Abstract
The generation and study of transgenic Aedes aegypti mosquitoes provides an essential tool for elucidating the complex molecular biology of this important vector. Within the field, genetic manipulation has now surpassed the proof of principle stage and is now utilised in both applied and theoretical vector control strategies. The application of new instruments, technologies and techniques allows ever more controlled experiments to be conducted. In this text we describe microinjection of Ae. aegypti embryos in the context of evaluating and performing genomic editing with transcription activator-like effector nucleases (TALENs).
Keywords: Mosquito, Aedes, TALEN, mutagenesis, HRMA, nuclease, gene editing
1. Introduction
Aedes aegypti mosquitoes are known vectors of a number of medically significant viruses and parasites including dengue viruses and yellow fever. Increased global travel, amongst other factors, has lead to the expansion of dengue to previously non-endemic regions of Europe, USA and China [1]. It is therefore imperative to understand the molecular biology of this complex vector, and over the last 20 years molecular techniques available to investigate this organism have advanced significantly. At present, the technology and techniques have advanced sufficently to generate transgenic lines or follow a reverse-engineered genetic approach.
One of the most exciting breakthroughs in the last two years has been the application of gene-editing techniques that can truly generate genetic knock-out mosquito lines. Transcription activator-like effector nucleases (TALENs) have been shown to work effectively in Ae. aegypti [2] and there is also evidence for successful homologous recombination [3, 4]. The adaptation of other technologies, such as high-resolution melt analysis (HRMA), are very useful in helping to characterize and track gene-editing events. To the uninitiated these experiments may appear time consuming and daunting but are actually straight-forward in their work-flow. In this chapter we have worked to create structured and robust protocols for the development of standard procedures building upon previous methods [5] to introduce and evaluate TALEN-induced genetic lesions in Aedes aegypti.
Assessment of gene editing activity using germline experiments is time consuming and labor intensive, requiring at least 2 months for Ae. aegypti. To this end we descrbie two rapid assays to make initial assessments of TALEN activity. Both assays have their own particular advantages and disadvantages. The single strand annealing (SSA) assay is the more sensitive of the two assays reviewed here. In this assay the TALEN target site is placed between two direct repeat sequences. The sequence upstream of target site is the first 300 bp of the firefly luciferase gene and the full length of firefly luciferase open reading frame (ORF) is located downstream of the target site. Successful double stranded DNA break induction can be repaired using the single stranded annealing pathway, resulting in the collapse of the direct homolog sequences and expression of the full length firefly luciferase ORF [2]. The disadvantage of this method is that it measures activity against an extrachromosomal (plasmid) target, not the endogenous target. Also, each unique target must first be cloned into a plasmid, limiting the ability of this method to scale. The second assay uses high resolution melt analysis (HRMA) [8] to determine differences in the melting temperature of PCR amplicons generated by primers flanking the targeted region of interest. Insertion/deletion events (indels) will alter the melt profile of the product in comparison to the wild-type control. The advantages of this method are that no cloning step is required, and that multiple TALENs or other gene-editing agents (CRISPRs) can be tested within the same injection mix as the assay allows for the same template to be employed in a number of different reactions. Two or three pairs of TALENs can be injected and assessed together, rather than individually (as in the case of the SSA assay). Disadvantages are that this method is not as sensitive as the SSA-based assay and is susceptible to false positive results if the amplified region contains natural sequence variant; this is easily avoided through careful pre-screening of potential target sites [7].
Once an active nuclease is identified in one or both transient assays, the described injection protocol can then be used for germline modification experiments. The final section describes our screening protocol to identify heteritable mutations in the absence of a phenotypic marker using HRMA.
2. Materials
All DNA plasmids for injection must be prepared using an endotoxin-free kit, we recommend using Nucleobond Xtra Midi EF Kit (catalog number 740420) (Macharey-Nagel, Macharey-Nagel, Bethlehem, PA). All insectary rearing should be conducted in standard Ae. aegypti laboratory conditions (28°C, 60–80% relative humidity, photoperiod of 16h light:8h dark) with all water required for mosquito rearing either reverse-osmosis treated or double-distilled.
2.1 Components for Microinjection of Ae. aegypti embryos
-
1
50 ml concial tube.
-
2
X-large cotton balls non-sterile (US Cotton, Gastonia, NC)
-
3
1.5″ hole puncher.
-
4
Chromatography paper (Fisher Scientific, Pittsburgh, PA).
-
5
Plastic coverslips (Thermo Fisher, Waltham, MA).
-
6
Double-sided adhesive non-toxic tape (catalog number #137) (Scotch St.Paul, MN).
-
7
100mm × 15mm petri dish.
-
8
Whatman paper Grade #40 (GE Healthcare, Bio-Sciences, Pittsburgh, PA).
-
9
Whatman paper Grade #50 (GE Healthcare, Bio-Sciences, Pittsburgh, PA).
-
10
40ml beaker (Fisherbrand, Fisher Scientific, Pittsburgh, PA).
-
11
Parafilm (Pechiney Plastic Packaging Company, Chicago, IL).
-
12
150mm × 15mm petri dish.
-
13
Adhesive putty (Scotch, St.Paul, MN).
-
14
Borosilicate microcapillaries, inner diameter 0.64mm, outer diameter 1.0mm, length 100mm (catalog number 1B100-4) (WPI Inc., Sarasota, FL).
-
15
Sutter P2000 Laser Based Micropipette Puller (catalog number P-2000) (Sutter Instrument Company, Novato, CA).
-
16
3% H2O2: 3% solution in DEPC treated-water.
-
17
Sutter Microelectrode Beveler (catalog number BV-10) with fine abrasive plate (catalog number 104D) (Sutter Instrument Company, Novato, CA).
-
18
10x Injection Buffer: 50mM KCl, 1mM NaH2PO4 in nuclease free water. Filter sterilize using 0.22μM syringe filter (adapted [6]).
-
19
Chilled centrifuge.
-
20
Eppendorf Microloaders (Eppendorf, Hauppauge, NY).
-
21
Flashlight aspirator (catalog number 2809C) (Bioquip Products, Compton, CA).
-
22
Leica MZ6 stereomicroscope (Leica Microsystems, Buffalo Grove, IL).
