (PIP3A AT4G35100, PIP2B AT2G37170) were identified with differential phosphorylation compared with wild type (Fig. 4B). These differences were particularly apparent in the C-terminus or N-terminus of the aquaporins where two serine residues are located which were frequently identified as phosphorylated under various conditions (28). In both mutants, phosphorylation at the second of two consecutive serine residues in the C-terminus of the aquaporins PIP3A, PIP2D, and PIP2B was strongly reduced (marked with asterisks in Fig. 4B). An increased phosphorylation in the ap-3β mutant compared with wild type was observed for the first of the two consecutive serines in PIP2F and for the N-terminal serine in PIP2E. Furthermore, in the ap-3β mutant reduced phosphorylation of the exocyst complex component SEC10 (AT5G12370) was found, as well as for syntaxins SYP121 (AT3G11820) and SYP132 (AT5G08080). Increase in phosphorylation was observed for NPSN13 (AT3G17440) and GOS12 (AT2G45200), both members of the plant SNARE (soluble N-ethylmaleimide-sensitive factor attachment receptor) protein family. In the ap-4β mutant, decreased phosphorylation was found for syntaxin SYP122 (AT3G52400) (supplemental Table S4). Interestingly, for the ap-3β mutant calcium ATPases (ACA10, ACA11, ACA9, ACA8, ACA4), calcium channels CAX1 (AT2G38170), TPC1 (AT4G03560) as well as calnexin1 (CNX1, AT5G61790) appeared with altered distribution profiles in at least one replicate experiment, and partly also showed differential phosphorylation (ACA10, CNX1). Calcium is known to play an important role during endocytosis and vesicle fusion in general (37).
Table IV. List of proteins with altered distribution profile across gradients and with differential phosphorylation levels in mutants compared to wild type.
| AGI | Description | Mapman | Bincode | Subcellular location |
|---|---|---|---|---|
| ap-3ß mutant | ||||
| at3g11820 | SYP121 | Cell.vesicle transport | 31.4 | PM |
| at3g17440 | NPSN13 | Cell.vesicle transport | 31.4 | PM |
| at5g08080 | SYP132 | Cell.vesicle transport | 31.4 | PM |
| at1g32400 | TOM2A | Development.unspecified | 33.99 | V |
| at3g60600 | VAP27–1 | DNA.unspecified | 28.99 | PM,ER |
| at3g19820 | DWF1 | hormone Metabolism.brassinosteroid.synthesis-degradation.sterols | 17.3.1.2.8 | PM,G |
| at1g45688 | unknown protein | Not assigned.unknown | 35.2 | PM |
| at1g64650 | Major facilitator superfamily | Not assigned.unknown | 35.2 | V |
| at1g70770 | unknown protein | Not assigned.unknown | 35.2 | PM,ER |
| at5g39570 | unknown protein | Not assigned.unknown | 35.2 | PM |
| at2g38280 | FAC1 | Nucleotide metabolism.degradation | 23.2 | PM |
| at2g01470 | STL2P | Protein.degradation | 29.5 | ER |
| at3g61260 | remorin family protein | RNA.regulation of Transcription.putative transcription regulator | 27.3.67 | PM |
| at4g29900 | ACA10 | Signalling.calcium | 30.3 | PM |
| at5g61790 | CNX1 | Signalling.calcium | 30.3 | ER |
