Abstract
Deterministic and stochastic processes jointly determine the community dynamics of forest succession. However, it has been widely held in previous studies that deterministic processes dominate forest succession. Furthermore, inference of mechanisms for community assembly may be misleading if based on a single axis of diversity alone. In this study, we evaluated the relative roles of deterministic and stochastic processes along a disturbance gradient by integrating species, functional, and phylogenetic beta diversity in a subtropical forest chronosequence in Southeastern China. We found a general pattern of increasing species turnover, but little-to-no change in phylogenetic and functional turnover over succession at two spatial scales. Meanwhile, the phylogenetic and functional beta diversity were not significantly different from random expectation. This result suggested a dominance of stochastic assembly, contrary to the general expectation that deterministic processes dominate forest succession. On the other hand, we found significant interactions of environment and disturbance and limited evidence for significant deviations of phylogenetic or functional turnover from random expectations for different size classes. This result provided weak evidence of deterministic processes over succession. Stochastic assembly of forest succession suggests that post-disturbance restoration may be largely unpredictable and difficult to control in subtropical forests.
A major challenge in studies of community assembly is to understand the relative importance of deterministic versus stochastic processes determining species composition1,2,3. Niche-based equilibrium theory assumes that local community dynamics are shaped by deterministic processes dictated by the relationship between species-specific traits and local abiotic and biotic factors1,3. In contrast, a stochastic view suggests that community dynamics are determined solely by demographic stochasticity and dispersal limitation2,4. Hitherto studies on the relative importance of these two sets of processes in forest ecology have mainly focused on mature species-rich forests rather than secondary forests. However, the directly observable rapid community dynamics and striking changes of the forest structure in disturbed, successional communities provide a useful laboratory for studying community assembly5,6.
Species beta diversity or species turnover measures the rate of community compositional changes between two or more locations7,8. Exploring patterns of species beta diversity can provide considerable insights into the relative roles of deterministic and stochastic processes during secondary forest succession. For example, based on Clement’s9 concept of succession, the deterministic view postulates that forest in all locations converge on a single optimal assemblage over succession. The convergence may be resulted from decreasing variation in environmental conditions among locations such as relatively homogeneous shade after canopy closure4,10. Thus, the deterministic view assumes a trend of decreasing species beta diversity over succession11,12. In contrast, the stochastic view proposes that dispersal limitation and demographical stochasticity across locations would result in increasingly divergent species composition, and suggests an increase in species beta diversity with succession2. Christensen and Peet11 tested these predictions by examining species beta diversity and have found that in forests of five age-classes, the mature hardwood stands had the highest beta diversity while young pine stands had the lowest beta diversity, thus failing to support Clement’s deterministic view9.
However, despite these predictions, deterministic and stochastic processes may potentially produce similar patterns of species beta diversity over succession4,13. For example, increasing species beta diversity created by stochastic processes may also arise from deterministic processes such as increasing variation in environmental conditions across locations. Severe disturbances such as forest clearing may homogenize environmental conditions like light and soil properties. The post-disturbance recovery of variation in environmental conditions such as understory light and soil nutrient levels, may lead to increasing species beta diversity when species with different traits colonize environmentally heterogeneous locations. The same species beta diversity pattern could be created simultaneously by both stochastic and deterministic processes. Thus, it is difficult to discern the relative roles of deterministic and stochastic processes, by solely using a species-based beta diversity approach.
Considering functional beta diversity may provide complementary information to species beta diversity, it may help to overcome this limitation. Environmental conditions determine the available niches and the functional composition that can fill them, thus community assembly is expected to be deterministic in functional composition. Moreover, disturbance regimes, i.e., different levels of disturbance intensity and frequency create various levels of variation in environmental conditions among locations and initial community composition14. Therefore, functional beta diversity should decrease (or increase) when variation in initial environmental conditions decrease (or increase) over time or along a disturbance gradient. In contrast, functional beta diversity should not change significantly if late-arriving species have similar traits with early-arriving species (priority effect) or community dynamics and species performance are not governed by species traits. Analyses integrating species, functional and phylogenetic beta diversity approaches may provide ecologists better understanding of multiple facets of community assembly over succession. For example, Fukami et al.15 experimentally demonstrated in a 9-year study that a decline in functional beta diversity and an constant species beta diversity over succession may arise from simultaneous operations of deterministic processes and priority effect.
Functional beta diversity is based on a limited set of measurable traits, whereas phylogenetic relatedness approximates multivariate trait similarity of species16. Therefore, phylogenetic beta diversity may potentially provide additional insight by synthesizing a greater amount of information than functional beta diversity. However, inferences of community assembly from community phylogenetic structures strongly depend on the niche conservatism assumption that closely related species have similar niches. If traits are not phylogenetically conserved, it is difficult to understand the community assembly processes from community phylogenetic structures.
In this study, we aimed to evaluate the relative roles of deterministic and stochastic processes in forest succession by using multiple axes of beta diversity. We used a dataset from nine 1-ha stem-mapped plots along a disturbance gradient in a subtropical forest chronosequence in southeastern China. As found in recent studies, strong environmental filtering governs forest succession in early stage, while interspecific competitions dictate community assembly in late stage17,18,19. We thus hypothesized that deterministic processes dominate the subtropical forest succession. On the other hand, the relative importance of local processes strongly depends on the chosen scales20,21. For example, neighborhood interaction operates at smaller spatial scales than environmental filtering22. Thus we tested our hypothesis at spatial scales of 10 × 10 m and 20 × 20 m.
Results
Phylogenetic signal in traits
The phylogenetic signal of six examined traits were quantified using Blomberg’s K23. Six traits had a weak phylogenetic signal with K values of less than one (Table B1), ranging from 0.105 to 0.441. However, all traits, except leaf area, had K values significantly higher than expected by chance, indicating a weak but significant phylogenetic signal. The significant phylogenetic signal in traits suggested that patterns of phylogenetic turnover may mirror the patterns of functional turnover.