-
23
Tweezers: Dumont #5 Inox 11cm (WPI Inc., Sarasota, FL).
-
24
Halocarbon oil 27 (Sigma-Aldrich, St. Louis, MO).
-
25
Leica DM1000 microscope (Leica Microsystems, Buffalo Grove, IL) and a Leica Micromanipulator (Leica Microsystems, Buffalo Grove, IL) to position the microinjector needle holder (catalog number MINJ-4FEM) (Eppendorf, Hauppauge, NY).
-
26
Femtojet, programmable microinjector (Eppendorf, Hauppauge, NY).
-
27
Fine-spray water bottle.
2.2 Components for Single Strand Annealing Assay
-
28
Dual-Luciferase Reporter Assay System (Promega, Madison, WI).
-
29
Glomax Multi+Microplate Multimode Instrument (Promega, Madison, WI).
-
30
Fine paintbrush size 3/0.
-
31
Long forceps 12″.
-
32
Microcentrifuge pestle for 1.5ml microcentrifuge tube.
-
33
Costar White Solid 96-well plate (Fisher Scientific, Pittsburgh, PA).
2.3 Components for High Resolution Melt Analysis Transient Assay
-
34
Lightscanner mastermix (Biofire Defense, Salt Lake City, UT).
-
35
Hard-shell black-white 96well plate (catalog number #HSP9665) (Biorad, Hercules, CA).
-
36
Mineral Oil (Fisher Scientific, Pittsburgh, PA).
-
37
Optical film (catalog #2239444) ) (Biorad, Hercules, CA).
-
38
Gradient thermocyler (Biorad, Hercules, CA).
-
39
1xTAE buffer: add together 4.84g Tris, 1.1ml Glacial Acetic Acid, 2ml 0.5M EDTA (pH8.0). Make upto 1 liter with reverse-osmosis treated water then autoclave.
-
40
TAE-agarose gel: 2% agarose in 1xTAE buffer. Dissolve 2g agarose in 100ml 1xTAE solution (described in previous step).
-
41
Lightscanner (catalog number LSCN-ASY-0011) (Biofire Defense, Salt Lake City, UT).
-
42
Nucleospin Tissue Kit (catalog number 740952) (Macharey-Nagel, Bethlehem, PA).
-
43
Pellet pestle cordless motor.
-
44
Nanodrop Spectrophotometer (Thermo Fisher, Waltham, MA).
2.4 Components for Germline experiment
-
45
Phire Animal Tissue Direct PCR kit (catalog number #F-140) (Thermo Scientific, Pittsburgh, PA).
-
46
70% ethanol: 70% ethanol solution in water.
-
47
LC Green Plus+ (catalog number BCHM-ASY-0005) (Biofire Defense, Salt Lake City, UT).
-
48
Fish food (Tetra, Blacksburg, VA).
-
49
Soup cup 16oz (The Webstaurant Store, Lancaster, PA).
-
50
Mesh fabric (Nylon Tulle).
-
51
15ml disposable beaker (Fisherbrand, Fisher Scientific, Pittsburgh, PA).
-
52
5ml disposable polystrene beakers (VWR, Radnor, PA).
-
53
Cardboard popcorn container 46oz (The Webstaurant Store, Lancaster, PA).
-
54
Glue-gun.
-
55
Staples and stapler.
-
56
Dynarex 4″cotton stockinette (Medex Supply, Monsey, NY).
-
57
14″ cable ties.
-
58
Wide polystrene drsosphila tube (28.5mm × 95mm) (Genesee, San Diego, CA).
-
59
Flug (Genesee, San Diego, CA).
-
60
Mosquito rearing pan - 28 quart storage box and lid (23″×16 ¼ ″×6″) (catalog number 1655) (Sterilite, Townsend, MA).
-
61
Disposable plastic Pasteur pipette.
-
62
Pyrex glass petri dish, 150mm × 20mm (Corning Glass Works, Corning, NY).
-
63
Small tub and lid (500ml volume).
-
64
Plastic sheeting for use with a 2mm Impulse Heat Sealer.
3. Methods
3.1 Microinjection of Aedes aegypti embryos
3.1.1 Preparation
-
1
A cage of mosquitoes should be blood fed at least 72 hrs before the first day of embryo injections (see Note 1).
-
2
Prepare the laying chambers/tubes by placing a ball of wet cotton in the cone of a 50 ml conical tube. Place a damp 1.5″ diameter circular chromatography paper gently on top of the cotton. The dampness of the cotton and paper should not be in excess; another cotton ball is used as the stopper at the open end (see Note 2).
-
3
Prepare 10–12 coverslips with double-sided adhesive tape along one edge. Trim the tape back to the edge of the coverslip using scissors.
-
4
Prepare a petri dish (100 mm diameter) with a circle of Whatman paper-grade #40. The upturned petri dish lid makes manipulation under the microscope easier as the field of view can quickly be adjusted by moving the dish. Saturate the paper with water then transfer it from the laying chamber-tube to the petri-dish mounted paper. The level of water saturation is critical for transferring the embryos as too little moisture will promote desiccation of embryos, whereas too much will make manipulation of the embryos problematic.
-
5
Prepare pieces of transfer embryo paper. The line of embryos for transfer is generated upon a different piece of Whatmann paper -grade# 50. The different grade ensures that less fibers are transferred to the adhesive tape upon the coverslip.
-
6
Prepare embryo recovery beakers. After injecting, the embryos are moved to a labelled 40 ml recovery beaker containing a ball of damp cotton and sealed with parafilm.
-
7
Needle manufacture (also summarised in [7])
Prepare a covered container for the needles to be stored in; a 100mm petri dish or tip box with two lines of adhesive putty is ideal.
Generate needles using borosilicate microcapillaries - inner diameter: 0.64mm, outer diameter: 1.0mm, length 100mm. Pull using Sutter P2000 Laser puller. Program: Heat=270, FIL=3, VEL=37, DEL=250, PUL=140 (see Note 3 and 4).
If the injection mix includes RNA components then use treated needles to minimize RNase contamination (see Note 5).
-
8
Bevel needle using Sutter BV-10. Our preffered angle of bevel is approximately: 22.5° (see Note 6).