| at5g49760 | leucine-rich repeat protein kinase, putative | Signalling.receptor kinases.leucine rich repeat VIII.VIII-1 | 30.2.8.1 | PM |
| at1g51800 | leucine-rich repeat protein kinase, putative | Signalling.receptor kinases.misc | 30.2.99 | PM |
| at1g59870 | PEN3, PDR8 | Transport.ABC transporters and multidrug resistance systems | 34.16 | PM |
| at2g39010 | PIP2E, PIP2;6 | Transport.Major Intrinsic Proteins.PIP | 34.19.1 | PM |
| at4g35100 | PIP3A, PIP2;7 | Transport.Major Intrinsic Proteins.PIP | 34.19.1 | PM |
| at5g60660 | PIP2F, PIP2;4 | Transport.Major Intrinsic Proteins.PIP | 34.19.1 | PM |
| at4g39080 | VHA-A3 | Transport.p- and v-ATPases | 34.1 | V |
| at5g17010 | sugar transporter family protein | Transport.sugars | 34.2 | V |
| at1g71880 | SUC1 | Transport.sugars.sucrose | 34.2.1 | PM |
| ap-4ß mutant | ||||
| at1g52780 | unknown protein | Not assigned.unknown | 35.2 | PM |
| at1g59610 | ADL3, CF1 | Misc.dynamin | 26.17 | PM |
| at2g37170 | PIP2B, PIP2;2 | Transport.Major Intrinsic Proteins.PIP | 34.19.1 | PM |
| at3g52400 | SYP122 | Cell.vesicle transport | 31.4 | PM |
| at4g08850 | kinase | Signalling.receptor kinases.leucine rich repeat XII | 30.2.12 | PM |
| at4g13510 | AMT1;1 | Transport.ammonium | 34.5 | PM |
| at4g23630 | BTI1 | Stress.biotic | 20.1 | PM |
| at4g35100 | PIP3A, PIP2;7 | Transport.Major Intrinsic Proteins.PIP | 34.19.1 | PM |
| at5g39570 | unknown protein | Not assigned.unknown | 35.2 | PM |
| at5g64410 | OPT4 | Transport.peptides and oligopeptides | 34.13 | PM |
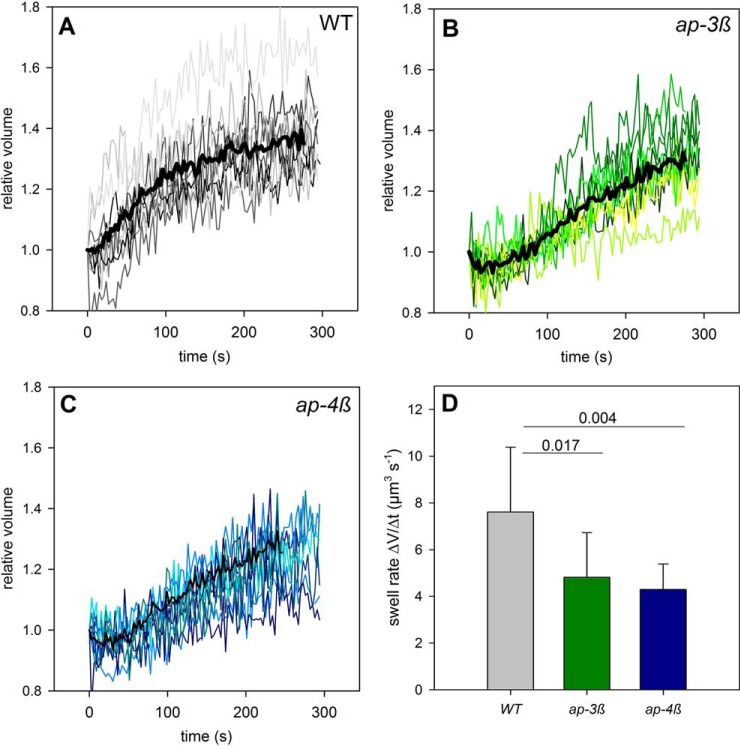
Altered Transport Activities in AP-complex Mutants
Because aquaporins were identified in both adaptor protein complex mutants as putative targets showing mis-localization during intracellular trafficking with possibly reduced abundance at the plasma membrane, we studied the water transport capacity by measuring volume changes of mesophyll protoplasts exposed to an osmotic shock (Fig. 5). Both mutants showed different swelling behavior compared with wild type protoplasts. Volume changes over time followed a sigmoidal curve in wild type (Fig. 5A). In contrast, volume changes were almost linear in the ap-3β mutant (Fig. 5B) and in the ap-4β mutant (Fig. 5C) resulting in much slower swelling rates for the mutants compared with wild type protoplasts (Fig. 5D). These findings suggest a lower amount of active water transporters, i.e. aquaporins, in the plasma membrane. Indeed, expression of N-teminal GFP-PIP2A constructs under 35S-promoter in mesophyll protoplasts in the ap-4β mutant resulted in a significantly (p < 0.01, t test) higher frequency of PIP2A being aggregated in internal compartments and a lower frequency of correct targeting to the plasma membrane (supplemental Fig. S3). This could be caused by disturbed vesicle and protein trafficking, thus failing to supply new molecules to the plasma membrane or by down-regulation of aquaporin activity by changing their phosphorylation state.