The effect of disturbance regimes on spatial turnover of diversity for all stems
In this study, linear mixed models were used to determine the effects of disturbance regime on three axes of turnover of diversity. Species, phylogenetic and functional turnover were quantified by Chao-Jaccard dissimilarity, τst and Bst, respectively. Subsequently, the turnover of three axes of diversity were plotted against disturbance regime (Figs 1 and 2) after controlling for the effect of habitat difference and other potentially important factors. The residuals of all linear mixed models had no significant spatial autocorrelation (illustrated in Fig. C1), suggesting that the spatial structure in the data had been accounted for in the models.
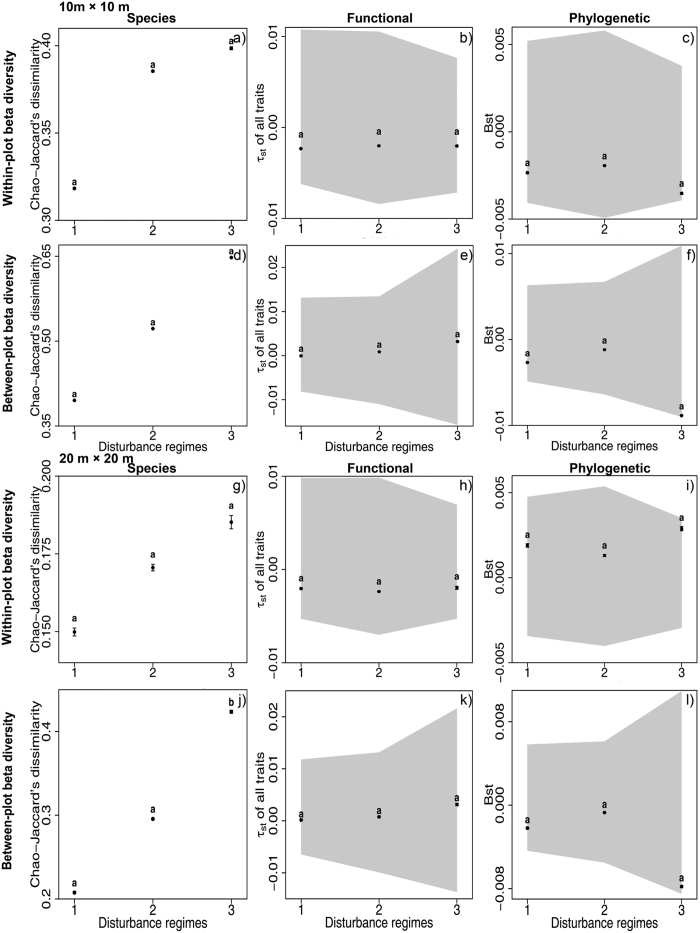
Figure 1. Species, functional and phylogenetic beta diversity for all subplot pairs (black square, mean ± SE) within different disturbance regimes at within- and between-plot level and scales of 10 × 10 m and 20 × 20 m.
(a,g) are species beta diversity at within-plot level, (b,h) for functional beta diversity at within-plot level, (c,i) for phylogenetic beta diversity at within-plot level, while (d,j) are species beta diversity at between-plot level, (e,k) for functional beta diversity at between-plot level, (f,l) for phylogenetic beta diversity at between-plot level. Letters indicate significant differences (P < 0.05) between disturbance regimes which was calculated by multiple comparison. The grey-shaded area represents the 95%-confidence interval for Bst and τst values from the 999 random communities. Bst and τst values inside the interval indicate phylogenetic or functional randomness, and Bst and τst values outside the interval indicate the significant phylogenetic or functional clustering or dispersion. Disturbance regimes: 1. young secondary forest: clear-cutting 50 years ago and selective cutting 20 years ago, 2. old secondary forest: clear-cutting 50 years ago, 3. old growth forest: without human disturbance more than 100 years.
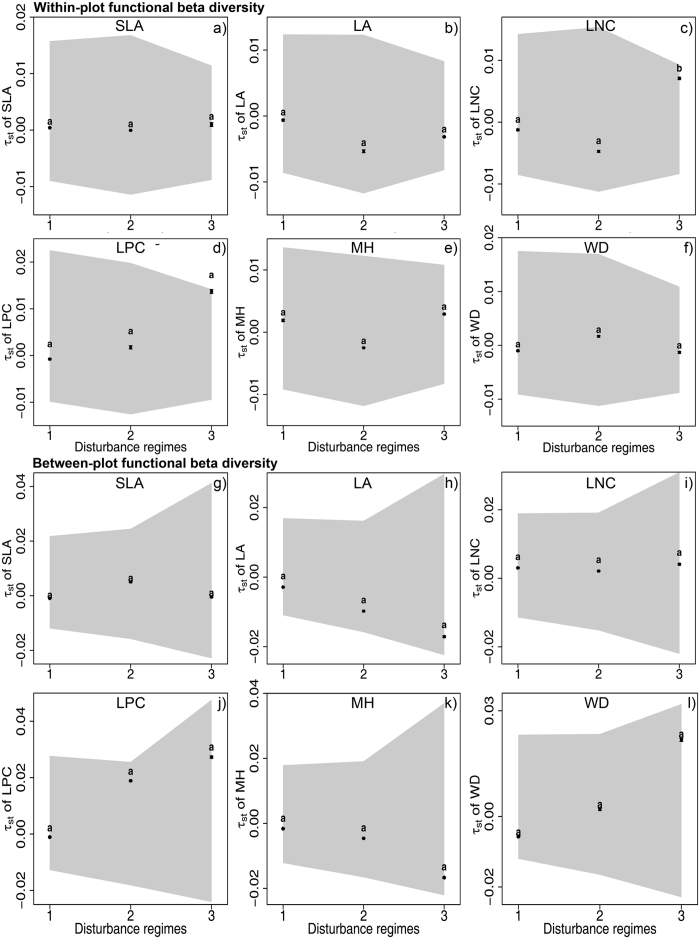
Figure 2. Functional beta diversity of individual traits for all subplot pairs (black square, mean ± SE) within different disturbance regimes at within-plot and between-plot levels and at a scale of 20 × 20 m.