-
9
Assemble the injection mix components in 1x injection buffer, if the mix includes RNA components then prepare needles and injection mix on that day. If the injection mix only includes DNA components then needles and injection mix can be prepared further ahead of time (see Note 7, 8, 9 and 10).
-
10
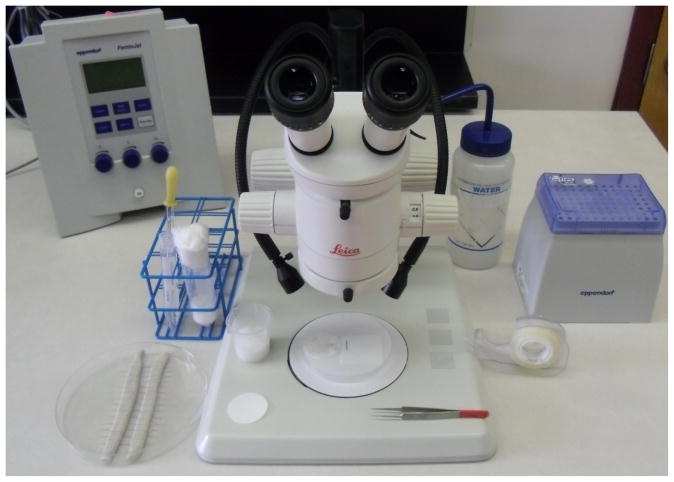
Figure 1 shows a prepared work station for microinjection with many of the described items.
Figure 1. A prepared work station ready for lining up embryos for microinjection.
Upon the desk from the left then proceeding clockwise: needles within their container, a laying chamber tube and a 15 ml tube of Halocarbon Oil with a glass Pasteur pipette ready for covering dessicated embryos, the Eppendorf Femtojet, a water bottle with a fine jet spray nozzle, Eppendorf microloaders (within their tip-rack) and double-sided adhesive tape. Upon the microscope stage (from left to right) are a small circle of paper for the laying chamber, a recovery beaker, a Whatman paper circle upon a petri dish lid showing a line of embryos ready for transfer, a pair of tweezers and three prepared coverslips.
3.1.2 Method
-
11
Within a pre-chilled centrifuge, spin the injection mix at >14000 rpm for >20 min at 4°C then maintain the injection mix chilled (on ice) while injecting (see Note 10).
-
12
Use Eppendorf Microloader tips to backfill the needle with injection mix (see Note 10).
-
13
Gently transfer 20–30 gravid females into the laying chamber using an aspirator. Leave the tube in darkness in insectary conditions for approximately 30 minutes (see Note 11 and 12). A small cardboard box works well.
-
14
Use tweezers (Dumont #5 Inox 11cm) to line up the embryos next to each other orientating the posterior pole in one direction. Line up 100–130 embryos in a single line that will span most of the width of the coverslip. When selecting embryos for lining up, observe and select embryos of a similar color. No matter how many embryos are lined up the embryos should be between 90–120 minutes old when injecting.
-
15
Dry the transfer paper by pressing down upon a folded kimwipe tissue before transfer to the coverslip. Be aware that the embryos will start to dessicate once the water is removed from around them (see Note 13).
-
16
Transfer the lined-up embryos to the prepared coverslip. Firmly but gently press the adhesive tape strip of the coverslip against the line of embryos. The edge of the coverslip is ideally only 2–3 mm away from the posterior poles of the lined up embryos (see Note 14).
-
17
Dessicate the embryos upon the coverslip. When they have achieved the desired desiccation level (first appearance of dimpling), cover in Halocarbon oil 27.
-
18
Using tweezers, remove any fibres that have also become stuck to the adhesive tape. This prevents the fibres obstructing or damaging the needle while injecting.
-
19
Transfer the coverslip to the injection microscope stage. Line up the edge of the coverslip closely with the edge of the injection microscope slide rostrum (see Fig. 2). Bring the bevel of the needle into focus using the micromanipulator then withdraw the needle from view and then focus the first embryo with the stage controls. Slowly bring the needle back into the microscope view until it is in the center of the image.
-
20
Using the Femtojet Injector, inject the embryos back to their full turgidity using the stage controls to pierce the embryos upon the fixed needle (see Notes 15, 16, 17, 18, 19, 20 and 21).
-
21
Drain the halocarbon oil from the injected embryos and using forceps transfer the embryos carefully to a damp paper surface for their development time to complete. Uninjected embryos are removed or squashed at this stage (see Note 22).
-
22
Embryos recover on a small piece of filter paper, Whatman paper grade #40 placed within a prepared recovery beaker. The number of transferred embryos are counted and noted then the recovery beaker is sealed with a stretched piece of parafilm over the top. The recovery beaker is placed back in the insectary for development under regular conditions.
-
23
If the injection mix includes RNA components then run a sample of the injection mix after injecting to observe if any RNA degradation occurred after the mix was assembled.
Figure 2. The microinjection setup.
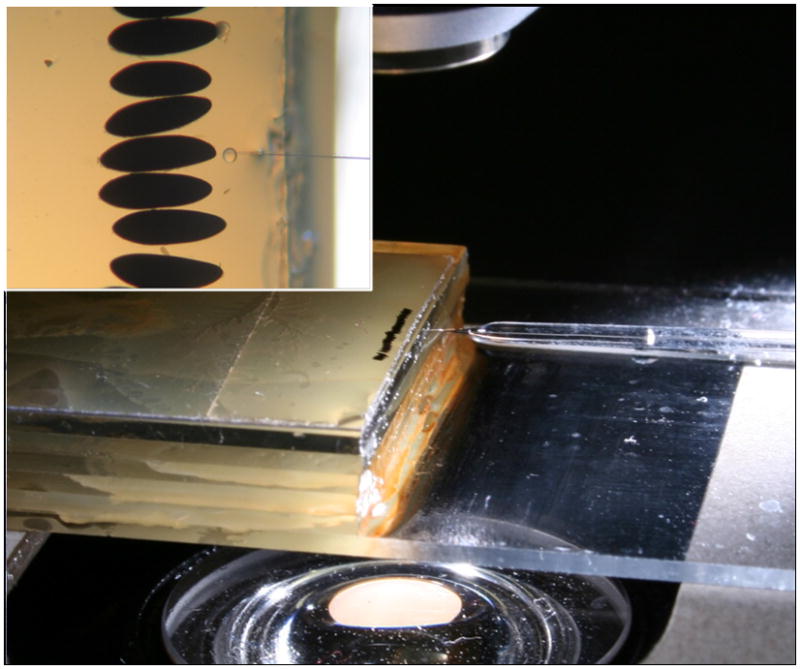
A coverslip with a line of embryos covered in Halocarbon oil placed upon the slide rostrum. The elevation allows the needle to easily access the posterior pole of the embryo without touching the stage.