Fig. 5.
Water transport measured by swelling rates of mesophyll cell protoplasts. Relative volumes of individual protoplasts (gray) and the resulting average swell rate of (A) wild type col-0, (B) ap-3β mutant, and (C) ap-4β mutant. D, Averaged volume increase rates with standard deviations of 10 protoplasts each.
The severe decrease of water transport capacity in the ap-4β mutant may result in the reduction in root growth observed especially in the ap-4β mutant (Fig. 6). However, in the ap-4β mutant the reduced root growth could also be connected to mis-targeting of uptake transporters for nitrogen compounds such as AMT1.1 (AT4G13510, Table IV); or altered phosphorylation of the N-terminal Ser11 in NRT2.1 (AT1G08090; supplemental Table S4). Interestingly, AP-2 assisted clathrin-mediated endocytosis of cellulose synthases seems to be important for root growth resulting in short root phenotypes also in AP-2 mutants (9). Thus, reduced root growth in the ap-4β mutant may also be connected to depletion of cell wall proteins (Table III) and/or altered phosphorylation patterns of pectin esterase proteins (supplemental Table S4).
Fig. 6.
Root length of wild type and AP-complex mutants. A, Images of seedlings of ap-3β, col-0 and ap-4β on JPL-3 medium + 1% w/v agar. After surface sterilization seeds were grown for 7 days at 22 °C under 16h light/8h dark. B, Root length measurement of WT and mutant seedlings treated as described in (A) Error bars represent S.D. (n = 30).
DISCUSSION
To identify putative targets of adaptor protein complexes AP-3 and AP-4, we characterized the protein distribution across density separated organelle membranes in ap-3β as well as in ap-4β mutants of Arabidopsis seedlings in comparison with wild type. Distribution of membrane proteins across density gradients is a powerful method that has found wide application in the definition of organelle membrane proteomes (35, 38, 39). Proteins of the endomembrane system are in constant movement. For example, plasma membrane (PM) proteins move through the endoplasmic reticulum (ER) and the Golgi apparatus before they reach their final destination and are re-cycled to endomembrane vesicles via endocytosis. Therefore, only a comprehensive view of many membrane compartments reflects the dynamic sub-cellular distribution of membrane proteins and thus, purification of single organelle-enriched fractions by other techniques such as the two-phase partitioning system gives only limited insights. The major advantage in using a sucrose step gradient is that all organelle fractions can be separated and enriched in a single preparative step. Here, we extend the application of such gradient preparations beyond the mere description of the organelle compositions to a comparative analysis with targeting mutants. Thereby, (i) gradient preparation turned out as very reproducible (Table I, supplemental Table S1) and (ii) the use of a metabolic labeling approach proved very powerful in detection of slight changes in abundance distribution profiles.
For efficient data analysis we followed a novel approach: in a first step, 14N peptide forms were identified using MaxQuant (40, 26), and 15N peaks were then assigned from unmatched features data within a narrow time and mass windows (supplemental Fig. S1) making advantage of retention time alignment features within MaxQuant (40). However, in contrast to classic 15N data analysis involving fragment spectra searches with 15N-parameters, high quantitation coverage by our approach depended on higher abundances of 14N over 15N peptides in the respective mixtures (Fig. 2) (34). Therefore, 1:3 mixtures of 15N:14N materials were used, yielding most reliable and reproducible results.
We identified two categories of putative cargo proteins: The first category included proteins with altered distribution profiles across the step gradient (Fig. 3), and the second category included proteins which were specifically absent in the mutants compared with wild type (Table III). The latter category consider the possibility that cargo proteins could be specifically degraded when components of the AP-complexes are missing. Among these cargo candidates, we especially found receptor kinases and cell wall proteins. The subunits AP-3β (AT3G55480), and AP-4β (AT5G11490) were identified in wild type only and not in their respective knock-out mutant. In addition, other subunits of the AP-4 complex, such as AP-4μ (AT4G24550) and AP-4σ (AT52G19790) were also found to be absent (not detected) in the ap-4β mutant. Because the mRNA at least of AP-4μ could be detected in the ap-4β mutant (data not shown), indeed degradation of the whole AP-complex and possibly also of its cargo seems to be occurring in the beta subunit knock out mutants.