Letters indicate significant differences (P < 0.05) between disturbance regimes which was calculated by multiple comparison. The grey-shaded area represents the 95%-confidence interval for the τst values from the 999 random communities. τst values inside the interval indicate phylogenetic or functional randomness, and τst values outside the interval indicate the significant phylogenetic or functional clustering or dispersion. SLA = specific leaf area, LA = leaf area, WD = wood density, LPC = leaf phosphorus content, LNC = leaf nitrogen content, MH = maximum height. Disturbance regimes: 1. young secondary forest: clear-cutting 50 years ago and selective cutting 20 years ago, 2. old secondary forest: clear-cutting 50 years ago, 3. old growth forest: without human disturbance more than 100 years.
At both scales of 10 × 10 m and 20 × 20 m, we detected increasing trends in species turnover with succession at within-plot or between-plot levels (Fig. 1a,d,g,j), one of which was significant (Fig. 1j). In contrast, functional and phylogenetic turnover (τst and Bst) of all traits showed no significant trends over succession at both within-plot and between-plot level (Fig. 1b,c,e,f,h,i,k,l). Moreover, τst and Bst values of all traits fall inside the 95%-confidence interval of random expectations (Fig. 1b,c,e,f,h,i,k,l), indicating that observed τst and Bst values of all traits were not significantly different from random expectations. Despite of insignificant effect of disturbance regime on τst and Bst, we found significant interaction terms between disturbance regimes and distance of habitat conditions in most phylogenetic and functional turnover models (Tables F1–F2). Functional turnover (τst) of individual traits showed consistent patterns with that of all traits at two spatial scales (Figs 2, D1).
The effect of disturbance regimes on spatial turnover of diversity for different size classes
The analyses of three axes of beta diversity for stems of different size classes revealed more details: at both scales of 10 × 10 m and 20 × 20 m, species turnover also tended to show increasing trends (Figs E1a–c, j–l, E2a–c, j–l) for three size classes over time at both within- and between-plot levels, some of which were significant (Figs E1a–c, l).
At a scale of 20 × 20 m, we detected no significant trends of Bst and τst along disturbance regimes (Fig. E1d–g,m,o,r) for most dbh size classes. Meanwhile, we found that Bst and τst values of all traits fall inside the 95%-confidence interval of random expectations (Fig. E1d–g,m,o,r) for most dbh size classes, indicating that observed Bst and τst values were not significant from random expectations. Exceptions included Bst for cohorts of 5–10 cm dbh at within-plot level and 1–5 cm and 5–10 cm dbh classes at between-plot level and τst of all traits for cohorts of 5–10 cm dbh class at between-plot level, which displayed significant increasing trends (Fig. E1h,n,p,q), and Bst for cohort of greater than 10 cm dbh at within-plot level which displayed significant decreasing trend (Fig. E1i). On the other hand, we detected significant interaction terms between disturbance regimes and distance of habitat conditions in most phylogenetic and functional dissimilarity models (not shown) for three size classes at the scale of 20 × 20 m. We found similar patterns of Bst and τst of all traits for cohorts of all size classes at a scale of 10 × 10 m (Fig. E2).
Discussion
The evidence of stochastic processes
At the spatial scale of 10 × 10 m and 20 × 20 m, species turnover showed overall increasing trends over succession (Fig. 1a,d,g,j) for all trees and three size classes, some of which were significant. Functional and phylogenetic turnover displayed little-to-no changes over succession (Fig. 1b,c,e,f,h,i,k,l and Figs 2, D1) for both within- and between-plot levels and for all trees and three size classes. An increasing species turnover but little-to-no variation in functional turnover could be resulted from stochastic processes due to the independence between colonization and species traits. Alternatively, this pattern may also be created by random arrival order and timing of species with similar traits (priority effects) under a constant environmental filtering24,25. Furthermore, functional and phylogenetic turnover did not significantly deviated from random expectations (Fig. 1b,c,e,f,h,i,k,l and Figs 2, D1), supporting the dominance of stochastic processes but rejecting the possibility of a priority effect under constant environmental filtering.
Our findings contradicted the general expectation from recent studies that deterministic processes dominate the forest succession17,18,19. There are some potential reasons for only little-to-no change in functional and phylogenetic turnover along the disturbance gradient. First, it is possible to miss the full gradient in this study as we did not include early successional forests before canopy closure and very old forests where succession may last for several centuries into our analyses6,26. Next, resprouting from both cut trees and shrubs may have major impacts on species composition in subtropical forests27,28,29. Some species such as Schima superba, Castanopsis fargesii, Camellia fraterna, and Eurya rubiginosa, could even colonize the forest from early to late successional stages. Their resprouting could impact on late-arriving species as demonstrated by Fukami et al.15. Finally, it is possible that random immigration may play an important role in community assembly over succession6. Therefore, it is likely that both initial community composition and random immigration may play important roles in community assembly. The mechanisms underlying random immigration may be resulted from weak trait-environment relationship in GNNR30,31. Letten et al.32 also found that phylogenetic and functional dissimilarity did not increase during the temporal succession in a fire-prone heathland in southeast Australia. They interpreted the stochastic functional and phylogenetic turnover as being due to changes in relative fitness of different traits through time, another possible mechanism underlying random immigration in GNNR.