3.2 Single Strand Annealing assay
3.2.1 Preparation
Clone target site (including both TALE binding regions and spacer) into the spacer region of the SSA test vector [7]. The activity of Firefly luciferase and Renilla luciferase can be measured by using the Dual-Luciferase Reporter Assay System with the Glomax Multi+Microplate Multimode instrument according to the manufacturer’s protocol. Different target sites can be assembled into a single SSA vector using gene synthesis technology. Numerous TALEN constructs can be tested within a single SSA test vector by using a multiple cloning site to allow insertion of the synthetic fragment.
3.2.2 Method
Assemble injection mix consisting of the SSA construct, a pair of TALEN expression plasmids and a control plasmid for normalization of the dual luciferase assay (see Note 23). Use 100–200 ng/μl of each TALEN expression construct, 200 ng/μl of SSA reporter plasmid and 200 ng/μl of the control plasmid. Add DEPC water to make a total volume of 50 μl.
Spin the injection mix down at the maximum speed for at least 10 min at 4°C.
Follow the injection protocol as stated above, ideally inject embryos in 3–6 replicates of 100–120 embryos each. Allow the embryos to recover for 24 hrs after injection, then transfer them into a labelled 1.5 ml microcentrifuge tube using a fine paintbrush. Snap-freeze the tubes in liquid nitrogen (see Note 24).
Remove the tubes from the liquid nitrogen one at the time using long forceps (see Note 25).
Dilute 5x Passive Lysis Buffer to 1x with nuclease-free molecular biology grade water (can be stored at 4°C for up to 1 month).
Prepare sufficient 1x Passive Lysis Buffer for 100 μl per tube of embryos.
Remove one tube at a time and crush within 50 μl of 1x Passive Lysis Buffer using a microcentrifuge pestle. Add an additional 50 μl of 1x Passive Lysis Buffer to the tube and place in a rack until all samples have been crushed (see Note 26).
Snap-freeze all samples again in liquid nitrogen then transfer them back to a rack to thaw.
Once thawed, spin down samples at ~6000 rpm for 10 sec at room temperature.
3.2.3 Luciferase assay
Prepare the Luciferase Assay Reagent II (LARII) by resuspending the lyophilized luciferase assay substrate in 10 ml of the supplied Luciferase Assay Buffer II (see Note 27).
Prepare the Stop & Glow substrate. Substrate is supplied as a 50x concentrate; dilute 1:50 into Stop &Glow buffer, each sample will require 80 μl per sample (see Note 28).
Equilibrate samples and reagent to ambient temperature.
Load 20 μl of each lysate into a Costar white solid 96-well plate.
Follow the manufacturer’s protocol Glomax Multi+Microplate Multimode instrument to inject 80 μl LARII for each sample using one injector, read the luminescence (delay 10s, integration 2s) then inject 80 μl Stop & Glo substrate using the second injector (delay 10s, integration 2s).
Create a ratio by normalizing the firefly RLU value using the renilla RLU value.
3.3 High Resolution Melt Ananlysis Transient assay
3.3.1 Preparation
Use the Lightscanner Primer design program to design a pair of primers for an assay. A good starting point for HRMA primer pairs design is a product length of 80–120 bp, primer Tm°C of 50–72°C, primer lengths of 19–30 nt and both 5′ and 3′ exclusion buffers set at 0 (see Note 29).
-
Set up the following reaction with the Lightscanner mastermix within a black-white 96 well plate.
Add 20 μl of mineral oil to each sample and 10 μl of the following mastermix reaction: 4 μl 2.5x Mastermix, 3 μl DEPC water, 1 μl of each primers (each to a final concentration of 250 μM).
Add the specific sample DNA (1 μl) to each single well; 10–20 ng of genomic DNA per 10 μl reaction is recommended (see Note 30).
Once the plate is loaded cover with optical film and ensure an airtight seal over each well and around the edge of the plate.
Centrifuge the plate in a swinging bucket rotor at 2000 rpm for 2 min at room temperature.
Perform gradient PCR using a standard thermocycler (95°C 2min, 40 cycles of [94°C 30 sec, 50–72°C 30 sec], 94°C 30 sec, 25°C 30 sec).
Load 7 μl of each sample into a TAE-agarose gel (2%) for analysis. Check the PCR product is the predicted size.
Thermal melt profiles can be generated using a Lightscanner (60°C–95°C, hold 57°C).
Using the Lightscanner Call-IT 2.0 software, examine the melt profiles within the annealing temperature determined by the gel image analysis. Find the temperature that has the greatest initial fluorescence tempered with the greatest difference in fluorescence change over the melt profile. This is the optimal annealing temperature for the reaction (see Note 31).
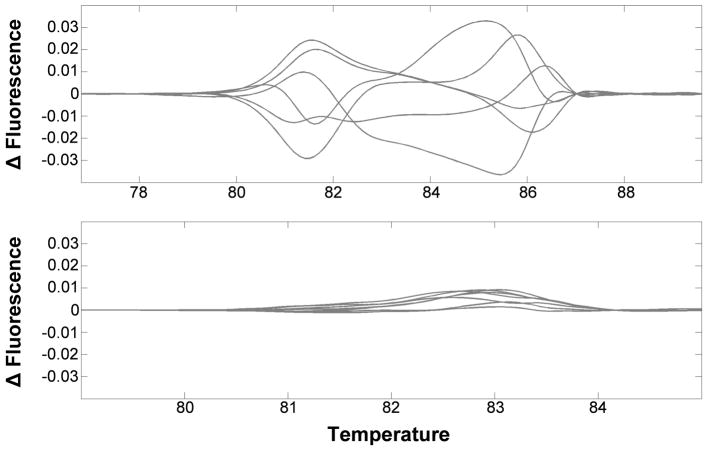
Perform the PCR reaction again (95°C 2 min, 40 cycles of [94°C 30 sec, x°C 30 sec], 94°C 30 sec, 25°C 30 sec), where ‘x’ is the optimized annealing temperature identified in the preparation steps, using DNA from 6–9 groups of 24 hr old embryos. Generate thermal melt profiles using a Lightscanner (50°C–95°C, hold 47°C). If excessive variation between samples is observed, primers should be redesigned and validated prior to proceeding (Fig. 3).