The putative cargo proteins of AP-3 and AP-4 of the first category contained several aquaporins and vesicle trafficking proteins of the SYP (syntaxin of plants) family. In the ap-3β mutant and ap-4β mutant, the same set of aquaporins and syntaxins were identified with altered distribution profiles. Except for PIP3A (AT4G35100) differential phosphorylation was also detected for distinct aquaporins and syntaxins. This suggests that, although in both mutants the same classes of proteins were affected, the AP-3 and AP-4 complexes could be delivering individual cargo proteins through distinct preferences for phosphorylated versions of aquaporins or syntaxins.
For aquaporins phosphorylation at the second of the two consecutive serines within the C-terminus is required for targeting of the protein to the plasma membrane (41) and especially for PIP2A (AT3G53420), phosphorylation at Ser283 decreased under osmotic stress (41). Phosphorylation at this site seems to favor an export from the ER. In both adaptor protein complex mutants, phosphorylation of this particular serine residue was strongly reduced (Fig. 4). Interestingly, syntaxins, such as SYP121, were shown to coordinate the trafficking of PIP3A to the plasma membrane (42). Furthermore, SYP121 was shown to not only be important for delivery of channel molecules to the membrane, but also for gating control of channels through specific interaction of the transporter with the SNARE protein (43). A recent study of interaction partners of different syntaxins revealed several aquaporins (PIP1, PIP2F, PIP3A), as well as plasma membrane H+-ATPase AHA2 as direct interaction partners to SYP132, but not to SYP121 and SYP122 (44). Members of adaptor protein complexes were not found as direct interactors of the SNARE proteins. SYPs are also regulated by phosphorylation as indicated to frequent identification of phosphorylation sites in the N-terminus under various stress conditions (28, 45). Recent work also showed an interaction of AP-4 with the vacuolar sorting receptor VSR1 (AT3G52850), which showed a distinct abundance profile in the ap-4β mutant (14).
Our findings suggest an involvement of AP-complexes in the process of targeting of PIPs together with other proteins to their destination, the plasma membrane, where they form active aquaporins. This involvement of the AP-complexes in the targeting of aquaporins could function through specific protein-protein interactions of the AP-complex and its cargo depending on modification status of the cargo proteins and other vesicle trafficking components. In this context, it is noteworthy, that particularly receptor kinases, for some of which the abundance was significantly depleted in the AP-complex mutants, are increasingly found to be involved in phosphorylation of transporters and channels within the membrane (46–48).
Targets for adaptor protein complex 3 were already suggested from cell biological experiments. Thereby, mis-localization of proteins in the ap-3β mutant was observed for PIN1, PIN2, PIN7, and BRI1, and C-terminal GFP-fusion of aquaporin PIP2A as well as vacuolar storage proteins when expressed in root tissues under a native promoter. This indicates that AP-3 is involved in correct delivery of plasma membrane located proteins (12, 13). Although we could not identify PIN1 and PIN2 in our data, we found the abundance profiles for PIN7, PIN3, and BRI1, to be different in the ap-3β as well as in the ap-4β mutants compared with wild type (supplemental Fig. S4). Also for vacuolar sorting receptor VSR1, a differential distribution profile was observed in the ap-4β mutant (supplemental Fig. S4). Most of these known candidates in our proteomic analysis were not detected in every fraction during the proteome analysis thus resulting in missing data values especially in the abundance profiles for the mutants. Nevertheless, the detection of shifted abundance distributions for the known ap-3β and ap-4β cargo does provide a validation of our approach to identify candidate cargo for AP-3 and AP-4 complexes. Also, the missing values in the mutant abundance distribution profiles could indicate higher protein degradation of the cargo protein in the mutants as discussed above. This further validates our criteria to include also proteins absent (not detected) in the mutants as putative cargo.