The weak evidence of deterministic processes
There were limited patterns showing significant changes of functional and phylogenetic turnover along disturbance gradient at two spatial scales and three size classes (Fig. E1e,h,i,n,p,q,E2i,p,r). The significant increase in phylogenetic turnover over time for smaller size classes (Fig. E1h,n,p,q,E2p) indicated higher level of phylogenetic relatedness within subplots at late-successional stage than at early stage. This suggested an increase of the variation in habitat conditions during post-disturbance recovery4. In contrast, the significant decline in phylogenetic turnover for larger dbh size classes (Fig. E1i,E2i,r) indicated lower level of phylogenetic relatedness within subplots at late-successional stage than early stage. This suggested the importance of interspecific interactions for larger size classes. However, the inconsistence between patterns of phylogenetic and functional turnover together with weak phylogenetic signal in traits suggested only weak evidences for above-described deterministic processes. These results were similar to the findings of Fukami et al.15 that increasing species turnover with decreasing functional turnover was likely produced by an interaction between priority effect and decreasing variation in environmental conditions. Purschke et al.19 found simultaneous declines in species, phylogenetic, and functional turnover with a decreasing variation in environmental conditions. Our result for the role of deterministic processes, was also consistent with those from analyses of phylogenetic and functional alpha diversity of the same chronosequence33.
The significant effect of the interaction term of disturbance regimes and distance of habitat conditions in most functional and phylogenetic dissimilarity models (e.g. Tables F1–F2) also provided evidence of deterministic processes. This result together with insignificant functional and phylogenetic turnover suggests an alternative explanation to the dominance of stochastic assembly, that the deterministic processes in habitats of different disturbance regimes may follow different trajectories and exhibit an overall stochastic assembly. This result also partly supported the importance of trait filtering during succession in GNNR34.
Despite a weak phylogenetic signal, phylogenetic turnover showed coherent patterns with functional turnover, suggesting trait conservatism in our study system. However, the hypothesis using community phylogenetic structure to infer the assembly processes is currently under debate35. As Mayfield and Levine35 noted, the hypothesis could be reinterpreted by Chesson’s framework how niche and competitive ability difference could influence species coexistence36, and evidence has recently begun to emerge from controlled experiments37,38,39. In this study, when functional and phylogenetic distances are correlated with competitive ability differences that generally exceed niche difference in a species pool, our inference of ecological processes from patterns of beta diversity should be robust. However, it could be difficult to infer ecological processes in this study when niche differences exceed competitive ability difference or phylogenetic and functional distances are correlated with niche differences. Our inference of ecological processes in this subtropical forest may be valid because the competitive ability differences likely exceed niche differences for a large proportion of species pool in such a heterogeneous landscape.
Conclusions
Disentangling the relative roles of deterministic and stochastic processes in shaping community dynamics over succession remains a major challenge in community ecology. Previous studies often hold that deterministic processes such as environmental filtering and competition dominate forest succession, leading to a transition from functional and phylogenetic clustering at earlier successional stages to functional and phylogenetic dispersion at later successional stages17,18. Contrary to this expectation, three lines of evidence from species, phylogenetic, and functional turnover revealed that community dynamics appear to be generally dominated by stochastic processes in this subtropical forest. Stochastic processes such as priority effect and random immigration, may play important roles in community assembly over succession. On the other hand, our finding of the weak evidence for deterministic processes may suggest an alternative explanation that deterministic processes in habitats of different disturbance regimes may follow different trajectories and exhibit an overall stochastic assembly. The dominance of stochastic processes over succession have important implications for subtropical forest management that the outcome of restoration would be largely unpredictable and difficult to control.
Methods
Study location and secondary forest plots
This study was conducted in the Gutianshan National Nature Reserve (GNNR) in the western part of Zhejiang Province, southeastern China (Fig. A1). The climate of GNNR is seasonal, and the vegetation represents a typical subtropical evergreen broad-leaved forest40. Detailed descriptions of climate, topography and flora can be found in Bruelheide et al.6 and Feng et al.33.
Forest plots were randomly selected and stratified by disturbance regimes in the GNNR. Three disturbance regimes of forests were estimated based on knowledge of local logging events, forest physiognomy (Fig. A3) and a dendrochronological study on the forest age41: 1) twice-cut forest: naturally restored forests after clear-cutting 50 years ago and selective cutting for forest tending 20 years ago (plots 1–3); 2) once-cut forest: naturally restored forests after clear-cutting 50 years ago (plots 4–6), and 3) old growth forest: forests without human disturbance for more than 100 years (plots 7–9). Three 1-ha plots (100 m in north-south direction by 100 m in west-east direction) were randomly selected within each disturbance regime, with a total of nine 1-ha forest plots established in the reserve in 200833 (see details on diversity and density in Appendix A).
All trees with diameter at breast height (dbh) ≥ 1 cm were tagged, identified and spatially mapped. Topographical variables were measured during the plot establishment in 2008. A total of 58,401 tree individuals belonging to 51 families and 183 species were recorded in these plots. All nomenclature in the datasets follows the Flora of China42. For the present study, the trees were grouped into subplots of 10 × 10 and 20 × 20 m2 in size, which allows for division of each plot into subplots of equal sizes.
Elevations for all of the intersections of the 20 m grid (324 intersections) were measured, and some of the intersections of the 10 m, 5 m or smaller grids (112 intersections) were also measured in some areas of rugged topography. Maximum elevation of the 9 plots ranged from 372.6 m to 772.9 m above sea level. Four topographic variables were calculated from elevation measurements: mean elevation, terrain convexity, slope and aspect as proxy variables to comprehensively portray the overall quality of habitat40,43.