Figure 3. Variability within the uninjected control samples reduces the sensitivity of HRMA analysis of gene editing events.
The HRMA melt profiles (top) show excessive variability between replicate control DNAs (n=6), compared to those (bottom) that show relatively little variation (n=7).
3.3.2 Method
Assemble injection mix consisting of the 300 ng/μl of each TALEN expression construct and 1× injection buffer. Add DEPC water to make a total volume of 50μl.
Follow the injection protocol as stated above. Inject embryos in three replicates of 100–120 embryos each. Allow the embryos to recover for 24 hours after injection then transfer the injected embryos into a 1.5ml microcentrifuge tube using a fine paintbrush. Snap-freeze labelled tubes in liquid nitrogen (see Note 25).
Also collect 6–9 samples of uninjected embryos of the same developmental time as these will serve as controls for the HRMA analysis.
-
Extract DNA from the experimental and control embryo samples using Macharey-Nagel Tissue Nucleospin Kit. Follow the stated protocol with the following considerations.
Homogenize the embryos (100–300) within a microcentrifuge tube using a microcentrifuge pestle in 180 ul of T1 buffer (see Note 26).
Incubate the proteinase K-T1 buffer homogenate 16 hours.
Before loading the homogenate into the spin column, spin the sample >13,000rpm for 2minutes at room temperature to pellet the embryo debris then transfer the solution to the column.
Elute in 100 ul Elution buffer and measure the DNA content using a Nanodrop spectrophotometer.
Follow the previous directions detailed in the transient assay preparation steps to set up the Lightscanner mastermix reactions within a hard-shell 96 well plate. Iinclude 6–9 control samples of uninjected embryos of the same developmental time period and at least one no-template control.
Perform the PCR reaction (95°C 2min, 40 cycles of [94°C 30 sec, x°C 30 sec], 94°C 30s, 25°C 30 sec), where ‘x’ is the optimized annealing temperature identified in the preparation steps. Thermal melt profiles are generated using a Lightscanner (50°C–95°C, hold 47°C).
Using the Lightscanner Call-IT 2.0 software, examine the melt profiles to determine whether indel events are occurring within the target region. After normalizing the data, set the uninjected controls as standards. Any indel events will cause a shift in melt profile of the PCR product from the uninjected controls. A difference curve of 0.1 or greater (from the set standards) is typically sufficent to justify a germline experiment; smaller differences may also be acceptable depnding on how much variation is present amongst the control samples.
3.4 Germline experiment
3.4.1 Preparation
-
Optimize PCR for your target sequence using the Phire Animal Tissue Direct PCR kit (see Fig. 4).
Set up a tube with 20 μl Dilution Buffer + 0.5μl DNA Release Additive.
Place one leg (or piece of leg) from a wild-type adult mosquito into the tube (see Note 32).
Incubate at room temperature for 2–5 min then 98°C (within a thermocycler) for 2 min. Allow to cool to room temperature while preparing PCR mix.
Using a multichannel pipette add 20 μl mineral oil to intended sample wells of a hard-shell 96 well plate.
Prepare the following: 19 μl Phire mastermix for each well across the gradient range; 10 μl 2x Phire Animal Tissue PCR Buffer, 1 μl primer mix (final concentration ~500nM), 0.4 μl Phire II polymerase, 2 μl LCGreen, 5.6 μl DEPC water. Pipette Phire master mix into the sample wells.
Transfer 1 μl of Phire-dilution sample to each well of the gradient range.
Once the plate is loaded, cover with optical film and ensure an airtight seal over each well and around the edge of the plate.
Centrifuge the plate in a swing bucket rotor for 1 min at 2000 rpm at room temperature.
Run Phire gradient PCR program (including a heteroduplex formation step) using a thermocycler (98°C 5min, 40 cycles of [98°C 5s, 60–72°C 5s, 72°C 20s], 72°C 1min, 94°C 30s, 25°C 30s, 4°C ∞).
Generate thermal melt profiles using a Lightscanner (60°C–95°C, hold 55°C).
Using the Lightscanner Call-IT 2.0 software, examine the melt profiles within the annealing temperature. Find the temperature that has the greatest initial fluorescence tempered with the greatest difference in fluorescence change over the melt profile. This will be the optimal annealing temperature for the assay.
Hatch wild type strain mosquitoes 4–7 days prior to hatching the injected embryos. This ensures that when G0 male and female have emerged the wild type females/males are ready to cross with them. Add a small amount of fish food in the pan.
-
Prepare between 80–100 soup-cup cages to house sibling-matings for the progeny of each G0 survivor using a 16 oz soup cup and the corresponding lid.
The cardboard inner top is punched out from the lid ring and discarded.
A mesh fabric (Nylon Tulle) is then placed over the cup opening and trapped down by replacing the ring of the adapted lid. The tension in the mesh needs to be sufficient to secure the mosquitoes inside and to support raisins and a small ball of damp cotton.
For each soup-cup cage prepare an oviposition beaker (15 ml disposable beaker) with a piece of chromatography paper curled around the inside.
After sex-separating the wild type pupae, place 30–35 individuals in 5 ml disposable polystyrene beakers into soup cup-cages. Provide a damp piece of the cotton wool and a number of raisins as sources of water and sugar.
-
Prepare 10–15 disposable popcorn-cages using a 46 oz cardboard containers:
Using a glue-gun, stick two 46 oz cups together, one inside the other.
A 10 cm diameter hole is cut through one side.