The lists of new candidate proteins identified here as cargo for AP-3 and AP-4 (Table III and Table IV; supplemental Table S3 and S4) show a large overlap. Out of 406 membrane located candidate cargo proteins for AP-3 and 273 candidate cargo proteins for AP-4, a total of 200 candidate cargo proteins were identified in both mutants. Among these were two remorin family proteins (49), syntaxins, several aquaporins, v- and p-ATPases, as well as receptor kinases. Given this large overlap of putative cargo proteins, the distinct role of the different AP-complexes does not become immediately apparent. However, although both AP-complexes may act on the same cargo molecule, their affinities or efficiencies of action may differ between tissues and specific cells leading to differences in the degree to which trafficking and targeting is disturbed in the ap-3β or ap-4β mutant. We could for example measure clear differences in water transport rates across the plasma membrane in ap-3β and ap-4β mutant (Fig. 5) indicating that although targeting and regulation of aquaporins is affected in both mutants, these proteins are affected with different severity which in turn, may cause the distinct difference in root growth. Thus, we conclude that adaptor protein complexes 3 and 4 provide a protein targeting system with some degree of redundancy allowing for dynamic, plastic reaction and fine-tuning of the regulatory network with which plants respond to different environmental challenges.
CONCLUSION
In conclusion our work provides insights into the cellular role of two adapter protein complexes, namely AP-3 and AP-4. We suggest that the action of cargo recognition by adaptor protein complexes requires a combination of protein-protein interactions and modification status of the cargo protein. We could further show that for one class of cargo proteins, the aquaporins, the disturbed abundance profiles and disturbed phosphorylation patterns in the adaptor protein complex mutants resulted in disturbed transport properties and even growth phenotypes. However, further research is required to unravel the precise mechanism by which the adapter protein complex recognizes and distinguishes the cargo proteins.
Acknowlegdements
We thank Sabine Schneider (Friedrich Alexander University Erlangen) for continuous and fruitful discussion. Ana Gimeno (Univ. Salzburg) is acknowledged for excellent technical assistance with the protoplast preparation and swelling assays.
Supplementary Material
Footnotes
Author contributions: H.P. and W.X.S. designed research; H.P., X.N.W., C.M., and G.O. performed research; X.N.W. and J.S. contributed new reagents or analytic tools; H.P., X.N.W., J.S., C.M., G.O., and W.X.S. analyzed data; H.P., G.O., and W.X.S. wrote the paper.
* This work was funded by the German Research Foundation (DFG) within Research Unit FOR1061.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- AP
- adapter protein
- AP-1
- adapter protein complex 1
- CME
- clathrin mediated endocytosis
- PM
- plasma membrane
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Peer W. A. (2011) The plant plasma membrane protein trafficking. Plant Cell Monographs 19, 31–57 [Google Scholar]
- 2. Zarsky V., Kulich I., Fendrych M., and Pecenkova T. (2013) Exocyst complexes multiple functions in plant cells secretory pathways. Curr. Opin. Plant Biol. 16, 726–733 [DOI] [PubMed] [Google Scholar]
- 3. Ding Y., Robinson D. G., and Liang L. (2014) Unconventional protein secretion (UPS) pathways in plants. Curr. Opin. Cell Biol. 29, 107–115 [DOI] [PubMed] [Google Scholar]
- 4. Nakatsu F., and Ohno H. (2003) Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct. Funct. 28, 419–429 [DOI] [PubMed] [Google Scholar]
- 5. Boehm M., and Bonifacino J. S. (2001) Adaptins: the final recount. Mol. Biol. Cell 12, 2907–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson T. (1998) Transport vesicles: coats of many colours. Curr. Biol. 8, R609–R612 [DOI] [PubMed] [Google Scholar]
- 7. Wang X., Cai Y., Wang H., Zeng Y., Zhuang X., Li B., and Jiang L. (2014) Trans-Golgi network-located AP1 gamma adaptins mediate dileucine motif-directed vacuolar targeting in Arabidopsis. Plant Cell 26, 4102–4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins B. M., McCoy A. J., Kent H. M., Evans P. R., and Owen D. J. (2002) Molecular architecture and functional model of the endocytic AP2 complex. Cell 109, 523–535 [DOI] [PubMed] [Google Scholar]