Phylogenetic tree construction
A molecular community phylogeny was reconstructed from DNA barcode for the tree community representing 177 species in the plots, and other 6 species were excluded from the analyses in this study. Leaf tissue was collected from three individuals for each species at GNNR and desiccated with silica gel. Total DNA was extracted from samples of leaf tissue using a standard CTAB protocol44. Three commonly used plant chloroplast DNA barcode loci (rbcL, matK and trnH-psbA) were amplified and sequenced using methods from Pei et al.45. The matK and rbcL loci were then globally aligned and the trnH-psbA sequences were aligned within orders using MUSCLE46, and three loci were assembled into a DNA supermatrix. Three-partition GTR + GAMMA models were applied to the three regions separately, and a maximum likelihood algorithm was subsequently used to reconstruct community phylogenies for the plots using RAxML47. A rapid bootstrapping analysis with 1000 replicates was conducted to assess the percentage support for each node. Finally, an ultrametric tree was obtained using the non-parametric rate smoothing approach in the r8s software package48.
Functional trait collection
We measured a total of six traits to quantitatively represent several important axes of plant functional strategy. Specific leaf area (SLA), leaf phosphorus content (LPC) and leaf nitrogen content (LNC) were measured to represent leaf economics spectrum49. Wood density (WD) was measured to represent wood economics spectrum50. Maximum height (MH) was used to represent adult light niche and competitive ability with neighbours49, and leaf area (LA) was measured to represent a balance between area deployed for light capture and leaf temperatures49. The trait collection protocols followed Cornelissen et al.51 with the exception of WD which followed the protocols of Wright et al.50.
Leaf area, SLA, LPC and LNC were measured using at least ten mature leaves collected from the tallest portion of the canopies of 5–10 of the largest individuals of each species randomly selected from the plots following the standard protocol51. The WD was calculated by collecting wood samples from 5 to 10 individuals for each species in the area surrounding the plots using methods described in Wright et al.50. The MH values were estimated using values reported in the Flora of China42. For each trait, a dendrogram was constructed using UPGMA clustering based on a Euclidean distance matrix representing interspecific trait similarity with species-level mean trait values of each species. In order to obtain a composite measure of functional similarity between species, a principal component analysis (PCA) was performed on a log-transformed and standardized trait data. Subsequently, a dendrogram of all traits was constructed by using UPGMA clustering based on a Euclidean distance of PCA axes52.
Phylogenetic signal in functional traits
To examine the extent to which the phylogenetic relatedness between species reflects ecological similarity, we used the K statistic of Blomberg et al.23 to quantify the phylogenetic signal of available functional traits in this study. The K statistic provides a comparison between observed and expected levels of a phylogenetic signal under an assumption of Brownian motion trait evolution for a given phylogenetic tree23. Values of K < 1 (or K > 1) indicate less (or more) phylogenetic signal in trait data than expected under Brownian motion trait evolution. We assessed the significance of the phylogenetic signal by randomly shuffling tip species 999 times and calculating 95% confidence intervals.
Quantifying the spatial turnover in species, phylogenetic and functional diversity
We quantified species beta diversity by calculating the Chao-Jaccard dissimilarity index53. Chao et al.53 showed that Jaccard’s dissimilarity index is systematically biased towards underestimation of similarity in hyperdiverse communities that are undersampled. The Chao-Jaccard dissimilarity index is an abundance-based dissimilarity index that incorporates the estimates of the numbers of unseen species and minimizes sampling biases53.
We used an index of abundance-weighted spatial partitioning of pairwise phylogenetic distance within- and between communities to measure phylogenetic beta diversity (Bst) and functional beta diversity (τst) between two communities. Bst and τst was calculated as54,55:
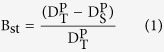 |
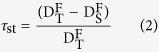 |
where 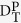 and
and 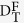 were the mean abundance-weighted phylogenetic and functional distance between community k and l respectively.
were the mean abundance-weighted phylogenetic and functional distance between community k and l respectively. 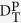 and
and 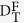 were measured as:
were measured as:
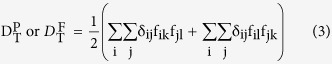 |
where 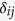 was phylogenetic or functional distance between species i and j, fik was the relative abundance of species i in community k and fjl was the relative abundance of species j in communities l.
was phylogenetic or functional distance between species i and j, fik was the relative abundance of species i in community k and fjl was the relative abundance of species j in communities l. 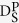 and
and 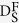 were the mean abundance-weighted phylogenetic and functional distance within community k and l respectively. Similarly,
were the mean abundance-weighted phylogenetic and functional distance within community k and l respectively. Similarly, 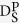 and
and 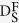 are measured as:
are measured as:
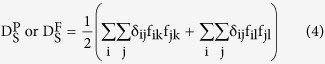 |
Within-plot and between-plot dissimilarity analysis
In order to determine the effect of disturbance regimes on dissimilarity within each plot, we calculated pairwise species, phylogenetic and functional dissimilarity between all subplot (10 × 10 or 20 × 20 m2) pairs within each plot using Chao-Jaccard’s dissimilarity, Bst and τst. We then used a linear mixed model to determine the effect of disturbance regimes on within-plot dissimilarity.
Studying the effect of disturbance regimes on forest succession has been criticized due to the unrealistic assumption that plots in each disturbance regime differ only in disturbance regime and have homogeneous habitat conditions13,56. We therefore removed the effect of within-plot (i.e. subplot-to-subplot) habitat difference by including the logarithm-transformed Euclidean distance of habitat variables and the logarithm-transformed spatial distance between subplots in the model. The distance of habitat conditions between each subplot pair was calculated as Euclidean distance of topographic variables after mean elevation, slope, and convexity were standardized to a zero mean and unit variance, and aspect was sine- and cosine-transformed as easting and northing. We included the mean elevation of each plot to control for the effect of elevational difference on diversity turnover at the plot scale due to elevational differences among plots (as shown in Fig. A2).