Cut an entry sock of ~25cm of Dynarex 4″ cotton stockinette.
Using a stapler secure the entry sock stockinette to the inside lip of the side hole.
Two cable ties (14″) are secured together to form a loop, these then trap a double layer of mesh (Nylon Tulle) to the top opening. The mesh is trapped to provide tension across the top of the opening allowing support of a blood feeder without excessive sagging.
Seal the mesh and cable ties in place using a hot glue gun.
-
Prepare laying tubes for single bloodfed female G0 mosquitoes.
Place a damp ball of cotton into the bottom of a wide polystrene drosophila tube.
Place a damp 1.5″ diameter circular chromatography paper gently on top of the cotton.
Use a tissue or paper towel to wipe dry the inside edge of the conical tube to discourage mosquitoes ovipositing eggs anywhere other than the circular paper. The dampness of the laying surface is crucial in that if the surface is too wet the mosquitoes will be trapped and prevent optimal laying. If too dry the mosquitoes will not lay.
Plug the opening with cotton or a flug.
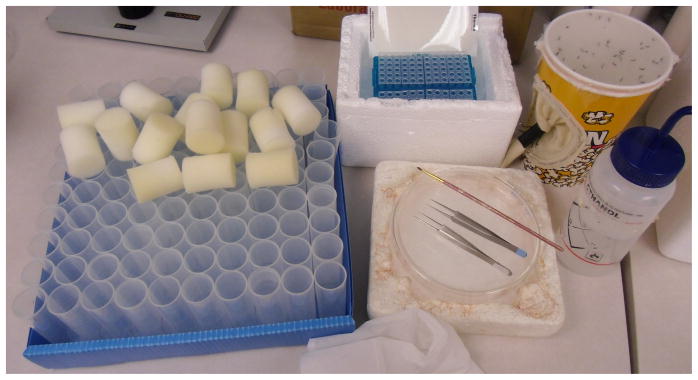
Figure 4. Preparation of materials for setting up a 96 well plate HRMA tracking experiment.
Clockwise from the left, the drosophila tubes ready for the adult mosquitoes with flugs to plug the tube, an ice-bucket with a 96 well plate already prepared with Phire mastermix in each sample well, a 46oz popcorn-cage with adult mosquitoes, an ethanol bottle to wipe the tweezer legs between samples, an ice-chilled glass petri dish with tweezers and a paintbrush to move and transfer the mosquitoes.
3.4.2 Method
-
1
Assemble injection mix with 300 ng/μl of each TALEN expression plasmid in 1x injection buffer.
-
2
Follow the injection protocol as stated above; inject approximately 1000 embryos.
-
3
At 5 days post-injection, prepare a pan containing 4 liters of reverse osmosis-purified water with an appropriate amount of ground fish food (see Note 33).
-
4
Hatch the injected embryos within the labelled pan (see Note 34 and 35).
Using a pair of tweezers, carefully transfer the paper containing injected eggs to the surface of water with the injected eggs face down.
-
5
Monitor the hatching pan daily for pupae, pick any developing pupae and separate the males and females into popcorn cages (see Note 36 and 37).
G0 females
-
6
Set up popcorn-cages with 20–30 wild type males each and a equal number of G0 females, anesthetize the adult mosquitoes using CO2 and then transfer them to 150 mm × 20 mm glass petri dishes where they can be examined and manipulated with ease (see Note 38).
-
7
Feed the G0 female cages 2–3 days after establishing the mating cross.
-
8
One day after blood feeding, anesthetize the mosquitoes, cut away the top mesh using a razor blade then transfer the mosquitoes (mated G0 females and wild type males) to a pre-chilled glass petri dish. Using a pair of tweezers, place a single bloodfed G0 female mosquito into a polystyrene drosophila laying tube then plug the tube (see Note 39).
-
9
Once the females have recovered, place the tubes gently in a horizontal orientation within a small cardboard box in the insectary. Keep 3–4 and record the number of tubes that show eggs have been laid. At this point the adult female mosquito can be discarded.
-
10
To hatch, gently fill the drosophila laying tube with water ensuring that the eggs are completely covered, along with a drop of very diluted fish-food solution.
-
11
After 2 to 3 days any viable G1 larvae will have hatched. At this point, larvae should be moved to an appropriately sized rearing pan.
-
12
Transfer G1 pupae from each founder female into a indivdual soup cup-cage allowing the sibling-adults to cross. At this time also add an oviposition beaker (see Note 40).
-
13
Blood feed the soup cup-cages 2–3 days after establishing the mating cross.
-
14
Two days after blood feeding, half-fill the oviposition beakers in the G1 inter-cross soup cup-cages with water by spraying/trickling water through the mesh.
-
15
After 3 or 4 days examine and record the number of soup cup-cages that show eggs have been laid.
-
16
When the G1 females have laid the eggs, anesthetize them using CO2 and transfer all the individuals to a labeled microcentrifuge tube. Snap-freeze and store at −80°C until ready for genomic DNA extraction.
-
17
Store the labeled G2 oviposition papers (see Note 41).
-
18
Extract genomic DNA from the G1 adult bodies. Use Macherey-Nagel Tissue Nucleospin kit. Elute DNA from spin column in 100 μl.
-
19
Use HRMA to analyse sequence at TALEN recognition site as detailed previously in HRMA Transient Assay section.
-
20
If indel mutation is identified by HRMA, hatch the corresponding G2 oviposition paper.
-
21
Place G2 male and female pupae into separate cages prior to genotyping by HRMA.
-
22
To identify individual adults with mutant alleles use Phire Animal Tissue Direct PCR kit:
Set up 96 well plate with 20 μl Dilution Buffer + 0.5μl DNA Release Additive in each well.
Place one leg (or piece of leg) from a wild-type adult mosquito within one well (see Note 32).
Seal plate with optical PCR film, incubate at room temperature for 2–5 min then 98°C (within a thermocycler) for 2 min. Allow to cool to room temperature while preparing PCR mix.
-
23
Follow the previous directions detailed in the preparation steps (Germline experiment, Preparation, Steps 2–3) to prepare the 96-well plate in which the Lightscanner reaction will be performed.