- 9. Bashline L., Li S., Zhu X., and Gu Y. (2015) The TWD40–2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 12870–12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gadeyne A., Sanchez-Rodriguez C., Vanneste S., Di Rubbo S., Zauber H., Vanneste K., Van Leene J., De Winne N., Eeckhout D., Persiau G., Van De Slijke E., Cannoot B., Vercruysse L., Mayers J. R., Adamowski M., Kania U., Ehrlich M., Schweighofer A., Ketelaar T., Maere S., Bednarek S. Y., Friml J., Gevaert K., Witters E., Russinova E., Persson S., De Jaeger G., and Van Damme D. (2014) The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691–704 [DOI] [PubMed] [Google Scholar]
- 11. Di Rubbo S., Irani N. G., Kim S. Y., Xu Z. Y., Gadeyne A., Dejonghe W., Vanhoutte I., Persiau G., Eeckhout D., Simon S., Song K., Kleine-Vehn J., Friml J., De Jaeger G., Van Damme D., Hwang I., and Russinova E. (2013) The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25, 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feraru E., Paciorek T., Feraru M. I., Zwiewka M., De Groodt R., De Rycke R., Kleine-Vehn J., and Friml J. (2010) The AP-3 beta adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22, 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zwiewka M., Feraru E., Moller B., Hwang I., Feraru M. I., Kleine-Vehn J., Weijers D., and Friml J. (2011) The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21, 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuji K., Shirakawa M., Shimono Y., Kunieda T., Fukao Y., Koumoto Y., Takahashi H., Hara-Nishimura I., and Shimada T. (2016) The Adaptor Complex AP-4 Regulates Vacuolar Protein Sorting at the trans-Golgi Network by Interacting with VACUOLAR SORTING RECEPTOR1. Plant Physiol. 170, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunkley T. P., Watson R., Griffin J. L., Dupree P., and Lilley K. S. (2004) Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell. Proteomics 3, 1128–1134 [DOI] [PubMed] [Google Scholar]
- 16. Borner G. H. H., Sherrier D. J., Weimar T., Michaelson L. V., Hawkins N. D., MacAskill A., Napier J. A., Beale M. H., Lilley K. S., and Dupree P. (2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lilley K. S., and Dupree P. (2007) Plant organelle proteomics. Curr. Opin. Plant Biol. 10, 594–599 [DOI] [PubMed] [Google Scholar]
- 18. Sadowski P. G., Groen A. J., Dupree P., and Lilley K. S. (2008) Sub-cellular localization of membrane proteins. Proteomics 8, 3991–4011 [DOI] [PubMed] [Google Scholar]
- 19. Groen A. J., Sancho-Andrés G., Breckels L. M., Gatto L., Aniento F., and Lilley K. S. (2014) Identification of trans-golgi network proteins in Arabidopsis thaliana root tissue. J. Proteome Res. 13, 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arrivault S., Guenther M., Florian A., Encke B., Feil R., Vosloh D., Lunn J. E., Sulpice R., Fernie A. R., Stitt M., and Schulze W. X. (2014) Dissecting the subcellular compartmentation of proteins and metabolites in Arabidopsis leaves using non-aqueous fractionation. Mol. Cell. Proteomics 13, 2246–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pertl H., Schulze W. X., and Obermeyer G. (2009) The pollen organelle membrane proteome reveals highly spatial-temporal dynamics during germination and tube growth of lily pollen. J. Proteome Res. 8, 5142–5152 [DOI] [PubMed] [Google Scholar]
- 22. Engelsberger W. R., Erban A., Kopka J., and Schulze W. X. (2006) Metabolic labeling of plant cell cultures with K15NO3 as a tool for quantitative analysis of proteins and metabolites. Plant Methods 2, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kierszniowska S., Walther D., and Schulze W. X. (2009) Ratio-dependent significance thresholds in reciprocal 15N-labeling experiments as a robust tool in detection candidate proteins responding to biological treatment. Proteomics 9, 1916–1924 [DOI] [PubMed] [Google Scholar]
- 24. Pertl H., Himly M., Gehwolf R., Kriechbaumer R., Strasser D., Michalke W., Richter K., Ferreira F., and Obermeyer G. (2001) Molecular and physiological characterisation of a 14–3-3 protein from lily pollen grains regulating the activity of the plasma membrane H+ ATPase during pollen grain germination and tube growth. Planta 213, 132–141 [DOI] [PubMed] [Google Scholar]
- 25. Wu X. N., and Schulze W. X. (2015) Phosphopeptide profiling of receptor kinase mutants. Methods Mol. Biol. 1306, 71–79 [DOI] [PubMed] [Google Scholar]