As forest plots were not equidistantly located in the reserve (Fig. A1), the logarithm-transformed spatial distance between each pair of subplots was also included as fixed effect variable in the model to remove the effect of physical proximity and other spatially autocorrelated unmeasured habitat factors such as soil nutrients. We used disturbance regimes (as a categorical variable), logarithm-transformed habitat distance and their interactions, mean elevation of plot, and logarithm-transformed spatial distance among subplots as fixed effect variables in this model. The pairwise diversity dissimilarity between subplots (level 1) was nested within plots (level 2), and each plot was, in turn, nested within a category of disturbance regime (level 3). Therefore, we included a plot-specific variable and a disturbance regime-specific variable, as random effect variables to remove the effect of plot and disturbance regime-specific conditions and the autocorrelation of spatial turnover within the same plot and the same disturbance regimes. We calculated the spatial correlogram of model residual against distance classes to check spatial autocorrelation of residuals of all models (Appendix C). Similar to the within-plot dissimilarity analysis, we used a linear mixed model to determine the effect of disturbance regime on phylogenetic and functional dissimilarity of subplots pairs between plots within each disturbance regime.
The significance of fixed effect variables were computed using F-tests for linear mixed models based on Satterthwaite’s approximation for denominator degrees of freedom57, and all insignificant terms of fixed effect variables were removed. A likelihood ratio test was used to compare the one level and two level random-effect variable models. Finally, because we were interested in the difference in dissimilarity between disturbance regimes, the appropriate linear contrasts were set up and tested, controlling for multiple comparisons, and the p-value of multiple comparisons was adjusted using the single-step method58.
Null model analysis
To examine whether species within communities were phylogenetically or functionally more (or less) than expected from a random assemblage from the pool of species in this study, we compared observed values of phylogenetic (Bst) and functional (τst) beta diversity with the null Bst and τst values from 999 random communities for each subplot pair. Random communities were generated by randomly shuffling the species names across the tips of phylogeny or functional dendrogram. Thus, the null model randomized the phylogenetic relatedness and functional similarity of species, while maintaining the observed species abundance, species richness, and species beta diversity in space. The above-described linear mixed model was used to control the effect of habitat difference and other potentially important factors on the observed and null Bst and τst values.
We also examined species, phylogenetic and functional beta diversity within each age category. We divided stems into three size classes: dbh 1 cm–5 cm, dbh 5 cm−10 cm, and dbh >10 cm to represent different regeneration classes during succession. We expected that phylogenetic and functional beta diversity changes across dbh size classes. This expectation follows because smaller stems in each plot were assumed to colonize at more recent successional stage within each plot, whereas larger stems established earlier in the process of succession.
Chao-Jaccard’s dissimilarity index was calculated using the R package ‘vegan’59. The Bst and τst values and null models of Bst and τst was calculated using the R package ‘spacodiR’60. Linear mixed models were fitted using the R package ‘lme4’61. The significance of fixed effect variables were computed using ‘lmerTest’ package57, and multiple comparisons were conducted using the ‘multcomp’ package58. All of the computations were conducted in R62.
Additional Information
How to cite this article: Mi, X. et al. Stochastic assembly in a subtropical forest chronosequence: evidence from contrasting changes of species, phylogenetic and functional dissimilarity over succession. Sci. Rep. 6, 32596; doi: 10.1038/srep32596 (2016).
Supplementary Material
Acknowledgments
This study was supported financially by National Key Research and Development Project of China (2016YFC0500202) the National Natural Science Foundation of China (31170401), and the Earthwatch Institute program “Quantify and monitor carbon pools and fluxes to assess the impact of climate change on subtropical forests under different anthropogenic disturbances”. The data analysis was conducted while XCM visited NGS’s lab in 2012. We thank the Administration Bureau of Gutianshan National Nature Reserve for logistic support. We also thank many HSBC climate champions and other field workers for their contributions to the establishment and census of the nine 1-ha forest plots. NGS was supported by two NSF USA-China Dimensions of Biodiversity Grants (DEB - 1046113; DEB - 1241136).
Footnotes
Author Contributions X.M., N.G.S. and K.M. designed the research. H.R., Q.J., G.F. and M.R. collected the data in the field. X.M. and N.G.S. conducted the data analyses. X.M., N.G.S., K.M. and D.P.B. wrote the manuscript. N.G.S. polished the English language. All authors discussed the results and revised the manuscript.
References
- Tilman D. Ressource Competition and Community Structure. (Princeton University Press, 1982). [PubMed] [Google Scholar]
- Hubbell S. P. The Unified Neutral Theory of Biodiversity and Biogeography. (Princeton University Press, 2001). [DOI] [PubMed] [Google Scholar]
- Chase J. M. & Leibold M. A. Ecological Niches: Linking Classical and Contemporary Approaches. (University of Chicago Press, 2003).