-
24
Transfer 1 μl of Phire-dilution sample to the 96-well plate, we recommend using 96 well plate plan to track samples.
-
25
Run Phire PCR at optimized temperature (including a heteroduplex formation step): (98°C 5min, 40 cycles of [98°C 5s, x°C 5s, 72°C 20s], 72°C 1min, 94 °C 30s, 25°C 30s, 4°C ∞).
-
26
Generate thermal melt profiles using a Lightscanner (60°C–95°C, hold 55°C) and analyze using the Lightscanner Call-IT 2.0 software.
-
27
Identify individual adult mosquitoes with mutant alleles.
G0 males
-
28
When crossing G0 males, first visually check the soup cups of virgin wild type females to see if there are any males in the group; these should be discarded. Anesthetize the adult mosquitoes using CO2 and then transfer them to 150 mm × 20 mm pre-chilled glass petri dishes. From the dishes the adults can be transferred using tweezers into the prepared soup cup-cages.
-
29
Put a single G0 male with 5 virgin females into each soup cup-cage. At this time also add an oviposition beaker into the soup cup cage (see Note 40).
-
30
Feed the soup cup-cages 2–3 days after establishing the mating cross.
-
31
Two days after blood feeding fill the oviposition cups in the male soup cups with water by spraying/trickling water through the mesh.
-
32
After 3 or 4 days examine and record the number of soup cup-cages that show eggs have been laid.
-
33
Using CO2, anesthetize the adults within those cages that have laid eggs. Those that have not can either be re-fed or discarded.
-
34
Using tweezers, tranfer the G1 oviposition paper into a small tub of water that has already been prepared with some fish-food. Alternatively the oviposition paper can be stored for screening later.
-
35
Continue the protocol as stated above for G1 progeny (of G0 females) by inter-crossing the adults within a soup cup-cage (see step 13), blood-feeding and collecting the G2 oviposition paper.
Acknowledgments
This protocol was developed with the support of grants [AI085091 and AI099843] from the NIAID. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID.
Footnotes
Preferentially the mosquitoes should be fed 96 hours in advance of the injection day. The number of females blood fed is dependent upon the number of injections needed for the experiment. For approximately 1000 injections, a minimum of 200 females is recommended.
Circular papers are manufactured using a hole puncher (1.5″) from chromatography paper. Use a tissue paper to wipe dry the inside edge of the conical tube to discourage mosquitoes ovipositing eggs anywhere other than the circular paper. The dampness of the laying surface is crucial in that if the surface is too wet the mosquitoes will be trapped and prevent optimal laying. If too dry the mosquitoes will not lay.
Handle the microcapillaries at the ends of the tubes to avoid oils or other contaminants being present at the heating point.
When pulling the needles, monitor the Heat-on-time value generated after every pull to ensure that the needles are consistent. Needles pulled outside a narrow variance should be discarded at this stage.
Handle microcapillaries with gloves. To treat needles for RNA work: soak all microcapillaries in 3% H2O2 solution overnight. Thoroughly rinse the microcapillaries numerous times in reverse osmosis treated water. Bake dry in an oven until all liquid has evaporated. Pull needles on the day of injections using standard embryo injection program.
To bevel microcappillaries, the 104D fine abrasive plate is recommended. Before use, blow any dust or fine particles away from the beveling surface using compressed air. The bevel of each needle can be checked using a microscope (magnification 100x–200x is ideal). The bore of the needle should also checked as if much greater than 1μm then the increased trauma to the embryo maybe lethal.
If the injection mix includes RNA components then normal RNA procedures should be followed (gloves, barrier tips, RNA treated needles).
Each component of the injection mix should be centrifuged at maximum for a minimum of 10 min at 4°C prior to pipeting to minimize debris carryover into the mix.
When assembling the injection mix, it is recommended use a maximum concentration of 800 ng/μl total nucleic acid as a higher concentration may block the needles.
Numerous steps can be taken to minimize clogging of the needle. The injection mix volume should be 30 μl or greaterl preferably in a 0.5 ml tube. This volume allows that the needle-backfiller can draw fluid from the uppermost level of the mix. When backfilling the needle, the end of the microloader should be as close to the needle point as possible. Dispense the injection mix gently while withdrawing the microloader to minimize the occurrence of bubbles within the needle. The injection mix can be left spinning at max (~14,000rpm) at 4°C within a benchtop centrifuge until it is needed for injection. While injecting, the injection mix should be spun down every 2 hours.
Ideally the aspirator can be adapted to have a narrow piece of tubing at the end (~5 mm inner diameter) that facilitates both the selection of gravid females and depositing them into the laying chamber, down the side of the stopper. Shake and blow the mosquitoes gently down the aspirator tube into the laying tube.
Check the laying chamber after 30 min for the appearance of embryos. If none can be observed then the oviposition time can be increased to 45–60 min. Multiple laying chambers can be setup simultaneously. The promptness of mosquitoes to oviposit varies based with each strain and time post-bloodmeal. We have found that most strains will lay eagerly after 4–6 days post blood meal but any further delay may result in reabsorption of the embryos or reduction in longevity.
Turn on the Femtojet Injector so that the injection pressure has already been achieved while the embryos are desiccating. This allows the operator to begin injecting shortly after moving the coverslip to the injection microscope rostrum.
Tamp down the embryos of the coverslip using an arm of the forceps to push the embryo more firmly into the adhesive tape before injection. Be aware that once the embryos are transferred they will begin to desiccate.
Place the coverslip upon the microscope stage or the microscope rostrum ready for injecting. This is manufactured by gluing 4 or 5 microscope slides together with the lower one offset to enable the microscope stage to grasp the edges while the light is not obscured by the specimen holder. Using a rostrum also allows greater variability in the angle of injection.
The angle of the injection can be accomplished between 0–20 degrees but we prefer approximately 5°.
The injection mix should be flowing freely from the tip of the needle. This allows the operator to inject the embryo without actually using the inject function of the Femtojet Injector. The pressure compensation value (pc) should be varied dependent upon the bore of the needle and the viscosity of the mix. The flow should be continuous but not too fast as the disruption to the embryo’s internal environment will increase mortality rates. If the yolk from the embryo is excessively spilling out then could indicate that the dessication stage was too short or that the needle is too blunt.