- 26. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 28. Durek P., Schmidt R., Heazlewood J. L., Jones A., MacLean D., Nagel A., Kersten B., and Schulze W. X. (2010) PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38, D828–D834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., and Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jouanneau J. P., and Peaud-Lenoel C. (1967) Growth and synthesis of proteins in cell suspensions of a kinetin dependent tobacco. Physiol. Plant. 20, 834–850 [Google Scholar]
- 31. Curtis M. D., and Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drechsel G., Bergler J., Wippel K., Sauer N., Vogelmann K., and Hoth S. (2011) C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J. Exp. Bot. 62, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abel S., and Theologis A. (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J. 5, 421–427 [DOI] [PubMed] [Google Scholar]
- 34. Arsova B., Zauber H., and Schulze W. X. (2012) Precision, proteome coverage and dynamic range of Arabidopsis proteome profiling using 15N metabolic labelling and label-free approaches. Mol. Cell. Proteomics 11, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunkley T. P., Hester S., Shadforth I. P., Runions J., Weimar T., Hanton S. L., Griffin J. L., Bessant C., Brandizzi F., Hawes C., Watson R. B., Dupree P., and Lilley K. S. (2006) Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nikolovski N., Rubtsov D., Segura M. P., Miles G. P., Stevens T. J., Dunkley T. P., Munro S., Lilley K. S., and Dupree P. (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 160, 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steer M. W. (2006) The role of calcium in exocytosis and endocytosis in plant cells. Physiol. Plant. 72, 213–220 [Google Scholar]
- 38. Gatto L., Vizcaino J. A., Hermjakob H., Huber W., and Lilley K. S. (2010) Organelle proteomics experimental designs and analysis. Proteomics 10, 3957–3969 [DOI] [PubMed] [Google Scholar]
- 39. Pertl H., Gehwolf R., and Obermeyer G. (2005) The distribution of membrane-bound 14–3-3 proteins in organelle-enriched fractions of germinating lily pollen. Plant Biol. 7, 140–147 [DOI] [PubMed] [Google Scholar]
- 40. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prak S., Hem S., Boudet J., Viennois G., Sommerer N., Rossignol M., Maurel C., and Santoni V. (2008) Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol. Cell. Proteomics 7, 1019–1030 [DOI] [PubMed] [Google Scholar]
- 42. Hachez C., Laloux T., Reinhardt H., Cavez D., Degand H., Grefen C., De Rycke R., Inze D., Blatt M. R., Russinova E., and Chaumont F. (2014) Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26, 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grefen C., Chen Z., Honsbein A., Donald N., Hills A., and Blatt M. R. (2010) A novel motif essential for SNARE interaction with the K(+) channel KC1 and channel gating in Arabidopsis. Plant Cell 22, 3076–30792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fujiwara M., Uemura T., Ebine K., Nishimori Y., Ueda T., Nakano A., Sato M. H., and Fukao Y. (2014) Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 55, 781–789 [DOI] [PubMed] [Google Scholar]
- 45. Van Wijk K. J., Friso G., Walther D., and Schulze W. X. (2014) Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 26, 2367–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fuglsang A. T., Kristensen A., Cuin T., Schulze W. X., Persson J., Thuesen K. H., Ytting C. K., Oehlschlaeger C., Mahmood K., Sondergaard T. E., Shabala S., and Palmgren M. G. (2014) Receptor kinase mediated control of primary active proton pumping at the plasma membrane. Plant J. 80, 951–964 [DOI] [PubMed] [Google Scholar]
- 47. Haruta M., Gray W. M., and Sussman M. R. (2015) Regulation of the plasma membrane proton pump (H-ATPase) by phosphorylation. Curr. Opin. Plant Biol. 28, 68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu X., Sanchez-Rodriguez C., Pertl-Obermeyer H., Obermeyer G., and Schulze W. X. (2013) Sucrose-induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Mol. Cell. Proteomics 12, 2856–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raffaele S., Mongrand S., Gamas P., Niebel A., and Ott T. (2007) Genome-wide annotation of remorins, a plant-specific protein family: Evolutionary and functional perspectives. Plant Physiol. 145, 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.