- Chazdon R. L. In Tropical Forest Community Ecology (eds Carson W. P. & Schnitzer S. A.) 384–408 (Blackwell, 2008). [Google Scholar]
- Norden N., Letcher S., Boukili V., Swenson N. & Chazdon R. Demographic drivers of successional changes in phylogenetic structure across life-history stages in plant communities. Ecology 93, S70–S82, doi: 10.1890/10-2179.1 (2012). [DOI] [Google Scholar]
- Bruelheide H. et al. Community assembly during secondary forest succession in a Chinese subtropical forest. Ecol. Monogr. 81, 25–41, doi: 10.1890/09-2172.1 (2011). [DOI] [Google Scholar]
- Anderson M. J. et al. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecol. Lett. 14, 19–28, doi: 10.1111/j.1461-0248.2010.01552.x (2011). [DOI] [PubMed] [Google Scholar]
- Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity. Ecography 33, 2–22, doi: 10.1111/j.1600-0587.2009.05880.x (2010). [DOI] [Google Scholar]
- Clements F. E. Plant Succession: An Analysis of the Development of Vegetation. (Carnegie Institution of Washington, 1916). [Google Scholar]
- Chazdon R. L. Second Growth: The Promise of Tropical Forest Regeneration in an Age of Deforestation. (The University of Chicago Press, 2014). [Google Scholar]
- Christensen N. L. & Peet R. K. Convergence during secondary forest succession. J. Ecol. 72, 25–36 (1984). [Google Scholar]
- Norden N., Chazdon R. L., Chao A., Jiang Y.-H. & Vílchez-Alvarado B. Resilience of tropical rain forests: tree community reassembly in secondary forests. Ecol. Lett. 12, 385–394, doi: 10.1111/j.1461-0248.2009.01292.x (2009). [DOI] [PubMed] [Google Scholar]
- Walker L. R., Wardle D. A., Bardgett R. D. & Clarkson B. D. The use of chronosequences in studies of ecological succession and soil development. J. Ecol. 98, 725–736, doi: 10.1111/j.1365-2745.2010.01664.x (2010). [DOI] [Google Scholar]
- Carreño-Rocabado G. et al. Effects of disturbance intensity on species and functional diversity in a tropical forest. J. Ecol. 100, 1453–1463, doi: 10.1111/j.1365-2745.2012.02015.x (2012). [DOI] [Google Scholar]
- Fukami T., Martijn Bezemer T., Mortimer S. R. & van der Putten W. H. Species divergence and trait convergence in experimental plant community assembly. Ecol. Lett. 8, 1283–1290, doi: 10.1111/j.1461-0248.2005.00829.x (2005). [DOI] [Google Scholar]
- Swenson N. G. The assembly of tropical tree communities – the advances and shortcomings of phylogenetic and functional trait analyses. Ecography 36, 264–276, doi: 10.1111/j.1600-0587.2012.00121.x (2013). [DOI] [Google Scholar]
- Letcher S. G. Phylogenetic structure of angiosperm communities during tropical forest succession. Proc. R. Soc. B 277, 97–104, doi: 10.1098/rspb.2009.0865 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Zang R., Letcher S. G., Liu S. & He F. Disturbance regime changes the trait distribution, phylogenetic structure and community assembly of tropical rain forests. Oikos 121, 1263–1270, doi: 10.1111/j.1600-0706.2011.19992.x (2012). [DOI] [Google Scholar]
- Purschke O. et al. Contrasting changes in taxonomic, phylogenetic and functional diversity during a long-term succession: insights into assembly processes. J. Ecol. 101, 857–866, doi: 10.1111/1365-2745.12098 (2013). [DOI] [Google Scholar]
- Graham C. H. & Fine P. V. A. Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol. Lett. 11, 1265–1277 (2008). [DOI] [PubMed] [Google Scholar]
- Smith A. B., Sandel B., Kraft N. J. B. & Carey S. Characterizing scale-dependent community assembly using the functional-diversity–area relationship. Ecology 94, 2392–2402, doi: 10.1890/12-2109.1 (2013). [DOI] [PubMed] [Google Scholar]
- Swenson N. G., Enquist B. J., Thompson J. & Zimmerman J. K. The influence of spatial and size scale on phylogenetic relatedness in tropical forest communities. Ecology 88, 1770–1780 (2007). [DOI] [PubMed] [Google Scholar]
- Blomberg S. P., Theodore Garland J. R. & Ives A. R. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 (2003). [DOI] [PubMed] [Google Scholar]
- Chase J. Community assembly: when should history matter? Oecologia 136, 489–498, doi: 10.1007/s00442-003-1311-7 (2003). [DOI] [PubMed] [Google Scholar]
- Fukami T. & Nakajima M. Community assembly: alternative stable states or alternative transient states? Ecol. Lett. 14, 973–984, doi: 10.1111/j.1461-0248.2011.01663.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegan B. Pattern and process in neotropical secondary rain forests: the first 100 years of succession. Trends Ecol. Evol. 11, 119–124, doi: 10.1016/0169-5347(96)81090-1 (1996). [DOI] [PubMed] [Google Scholar]
- Wang X.-H., Kent M. & Fang X.-F. Evergreen broad-leaved forest in Eastern China: Its ecology and conservation and the importance of resprouting in forest restoration. For. Ecol. Manage. 245, 76–87, doi: 10.1016/j.foreco.2007.03.043 (2007). [DOI] [Google Scholar]
- Li X., Wilson S. D. & Song Y. Secondary succession in two subtropical forests. Plant Ecol. 143, 13–21, doi: 10.1023/A:1009806512601 (1999). [DOI] [Google Scholar]
- Tang C., Zhao M.-H., Li X.-S., Ohsawa M. & Ou X.-K. Secondary succession of plant communities in a subtropical mountainous region of SW China. Ecol. Res. 25, 149–161, doi: 10.1007/s11284-009-0644-z (2010). [DOI] [Google Scholar]
- Liu X. et al. Covariation in Plant Functional Traits and Soil Fertility within Two Species-Rich Forests. PLoS ONE 7, e34767, doi: 10.1371/journal.pone.0034767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnke M., Kröber W., Welk E., Wirth C. & Bruelheide H. Maintenance of constant functional diversity during secondary succession of a subtropical forest in China. J Veg Sci 25, 897–911, doi: 10.1111/jvs.12114 (2014). [DOI] [Google Scholar]