Occasionally, it can be observed that embryos are being displaced from their adhesive tape position rather than pierced by the needle. This can occur when excess water from the transfer step causes the adhesive tape to soften, the pressure to stick them to the coverslip was not sufficent, when the needle is too blunt or when the embryos have darkened too much.
If the embryos are invaginating excessively before the needle is piercing the embryo then the needle maybe too blunt or the desiccation was too long.
If the needle becomes clogged with debris then the ‘Clean’ function may be used to unblock the aperture. If this does not resolve the problem (very slow or no injection mix exiting the needle) then replace the needle.
After repeated use the needle will blunt and should be replaced. Replacement is dependent upon the actual injections but ideally after 2 or 3 lines when using borosilicate glass.
Removal of halocarbon oil can be achieved in different ways. We have found the easiest to be gently spraying the coverslip with a water spray bottle. The fine jet can be directed just above the line of embryos as the coverslip is held perpendicularly to some paper towels. This encourages the oil to drain off the embryos and also softens the adhesion of the tape easing their removal with forceps.
It is advised that the experimental and control plasmids use different promoters to express luciferase to minimize any promoter cross-talk between them. As a normalization control plasmid, either pkhsp82-Renilla or pIE1-Renilla can be used with good effect to express Renilla luciferase [7].
It is advisable to also collect up the non-transferred uninjected embryos to act as a negative control; this hatch rate will give a baseline for the survival rate. An additional more stringent control for the injection process may include the recovery of lined-up but uninjected embryos to assess the potential mortality caused by the physical lining up and recovery stages.
Samples can be stored at −80°C at this stage. If samples have been stored then transfer them back into liquid nitrogen ready for homogenizing.
When using a pestle motor, add an initial volume of 30 μl and then crush as this will minimize liquid spraying out of the sample tube. Once the sample has been homogenized add the additional volume to make up the desired total.
LARII is stable for 1 month at −20°C and 1 year at −80°C. Make 1 ml aliquots of LARII and use 80 μl per sample.
Make up the 1x Stop & Glow buffer in a glass vial or siliconized polypropylene tube (a culture tube works well).
One obvious source of variation within the HRMA output can originate from SNPs within the sequence. SNPs can be accounted for if known samples are considered as standards; therefore sequence data of the target region for a number of individuals is advised. Where possible, the assay primers and the gene-editing elements should be designed around an invariable region, therefore any indel events within the target region are more easily recognized.
One of the most crucial preparation steps is to identify an optimal annealing temperature for the PCR reaction that yields the product for analysis. It is therefore imperative that the template preparation is standardized to minimize variability.
Check the gel image of the gradient PCR products to ensure that the optimal temperature identified by the melt profile results in a specific PCR product of the predicted size.
Ideally the entire leg is immersed in the reaction volume. Be sure to wipe the forcep tips and legs with 70% ethanol between individual samples to limit contamination.
To reduce chance of cross-contamination, maintain separate labeled disposable transfer Pasteur pipettes and small beakers (for food solutions) for each line.
If some eggs float on the surface of water, then trap them under a small piece of tissue. This prevents them from sticking to the side of the pan where a drop in water level through evaporation might cause them to become non-viable. It also helps ensure any remaining oil is siphoned away from the embryo surface.
Add a small amount of fish food in the pan. The pan with the injected-recovered egg papers should be maintained for at least two weeks to ensure all viable eggs will hatch. During this time the water should not be changed. If too much food is present then the water may turn cloudy. Only add more food when all has been consumed.
We recommend to have 25–30 G0 females in each medium-sized cage.
If there is uncertainty about the size of male and female pupae (as the G0 pupae sometimes differentiate less obviously than the wild type), place each pupa in a drosophila tube (28.5 mm × 95 mm) in a small amount of water and stopper the tube with a flug or cotton. Check the tubes every day and as soon as they emerge, release them in the male and females popcorn-cages.
Anesthetize adult mosquitoes using CO2 then transfer them to an pre-chilled glass petri dish (150 mm × 20 mm) on ice. Be aware of the amount of time the mosquitoes are knocked out as extended period of times anesthetized may have an effect on their longevity. Check for any contaminating adults of the other sex then transfer adults into the appropriate cage.
Place the female mosquito gently inside the tube as it is laid flat (rather than standing). This will allow the female to recover on a dry surface rather than against the damp paper.
Placing the dry oviposition beaker in the soup cup-cage at this same time saves repeatedly anesthetizing the adult mosquitoes after the mated females have blood fed. Also a punch a hole in the side of the container to allow the addition of more pupae if necessary.
To store Aedes aegypti oviposition papers first gently but firmly blot the oviposition paper upon a paper towel or tissue. The amount of moisture within the paper is crucial, too much will encourage mold growth but too little will result in very few or no eggs hatching. Place the dried (but not desiccated) paper within a plastic pouch then seal the edges around the enclosed paper using a 2 mm Impulse Heat Sealer. In this manner the oviposition papers can be stored for up to 4 months at room temperature.
References
- 1.WHO. Dengue and severe dengue - Fact sheet No.117. World Health Organization; 2012. (updated 2014) [Google Scholar]
- 2.Aryan A, et al. TALEN-based gene disruption in the dengue vector Aedes aegypti. PLoS One. 2013;8(3):e60082. doi: 10.1371/journal.pone.0060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liesch J, Bellani LL, Vosshall LB. Functional and Genetic Characterization of Neuropeptide Y-Like Receptors in Aedes aegypti. PLoS Negl Trop Dis. 2013;7(10):e2486. doi: 10.1371/journal.pntd.0002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMeniman CJ, et al. Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell. 2014;156(5):1060–71. doi: 10.1016/j.cell.2013.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jasinskiene N, Juhn J, James A. Microinjection of A. aegypti embryos to obtain transgenic mosquitoes. J Visual Exp. 2007;5:219. doi: 10.3791/219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coates CJ, et al. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95(7):3748–51. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aryan A, Myles KM, Adelman ZN. Targeted genome editing in Aedes aegypti using TALENs. Methods. 2014 doi: 10.1016/j.ymeth.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassett Andrew R, et al. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Reports. 2013;4(1):220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]