- Letten A. D., Keith D. A. & Tozer M. G. Phylogenetic and functional dissimilarity does not increase during temporal heathland succession. Proc. R. Soc. B, 281, doi: 10.1098/rspb.2014.2102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G. et al. Anthropogenic disturbance shapes phylogenetic and functional tree community structure in a subtropical forest. For. Ecol. Manage. 313, 188–198, doi: 10.1016/j.foreco.2013.10.047 (2014). [DOI] [Google Scholar]
- Kröber W., Böhnke M., Welk E., Wirth C. & Bruelheide H. Leaf trait-environment relationships in a subtropical broadleaved forest in South-East China. PLoS ONE 7, e35742, doi: 10.1371/journal.pone.0035742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield M. M. & Levine J. M. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 13, 1085–1093, doi: 10.1111/j.1461-0248.2010.01509.x (2010). [DOI] [PubMed] [Google Scholar]
- Chesson P. L. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Evol. Syst. 31, 343–366 (2000). [Google Scholar]
- Godoy O., Kraft N. J. B. & Levine J. M. Phylogenetic relatedness and the determinants of competitive outcomes. Ecol. Lett. 17, 836–844, doi: 10.1111/ele.12289 (2014). [DOI] [PubMed] [Google Scholar]
- Bennett J. A., Lamb E. G., Hall J. C., Cardinal-McTeague W. M. & Cahill J. F. Increased competition does not lead to increased phylogenetic overdispersion in a native grassland. Ecol. Lett. 16, 1168–1176, doi: 10.1111/ele.12153 (2013). [DOI] [PubMed] [Google Scholar]
- Narwani A., Alexandrou M. A., Oakley T. H., Carroll I. T. & Cardinale B. J. Experimental evidence that evolutionary relatedness does not affect the ecological mechanisms of coexistence in freshwater green algae. Ecol. Lett. 16, 1373–1381, doi: 10.1111/ele.12182 (2013). [DOI] [PubMed] [Google Scholar]
- Legendre P. et al. Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 90, 663–674, doi: 10.1890/07-1880.1 (2009). [DOI] [PubMed] [Google Scholar]
- Xing P., Zhang Q.-B. & Baker P. Age and radial growth pattern of four tree species in a subtropical forest of China. Trees 26, 283–290, doi: 10.1007/s00468-011-0590-6 (2012). [DOI] [Google Scholar]
- Wu Z., Raven P. H. & De-Yuan Hong. Flora of China. (Beijing: Science Press & St Louis: Missouri Botanical Garden Press, 1994-2009). [Google Scholar]
- Harms K. E., Condit R., Hubbell S. P. & Foster R. B. Habitat associations of trees and shrubs in a 50-ha neotropical forest plot. J. Ecol. 89, 947–959, doi: 10.1111/j.1365-2745.2001.00615.x (2001). [DOI] [Google Scholar]
- Doyle J. J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15 (1987). [Google Scholar]
- Pei N. et al. Exploring tree-habitat associations in a chinese subtropical forest plot using a molecular phylogeny generated from DNA barcode loci. PLoS ONE 6, e21273, doi: 10.1371/journal.pone.0021273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797, doi: 10.1093/nar/gkh340 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P. & Rougemont J. A Rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771, doi: 10.1080/10635150802429642 (2008). [DOI] [PubMed] [Google Scholar]
- Sanderson M. J. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302, doi: 10.1093/bioinformatics/19.2.301 (2003). [DOI] [PubMed] [Google Scholar]
- Westoby M., Falster D. S., Moles A. T., Vesk P. A. & Wright I. J. Plant ecological strategies: some leading dimensions of variation between species. Annu. Rev. Ecol. Syst. 33, 125–159, doi: 10.1146/annurev.ecolsys.33.010802.150452 (2002). [DOI] [Google Scholar]
- Wright S. J. et al. Functional traits and the growth–mortality trade-off in tropical trees. Ecology 91, 3664–3674, doi: 10.1890/09-2335.1 (2010). [DOI] [PubMed] [Google Scholar]
- Cornelissen J. H. C. et al. A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust. J. Bot. 51, 335–380, doi: 10.1071/BT02124 (2003). [DOI] [Google Scholar]
- Legendre P. & Legendre L. Numerical Ecology. 3rd English edn, (Elsevier Science BV, 2012). [Google Scholar]
- Chao A., Chazdon R. L., Colwell R. K. & Shen T.-J. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8, 148–159, doi: 10.1111/j.1461-0248.2004.00707.x (2005). [DOI] [Google Scholar]
- Hardy O. J. & Senterre B. Characterizing the phylogenetic structure of communities by an additive partitioning of phylogenetic diversity. J. Ecol. 95, 493–506 (2007). [Google Scholar]
- Hardy O. J. & Jost L. Interpreting and estimating measures of community phylogenetic structuring. J. Ecol. 96, 849–852, doi: 10.1111/j.1365-2745.2008.01423.x (2008). [DOI] [Google Scholar]
- Johnson E. A. & Miyanishi K. Testing the assumptions of chronosequences in succession. Ecol. Lett. 11, 419–431, doi: 10.1111/j.1461-0248.2008.01173.x (2008). [DOI] [PubMed] [Google Scholar]
- Kuznetsova A., Brockhoff P. B. & Christensen R. H. B. lmerTest: Tests for random and fixed effects for linear mixed effect models (lmer objects of lme4 package). version 2.0-0. URL http://CRAN.R-project.org/package=lmerTest. (2013).
- Bretz F., Hothorn T. & Westfall P. Multiple Comparisons Using R. (CRC Press, 2010). [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., Stevens M. H. H. & Wagner H. vegan: community ecology package. R package version 2.0–4. URL: http://cran.r-project.org/ (2012).
- Eastman J. M., Paine C. E. T. & Hardy O. J. SpacodiR: structuring of phylogenetic diversity in ecological communities. Bioinformatics 27, 2437–2438, doi: 10.1093/bioinformatics/btr403 (2011). [DOI] [PubMed] [Google Scholar]
- Bates D. M. & Maechler M. lme 4: Linear mixed-effects models using S4 classes. R package version 0.999999–0, URL https://cran.r-project.org/package=lme4 (2012).
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/ (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.